Abstract
Background
The parasitic nematodes Ascaris lumbricoides and A. suum are of great public health and economic significance, and the two taxa were proposed to represent a single species. miRNAs are known with functions of gene regulations at post-transcriptional level.
Results
We herein compared the miRNA profiles of A. lumbricoides and A. suum female adults by Solexa deep sequencing combined with bioinformatics analysis and stem-loop real-time PCR. Using the A. suum genome as the reference genome, we obtained 171 and 494 miRNA candidates from A. lumbricoides and A. suum, respectively. Among which, 74 miRNAs were shared between the two taxa, 97 and 420 miRNAs were A. lumbricoides and A. suum specific. Target and function prediction revealed a significant set of targets which are related to ovarian message protein, vitellogenin and chondroitin proteoglycan of the two nematodes. Enrichment analysis revealed that the percentages of most predicted functions of the miRNA targets were similar, with some taxon specific or taxon enhanced functions, such as different target numbers, specific functions (NADH dehydrogenase and electron carrier functions), etc.
Conclusions
This study characterized comparatively the miRNAs of adult A. lumbricoides and A. suum, and the findings provide additional evidence that A. lumbricoides and A. suum represent a single species. Due to the fast evolution nature of miRNAs and the different parasitic living conditions of humans and pigs, the phenomenon above might indicate a fast evolution of miRNAs of Ascaris in humans and pigs.
Keywords: MicroRNA (miRNA), Ascaris lumbricoides, Ascaris suum, Comparative analysis
Background
Roundworms Ascaris lumbricoides and A. suum are important parasites of human and pig health and socio-economic significance, with global distribution [1,2]. Ascaris infects approximately 1.2 billion people globally and has been associated with intestinal pathology, respiratory symptoms and malnutrition in children from endemic areas [3]. It was reported that humans and pigs can both be infected with the two nematodes [2]. There are very limited nucleotide differences in the sequences of internal transcribed spacers (ITS) of ribosomal DNA between the two ascarid species [4]. A. lumbricoides and A. suum are now proposed to represent a single species, and A. suum is considered a synonym of A. lumbricoides[5]. The genome and transcripts of A. suum were recently available, which provided valuable resources for better understanding and further studies of the biology of the Ascaris parasites [6-8].
MicroRNAs (miRNAs) are non-coding small RNA of 18–24 nt in length. They are considered as key regulators for gene expression at the post-transcriptional level [9-12]. Due to their key regulating functions in growth, metabolism, development and cell differentiation and their ability to respond to environmental and developmental signals, miRNA is essential for the complex life cycles of human and animal parasites [13-22]. miRNAs are also important for pathogen-host interactions [23-26]. Recently studies have indicated that miRNAs may represent potential biomarkers for various biomedical problems, such as the differentiation of different tumors and muscular tissues [27-29], and they may provide biomarkers for the characterization of different genotypes of Toxoplasma gondii[30].
There might be differences in gene expression and regulation given the different living environments of A. lumbricoides and A. suum, although they are considered to represent the same species. Therefore, it would be interesting to characterize the expression profiles of miRNAs in the two taxa. Herein the objective of the present study was to examine and compare the miRNA profiles of A. lumbricoides and A. suum using an integrative approach combining Solexa deep sequencing combined with bioinformatics analysis and stem-loop real-time PCR.
Methods
Ethics statement
The present study was approved by the Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Approval No. LVRIAEC2011-006), and the A. lumbricoides and A. suum samples were collected strictly according to the requirements of the Ethics Procedures and Guidelines of the People's Republic of China.
Nematodes
Female adults of A. suum roundworms were obtained from slaughtered pigs in Shenzhen, China. Female adults of A. lumbricoides were obtained from a patient with ascariasis after being treated with piperazine in Zhanjiang, Guangdong Province, China. Worms were incubated in physiological saline for 3 h at 37°C and then washed 3 times to get rid of contamination from hosts. Female adults were identified morphologically and their identity was further ascertained by PCR amplification and sequencing of the first internal transcribed spacer (ITS-1) of rDNA [4]. The nematodes were stored at −80°C until use.
Total RNA isolation, small RNA preparation and deep sequencing
Total RNA was prepared from a whole single adult of A. lumbricoides and A. suum respectively, using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Ten μg total RNA and Novex 15% TBE-Urea gel (Invitrogen) were used for small RNA isolation. The RNA fragments of 18–30 bases long were added with 5’ and 3’ adaptors (Illumina) to the both ends, reverse transcripted with a RT-PCR kit (Invitrogen), and sequenced employing a Solexa sequencer at Huada Genomics Institute Co. Ltd, China.
Computational analysis
Adaptors, low quality sequences and reads smaller than 18 nt were removed from the raw sequencing data. The reads were then searched against GenBank and Rfam database ( http://www.sanger.ac.uk/software/Rfam) to identify non-coding RNA. The remaining reads were mapped onto the A. suum genome by using the SOAP [31] with the sequences of pre-miRNA meeting the three criteria: 1) there was a standard stem-loop structure of pre-miRNA; 2) mature miRNAs were present in one arm instead of the loop of hairpin precursors; and 3) the free energy hybridization was lower than −18 kcal/mol. The identified miRNA candidates were searched against the known miRNAs of A. suum deposited in the Sanger miRBase with Blast [32]. Unmatched miRNA candidates were marked as novel miRNA. Targets of miRNA candidates were predicated with RNAhybrid [33] with the following extra parameters: A) the △△G was set as lower than −25 kcal/mol; B) P-value was set as ≤ 0.01. The Gene Ontology database (GO, http://www.geneontology.org/), Blast and Interproscan ( http://www.ebi.ac.uk/Tools/pfa/iprscan/) were used for prediction of functions of predicted targets.
Analysis of novel miRNA expression
The representative novel miRNA in the two nematodes were certified using stem-loop real-time reverse transcription polymerase chain reaction (RT-PCR) with SYBR Green [34]. The house keeping gene β-actin of A. suum (GenBank accession no. BI594141) was used as the endogenous control with primers as follows: forward primer (5′-CTCGAAACAAGAATACGATG-3′) and reverse primer (5′-ACATGTGCCGTTGTATGATG-3′). Primers were synthesized by Shenggong Co, Ltd., China. The cycle conditions were as follows: 94°C for 30s, 52°C for 30s, 72°C for 30s and finally with a single extension at 72°C for 10 min. The quantification of each miRNA relative to β-actin gene was calculated using the equation: N = 2-ΔCt, ΔCt = CtmiRNA-Ctacin[35].
Results
Profile differences in short RNAs between the two Ascaris taxa
Deep sequencing yielded 18.29 and 11.72 million raw reads in adult of A. lumbricoides and A. suum, respectively, with 14.69 and 9.76 million high quality reads that longer than 18 nt. Length distribution analysis showed that the reads of A. lumbricoides and A. suum were focused on 21–23 nt. Among the clean reads, 46.11% and 39.97% were identified as non-coding RNA (ncRNA) in A. lumbricoides and A. suum respectively, including tRNA, rRNA, snRNA and snoRNA, which were at near the same level. Among the high quality reads, 72.47% of the total reads were shared by the two parasites (Table 1), while 1.71 and 1.08 million was A. lumbricoides and A. suum specific.
Table 1.
Common and taxon-specific reads of Ascaris lumbricoides and Ascaris suum
| Classification | Unique sRNAs (%) | Total sRNAs (%) |
|---|---|---|
| Total sRNAs |
2917391 (100%) |
24449036 (100%) |
| Common readsa |
126807 (4.35%) |
17718517 (72.47%) |
|
A. lumbricoides specificb |
1711536 (58.67%) |
4379273 (17.91%) |
| A. suum specificc | 1079048 (36.99%) | 2351246 (9.62%) |
Note: areads that were shared by the two taxa; breads found in A. lumbricoides, but not found in A. suum; creads found in A. suum, but not found in A. lumbricoides.
miRNA profiles of the two taxa
By mapping onto the A. suum genome, we obtained 171 and 494 miRNA candidates, with precursors meeting the criteria listed in the method and having standard stem-loop structures (Table 2, Additional file 1: Table S1). Among the miRNA candidates, 68 and 87 miRNAs were matched with the known A. suum miRNAs deposited in the miRBase database, and 63 of them were shared. The left miRNA candidates were marked as novel miRNAs, and among which only 11 miRNAs were shared, therefore, 92 and 396 miRNAs were A. lumbricoides and A. suum specific. Totally, there were 74 miRNAs shared by the two nematodes, including 63 known miRNA and 11 novel ones; and there were 97 and 420 miRNAs being A. lumbricoides and A. suum specific.
Table 2.
Comparison of miRNA profiles in Ascaris lumbricoides (Alu) and A. suum (Asu)
| Shared | Asu-specific | Alu-specific | Asu-total | Alu-total | |
|---|---|---|---|---|---|
| Novel |
11 |
396 |
92 |
407 |
103 |
| Known |
63 |
24 |
5 |
87 |
68 |
| Total | 74 | 420 | 97 | 494 | 171 |
Target prediction and the functional prediction of the predicted targets
A total of 57,359 mRNA and EST items of Ascaris deposited in NCBI were downloaded and used for target prediction. Under the stringent matching criteria, it was found that the target numbers of both nematodes ranged from one to thousands. For A. lumbricoides, the target number ranged from one (Alu-miR-novel-012-3p, Alu-miR-novel-066-5p, Alu-miR-novel-102-3p) to 3,562 (Alu-miR-novel-063-3p), 255 in average. For A. suum, the target number ranged between one (asu-miR-novel-027-3p, asu-miR-novel-044-5p, and asu-miR-novel-068-5p) and 3,343 (asu-miR-novel-039-3p), 168 in average.
Functional prediction revealed a significant set of targets which are related to ovarian message protein in A. lumbricoides (n = 9) and A. suum (n = 10), a set of vitellogenin (5 in A. lumbricoides and 18 in A. suum), and a set of chondroitin proteoglycan (5 in A. lumbricoides and 7 in A. suum). The same phenomenon is found in NADH dehydrogenase (n = 7 in A. suum, n = 3 in A. lumbricoides), 40s ribosomal protein (n = 14 in A. suum, n = 2 in A. lumbricoides), and 60s ribosomal protein (n = 11 in A. suum, n = 7 in A. lumbricoides). As a distinguished character, there were more targets related to movement in A. suum, such as actin (n = 5) and tubulin (n = 6), while such targets were very few in A. lumbricoides.
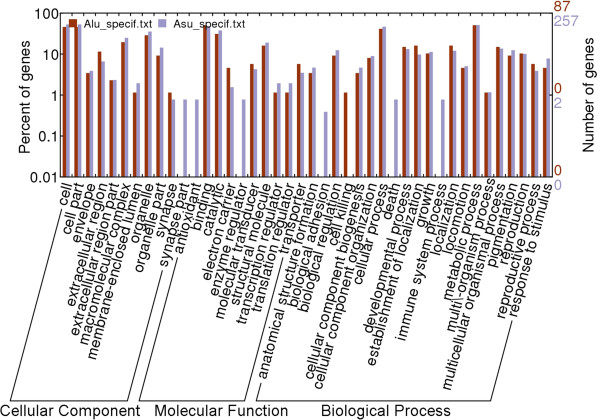
Gene Ontology (GO) analysis of the targets of miRNAs resulted in 3 respect outcomes, including cellular component, molecular function and biological process. Enrichment analysis showed that percentages of most targets functions of A. suum and A. lumbricoides were similar, except a few items (Figure 1), which indicated a very closely metabolism pattern of the two parasites. However, for cell component, miRNA targets of A. suum had an extra cellular component part named as “synapse part”; for molecular function, one function named “electron carrier” was only found in targets of A. suum. In addition, three biological processes named biological adhesion, death, and immune system process were only found in the miRNA targets of A. suum.
Figure 1.
Enrichment analysis of the functions of strain-specific miRNAs targets of Ascaris suum and A. lumbricoides. The horizontal axis: Gene Ontology analysis to the targets, including cellular component, molecular function, and biological process. The vertical axis: percentage of genes in total targets.
Quantification of A. lumbricoides and A. suum miRNAs
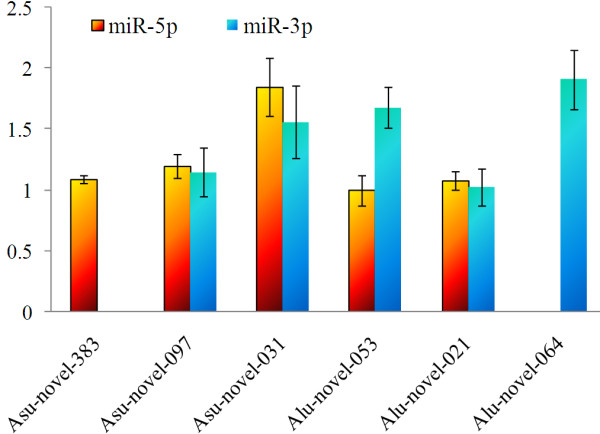
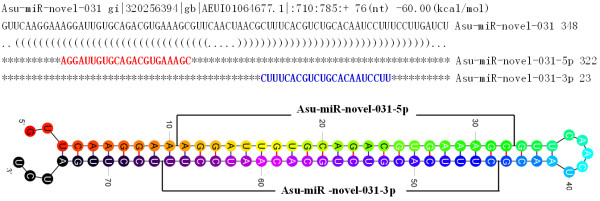
Three A. suum and A. lumbricoides specific novel miRNAs, including asu-miR-novel-383, asu-miR-novel-097 and asu-miR-novel-031 in A. suum, and alu-miR-novel-053, alu-miR-novel-021, and alu-miR-novel-064 in A. lumbricoides, were representatively selected for quantification using modified stem-loop qRT-PCR (Figure 2). These 6 miRNAs had the lowest △G energy, and/or more matched variants, and had mature miRNAs at both of the arms of the stem-loop precursors. The representative blast and standard stem-loop structure as asu-miR-novel-031 was shown in Figure 3. Detailed blast information with variants of others was shown in Additional file 2.
Figure 2.
The main blast result and standard stem-loop structure of precursors of one of the representative miRNAs. Upper: plain text sequence of precursor of asu-miR-share-031 and the main blast result with mature miRNA matched; Down: stem-loop structure of the precursor. Detailed blast information was shown in (Additional file 2: Figure S1).
Figure 3.
Quantification of representative miRNAs of Ascaris lumbricoides and A. suum. Three novel miRNAs of each nematode were detected. Two miRNAs named asu-miR-novel-031 and alu-miR-novel-53 had significant expression differences of 5p and 3p mature miRNAs. The alu-miR-novel-64-5p and asu-miR-novel-383-3p were not successfully amplified, which might due to their low expression levels.
Both of the mature miRNAs of the 6 selected miRNAs can be successfully detected, except the alu-miR-novel-64-5p and asu-miR-novel-383-3p. Two miRNAs named asu-miR-novel-031 and alu-miR-novel-53 had significant expression differences at 5p and 3p. For alu-miR-novel-53, the expression levels were 1.00 ± 0.12 at 5p, and it was 1.68 ± 0.17 at 3p. And for asu-miR-novel-031, the relative expression level was 1.85 ± 0.24 at 5p, while it was 1.56 ± 0.30 at 3p. The alu-miR-novel-64-5p and asu-miR-novel-383-3p were not successfully amplified, which could be attributed to the very low expression levels, despite that the primers and reaction mixture were modified several times.
Discussion
A previous study revealed that A. lumbricoides and A. suum have identical 5.8S and ITS-2 rDNA sequences [4]. There were only six (1.3%) nucleotide differences in ITS-1. All these genetic evidences supported the proposal that A. lumbricoides and A. suum represent a single species [4,5].
Due to their specific expression in different organisms, tissues and cells, miRNAs may provide potential novel biomarkers [27-29]. Our previous study indicated that different T. gondii genotypes have their unique miRNA profiles [30]. In the present study, the two nematode taxa shared very high percentage of total reads (72.47%), while having very small percentage of unique reads (4.35%), and the total percentage of ncRNA was at similar level (46.11% and 39.97% in A. lumbricoides and A. suum, respectively), which indicated huge redundance of some ncRNA of the two nematodes. When parasitic environments change, the gene expression of parasites will be correspondingly changed, and regulators such as miRNAs will also be changed, therefore the death and new creation of miRNAs are very common [36]. For the different parasitic environment of pigs and humans, adaptive modifications should have occurred. So although the sRNA/shared-miRNAs for fundamental metabolism were similar in the two taxa, there still are some specific miRNAs in each nematode taxon. We obtained 171 and 494 miRNA candidates as expression profiles, with 74 miRNAs being shared by the two nematode taxa, and 97 and 420 miRNAs being A. lumbricoides and A. suum specific. The same precursors of a miRNA can be found in the genome of A. lumbricoides or A. suum, however, we obtained fewer miRNAs from A. lumbricoides than from A. suum, which indicated that some miRNAs in A. lumbricoides were not expressed. This difference might be resulted from the different parasitic living environment in pigs and humans.
Function prediction and enrichment analysis showed that targets of the miRNAs of the two parasites had similar metabolism patterns, including cellular component, molecular function and biological process, with some specific differences, such as different target number, specific functions (NADH dehydrogenase and electron carrier functions), etc.
Conclusion
The present study characterized comparatively the miRNAs of adult A. lumbricoides and A. suum, and the findings support the recent proposal that A. lumbricoides and A. suum represent a single species [5]. Due to the fast evolution nature of miRNAs and the different parasitic living conditions of humans and pigs, the phenomenon above might indicate a fast evolution of miRNAs of Ascaris.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XQZ, MJX and JPT conceived and designed the experiment, and critically revised the manuscript. CCS, MJX, SA and HQS performed the experiments, analyzed the data, and drafted the manuscript. LP helped in the study design, study implementation, and manuscript revision. All authors read and approved the final manuscript.
Supplementary Material
Shared and strain-specific miRNA with their predicated target of Ascaris lumbricoides and A. suum.
Detailed blast results of variants and precursors of representative miRNAs in Ascaris lumbricoides and A. suum.
Contributor Information
Chang-Chun Shao, Email: 7330968@163.com.
Min-Jun Xu, Email: xuminjun@caas.cn.
Samer Alasaad, Email: samer@ebd.csic.es.
Hui-Qun Song, Email: songhuiqun@scau.edu.cn.
Lifei Peng, Email: pengjida@gdmc.edu.cn.
Jian-Ping Tao, Email: yzjptao@126.com.
Xing-Quan Zhu, Email: xingquanzhu1@hotmail.com.
Acknowledgements
This work was supported in part by the International Science & Technology Cooperation Program of China (Grant No. 2013DFA31840), the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006) to XQZ, and the China Postdoctoral Science Foundation (Grant No. 201104363) to MJX.
References
- Dold C, Holland CV. Ascaris and ascariasis. Microbes Infect. 2011;13:632–637. doi: 10.1016/j.micinf.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Crompton DW. Ascaris and ascariasis. Adv Parasitol. 2001;48:285–375. doi: 10.1016/s0065-308x(01)48008-0. [DOI] [PubMed] [Google Scholar]
- Hagel I, Giusti T. Ascaris lumbricoides: an overview of therapeutic targets. Infect Disord Drug Targets. 2010;10:349–367. doi: 10.2174/187152610793180876. [DOI] [PubMed] [Google Scholar]
- Zhu X, Chilton NB, Jacobs DE, Boes J, Gasser RB. Characterisation of Ascaris from human and pig hosts by nuclear ribosomal DNA sequences. Int J Parasitol. 1999;29:469–478. doi: 10.1016/S0020-7519(98)00226-4. [DOI] [PubMed] [Google Scholar]
- Leles D, Gardner SL, Reinhard K, Iniguez A, Araujo A. Are Ascaris lumbricoides and Ascaris suum a single species? Parasit Vectors. 2012;5:42. doi: 10.1186/1756-3305-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, Yang L, Zeng N, Xu X, Xiong Z, Chen F, Wu X, Zhang G, Fang X, Kang Y, Anderson GA, Harris TW, Campbell BE, Vlaminck J, Wang T, Cantacessi C, Schwarz EM, Ranganathan S, Geldhof P, Nejsum P, Sternberg PW, Yang H, Wang J, Wang J, Gasser RB. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Zou FC, Hall RS, Zhong W, Jex AR, Campbell BE, Ranganathan S, Sternberg PW, Zhu XQ, Gasser RB. Bioinformatic analysis of abundant, gender-enriched transcripts of adult Ascaris suum (Nematoda) using a semi-automated workflow platform. Mol Cell Probes. 2009;23:205–217. doi: 10.1016/j.mcp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Huang CQ, Gasser RB, Cantacessi C, Nisbet AJ, Zhong W, Sternberg PW, Loukas A, Mulvenna J, Lin RQ, Chen N, Zhu XQ. Genomic-bioinformatic analysis of transcripts enriched in the third-stage larva of the parasitic nematode Ascaris suum. PLoS Negl Trop Dis. 2008;2:e246. doi: 10.1371/journal.pntd.0000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tuo W, Gao H, Zhu XQ. MicroRNAs of parasites: current status and future perspectives. Parasitol Res. 2010;107:501–507. doi: 10.1007/s00436-010-1927-6. [DOI] [PubMed] [Google Scholar]
- Lin WC, Li SC, Lin WC, Shin JW, Hu SN, Yu XM, Huang TY, Chen SC, Chen HC, Chen SJ, Huang PJ, Gan RR, Chiu CH, Tang P. Identification of microRNA in the protist Trichomonas vaginalis. Genomics. 2009;93:487–493. doi: 10.1016/j.ygeno.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Wang CR, Xu MJ, Fu JH, Nisbet AJ, Chang QC, Zhou DH, Huang SY, Zou FC, Zhu XQ. Characterization of microRNAs from Orientobilharzia turkestanicum, a neglected blood fluke of human and animal health significance. PLoS ONE. 2012;7:e47001. doi: 10.1371/journal.pone.0047001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Ai L, Fu JH, Nisbet AJ, Liu QY, Chen MX, Zhou DH, Zhu XQ. Comparative characterization of microRNAs from the liver flukes Fasciola gigantica and F. hepatica. PLoS ONE. 2012;7:e53387. doi: 10.1371/journal.pone.0053387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Fu JH, Nisbet AJ, Huang SY, Zhou DH, Lin RQ, Song HQ, Zhu XQ. Comparative profiling of microRNAs in male and female adults of Ascaris suum. Parasitol Res. 2013;112:1189–1195. doi: 10.1007/s00436-012-3250-x. [DOI] [PubMed] [Google Scholar]
- Xu MJ, Liu Q, Nisbet AJ, Cai XQ, Yan C, Lin RQ, Yuan ZG, Song HQ, He XH, Zhu XQ. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;11:521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Wang CR, Huang SY, Fu JH, Zhou DH, Chang QC, Zheng X, Zhu XQ. Identification and characterization of microRNAs in the pancreatic fluke Eurytrema pancreaticum. Parasit Vectors. 2013;6:25. doi: 10.1186/1756-3305-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Cannella D. Apicomplexan parasites and subversion of the host cell microRNA pathway. Trends Parasitol. 2011;27:481–486. doi: 10.1016/j.pt.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Zhao GH, Xu MJ, Zhu XQ. Identification and characterization of microRNAs in Baylisascaris schroederi of the giant panda. Parasit Vectors. 2013;6:216. doi: 10.1186/1756-3305-6-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Jia B, Lu H, Peng S, Piao X, Hou N, Cai P, Yin J, Jiang N, Chen Q. A comparative study of small RNAs in Toxoplasma gondii of distinct genotypes. Parasit Vectors. 2012;5:186. doi: 10.1186/1756-3305-5-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Zhai H, Fesler A, Ju J. MicroRNA: a third dimension in autophagy. Cell Cycle. 2013;12:246–250. doi: 10.4161/cc.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall ES, Elshekiha HM, Hakimi MA, Flynn RJ. Toxoplasma gondii peroxiredoxin promotes altered macrophage function, caspase-1-dependent IL-1beta secretion enhances parasite replication. Vet Res. 2011;42:80. doi: 10.1186/1297-9716-42-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Lan J, Wu X, Yang D, Zhang Z, Nie H, Hou R, Zhang R, Zheng W, Xie Y, Yan N, Yang Z, Wang C, Luo L, Liu L, Gu X, Wang S, Peng X, Yang G. Identification of Dirofilaria immitis miRNA using Illumina deep sequencing. Vet Res. 2013;44:3. doi: 10.1186/1297-9716-44-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søkilde R, Vincent M, Møller AK, Hansen A, Høiby PE, Blondal T, Nielsen BS, Daugaard G, Møller S, Litman T. Efficient identification of miRNAs for classification of tumor origin. J Mol Diagn. 2014;16:106–115. doi: 10.1016/j.jmoldx.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Endo K, Weng H, Naito Y, Sasaoka T, Takahashi A, Fukushima Y, Iwai N. Classification of various muscular tissues using miRNA profiling. Biomed Res. 2013;34:289–299. doi: 10.2220/biomedres.34.289. [DOI] [PubMed] [Google Scholar]
- Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014. doi:10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed]
- Xu MJ, Zhou DH, Huang SY, Zhao FR, Nisbet AJ, Lin RQ, Song HQ, Zhu XQ. Comparative characterization of microRNA profiles of different genotypes of Toxoplasma gondii. Parasitology. 2013;140:1111–1118. doi: 10.1017/S003118201300053X. [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Mount DW. Using the Basic Local Alignment Search Tool (BLAST) CSH Protoc. 2007;2007:p17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shared and strain-specific miRNA with their predicated target of Ascaris lumbricoides and A. suum.
Detailed blast results of variants and precursors of representative miRNAs in Ascaris lumbricoides and A. suum.