Abstract
Amnestic mild cognitive impairment (aMCI) often is an early stage of Alzheimer’s disease (AD). MCI is characterized by cognitive decline departing from normal cognitive aging but that does not significantly interfere with daily activities. This study explores the potential of scalp EEG for early detection of alterations from cognitively normal status of older adults signifying MCI and AD. Resting 32-channel EEG records from 48 age-matched participants (mean age 75.7 years)–15 normal controls (NC), 16 early MCI, and 17 early stage AD–are examined. Regional spectral and complexity features are computed and used in a support vector machine model to discriminate between groups. Analyses based on three-way classifications demonstrate discrimination accuracies of 83.9%–96.8% for MCI vs. NC (p-value<0.0029), 71.9%–96.9% for AD vs. NC (p-value<0.0333), and 87.9%–90.9% for AD vs. MCI (pvalue<0.0136), depending on the EEG protocol condition employed. These results demonstrate the great promise for scalp EEG spectral and complexity features as noninvasive biomarkers for detection of MCI and early AD.
Keywords: EEG-based diagnosis, early Alzheimer’s disease, mild cognitive impairment, spectral, entropy
INTRODUCTION
Mild cognitive impairment (MCI), especially the amnestic type, is a degenerative neurological disorder characterized by cognitive decline which is greater than expected for an individual’s age, but does not necessarily interfere with daily activities [1]. Early stages of Alzheimer’s disease (AD) are characterized by progressive memory loss, diminished vocabulary, and lowered ability to execute precise motor movements, together with impaired activities of daily living. Previous research has shown that MCI patients progress to AD at a rate of approximately 10% to 15% of patients per year [2],[3]. The prevalence of MCI and AD increases with age, making MCI/AD research of significant interest in geriatric medicine. Current interest in the field is focused on detection of MCI at the earliest possible time for purposes of instituting medication and lifestyle changes to slow or halt progression of further cognitive decline. However, there is no biomarker-based test or screening tool of utility in the primary care setting.
Several causes of MCI and AD have been hypothesized based on pathologic features observed in the brains of AD patients. The current leading hypothesis for the cause of AD is abnormal deposition of amyloid beta protein in the brain together with a second abnormal protein, microtubule-associated tau [4],[5]. No single root cause can be attributed to MCI. However, given the high number of MCI patients who progress to AD, the fundamental cause of both MCI and AD is likely to be the same in many cases [6]. In the current study, the MCI participants were all diagnosed as amnestic MCI.
The earliest stage of MCI and AD diagnosis is often based on neuropsychological tests and patient history evaluations [7]. Once it is determined that the subject is experiencing abnormal cognitive decline, physicians in advanced memory disorders clinics employ more quantitative diagnostic tools. One such tool is the analysis of cerebrospinal fluid (CSF) biomarkers. The most common CSF biomarkers used for AD diagnosis are amyloid beta and tau. These biomarkers provide a reliable means to distinguish AD from other forms of dementia and appear in characteristically abnormal levels at the earliest onset of the disease [8],[9]. Published findings suggest that CSF examination can identify abnormal proteins in patients who went on to develop AD later [10].
Imaging in AD has been studied intensively in recent years [11]. Positron emission tomography (PET) detects fibrillar amyloid protein in living subjects using amyloid-binding tracer, Pittsburgh Compound B (PiB)14. The regional variations in PiB binding in vivo are strikingly similar to the deposits of amyloid beta and tau neurofibrillary tangle distributions post-mortem [12]. PET scans have been able to measure the PiB uptake in the neocortex and identify the regional distribution of amyloid beta plaque burden with high specificity in AD. Additionally, structural magnetic resonance imaging (sMRI) studies have shown reductions in volume of the hippocampus, one of the key areas of the brain affected by amyloid beta and tau deposition early in the disease [13]. Diffusion tensor MRI can measure white matter integrity in the brain and may be more useful as an earlier biomarker of AD than sMRI [14]. Structural MRI and amyloid-PET scans may be used in combination in AD, but they are complex, expensive, require radiation exposure in the case of PET, and are not yet standardized tools for AD detection and diagnosis.
EEG is a noninvasive method indirectly measuring brain neural electric activity from the scalp of the head. Even though it has been around for decades, using EEG as cognitive biomarker to detect and predict MCI and AD in individuals is a relatively new effort [15]. Scalp EEG has the potential to play a significant role as one of the earliest biomarkers for MCI and early-stage AD, before clinical diagnosis. EEG operates through the recording of oscillations of brain electric potentials from electrodes attached to the scalp. EEG data from MCI and AD patients have been shown to have lower mean levels of channel-to-channel synchronization than those of healthy controls [16],[17]. The effects of cortical neurons’ deaths, axonal pathology, cholinergic deficits, and other neural network disconnections as concomitants of the disease are manifested by multiple changes in EEG characteristics [18]–[21]. Cognitive impairment as a result of AD is correlated with a reduction in dominant posterior rhythm while dementia in general correlates well with a rise in theta activity [19]. Decreased synchronization in local and global networks is also observed with AD [17],[22],[23]. Other potential differences in spectral features of EEG for MCI/AD have also been noted [24]–[28]. Experimental results using EEG to correctly distinguish AD patients from healthy controls have proven promising; however, researchers have encountered difficulty in distinguishing between different stages of dementia (e.g., MCI vs. AD) [29],[30]. In many cases, group differences can be demonstrated, but group means are not sufficiently different to allow for diagnostic identification of an individual with a specific group [31]–[37]. If EEG technology can be developed to discriminate MCI from normal EEG for individuals and to show changes over relatively short time periods in and individual, it will be transformative in the early detection of MCI, AD, and other dementias, as it is more sensitive than behavioral testing, inexpensive, noninvasive, and can be made practical for primary care settings. In this study we focus on the spectral and complexity features of EEG as distinctions between normal older, MCI, and AD subjects.
MATERIALS AND METHODS
Participants
The EEG data used in this study were recorded in the Aging Brain and Cognition laboratory in the Behavioral Science Department and Sanders-Brown Center on Aging at the University of Kentucky (UK) College of Medicine. Participants between the ages of 60 and 90 years were recruited from a study cohort of cognitively normal older adults identified by the Alzheimer’s Disease Center (ADC) of the UK College of Medicine. The normal older participants are screened annually and were followed into likely MCI or AD stage till autopsy. MCI and AD participants were diagnosed and recruited by cognitive neurologists Drs. C. Smith and G. Jicha at the UK ADC Clinical Core and from its Research Memory Disorders Clinic. All MCI participants belonged to the amnestic MCI subtype. A list of neurological assessments used to make MCI/AD diagnoses is provided in Table 1 [26]. Means and standard deviations for MMSE scores for the three groups are presented in Table 2. The MCI and early AD participants’ EEG data were recorded as soon as possible after diagnoses were made.
Table 1.
UK-ADC Uniform Research Battery of Cognitive Tests and Other Evaluations
| General Cognitive Measures |
| MMSE; Kokmen Test |
| Clinical Dementia Rating (CDR) |
| Memory Domain Measures |
| WMS Logical Memory I & II |
| California Verbal Learning Test* |
| Attention/Executive Domain Measures |
| Trail Making Tests A & B |
| WAIS-R Digit Span & Digit Symbol |
| Language Domain Measures |
| COWAT* |
| Animal & Vegetable Fluency |
| Boston Naming |
| Visual/Spatial Domain Measures |
| CERAD Figures |
| Baseline Only |
| National Adult Reading Test* |
| Medical Evaluation |
| Physical exam* |
| Neurological exam* |
| Medical history* |
| Medications |
| Nutritional supplements |
| Food Frequency Questionnaire (FFQ) |
| Psychiatric Evaluation |
| Neuropsychiatric Inventory Questionnaire (NPI-Q) |
| Geriatric Depression Scale (GDS) |
| Functional Ability Measures |
| Functional Assessment Questionnaire (FAQ) |
| SF-36* |
| ADCS-ADL* |
Additional test measures or expanded content beyond UDS core measures
Table 2.
MMSE Scores for Groups
| AD | MCI | NC |
|---|---|---|
| 24.26 (2.42) | 27.40 (2.00) | 29.43 (0.73) |
Group means for Mini Mental; standard deviations in parentheses
All participants were screened to exclude active or unstable medical conditions, depression, and other psychiatric disorders, or history of neurologic or neurosurgical conditions. No participants were to have the ApoE4 allele for Alzheimer’s risk or to be on any psychoactive medication other than antidepressants. For cognitively normal individuals, we excluded those with the ApoE4 genes risk factor in order to have low-risk normal control (NC) group. Systolic hypertension, donepezil, and anti-depressants are known to modulate EEG markers including event-related potentials associated with cognitive functions. These conditions are very common in the population of interest, complicating outright exclusion; however, uncontrolled hypertension or use of sedatives such as the benzodiazepines were bases for exclusion from the study.
Participants were well-matched with regards to age, with normal controls, MCI, and AD participants having mean ages of 75.7 years (SD 5.5 years), 74.6 years (SD 9.0 years), and 76.7 years (SD 5.2 years). NC and AD participants were also well-matched in regards to gender, with NC and AD participants being comprised of 60% females. MCI participants were only 25% female. Difference in MCI gender was likely due to recruiting and does not reflect population trends.
EEG Protocols
Participants were connected to 64- or 32-channel EEG caps using a Neuroscan II system (10–20 montage). In either case, only the 32 common channels were recorded. EEG data were recorded under a protocol using three different non-memory-task conditions. These included: (1) resting with eyes open for 5 minutes, (2) resting with eyes closed while counting backwards by ones for 10 minutes while tapping a finger, and (3) resting with eyes closed for 10 minutes, followed by another 5 minutes of eyes open while resting. EEG recordings were performed without interruption at the same appointment for each participant. EEG data were acquired at 500 Hz. The 32 EEG channels included 2 ocular channels that were used to determine the dominant eye blink frequency. A simple 2nd order Butterworth filter was used to attenuate frequencies greater than 200 Hz. Notch filters were used to remove dominant eye blink frequencies and to remove 60 Hz frequencies, which may have been amplified by background electronic devices. In addition, localized subtraction of baseline for identified muscular and other low frequency artifacts was performed. Analysis of EEG data examined only frequency components less than 40 Hz.
Feature Extraction
Spectral and complexity features were computed for the preprocessed EEG based on oscillations. A few spectral and complexity features were also computed for the first time derivative of the EEG. Investigation of spectral and complexity features of the first time derivatives was motivated by the fact that time derivatives of the EEG voltages (V′(t)) contain indirect information regarding time-dependent rate of change of impedance (Z′(t)) and distribution of charge (rate of change of current flow, l′(t)) in the brain, both of which may be affected by disruption of neural pathways due to disease pathology (V′(t) = Z′(t)l(t) + l′(t)Z(t)). A total of 24 features were calculated for each channel: 18 for the preprocessed EEG and 6 for the time derivatives; see Table 3 for a list of features.
Table 3.
Features
| Relative Spectral Power Features |
| (1) : relative power in θ band |
| (2) : relative power in α1 band |
| (3) : relative power in α2 band |
| (4) : relative power in β1 band |
| (5) : relative power in β2 band |
| (6) : relative power in γ band |
| Additional Spectral Features |
| (7) Ptotal: total spectral power |
| (8) : peak α band freq. |
| (9) fmed: median freq. |
| (10) Sspec: spectral entropy |
| Spectral Power Ratios |
| (11) R1: first ratio |
| (12) R2: second ratio |
| (13) R3: third ratio |
| Entropy/Complexity Features |
| (14) A: activity |
| (15) M: mobility |
| (16) C: complexity |
| (17) Ssamp: sample entropy |
| (18) CLZ: Lempel-Ziv complexity |
| First Derivative Features |
| (19) (Ptotal)′: total spectral power of first derivative |
| (20) : peak α band freq. of first derivative |
| (21) (fmed)′: median freq. of first derivative |
| (22) (Sspec)′: spectral entropy of first derivative |
| (23) (Ssamp)′: sample entropy of first derivative |
| (24) (CLZ)′: Lempel-Ziv complexity of first derivative |
Definitions for features and descriptions of how they were computed are provided in the Spectral Features and Entropy and Complexity Features sections.
Spectral Features
Relative spectral power in the θ band (, 3.5–7.5 Hz), α1 band (, 7.5–9.5 Hz), α2 band (, 9.5–12.5 Hz), β1 band (, 12.5–17.5 Hz), β2 band (, 17.5–25 Hz), and γ band (, 25–40 Hz) were computed. Additional spectral features included total spectral power (Ptotal, 3.5–40 Hz), peak α band frequency (), median frequency (fmed), and spectral entropy (Sspec). Peak α band frequency is defined as the frequency in the range 7.5–12.5 Hz at which the power spectral density curve is maximized. Median frequency is defined as the frequency at which the total spectral power is halved. Spectral entropy is defined as Shannon entropy computed over the normalized power spectral density curve. The mathematical definition for Sspec is given by Equation (1), where p(fi) is the probability of occurrence of frequency fi, P(fi) is the spectral power at frequency fi, and Ptotal is the total spectral power (3.5–40 Hz).
| (1) |
Three spectral power ratios were also computed. These ratios were used based on previous observations of increased power in the θ band of EEG taken from MCI and AD individuals [30],[38]. The definitions for these ratios are presented in Equations (2), (3), and (4).
| (2) |
| (3) |
| (4) |
Spectral features were computed using the power spectral density of individual channels’ records as determined by the Welch Modified Averaged Periodogram Method [39]. In applying Welch’s method, a signal is split into overlapping segments of a given length. The overlapping segments have a tapering window applied to them in the time domain which tapers the data at either end. Periodograms are then calculated for each segment by computing the discrete Fourier transform and the squared magnitude of the result. The periodograms are then averaged. In calculating spectral features, two-second segments with 50% overlap were used. Each two-second segment was modified by a Hanning window (50% cosine taper). Periodograms were then calculated for each two-second segment and averaged. Choice of window length and tapering window were based on methods for computing other common spectral features of EEG presented by previous researchers [30].
Entropy and Complexity Features
Entropy and complexity measures were also computed, including activity (A), mobility (M), complexity (C), sample entropy (Ssamp), and Lempel-Ziv complexity (CLZ). The definitions for activity, mobility, and complexity are presented in Equation (5), where σ0 is the variance of the signal, σ1 is the variance of the first derivative of the signal, and σ2 is the variance of the second derivative of the signal.
| (5) |
Sample entropy is defined as the negative natural log of the conditional probability that time series of length N, having repeated itself within a tolerance of r for m data points, will also repeat itself for m + 1 points. Note that this definition excludes self-matches. One can define Nm as the number of matches of length m and as the subset of Nm that also matches for length m + 1. Sample entropy can then be defined mathematically by Equation (6). In our analyses, we chose m = 2 and r = 0.2 times the standard deviation of the signal. Choice of m and r were based on approaches presented by previous researchers [30].
| (6) |
Lempel-Ziv complexity CLZ is a commonly-used measure for characterizing the randomness of biomedical signals. To compute CLZ, the numerical sequence (signal) must first be transformed into a symbolic sequence S. The most usual means to do this is to convert the signal into a 1/0 symbolic sequence by comparing the signal to a threshold value, usually the median value of the signal. For example, whenever the signal is larger than the median value, one maps the signal to 1, otherwise, to 0. After mapping the signal into its symbolic 1/0 sequence, the sequence can be parsed to obtain distinct “words”, and the “words” can be encoded. Lempel-Ziv complexity can then be defined as the length of the encoded sequence L divided by the length of the signal n.
In order to parse the signal one can sequentially scan the sequence and S = s1s2 … sn rewrite it as a concatenation W = w1−w2 … wk of k “words” chosen such that w1 = s1 and wj is the shortest “word” that has not appeared previously. Thus, wj = wi0 or wi1, where 1 ≤ i ≤ j−1. For example, for the sequence S = 1011010100010, W = 1-0-11-01-010-00-10. The “word” sequence W must then be encoded. This can be done as follows. For each “word” use log2 k bits to describe the location of the prefix to each “word” and 1 bit to describe the last bit of the “word”. For this example, k = 7 words. Thus, one would use 3 bits to describe the prefix location for each word, with 000 being the prefix for an empty set. The encoded sequence would then be (000,1)-(000,0)-(001,1)-(010,1)-(100,0)-(010,0)-(001,0). The length of the encoded sequence L is then equal to k(log2 k + 1). Lempel-Ziv complexity can then be defined mathematically by Equation (7). When the length of the signal n is very large, k ≤ n/log2 n and Equation (7) reduces to Equation (8) [30].
Entropy and complexity features were computed using five-second windows with 50% overlap and then averaged. Choice of window length was based on methods presented by previous researchers [30].
| (7) |
| (8) |
Regionally-Averaged Features
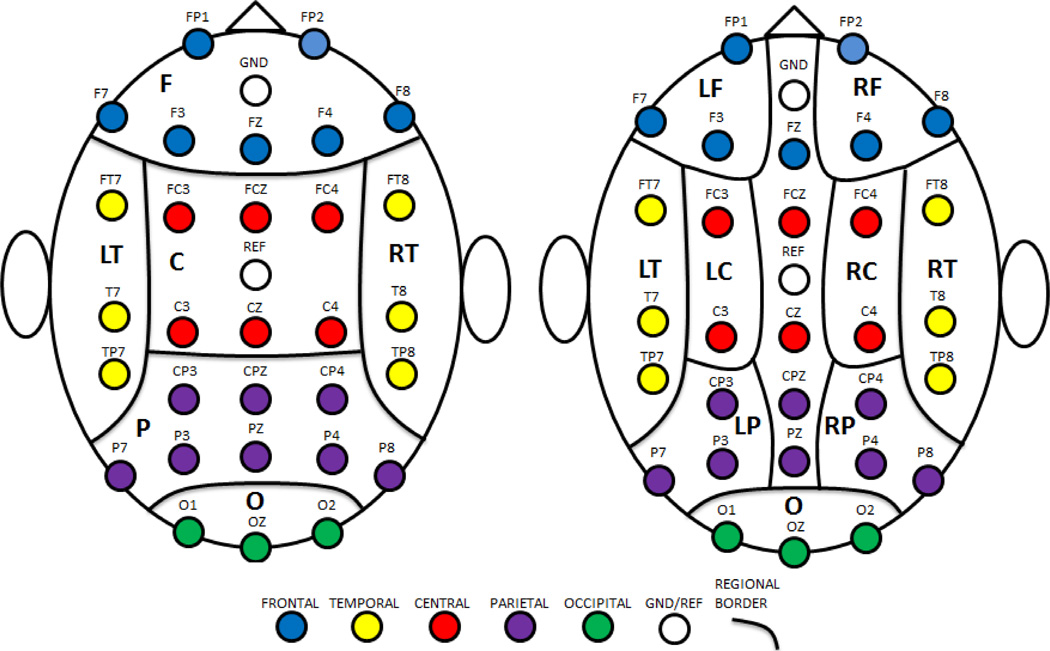
The 24 features described above and presented in Table 3 were calculated for the first 2 minutes of data for each protocol condition. Features were computed for each channel used in analyses. The 30 channels were grouped into 12 scalp regions based on their arrangement and location on the scalp. The regions included: (1) left frontal (LF); (2) right frontal (RF); (3) frontal (F = LF + RF + channel FZ); (4) left temporal (LT); (5) right temporal (RT); (6) left central (LC); (7) right central (RC); (8) central (C = LC + RC + channels FCZ and CZ); (9) left parietal (LP); (10) right parietal (RP); (11) parietal (P = LP + RP + channels CPZ and PZ); and (12) occipital (O). Note that left and right regions do not include central line channels; see Fig. 1 for regional boundaries. A global region (G) comprised of all channels was also considered. Each of the 24 features was averaged over each region (including G) for a total of 13 regional average feature groups.
Fig. 1.
Regional Boundaries. Left: major regions; right: subregions. LF=left frontal; RF=right frontal; F=frontal; LT=left temporal; RT=right temporal; LC=left central; RC=right central; C=central; LP=left parietal; RP=right parietal; P=parietal; O=occipital.
Feature Selection
The three groups of participants (NC, MCI, and AD) provided three binary discrimination problems: (1) MCI vs. NC, (2) AD vs. NC, and (3) MCI vs. AD. Twenty-four features were computed for each of the 13 regions described above for each subject for each protocol condition. Feature selection was performed as follows. For each discrimination problem and each condition, combinations of up to eight features were tested using support vector machine (SVM) functions in MATLAB™ [39]. Quadratic kernel functions were used in all discriminations and the cost coefficient was held constant at unity. To help avoid overfitting, nested leave-one-out cross-validation loops were used to suggest and test different combinations of features. The inner loop was used to generate a list of suggested combinations of features using a forward, supervised, high-score feature selection method where combinations were scored using leave-one-out cross-validation accuracy of SVM model predictions based on a smaller, randomized, subset of records [24]. The outer loop determined the leave-one-out cross-validation accuracy of the combinations of features suggested by the inner loop for all available records. The contribution of individual features was then assessed based on how often they appeared in the 200 best performing feature combinations tested in the outer loop simulations. Ultimately, the eight features that appeared most often were selected for each protocol condition and binary classifier.
Statistical Significance of Binary Classifiers
The statistical significance of the leave-one-out cross-validation accuracies of the binary classifiers was assessed using Monte Carlo permutation testing [40]. Specifically, a random sample of 10,000 permutations of shuffled labels indicating groups (NC, MCI, or AD) was used to estimate a 95% confidence interval for the probability that the leave-one-out cross-validation accuracies obtained were due to chance. The p-values presented were determined using this method.
Three-way Classification via Binary Classifiers
For each protocol condition, a three-way classification scheme was constructed using the pairwise coupling approach proposed by Hastie and Tibshirani [41]. For a given record, each binary SVM classifier was trained using all other available records and was applied to the given record. If two out of three of the SVM binary classifiers classified a record as belonging to class i, then the final decision of the classifier was to classify the record as belonging to class i. If instead, each of the three binary classifiers classified the record as a different one of the three possible classes, then the record was categorized as undetermined. For each undetermined record, the probability of that record belonging to each class, pi, i = 1, 2, 3, was then estimated. The final decision for an undetermined record was chosen as the class corresponding to the largest probability, argmaxi(pi)[41].
RESULTS
For each protocol condition, the selected features for each binary classifier were selected for further analyses. Two-sample Student t-distribution tests (unequal variance) were performed on group means of individual features in order to determine whether linear group differences based on the given samples were great enough to statistically infer differences in the groups’ populations. The results of the t-distribution tests thus provided a measure of linear separability of groups for individual features. Features for which significant linear separation existed, and the significance of the separability, are indicated in Table 4. Given the small sample sizes, such inference required large differences between group means and small variation within groups. It should be noted that such tests are dependent on the assumption of representative samples.
Table 4.
Group Comparisons of Selected Features for Binary Classifiers
| Condition | MCI vs. NC | AD vs. NC | AD vs. MCI | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
Selected Features |
Group Means |
Selected Features |
Group Means |
Selected Features |
Group Means |
||||
|
Resting (eyes open) |
MCI>NC | AD<NC | F-(Ptotal)′ | AD>MCI*** | |||||
| C-Ptotal | MCI>NC*** | RF-fmed | AD<NC | LF-(Ssamp)′ | AD>MCI** | ||||
| RC-Ssamp | MCI<NC | F-Ssamp | AD>NC | AD<MCI | |||||
| RF-fmed | MCI<NC | F-(Ptotal)′ | AD>NC*** | AD<MCI | |||||
| RF-(fmed)′ | MCI<NC*** | AD>NC | LT-fmed | AD<MCI | |||||
| MCI<NC** | AD<NC | AD>MCI | |||||||
| RP-(Ptotal)′ | MCI<NC | LT-C | AD>NC* | AD>MCI | |||||
| G-Sspec | MCI<NC | LP-(Sspec)′ | AD<NC** | RT-Ptotal | AD>MCI*** | ||||
| acc. (sens., spec.): | acc. (sens., spec.): | acc. (sens., spec.): | |||||||
| 96.8% (93.8%, 100%) | 84.4% (88.2%, 80.0%) | 90.9% (82.4%, 100%) | |||||||
| 95% CI for p-value: | 95% CI for p-value: | 95% CI for p-value: | |||||||
| (0.0002, 0.0014) | (0.0021, 0.0043) | (0.0094, 0.0136) | |||||||
|
Selected Features |
Group Means |
Selected Features |
Group Means |
Selected Features |
Group Means |
||||
|
Counting (eyes closed) |
MCI<NC | AD<NC | AD>MCI | ||||||
| C-Ptotal | MCI>NC*** | F-Ptotal | AD>NC*** | AD>MCI | |||||
| MCI>NC | F-A | AD>NC*** | RC-Ptotal | AD>MCI*** | |||||
| C-A | MCI>NC*** | LF-Ptotal | AD>NC*** | RC-A | AD>MCI*** | ||||
| LC-Ptotal | MCI>NC*** | AD>NC | RC-Ssamp | AD<MCI | |||||
| RC-R3 | MCI>NC | RF-(Ptotal)′ | AD>NC*** | AD<MCI | |||||
| MCI<NC** | LT-R3 | AD>NC | O-CLZ | AD<MCI | |||||
| O-(fmed)′ | MCI<NC*** | P-R1 | AD>NC | P-A | AD>MCI*** | ||||
| acc. (sens., spec.): | acc. (sens., spec.): | acc. (sens., spec.): | |||||||
| 83.9% (93.8%, 73.3%) | 96.9& (100%, 93.3%) | 90.9% (88.2%, 93.8%) | |||||||
| 95% CI for p-value: | 95% CI for p-value: | 95% CI for p-value: | |||||||
| (0.0011, 0.0029) | (0, 0.0003) | (0.0049, 0.0081) | |||||||
|
Selected Features |
Group Means |
Selected Features |
Group Means |
Selected Features |
Group Means |
||||
|
Resting (eyes closed) |
C-R1 | MCI>NC | C-Ptotal | AD>NC*** | C-Ptotal | AD>MCI*** | |||
| C-(Ssamp)′ | MCI<NC*** | C-R1 | AD>NC | LC-Ptotal | AD>MCI*** | ||||
| LC-Ptotal | MCI>NC*** | C-R2 | AD>NC | AD<MCI | |||||
| MCI<NC | C-A | AD>NC*** | AD<MCI | ||||||
| LF-R2 | MCI<NC | LC-Ptotal | AD>NC*** | AD<MCI | |||||
| LT-R2 | MCI>NC | RC-Ptotal | AD>NC*** | AD>MCI | |||||
| LP-Ptotal | MCI<NC*** | O-Ssamp | AD<NC | LT-A | AD>MCI*** | ||||
| MCI>NC*** | RT-(Ptotal)′ | AD<NC*** | LP-(Ssamp)′ | AD>MCI | |||||
| acc. (sens., spec.): | acc. (sens., spec.): | acc. (sens., spec.): | |||||||
| 93.6% (100%, 86.7%) | 71.9% (64.7%, 80.0%) | 87.9% (88.2%, 87.5%) | |||||||
| 95% CI for p-value: | 95% CI for p-value: | 95% CI for p-value: | |||||||
| (0, 0.0003) | (0.0314, 0.0333) | (0.0035, 0.0063) | |||||||
p< 0.05
p<0.01,
p<0.001.
Feature designations are preceded by regional indices: LF=left frontal; RF=right frontal; F=frontal; LT=left temporal; RT=right temporal; LC=left central; RC=right central; C=central; LP=left parietal; RP=right parietal; P=parietal; O=occipital; G=global. See Table 2 for feature designations; see Fig. 1 for regional boundaries.
Leave-one-out binary classification accuracies were computed for each binary classifier for each protocol condition. In each case, all eight of selected features listed in Table 4 were employed in combination using a quadratic SVM model. The results are summarized in Table 4 where 95% confidence intervals for corresponding p-values of the resulting leave-one-out cross-validation accuracies were assessed using Monte Carlo permutation testing.
MCI vs. NC Binary Classification
For both the eyes open, resting protocol condition and the counting task, MCI patients demonstrated a significant increase in total power in the central region (C-Ptotal) compared to normal controls. While resting with eyes open, the MCI group’s data show a decreased in alpha and beta activity in the first derivative for the right frontal (RF) and parietal (P) regions, as evidenced by a significant decrease in the median frequency in the right frontal region (RF-fmed) and peak alpha frequency in the parietal region (). A significant decrease in the alpha and beta activity of the first derivative was also observed in the right frontal region (decreased RF-(fmed)′) during the counting task. MCI participants also demonstrated a significant increase in the activity measure (A) in the central region (C) during the counting task. When resting with eyes closed, MCI participants exhibited increased theta activity in the central (C) and left temporal (LT) regions coupled with decreased higher beta band activity in the left frontal (LF) region compared to NC participants, although these differences were not substantial enough to allow statistical inferences regarding population trends. Significant differences observed for MCI participants while resting with eyes closed included a decrease in the sample entropy of the first derivative (C-(Ssamp)′), a decrease in total power in the left parietal regions (LP-Ptotal), and an increase in gamma band activity on a global level ().
Leave-one-out cross-validation discrimination accuracies of 96.8% (p<0.0014), 83.9% (p<0.0029), and 93.6% (p<0.0003) were achieved for MCI vs. NC discrimination for the eyes open condition, counting task, and eyes closed while resting conditions, respectively.
AD vs. NC Binary Classification
A leave-one-out cross-validation accuracy of 84.4% (p<0.0043) was achieved for AD vs. NC discrimination during the eyes open, resting condition. Group differences in selected features indicate a significant increase in the total power of the first derivative in the frontal region (F-(Ptotal)′) and a decrease in spectral entropy of the first derivative in the left parietal region (LP-(Sspec)′) for AD participants. AD participants also demonstrated an increase in complexity in the left temporal region (LT-C). Increases in theta activity and decreases in alpha and beta activity were also observed in the frontal (F) regions; however, groups were not linearly separable at a significant level for these features.
While counting backwards with eyes closed, AD patients were observed to have a significant increase in total power (Ptotal) and activity (A) in the frontal region (F) compared to normal controls, including a significant increase in total power of the first derivative in the right frontal region (RF-(Ptotal)′). Increases in theta activity were also observed in the left temporal (LT) and parietal (P) regions; however group differences were not significantly linearly separable. A leave-one-out cross-validation accuracy of 96.9% (p-value<0.0003) was achieved for AD vs. NC discrimination during the counting task.
For resting with eyes closed, the AD group data show significant increases in the total power (LC-Ptotal) and activity (C-A) in the central region compared to normals. In addition, AD participants were also observed to have a significant decrease the total power of the first derivative in the right temporal region (RT-(Ptotal)′). Increases in theta activity in the central (C) region and a decrease in sample entropy in the occipital (O) region were also observed in AD data, although differences were not linear. A leave-one-out cross-validation accuracy of only 71.9% (p-value<0.0333) was achieved for the eyes closed, resting condition.
AD vs. MCI Binary Classification
Differences between MCI and AD patients while resting with eyes open included a significant increase in total power in the right temporal region (RT-Ptotal), total power of the first derivative in the frontal region (F-(Ptotal)′), and sample entropy of the first derivative in the left frontal region (LF-(Ssamp)′) for AD participants. In addition, AD participants were observed to have lower gamma activity in the right frontal (RF) and left temporal (LT) regions, decreased alpha and beta activity in the left temporal (LT) region, and increased alpha and beta activity in the right parietal (P) and occipital (O) regions. These differences, however, did not significantly deviate in a linear manner from patterns observed for MCI participants. During the counting task, AD participants demonstrated significant increases in total power in the right central region (RC-Ptotal) and activity (A) in the right central (RC) and parietal (P) regions. Less significant differences included increased alpha activity in the central regions and decreases in alpha activity and complexity in the left frontal and occipital regions, respectively. While resting with eyes closed, AD participants again demonstrated significant increases in total power (Ptotal) in the central (C) regions. This was accompanied by a significant increase in activity in the left temporal region (LT-A). AD participants also had decreased beta activity in the right central (RC) and frontal (F) regions.
Leave-one-out cross-validation discrimination accuracies of 90.9% (p<0.0136), 90.9% (p<0.0081), and 87.9% (p<0.0063) were achieved for AD vs. MCI discrimination for the eyes open condition, counting task, and eyes closed while resting conditions, respectively.
Three-way Classification Results
Three-way classification results for eye open condition are presented in Table 5. Of those records classified as MCI, 100% were MCI; and of those records classified as AD, 81.3% were AD. Normal predictions were not as accurate, with only 70.6% of those records classified as being normal actually being normal. On the other hand, of all the true normal participants, 80.0% of them were accurately identified; of all the true MCI participants, 93.8% were accurately identified; and of all the true AD participants, 76.5% were accurately identified. Overall, the prediction accuracy was 83.3%.
Table 5.
Confusion Table for Resting, Eyes Open 3-Way Classification
| True Classes | |||||
|---|---|---|---|---|---|
| Predicted Classes | NC | MCI | AD | ||
| NC | 12 | 1 | 4 | 70.6% | |
| MCI | 0 | 15 | 0 | 100% | |
| AD | 3 | 0 | 13 | 81.3% | |
| 80.0% | 93.8% | 76.5% | Overall Acc.: 83.3% | ||
Three-way classification results for the counting task are presented in Table 6. Of those records classified as normal, 100% were normal; of those classified as MCI, 75% were MCI; and of those classified as AD, 88.2% were AD. On the other hand, of all the true normal participants, 73.3% of them were accurately identified; of all the true MCI participants, 93.8% were accurately identified; and of all the true AD participants, 88.2% were accurately identified. Overall, the prediction accuracy was 85.4%.
Table 6.
Confusion Table for Counting, Eyes Closed 3-Way Classification
| True Classes | |||||
|---|---|---|---|---|---|
| Predicted Classes | NC | MCI | AD | ||
| NC | 11 | 0 | 0 | 100% | |
| MCI | 3 | 15 | 2 | 75.0% | |
| AD | 1 | 1 | 15 | 88.2% | |
| 73.3% | 93.8% | 88.2% | Overall Acc.: 85.4% | ||
Three-way classification results for the resting with eyes closed condition are presented in Table 7. Of those records classified as normal, 78.6% were normal; of those classified as MCI, 82.4% were MCI; and of those classified as AD, 76.5% were AD. On the other hand, of all the true normal participants, 73.3% of them were accurately identified; of all the true MCI participants, 87.5% were accurately identified; and of all the true AD participants, 76.5% were accurately identified. Overall, the prediction accuracy was 79.2%. Among all the protocol conditions, the resting with eyes closed three-way classifier performed most poorly in overall accuracy. This is not surprising, however, given the resting with eyes closed condition resulted in the poorest binary classifier performance for distinguishing AD from normal or MCI participants.
Table 7.
Confusion Table for Resting, Eyes Closed 3-Way Classification
| True Classes | |||||
|---|---|---|---|---|---|
| Predicted Classes | NC | MCI | AD | ||
| NC | 11 | 0 | 3 | 78.6% | |
| MCI | 2 | 14 | 1 | 82.4% | |
| AD | 2 | 2 | 13 | 76.5% | |
| 73.3% | 87.5% | 76.5% | Overall Acc.: 79.2% | ||
DISCUSSION
In this study, regional spectral and complexity features of scalp EEG recorded during resting state conditions and a simple cognitive task of counting backwards by ones are explored as potential biomarkers for discriminating among NC, MCI and AD groups. Resting state protocols and a simple cognitive task (i.e., counting backwards) eliminates the need for extra training of primary care personnel in administering complex cognitive tasks. We have also demonstrated good result from analyzing short data segments (two minutes), practical for a primary care setting.
Previous studies have explored various EEG features for the discrimination of MCI/AD from normal individuals. Several of these researchers have observed that features which appear to discriminate AD from NC well, do not necessarily perform well when applied to MCI data [4],[5],[26]. While it would be convenient to have features that operate with normals at one extreme value, AD at the other extreme, and MCI somewhere in the middle, thus making the three groups clearly and linearly separable with a clear linear pattern of progression, that is often not the case. Instead, MCI individuals often appear to be spread across boundaries that otherwise clearly discriminate AD from normal participants. A different set of features may, therefore, be required for discriminating between MCI vs. NC, AD vs. NC, and MCI vs. AD [42].
Three generally accepted trends in the literature for MCI/AD discrimination via EEG are: (1) a shift in the power spectrum toward the lower frequencies (delta and theta band) coupled with a decrease in alpha and beta band activity; (2) decreased complexity; and (3) decreased synchronization [17],[23],[22],[43]. These trends, however, are generally reported as global trends observed during eyes closed, resting protocol conditions. Trends in the complexity and spectral power distribution of the first derivative of EEG for MCI and AD are less well established. Furthermore, patterns may differ at different parts of the brain. The features selected to serve as discriminating features for the binary classifiers were examined for trends between groups. For the eyes closed, resting condition, we have consistent results (i.e., decreased complexity, decreased alpha and beta band activity), however these trends were not statistically significant for our participants.
As stated in [44], evidence of the diagnostic utility of resting EEG in dementia and mild cognitive impairment (MCI) is still not sufficient to establish this method for the initial evaluation of subjects with cognitive impairment in the routine clinical practice. In addition, AD EEG studies have been done at the group level. Much work has been done showing group differences, i.e. AD brains differ from those of the normal brains, but differences were not sufficient to categorize individuals. In this work, participants are classified at the individual level, which is the case when a patient comes into a doctor’s office. Individualized analysis allows these EEG markers to be tested against known biomarkers for Alzheimer's disease and with cognitive tests. It is our goal to be able to use easy neural indicators to diagnose and predict cognitive decline pre-clinically, such as among people with subjective memory complaints.
Dementia can cause different neurological changes in different individuals–especially in the case of more enigmatic diagnoses such as MCI [45],[46]. Such differences may lead to the failure in the generalization of globally-averaged spectral measures to other data sets. While most of the features employed have been used before on either a global or individual channel basis, we examine how the features' values change on a topographical regional basis. The topographical regions do not necessarily correspond to the cortical location of the source of the brain activity; e.g., activity recorded at a central electrode site may reflect a source from the frontal cortex. One goal of this work was to discover regional differences in common EEG features to be potential indicators for cognitive degeneration. In addition, we examined the discriminating power of different features for different protocols, including resting protocols—eyes open and eyes closed—and a simple cognitive task.
This study explores differences in common spectral and complexity measures of EEG on a regional and global basis for the discrimination of preclinical dementia from the normal aging condition. The successful discrimination between the three groups of EEG records (NC, MCI, and AD) are the result of differences in regional electrical activity in specific frequency bands in resting states and during a simple cognitive task. The current work does not distinguish between different subtypes of MCI such as single- vs. multiple-domain deficits. Such questions are potential topics for future research.
The results for the three-way discrimination presented here are encouraging, especially those for MCI, as detection of MCI at the earliest possible stage is currently of great interest in the field. Precisions of 100%, 75%, and 82.4% were achieved for MCI participants for the eyes open condition, counting task, and eyes closed condition, respectively. The highest precisions for NC and AD participants were achieved during the counting task, which suggests that a simple cognitive task may be a reliable protocol for detection of characteristic EEG differences associated with AD. The three-way classification scheme presented here allows for the differentiation of the three groups without a priori knowledge that individuals fall into two of the three groups. One limitation of this study is the small number of participants. Future work will increase the sample size, and examine our EEG indicators against AD biomarkers (e.g., CSF) to test the robustness and generality of the results here. Future work will also examine whether longitudinal changes over relatively short time periods can be detected at the individual level.
ACKNOWLEDGEMENTS
Research was sponsored in part by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the US Department of Energy under Contract No. DE-AC05-00OR22725; by the NSF under grant numbers CMMI-0845753 and CMMI-1234155; and in part by the NIH under grants NIH P30 AG028383 to UK Sanders-Brown Center on Aging, NIH AG00986 to YJ, and NIH NCRR UL1RR033173 to UK Center for Clinical and Translational Science. The contributions to this paper by two of the authors (N. B. Munro and L. M. Hively) was prepared while acting in their own independent capacities and not on behalf of UT-Battelle, LLC, or its affiliates or successors, or Oak Ridge National Laboratory, or the US Department of Energy.
We deeply thank Dr. David Wekstein of the UK Alzheimer’s Research Center for his key role in getting the collaboration between ORNL and UK in place to make the pilot study possible. We thank A. Lawson, E. Walsh, J. Lianekhammy, S. Kaiser, C. Black, K. Tran, and L. Broster at the University of Kentucky for their assistance in data acquisition and database management, and Schmitt, F, Kryscio, R, and Abner, E at the Biostatics Core at the UK aging center for providing MMSE scores of some participants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K, Belleville S, Brodaty H, Bennett D, Chertkow H, Cummings JL, de Leon M, Feldman H, Ganguli M, Hampel H, Scheltens P, Tierney MC, Whitehouse P, Winblad B. Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- 2.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Archives of Neurology. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 3.Wee CY, Tap PT, Li W, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Enriched white matter connectivity networks for accurate identification of MCI patients. NeuroImage. 2011;54:1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iqbal K, Alonso AC, Chen S, Chohan MO, El-Akkad E, Gong CX, Khatoon S, Li B, Liu F, Rahman A, Tanikuai H, Grundke-Iqbal I. Tau pathology in Alzheimer’s disease and other tauopathies . Biochimica et Biophysica Acta. 2005;1793:198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Wong DF, Rosenberg PB, Zhou Y, Jumar A, Raymont V, Ravert HT, Dannals RF, Nandi A, Brasić JR, Ye W, Hilton J, Lyketsos C, Kung HF, Joshi AD, Skovronsky DM, Pontecorvo MJ. In vivo imaging of amyloid deposition in Alzheimer’s disease using the novel radioligand [18F]AV-45 (florbetapir F 18) Journal of Nuclear Medicine. 2010;51:913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossini PM, Del Percio C, Pasqualetti P, Cassetta E, Binetti G, Dal Forno G, Ferreri F, Frisoni G, Chiovenda P, Miniussi C, Paris L, Tombini M, Vecchio F, Babiloni C. Conversion from mild cognitive impairment to Alzheimer’s disease is predicted by source and coherence of brain electroencephalography rhythms. Neuroscience. 2006;143:793–803. doi: 10.1016/j.neuroscience.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt FA, Nelson PT, Abner E, Scheff S, Jicha GA, Smith C, Cooper G, Mendiondo M, Danner DD, Van Eldik LJ, Caban-Holt A, Lovell MA, Kryscio RJ. University of Kentucky Sanders-Brown healthy brain aging volunteers: donor characteristics, procedures and neuropathology. Current Alzheimer Research. 2012;9:724–733. doi: 10.2174/156720512801322591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anoop A, Singh PK, Jacob RS, Maji SK. CSF biomarkers for Alzheimer’s disease diagnosis. International Journal of Alzheimer’s Disease. 2010;2010:1–12. doi: 10.4061/2010/606802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zetterberg H, Mattsson N, Blennow K. Cerebrospinal fluid analysis should be considered in patients with cognitive problems. International Journal of Alzheimer’s Disease. 2010;2010:1–4. doi: 10.4061/2010/163065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, Coart E, Hansson O, Minthon L, Zetterberg H, Blennow K, Shaw L, Trojanowksi JQ. Diagnosis-independent Alzheimer’s disease biomarker signature in cognitively normal elderly people. Archives of Neurology. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Aykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toqa AW, Trojanowksi JQ. The Alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimer’s & Dementia. 2012;8:S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikonomovic MD, Klunk WE, Abrahamson EE, Mathis CA, Price JC, Tsopelas ND, Lopresti BJ, Ziolko S, Bi W, Paljug WR, Debnath ML, Hope CE, Isanski BA, Hamilton RL, DeKosky ST. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer’s disease. Brain. 2008;131:1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jack CR, Jr, Lowe VJ, Senjem ML, Wigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of AD and amnestic MCI. Brain. 2008;131:665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartzokis G, Lu PH, Mintz J. Quantifying age-related myelin breakdown with MRI: novel therapeutic targets for preventing cognitive decline and Alzheimer’s disease. Journal of Alzheimer’s Disease. 2004;6:S53–S59. doi: 10.3233/jad-2004-6s604. [DOI] [PubMed] [Google Scholar]
- 15.Olichney JM, Yang J, Taylor J, Kutas M. Cognitive event-related potentials: biomarkers of synaptic dysfunction across the stages of Alzheimer’s disease. Journal of Alzheimer’s Disease. 2011;26:215–228. doi: 10.3233/JAD-2011-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenig T, Prichep L, Dierks T, Huhl D, Wahlund LO, John ER, Jelic V. Decreased EEG synchronization in Alzheimer’s disease and mild cognitive impairment. Neurobiology of Aging. 2005;26:165–171. doi: 10.1016/j.neurobiolaging.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Stam CJ, Montex T, Jones BF, Rombouts SA, van der Made Y, Pijnenburg YA, Scheltens P. Disturbed fluctuations of resting state EEG synchronization in Alzheimer’s disease. Clinical Neurophysiology. 2005;116:708–715. doi: 10.1016/j.clinph.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Brenner RP, Ulrich RF, Spiker DG, Sclabassi RJ, Reynolds 3rd CF, Marin RS, Boller F. Computerized EEG spectral analysis in elderly normal, demented and depressed subjects. Electroencephalography and Clinical Neurophysiology. 1986;64:483–492. doi: 10.1016/0013-4694(86)90184-7. [DOI] [PubMed] [Google Scholar]
- 19.Jeong J. EEG dynamics in patients with Alzheimer’s disease. Clinical Neurophysiology. 2004;115:1490–1505. doi: 10.1016/j.clinph.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Signorino M, Pucci E, Belardinelli N, Nolfe G, Angeleri F. EEG spectral analysis in vascular and Alzheimer’s dementia. Electroencephalography and Clinical Neurophysiology. 1995;94:313–325. doi: 10.1016/0013-4694(94)00290-2. [DOI] [PubMed] [Google Scholar]
- 21.Soininen H, Partanen J, Laulumaa V, Helkala EL, Laasko M, Riekkinen PJ. Longitudinal EEG spectral analysis in early stage of Alzheimer’s disease. Electroencephalography and Clinical Neurophysiology. 1989;72:290–297. doi: 10.1016/0013-4694(89)90064-3. [DOI] [PubMed] [Google Scholar]
- 22.Stam CJ, van der Made Y, Pijnenburg YA, Scheltens P. EEG synchronization in mild cognitive impairment and Alzheimer’s disease. Acta Neurologica Scandinavica. 2003;108:90–96. doi: 10.1034/j.1600-0404.2003.02067.x. [DOI] [PubMed] [Google Scholar]
- 23.Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Small-world networks and functional connectivity in Alzheimer’s disease. Cerebral Cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- 24.Besthorn C, Cursi M, Liberati D, Franceschi M, Comi G. EEG coherence in Alzheimer’s disease. Electroencephalography and Clinical Neurophysiology. 1993;90:242–245. doi: 10.1016/s0013-4694(97)00129-6. [DOI] [PubMed] [Google Scholar]
- 25.Hermann CS, Demiralp T. Human EEG gamma oscillations in neuropsychiatric disorders. Clinical Neurophysiology. 2005;116:2719–2733. doi: 10.1016/j.clinph.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Huang C, Wahlund L, Dierks T, Julin P, Winblad B, Jelic V. Discrimination of Alzheimer’s disease and mild cognitive impairment by equivalent EEG sources: a crosssectional and longitudinal study. Clinical Neurophysiology. 2000;111:1961–1967. doi: 10.1016/s1388-2457(00)00454-5. [DOI] [PubMed] [Google Scholar]
- 27.Jelic V, Johansson SE, Almkvist O, Shigeta M, Julin P, Nordberg A, Winblad B, Wahlund LO. Quantitative electroencephalography in mild cognitive impairment: longitudinal changes and possible prediction of Alzheimer’s disease. Neurobiology of Aging. 2000;21:533–540. doi: 10.1016/s0197-4580(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 28.Prichep LS, John ER, Ferris SH, Reisberg B, Almas M, Alper K, Cancro R. Quantitative EEG correlates of cognitive deterioration in the elderly. Neurobiology of Aging. 1994;15:85–90. doi: 10.1016/0197-4580(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 29.He B. Neural signal processing, in Neural Engineering. New York, NY: Kulwer Academic/Plenum Publishers; 2005. pp. 193–321. [Google Scholar]
- 30.Snaedal J, Johannesson GH, Gudmundsson TE, Gudmundsson S, Pajdak TH, Johnsen K. The use of EEG in Alzheimer’s disease, with and without scopolamine-a pilot study. Clinical Neurophysiology. 2010;121:836–841. doi: 10.1016/j.clinph.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 31.McBride J, Zhao X, Munro N, Smith C, Jicha G, Jiang Y. Resting EEG Discrimination of early stage Alzheimer's disease from normal aging using inter-channel coherence network graphs. Annals of Biomedical Engineering. 2013;41:1233–1242. doi: 10.1007/s10439-013-0788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride J, Zhao X, Nichols T, Vagnini V, Munro N, Berry D, Jiang Y. Scalp EEG-based discrimination of cognitive deficits after traumatic brain injury using eventrelated tsallis entropy analysis. IEEE Transaction on Biomedical Engineering. 2013;60:90–96. doi: 10.1109/TBME.2012.2223698. [DOI] [PubMed] [Google Scholar]
- 33.Zavala-Fernandez H, Orglmeister R, Trahms L, Sander TH. Identification enhancement of auditory evoked potentials in EEG by epoch concatenation and temporal decorrelation. Computer Methods and Programs in Biomedicine. 2012;108:1097–1105. doi: 10.1016/j.cmpb.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Siadat M-R, Soltanian-Zadeh H, Fotouhi F, Elisevich K. Content-based image database system for epilepsy. Computer Methods and Programs in Biomedicine. 2005;79:205–226. doi: 10.1016/j.cmpb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Shah EN, Reddy NP, Rothschild BM. Fractal analysis of acceleration signals from patients with CPPD, rheumatoid arthritis, and spondyloarthroparthy of the finger joint. Computer Methods and Programs in Biomedicine. 2005;77:233–239. doi: 10.1016/j.cmpb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Gómez C, Hornero R, Abásolo D, Fernández A, Escudero J. Analysis of the magnetoencephalogram background activity in Alzheimer's disease patients with auto-mutual information. Computer Methods and Programs in Biomedicine. 2007;87:239–247. doi: 10.1016/j.cmpb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Poza J, Hornero R, Abásolo D, Fernández A, Mayo A. Evaluation of spectral ratio measures from spontaneous MEG recordings in patients with Alzheimer's disease. Computer Methods and Programs in Biomedicine. 2008;90:137–147. doi: 10.1016/j.cmpb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Rossini PM, Parisi L. ch. 6. In: Abou-Saleh MT, Katona C, Kumar A, editors. Neurophysiology of the ageing brain, in Principles and Practice of Geriatric Psychiatry. 3rd ed. Chichester, UK: John Wiley & Sons, Ltd.; 2011. [Google Scholar]
- 39.MathWorks. [Accessed 5 May. 2012];MATLAB. Available at mathworks.com/products/matlab.
- 40.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human Brain Mapping. 2001;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hastie T, Tibshirani R. Classification by pairwise coupling. Annals of Statistics. 1998;26:451–471. [Google Scholar]
- 42.Baker M, Akrofi K, Schiffer R, O'Boyle MW. EEG patterns in mild cognitive impairment (MCI) patients. The Open Neuroimaging Journal. 2008;2:52–55. doi: 10.2174/1874440000802010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dauwels J, Vialatte F, Cichocki A. Diagnosis of Alzheimer’s disease from EEG signals: where are we standing. Current Alzheimer’s Research. 2010;7:487–505. doi: 10.2174/156720510792231720. [DOI] [PubMed] [Google Scholar]
- 44.Jelic V, Kowalski J. Evidence-based evaluation of diagnostic accuracy of resting EEG in dementia and mild cognitive impairment. Clinical EEG Neuroscience. 2009;40:129–142. doi: 10.1177/155005940904000211. [DOI] [PubMed] [Google Scholar]
- 45.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Archives of Neurology. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 46.Petersen RC. Mild cognitive impairment as a diagnostic entity. Journal of Internal Medicine. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]