Abstract
Background
Abnormalities in pyloric development or in contractile function of the pylorus cause reflux of duodenal contents into the stomach and increase the risk of gastric metaplasia and cancer. Abnormalities of the pyloric region are also linked to congenital defects such as the relatively common neonatal hypertrophic pyloric stenosis, and primary duodenogastric reflux. Therefore, understanding pyloric development is of great clinical relevance. Here, we investigated the role of the LIM homeodomain transcription factor Isl1 in pyloric development.
Results
Examination of Isl1 expression in developing mouse stomach by immunohistochemistry, whole mount in situ hybridization and real-time quantitative PCR demonstrated that Isl1 is highly expressed in developing mouse stomach, principally in the smooth muscle layer of the pylorus. Isl1 expression was also examined by immunofluorescence in human hypertrophic pyloric stenosis where the majority of smooth muscle cells were found to express Isl1. Isl1 function in embryonic stomach development was investigated utilizing a tamoxifen-inducible Isl1 knockout mouse model. Isl1 deficiency led to nearly complete absence of the pyloric outer longitudinal muscle layer at embryonic day 18.5, which is consistent with Gata3 null mouse phenotype. Chromatin immunoprecipitation, luciferase assays, and electrophoretic mobility shift assays revealed that Isl1 ensures normal pyloric development by directly targeting Gata3.
Conclusions
This study demonstrates that the Isl1-Gata3 transcription regulatory axis is essential for normal pyloric development. These findings are highly clinically relevant and may help to better understand pathways leading to pyloric disease.
Keywords: α-smooth muscle actin, Gata3, Isl1, Pylorus
Background
The vertebrate gut is a remarkable structure that ingests and digests food, absorbs nutrients, and removes waste products. The gut originates from a simple tubular structure composed of three germ layers including an underlying endoderm, a surrounding splanchnic mesoderm, and an ectoderm [1-3]. In mouse embryos, the gut becomes patterned along the anterior-posterior, dorsal-ventral, left-right, and radial axes. The gut tube consists of the foregut, midgut, and hindgut along its anterior-posterior axis [4,5]. As development progresses, the foregut gives rise to the esophagus, stomach, liver, lungs, and pancreas. The midgut forms the small intestine and the hindgut develops into the large intestine [1,5-8].
The stomach is derived from the posterior foregut. The stomach morphologically differentiates from the foregut tube around embryonic day 9.5 (E9.5) and the expansion of the pre-gastric mesenchyme allows the domain of the stomach to be visible beginning at E10.5 [9]. Mesenchymal cells of stomach differentiate into four distinct concentric layers, including lamina propria, muscularis mucosae, and circular and longitudinal smooth muscle at different stages of embryonic development [10]. By E11.5, the stomach is distinctly enlarged. The stomach smooth muscle differentiates at E13, with a distinct layer of α-smooth muscle actin (α-SMA)-positive cells appearing and a circular muscle layer forming throughout the stomach [11]. The smooth muscle layer thickens in the constricted prospective pyloric sphincter region at about E14.5 [2,9]. At E18.5, the pyloric sphincter begins to function in preventing the reflux of duodenal contents into the stomach [9].
The posterior or pylorus portion of the stomach is the anatomical junction between the stomach and the duodenum. At the terminus of the pylorus, the distinct valvular flaps of the pyloric sphincter can be seen [2]. Under normal physiological conditions, the stomach depends on its peristaltic contraction to grind and thrust the partially digested food, and the pylorus relies on its thickened pyloric sphincter to control the flow of food into the small intestine. Abnormalities in pyloric development or in the contractile function of the pylorus cause reflux of duodenal contents into the stomach and increase the risk of gastric metaplasia and cancer [12,13]. Abnormalities of the pylorus are related to congenital defects [14-16]. Therefore, much attention has been given to the regulating elements and pathways of stomach development, especially pylorus and pyloric sphincter development.
Previous data in chick suggested that bone morphogenetic protein (BMP) signaling regulates mesenchymal expression of Nkx2.5 and Sox9, which affects the character of the pyloric epithelium but has no effect on pyloric smooth muscle [5,17], suggesting that mesenchymal signaling by unknown factors affects the pyloric epithelial phenotype. In the mouse, molecular mechanisms of pyloric formation are little understood, with relatively few of the factors required for normal pyloric development having been identified. Those that have been include Sox9[17], Six2[9], Bapx1[18], Nkx2.5[3,17], Gremlin[9], and Gata3[19,20]. Ablation of the homeodomain transcription factor, Six2, expressed in posterior stomach, disrupts thickening of the pyloric smooth muscle layer and attenuates constriction of the pylorus sphincter. In addition, loss of Six2 eliminates Sox9 expression, and reduces Nkx2.5 and Gremlin expression in the pylorus, although this expression later recovers [9], suggesting that Six2, Sox9, Nkx2.5, and Gremlin are required for pyloric development. In addition, Nkx2.5, Sox9, and Gata3 are co-expressed in the dorsal pyloric outer longitudinal muscle (OLM) layer that matures between E14.5 and E16.5. Following deletion of Nkx2.5 or Gata3, the dorsal pyloric OLM is almost absent and constriction of the pylorus sphincter is attenuated [20].
The LIM homeodomain (LIM-HD) transcription factor Isl1 was originally found to function as an insulin gene enhancer binding protein [21]. Isl1 is comprised of two tandem LIM domains and a homeodomain. The homeodomain, with its helix-turn-helix structure, binds to regulatory DNA sequences of target genes, while the LIM domains are mainly involved in protein-protein interactions that regulate the activity of the LIM-HD [22]. Isl1 plays critical roles in cell determination, proliferation, and differentiation in the nervous system [23,24], heart [25], and pituitary gland [26]. In addition, Isl1 expression has been detected in gastric mesenchyme [27,28] and gastrointestinal epithelium in both embryonic and adult mice [29]. However, the role of Isl1 in stomach development has yet to be explored. In the present study, we examined Isl1 expression in the stomach. Isl1 was highly expressed in the posterior stomach in early stages of development and was mainly restricted to the smooth muscle cells of the pylorus. To examine Isl1 function in stomach development, we utilized a tamoxifen-inducible knockout mouse model. An inducible model was needed because Isl1-/- mutants die at approximately E10.5 owing to defects in heart formation. Our results show that Isl1 is vital for formation of the pyloric OLM layer during stomach organogenesis.
Results
Isl1 is expressed in embryonic mouse stomach
We examined Isl1 mRNA levels in embryonic mouse stomach by real-time quantitative PCR (RT-qPCR) and whole mount in situ hybridization (WISH). Isl1 mRNA was initially detected at E11.5 by RT-qPCR. Isl1 reached the highest level at E13.5, followed by a sharp decline at E14.5, and had no significant changes into adulthood (Additional file 1: Figure S1a). This result was similar to a previous report [29]. The localization of Isl1 mRNA expression was investigated using WISH. We performed Isl1 WISH in embryonic stomach at E11.5, E13.5, and E14.5. At E11.5, Isl1 was localized to the posterior half of the stomach (Additional file 1: Figure S1b). However, the Isl1 WISH signal was much stronger and condensed in the pylorus by E13.5 (Figure 1A). As stomach development progressed, the pylorus continued to express Isl1 and expression of Isl1 extended to the prospective pyloric sphincter at E14.5 (Additional file 1: Figure S1b). However, the Isl1 WISH signal weakened considerably from E13.5 to E14.5. These Isl1 WISH results concurred with Isl1 RT-qPCR results.
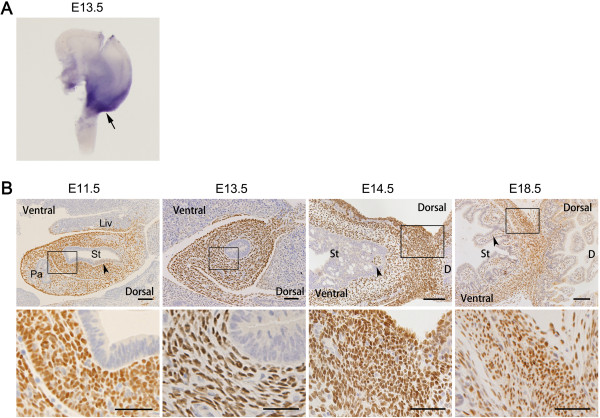
Figure 1.
Isl1 is expressed in developing mouse stomach. (A) Embryonic stomach WISH analysis demonstrated that Isl1 expression was limited to the pylorus at E13.5 (arrow). (B) Immunohistochemical staining for Isl1 in stomach. Sections from embryos were arranged in rostral to caudal sequence at E11.5 and E13.5, and Isl1 expression was mainly localized to the mesenchyme of the posterior stomach. At E14.5 and E18.5, Isl1 was highly expressed in smooth muscle cells of the pylorus, although there were some Isl1-positive cells in the lamina propria (arrowheads). Enlarged images in boxed regions are shown below original images. D, duodenum; Liv, liver; Pa, pancreas; St, stomach. Scale bars of original images: 100 μm; scale bars of enlarged images: 50 μm.
We then assessed Isl1 protein expression by immunohistochemistry. Results demonstrated that Isl1 was mainly localized to the posterior stomach mesenchyme from E11.5 to E13.5, then Isl1 was expressed in smooth muscle cells of the pylorus (Figure 1B and Additional file 1: Figure S1c), and was also detectable in the lamina propria (Figure 1B, arrowheads). In adult mouse stomach, only a few Isl1-positive cells were observed in the smooth muscle layer (Additional file 1: Figure S1c).
Isl1-positive cells are co-expressed with α-smooth muscle actin in embryonic mouse stomach
To see whether Isl1 expression was related to smooth muscle development of the pylorus, we examined the expression of Isl1 and the earliest smooth muscle marker α-SMA using immunofluorescence. Results demonstrated that the proportion of Isl1-positive cells expressing α-SMA gradually increased (Figure 2). At E11.5, no α-SMA-positive cells were detected, although Isl1 was highly expressed in the mesenchyme of the posterior stomach. At E14.5, a subset of Isl1-positive cells, mainly those in the inner circular muscle (ICM) of the pylorus, were α-SMA positive. By E16.5, pyloric OLM has already undergone differentiation [20]. At E18.5, the majority of Isl1-positive cells in the pylorus were α-SMA positive. Isl1 expression persisted in mature pyloric ICM and OLM, and lamina propria cells also expressed Isl1 (Additional file 1: Figure S2). In addition, Isl1 expression was examined in human samples of hypertrophic pyloric stenosis by immunofluorescence, and results demonstrated that Isl1 was also expressed in human smooth muscle cells of the pylorus (Additional file 1: Figure S3). Therefore, these results suggest that Isl1 may participate in the formation of pyloric sphincter.
Figure 2.
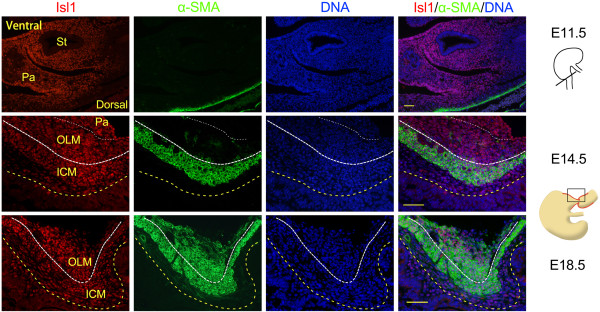
Double immunostaining for Isl1 and α-smooth muscle actin in mouse smooth muscle cells of the dorsal pylorus. Isl1 and α-SMA co-expression in smooth muscle cells at E11.5, E14.5, and E18.5. Yellow dotted lines mark the epithelial basement membrane, white thick dotted lines indicate ICM and OLM boundary, and white dotted lines indicate OLM and pancreas boundary. Red staining is Isl1, green staining is α-SMA, and DAPI nuclear counterstaining (DNA) is blue. α-SMA, α-smooth muscle actin; ICM, inner circular muscle; OLM, outer longitudinal muscle; Pa, pancreas; St, stomach. Scale bars: 50 μm.
Isl1 expression is effectively ablated in Isl1MCM/F-inducible knockout mice
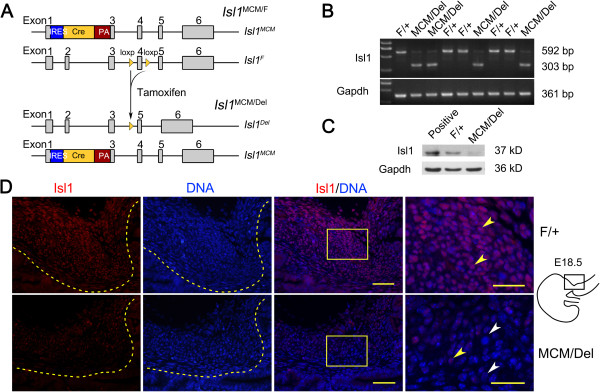
To investigate effects of Isl1 ablation on stomach development, we utilized Isl1MCM/F-inducible Cre (Isl1MCM/Del) mice (Figure 3A) and Isl1F/+mice were used as controls [30,31]. Embryos were genotyped by PCR at E18.5 (Additional file 1: Figure S4) and intact or mutant Isl1 mRNA was distinguished by semi-quantitative PCR (Figure 3B).
Figure 3.
Efficiency of Isl1 ablation in stomachs of Isl1MCM/Del mutant mouse stomachs at E18.5. (A) Tamoxifen-inducible Cre recombinase excised DNA sequences flanked by two loxP sites. (B)Isl1 RNA levels were ablated in Isl1MCM/Del mutant stomachs as seen by semi-quantitative PCR. Isl1F/+mice showed a 592 base pair product whereas Isl1MCM/Del mice generated a 303 base pair product. (C) Isl1 was significantly down-regulated at the protein levels in Isl1MCM/Del mutant stomachs as shown by western blot. Expression of embryos at E11.5 was used as positive control. (D) Isl1 protein expression in Isl1F/+and Isl1MCM/Del embryonic pylorus. Isl1 expression was significantly reduced in Isl1MCM/Del embryonic stomachs, as seen by immunofluorescence. Images in Isl1F/+and Isl1MCM/Del were processed on the same slide and photographed at the same exposure. Enlarged images of the boxed areas are shown on the right side of the merged pictures. Yellow arrowheads show representative Isl1-positive cells, and white arrowheads show representative Isl1-negative cells. Yellow dotted lines mark the epithelial basement membrane. Scale bars: 50 μm.
Western blot analyses showed that Isl1 protein levels in embryonic stomach of Isl1MCM/Del mice were significantly lower than those in Isl1F/+mice (Figure 3C). Immunofluorescence results demonstrated significantly reduced Isl1 staining in pylorus of Isl1MCM/Del mice as compared to controls (Figure 3D). These data demonstrate that Isl1 expression was effectively down-regulated in Isl1MCM/Del mutant stomachs.
Pyloric abnormalities in Isl1MCM/F mutants
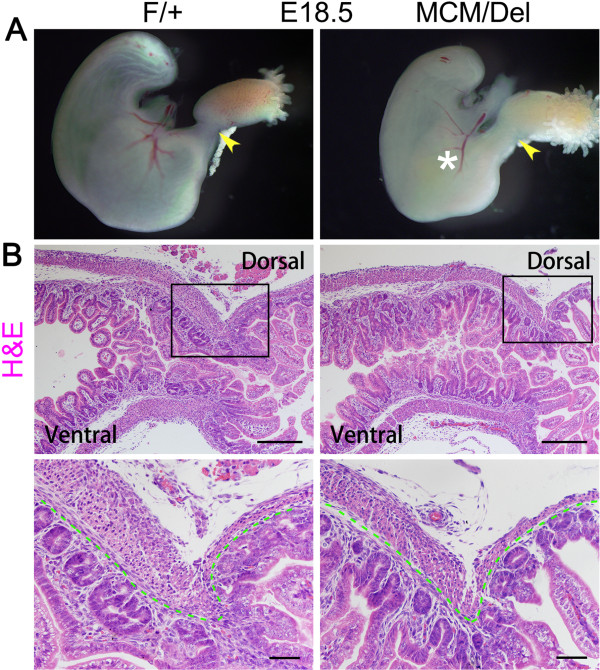
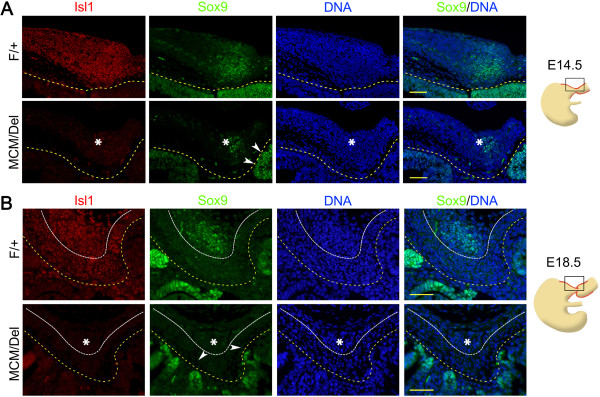
To investigate effects of Isl1 ablation on stomach development, we compared morphological and histological differences between Isl1MCM/Del and Isl1F/+stomachs at E18.5. At E18.5, yellow fluid was observed in Isl1MCM/Del stomachs but not in stomachs of Isl1F/+littermates (Figure 4A, asterisk). Histological examination demonstrated that the dorsal pyloric smooth muscle layer was much thinner in the pylorus of Isl1MCM/Del mice compared with controls (Figure 4B). We examined expression and distribution of α-SMA in both Isl1MCM/Del mutants and Isl1F/+pylorus. Immunofluorescence results demonstrated that Isl1 deficiency led to nearly complete absence of the pyloric OLM layer at E18.5, and remaining cells were loosely organized (Figure 5A, asterisks). In addition, constriction of the pyloric sphincter was attenuated in Isl1MCM/Del mutant stomachs when compared with constriction in Isl1F/+stomachs (Figure 5B). Furthermore, we analyzed expression of the smooth muscle specific protein Calponin-1 at E18.5, and immunofluorescence results demonstrated that loss of Isl1 also resulted in near absence of Calponin-1 expression in the dorsal pyloric OLM layer, similar to result with α-SMA (Additional file 1: Figure S5). Sox9 is expressed in both epithelium and mesenchyme [9] and is required for development of OLM and formation of pyloric sphincter constriction [20]. Our immunofluorescence results showed that Sox9 remained at normal levels in stomach epithelium of Isl1MCM/Del mice at E14.5 and E18.5 (Figure 6, arrowheads), but the area of pyloric smooth muscle expressing Sox9 was significantly reduced in Isl1MCM/Del mutants at E14.5 (Figure 6A, asterisks) and absent at E18.5 (Figure 6B, asterisks). Thus, Isl1 was required for Sox9 expression in dorsal pyloric OLM cells. These results indicate that Isl1 is critical for regulating development of mouse pyloric smooth muscle.
Figure 4.
Morphological and histological changes in developing stomach of Isl1MCM/Del mutants. (A) Gross and microscopic evidence for stomach defects in Isl1MCM/Del mice. Whole mount views at E18.5 in Isl1F/+and Isl1MCM/Del mouse stomachs. Isl1MCM/Del mutant stomachs lacked a functional pyloric sphincter (arrowhead), thereby allowing reflux of fluid as observed in mutant embryos. Yellow fluid is denoted by asterisk. (B) Hematoxylin and eosin staining of Isl1F/+and Isl1MCM/Del mouse pylorus at E18.5. The dorsal pyloric smooth muscle (black boxed region) was prominent in Isl1F/+embryos, but was much thinner in Isl1MCM/Del embryos. The remainder of the pylorus was histologically normal. Green dotted lines mark the epithelial basement membrane. Enlarged images in boxed regions are shown below original photos. Scale bars of original photos: 200 μm; scale bars of enlarged images: 50 μm. H&E, hematoxylin and eosin.
Figure 5.
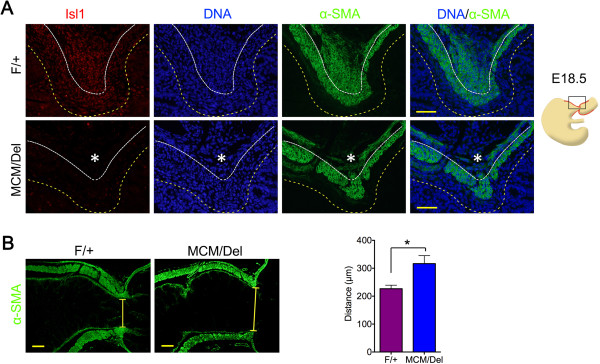
Loss of Isl1 disrupts formation of the dorsal pyloric outer longitudinal muscle. (A) Immunofluorescence of Isl1 and α-SMA in Isl1F/+and Isl1MCM/Del embryonic pylorus at E18.5. Loss of Isl1 resulted in nearly complete loss of α-SMA-positive cells in the dorsal pyloric OLM (asterisks). Yellow dotted lines mark the epithelial basement membrane and white dotted lines indicate ICM and OLM boundary. Red staining is Isl1, green staining is α-SMA, and DAPI nuclear counterstaining (DNA) is blue. Scale bars: 50 μm. (B) α-SMA immunofluorescence of Isl1F/+and Isl1MCM/Del embryonic pylorus at E18.5. Compared with Isl1F/+embryos, the pyloric sphincter constriction was wider in Isl1MCM/Del animals. Sphincter constriction measurements are shown. Yellow bars demarcate the pylorus and highlight the marked difference in width between Isl1F/+and Isl1MCM/Del samples. Data are mean ± SEM (n = 6 mice per group), *P <0.05 (Student’s t-test). Scale bars: 50 μm. α-SMA, α-smooth muscle actin; ICM, inner circular muscle; OLM, outer longitudinal muscle.
Figure 6.
Loss of Isl1 eliminates the dorsal pyloric outer longitudinal muscle Sox9 expression. (A) Double immunostaining for Isl1 and Sox9 in the dorsal pylorus at E14.5. In the absence of Isl1, the domain of mesodermal cells (asterisks) expressing Sox9 was smaller. (B) Double immunostaining for Isl1 and Sox9 in the dorsal pylorus at E18.5. Inducible Isl1 knockout effectively eliminated Isl1 expression, with concomitant loss of Sox9 expression in the dorsal OLM cells (asterisks). Sox9 expression was normal in the stomach epithelium at both stages (arrowheads). Yellow dotted lines mark the epithelial basement membrane and white dotted lines separate ICM and OLM. Red staining is Isl1, green staining is Sox9, and DAPI nuclear counterstaining (DNA) is blue. Scale bars: 50 μm. ICM, inner circular muscle; OLM, outer longitudinal muscle.
Expression and distribution of protein gene product 9.5 (PGP9.5), an enteric nervous system marker [32], was intact at E18.5 in Isl1MCM/Del mutant stomachs (Additional file 1: Figure S6). Pancreatic and duodenal homeobox gene 1 (Pdx1) is expressed in epithelial cells of the antral-pyloric segment and the rostral duodenum [33]. Our immunofluorescence results showed that Pdx1 expression was similar in Isl1MCM/Del mice when compared with controls at E18.5 (Additional file 1: Figure S7). In addition, the mouse stomach and duodenal epithelial boundary was established between E14.5 and E16.5 [34], this period coinciding with development of the OLM layer [20]. We tested the integrity of the stomach-small intestine epithelial pyloric border at E18.5 by examining expression of an intestine-specific epithelial marker Cdx2 [19]. Our immunohistochemistry results demonstrated that the position of the epithelial pyloric border in Isl1MCM/Del mice was similar to that of controls (Additional file 1: Figure S8). These results indicate that loss of Isl1 does not affect innervation or epithelial development of the pylorus.
Loss of Isl1 does not affect proliferation or apoptosis of pyloric inner circular muscle and outer longitudinal muscle cells
To see whether Isl1 expression was related to cell proliferation of the pylorus, we examined co-localization of Isl1 and the proliferative marker bromodeoxyuridine (BrdU) using immunofluorescence in Isl1F/+mice. Our results showed that BrdU-positive cells were dense at E11.5 and scattered throughout the ICM and OLM regions at E14.5 and E18.5 (Additional file 1: Figure S9a). In addition, the proportion of proliferating ICM and OLM cells was not significantly different between Isl1MCM/Del and Isl1F/+mice at E18.5 (Additional file 1: Figure S9b).
To assess a potential impact on apoptosis, we examined cleaved Caspase 3 expression at E18.5, and our immunofluorescence results showed there were no Caspase 3-positive cells in pyloric ICM or the OLM layer of Isl1MCM/Del and Isl1F/+mice (Additional file 1: Figure S10). These data indicate that Isl1 ablation does not affect proliferation or apoptosis of pyloric ICM and OLM cells.
Loss of Isl1 results in decreased expression of Gata3, Gremlin, and Nkx2.5
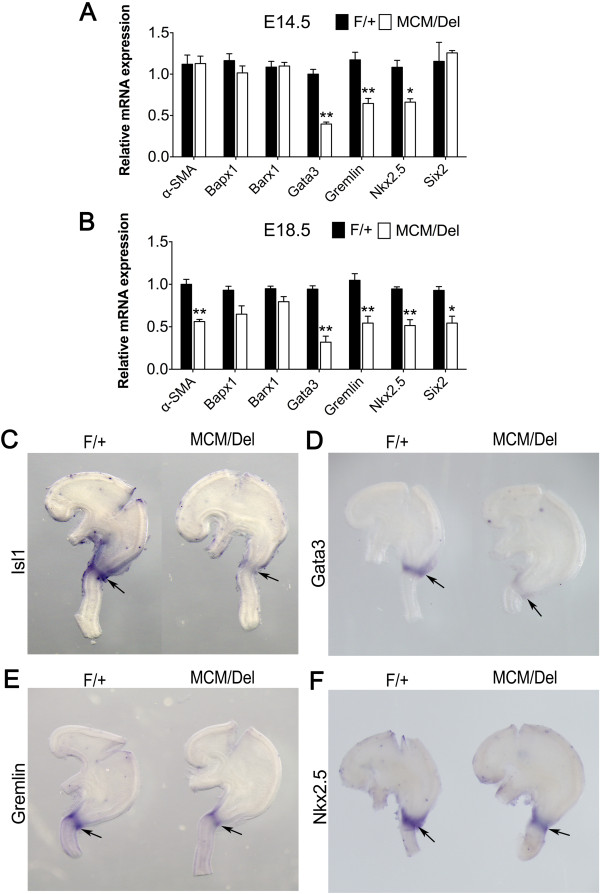
Since a numbers of factors, including Bapx1[18], Barx1[35], Gata3[19,36], Gremlin[5], Nkx2.5[2], and Six2[9], have been reported to be involved in regulation of pyloric development, we examined the effects of loss of Isl1 on expression of these genes at E14.5 and E18.5. RT-qPCR results showed that loss of Isl1 did not affect expression of Bapx1 or Barx1 at either E14.5 or E18.5 (Figure 7A,B). At E18.5, α-SMA and Six2 mRNA levels were significantly lower in Isl1MCM/Del mice as compared with controls (Figure 7B). At both E14.5 and E18.5, Nkx2.5, Gata3, and Gremlin mRNA levels in the stomach of Isl1MCM/Del mice were lower than controls (Figure 7A,B). Gata3 mRNA levels were approximately 70% decreased at both stages examined (Figure 7A,B). Based on these results, we investigated Isl1, Gata3, Gremlin, and Nkx2.5 expression in Isl1MCM/F mutant and Isl1F/+stomachs using WISH. Results demonstrated that expression of each of these genes was mainly confined to the pyloric region, as expected; Gata3 expression was more reduced in mutant stomachs; and Gremlin and Nkx2.5 only had subtle changes (Figure 7E,F). Isl1 and Gata3 expression were the most strongly affected (Figure 7C,D). These results were consistent with RT-qPCR data and suggest that Isl1 regulates expression of Gata3, Gremlin, and Nkx2.5.
Figure 7.
Aberrant gene expression in hindstomach in Isl1MCM/Del mutants. (A) RT-qPCR analysis of mRNA levels of hindstomach-enriched transcription factors at E14.5 indicates significant reduction of α-SMA, Six2 Nkx2.5, Gata3, and Gremlin mRNA in Isl1MCM/Del mutant stomachs (n = 4). All results were normalized to levels of Gapdh mRNA. (B) RT-qPCR analysis of mRNA levels of hindstomach-enriched transcription factors at E18.5 indicates a significant reduction of Nkx2.5, Gata3, and Gremlin mRNA in the Isl1MCM/Del mutant stomachs (n = 4). All results were normalized to levels of Gapdh mRNA. Data are mean ± SEM (n = 6 mice per group). *P <0.05 versus Isl1F/+; **P <0.01 versus Isl1F/+ (Student’s t-test). (C-F) WISH mRNA analysis confirmed loss of Isl1, Gata3, Gremlin, and Nkx2.5 mRNA expression at E14.5 in the Isl1MCM/Del mutant stomachs. Isl1 and Gata3 mRNA were severely down-regulated in Isl1MCM/Del mice, whereas Gremlin and Nkx2.5 expression were slightly reduced. Arrows point to the pyloric sphincter.
Isl1 targets Gata3 and activates its transcription
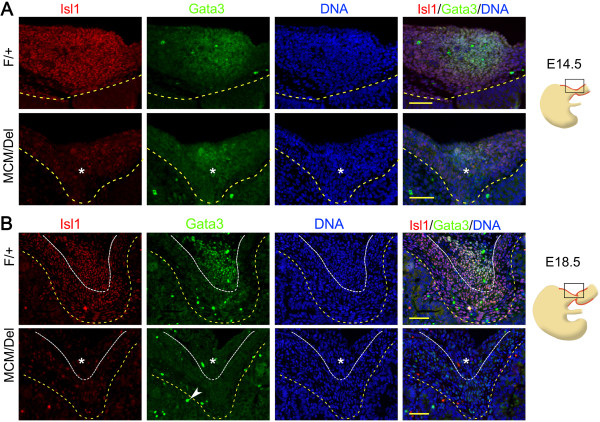
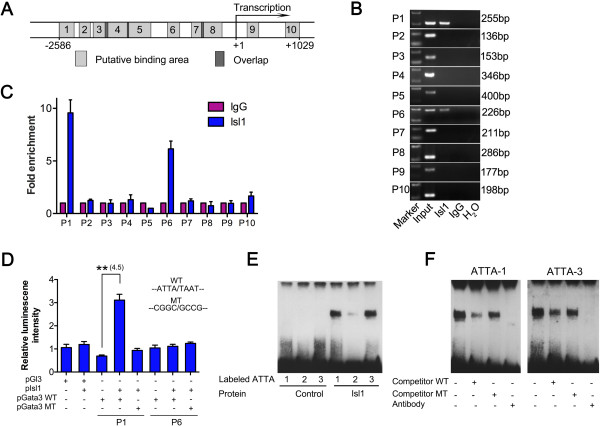
Gata3 is selectively expressed in the pylorus of the developing mouse embryo [19,20]. Expression of both Isl1 and Gata3 mRNA was observed in the pylorus at E14.5, but whether Gata3 and Isl1 are expressed in the same cells has not been explored. Therefore, we examined expression of Isl1 and Gata3 by immunofluorescence analyses. Results demonstrated that Isl1 and Gata3 proteins were co-expressed within the same cells of the pylorus at E14.5 and E18.5 in Isl1F/+control stomachs (Figure 8). In addition, the area expressing Gata3 was significantly smaller in Isl1MCM/Del mutant pyloric smooth muscle layer at E14.5 (Figure 8A) and it was lost at E18.5 in the pyloric OLM layer (Figure 8B). Thus, Isl1 was required for Gata3 expression in the dorsal pyloric OLM layer. To investigate whether Isl1 regulates pyloric development by directly regulating Gata3, we performed bioinformatics analysis of the Gata3 genomic locus. The mouse Gata3 gene contains several putative Isl1 response elements (ATTA/TAAT) at -2,832 base pairs (bp) to +1,002 bp from the transcription initiation sites [37]. We identified 10 areas that contained a putative Isl1 binding site (Figure 9A), and 10 pairs of corresponding primers were designed to amplify these regions following chromatin immunoprecipitation (ChIP) studies utilizing antibody to Isl1. Immunoprecipitated genomic DNA was obtained from pyloric regions of mouse embryos at E14.5. Of the 10 putative Isl1 binding areas, two discrete regions, in the -2,558 bp to -2,303 bp (P1 region) and -1,081 bp to -855 bp (P6 region), were occupied by Isl1 protein. This result was confirmed by semi-quantitative PCR (Figure 9B) and the fold enrichment method (Figure 9C).
Figure 8.
Loss of Isl1 eliminates the dorsal pyloric outer longitudinal muscle Gata3 expression. (A) Double immunostaining for Isl1 and Gata3 in the dorsal pylorus at E14.5. The region of mesodermal cells (asterisks) expressing Gata3 was smaller in the Isl1MCM/Del pylorus than Isl1Fl+. (B) Double immunostaining for Isl1 and Gata3 in the dorsal pylorus at E18.5. Inducible Isl1 knockout effectively eliminated Isl1 expression, with concomitant loss of Gata3 expression in the dorsal OLM cells (asterisks). Yellow dotted lines mark the epithelial basement membrane and white dotted lines indicate the ICM and OLM boundary. White arrowhead indicates non-specific stain. Red staining is Isl1, green staining is Gata3, and DAPI nuclear counterstaining (DNA) is blue. Scale bars: 50 μm. ICM, inner circular muscle; OLM, outer longitudinal muscle.
Figure 9.
Isl1 directly binds to Gata3 enhancer regions and regulates the Gata3 enhancer activity. (A) A schematic representation of the Gata3 gene surrounding the transcription start site. Putative Isl1 binding sequences (containing the ATTA/TAAT sequence) are shown as grey rectangles. (B) ChIP-PCR amplification was obtained using P1 to P10 primers which would amplify Isl1 consensus-containing fragments in the vicinity of the Gata3 transcription start site. ChIP with Isl1 antibody and amplification of fragments using the indicated primers (Additional file 2: Table S3) demonstrated binding of Isl1 to the Gata3 promoter regions in pylorus of wild-type mouse embryos at E14.5. A cell aliquot before precipitation was designated as the input sample. IgG was a negative control provided by the kit. (C) Fold change of enriched DNA fragment from ChIP detected by qPCR. (D) Effects of an Isl1 expression vector on the transiently transfected Gata3 gene enhancers (P1 and P6 regions) fused to luciferase reporter genes in 293FT cells. Data are mean ± SEM (n = 4). **P <0.01 (Student’s t-test). (E) EMSA were performed with in vitro translated pcDNA3.1-Isl1 and control vector respectively. Isl1 efficiently bound to oligonucleotides representing number 1 and 3 sites of the Gata3-P1 enhancer region. (F) Labeled ATTA number 1 and 3 probes of the P1 region were incubated with in vitro translated pcDNA3.1-Isl1 protein and assayed by EMSA. Specificity of protein-DNA binding was determined by competition with excess unlabeled wild-type or mutant competitor oligonucleotides. Additionally, Isl1 binding to oligonucleotide probes was blocked by antibodies to Isl1. bp, base pairs; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assays; IgG, immunoglobulin G; MT, mutant type; WT, wild type.
Luciferase assays were also performed to investigate the ability of Isl1 to regulate the Gata3-P1 or Gata3-P6 enhancer regions. Results of these luciferase reporter assays demonstrated that Isl1 overexpression enhanced activity of the Gata3-P1-wild-type luciferase reporter approximately 4.5-fold (Figure 9D). Site-directed mutagenesis revealed that mutation of the Isl1 consensus site within the P1 enhancer selectively decreased the ability of Isl1 co-transfection to activate the reporter. Isl1 expression did not affect luciferase activities of Gata3-P6-wild-type, Gata3-P6-mutant-type and pGL3.0-basic (Figure 9D). Together, the data strongly suggest that Isl1 regulates Gata3 transcription by binding to the Gata3-P1 element at the -2,558 bp to -2,303 bp region.
To further investigate this, electrophoretic mobility shift assays (EMSA) were performed with in vitro translated pcDNA3.1-Isl1 and control vector respectively. The Gata3-P1 enhancer region included three putative ATTA binding sites, and Isl1 efficiently bound to oligonucleotides representing number 1 and 3 sites (Figure 9E). Binding of Isl1 to number 1 and 3 sites was specifically competed for by excess unlabeled probes but not by excess unlabeled probes containing mutations within the Isl1 consensus binding sites (Figure 9F). Additionally, binding to Isl1 consensus site containing oligonucleotides was blocked by Isl1 antibody. Collectively, these data demonstrate that Isl1 is a direct regulator of Gata3 transcription.
Discussion
The presented results show that Isl1 is highly expressed in early stages of stomach development in mouse embryos, being confined at later stages to the muscle layer of the pylorus. Previous results demonstrated that Isl1 expression in the developing stomach is restricted to the ventral gastric mesenchyme at E9.5 [29], and sharply increases until E13.5. During this period of time, the mouse stomach undergoes expansion from the foregut tube [9], and the circular muscle layer of the stomach forms [11]. Our results further demonstrate that Isl1 expression is localized to the posterior stomach mesenchyme from E11.5 to E13.5, and is concentrated in the smooth muscle cells of the pylorus at later stages of stomach development, although Isl1-positive cells are also detectable in the lamina propria. These results suggest that Isl1 might be involved in the regulation of stomach organogenesis and in development of the pyloric smooth muscle layer, which is derived from stomach mesenchyme. In support of this, ablation of Isl1 led to nearly complete absence of the pyloric OLM layer at E18.5.
Stomach organogenesis occurs after E9.5 during mouse development [9]. Isl1 null mouse embryos show developmental anomalies at E9.5 and die at E10 [24]. To prolong the life of the embryos, we adopted a delayed knockout strategy using a tamoxifen-inducible mutated estrogen receptor ligand-binding domain (mER)-Cre-mER recombinase targeted to the Isl1 locus, administering tamoxifen at E11.5. Our results are in agreement with a previous report that showed that the Isl1MCM/Del mice died in the perinatal period [30]. We thus examined effects of Isl1 ablation beginning at E18.5 on mouse stomach development during the subsequent embryonic development period. We found that Isl1 expression was effectively down-regulated at both gene and protein levels. Further morphological and histological results demonstrated that the dorsal pyloric smooth muscle layer was much thinner in the pylorus of Isl1MCM/Del mice when compared with that of Isl1F/+mice. Further evidence that Isl1 is required for formation and growth of the pylorus was that duodenogastric reflux, which results from reduced contractile activity of the pyloric sphincter [9,18], was clearly observed in Isl1MCM/Del stomachs.
To investigate the cellular mechanisms by which loss of Isl1 resulted in underdevelopment of the pylorus, we tested effects of Isl1 ablation on pyloric cell differentiation, proliferation, and apoptosis. Loss of Isl1 had no significant effects on pyloric cell proliferation or apoptosis. These results are consistent with previous results suggesting that Isl1 is not likely to be involved in promoting proliferation of gastrointestinal epithelium [29]. α-SMA is essential for muscle differentiation, and widely used as a smooth muscle marker [9]. The proportion of cells expressing α-SMA among Isl1-positive cells significantly increased from E11.5 to E18.5. Isl1 ablation resulted in loss of the dorsal pyloric OLM layer and decreased α-SMA expression in Isl1MCM/Del stomachs when compared to Isl1F/+at E18.5. Therefore, we suggest that Isl1 affects pyloric development mainly by regulating dorsal pyloric OLM layer formation.
To reveal the molecular mechanisms by which Isl1 regulates pyloric development, we assessed the relationship between Isl1 and genes that are required for pyloric development, including Bapx1, Barx1, Nkx2.5, Gremlin, Six2, and Gata3. Isl1MCM/Del mutants exhibited somewhat decreased expressions of Nkx2.5 and Gremlin. Subtle changes in Nkx2.5 and Gremlin expression may be owing to the loss of some muscle, where these genes were expressed. However, expression of Gata3 was most dramatically down-regulated. Furthermore, Gata3 deletion also abrogated development of the OLM layer, leading to loss of Sox9 expression and pyloric constriction [20]. These results in Gata3 null mice demonstrate that Gata3 is required for the survival of these smooth muscle cells, and stomachs are phenotypically similar to those observed in Isl1MCM/Del mutants.
To investigate whether Gata3 is a direct downstream target of Isl1 in stomach, we performed ChIP assays utilizing Isl1 antibody and chromatin from embryonic stomach, and EMSA assays with in vitro translated Isl1 protein. We found direct binding of Isl1 to several consensus Isl1 response elements in regions surrounding the Gata3 transcription start site. In addition, co-transfection studies demonstrated the ability of an Isl1 expression vector to activate expression of the defined Gata3 enhancer element. Collectively, our data demonstrate that Isl1 directly interacts with enhancer elements in the Gata3 promoter region in stomach to activate Gata3 expression at the transcriptional level.
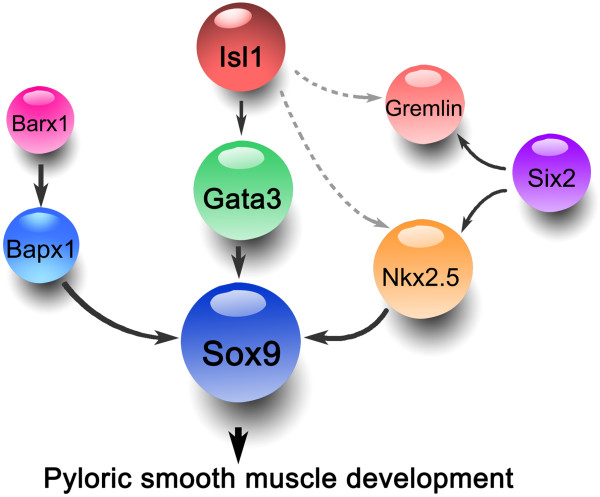
Based on results presented here and previously published for mouse pyloric development, we propose a model for a molecular interaction network controlling pyloric development (Figure 10). Bapx1 expression is lost in Barx1-null stomachs, and loss of Bapx1 does not affect Nkx2.5 expression, but gene expression microarrays show decreased Sox9 [18,38]. Thus, Barx1 may regulate Sox9 through Bapx1. Loss of Six2 reduces Nkx2.5, Gremlin, and Sox9 expression in pylorus [9], and Nkx2.5 null stomachs also lead to loss of Sox9 expression [20]; so, it is possible that Sox9 is regulated by Six2 through Nkx2.5. In addition, Sox9 is absent after deletion of Gata3, and there is no direct relationship between Gata3 and Nkx2.5[20], and our results demonstrate that Isl1 directly regulates Gata3, which suggests that Sox9 is regulated by Isl1 via Gata3. Thus, all of these pathways converge on Sox9 and confirm the critical role of Sox9 in pyloric development.
Figure 10.
Model of Isl1 function in mouse developing pyloric muscle. Bapx1 is lost in Barx1-null stomachs, Barx1 functions upstream of Bapx1, and loss of Bapx1 down-regulates Sox9 expression. We therefore suggest that Barx1 regulates Sox9 through Bapx1. Loss of Six2 reduces Nkx2.5, Gremlin, and Sox9 expression, and loss of Nkx2.5 also leads to loss of Sox9 expression. In addition, Sox9 is absent after deletion of Gata3. Our results demonstrate that Isl1 directly regulates Gata3, which suggests that Sox9 is regulated by Isl1 via Gata3. Dotted lines indicate that Nkx2.5 and Gremlin are down-regulated in Isl1MCM/Del stomachs, but specific regulatory mechanisms still remain unclear.
Our study demonstrated that Isl1 is highly expressed in the developing mouse stomach and in particular in the pylorus. Functionally, Isl1 is required for pyloric OLM layer development. We have further shown that Isl1 directly targets Gata3. Reduced expression of Gata3 can account for the pyloric phenotype observed in Isl1 mutants. In light of the results presented here, Isl1 is critical for stomach organogenesis and pyloric OLM development. These findings are important for our understanding of diseases resulting from abnormalities of pyloric sphincter development.
Conclusions
This work sheds new light on Isl1 expression and gives mechanistic insight into Isl1 function in developing pylorus of mouse embryos. We found that Isl1 was strongly expressed in the posterior stomach of mouse embryos and mainly confined to the muscle layer of the pylorus. In addition, the proportion of Isl1-positive cells expressing α-SMA gradually increased in the pylorus as development progressed and loss of Isl1 resulted in loss of the dorsal pyloric OLM layer in Isl1MCM/Del stomachs at E18.5. These new findings demonstrate that Isl1 is involved in regulating pyloric OLM development. Subsequent analysis further revealed that Isl1 ensures normal stomach pyloric development via directly targeting Gata3. These findings are highly clinically relevant and will help us to better understand the cause of related diseases such as hypertrophic pyloric stenosis resulting from smooth muscle hypertrophy in the pylorus.
Methods
Animals
Adult (6- to 8-week-old) male and female C57BL/6 mice were used for this study. All animal studies were approved by the Chinese Association for Laboratory Animal Sciences. The age of mouse embryos was determined by the appearance of the vaginal plug, which was taken to be E0.5. The birth day of the pup was marked as P1 for these experiments. Generations of Isl1MCM/+and Isl1F/F mice have been reported previously [30,31]. In brief, we used a ‘floxed’ Isl1 allele (Isl1F) in which loxP sites were inserted into the introns flanking exon 4 of the Isl1 locus [30], and a tamoxifen-inducible knockin Isl1 mER-Cre-mER allele [31,39]. Isl1F/F mice were mated with Isl1MCM/+mice to generate litters with equal numbers of Isl1MCM/F-inducible knockouts (Isl1MCM/Del) and Isl1F/+controls. To induce excision in Isl1MCM/F embryos, pregnant females were administered an oral gavage of 300 μl of tamoxifen (T5648; Sigma, St. Louis, MO, USA) in sesame oil (10 mg/ml) at E11.5 for three consecutive days just before Isl1 expression sharply increased, and the embryos were harvested at E14.5 or E18.5.
Patient material
Two patients with hypertrophic pyloric stenosis were selected from the 306th Hospital of People’s Liberation Army, Beijing. Pyloric tissue stored in the 4% Paraformaldehyde buffered in 0.01M PBS were selected from excess material collected from patients undergoing operations to retrieve surgical specimens. The study on human material was performed according to the instructions and guidelines of the 306th Hospital Ethics Committee. Approval of this study was granted by the Chinese Association for Laboratory Animal Sciences and the 306th Hospital Ethics Committee.
PCR, semi-quantitative PCR and real-time quantitative PCR
Genomic DNA was isolated from tail biopsies following the HotSHOT method [40] and genotyping was performed using standard PCR methods with sequence-specific primers (Additional file 2: Table S1). Total RNA was extracted from the pyloric regions of stomachs at E14.5 and E18.5 using commercial reagents (12183–016; Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was converted to cDNA using M-MLV reverse transcription reagents (M170A; Promega, Madison, WI, USA). RT-qPCR was performed using SYBR Green master mix (DRR420A; TaKaRa, Dalian, China) in the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and reactions were done in triplicate. RT-qPCR conditions were as follows: 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Relative RNA quantifications were normalized to endogenous control Gapdh. PCR and semi-quantitative PCR was performed in the PCR instrument (Bio-Rad Laboratories, Hercules, CA, USA) as follows: 94°C for 5 minutes (one cycle); 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds (32 cycles); 72°C for 10 minutes; and 4°C holding. PCR products were visualized on a 2% agarose gel with added ethidium bromide. Primers for detecting Isl1 knockdown efficiency and identifying gene expression change in Isl1MCM/Del mouse embryos are listed in Additional file 2: Table S1.
Western blot
Embryonic stomachs were lysed with RIPA buffer (9806; Cell Signaling, Danvers, MA, USA) containing 1 mM phenylmethylsulfonyl fluoride (8553S; Cell Signaling). The protein concentration of each group was determined using a bicinchoninic acid assay reagent (Vigorous Biotechnology, Beijing, China) according to the manufacturer’s recommendations. Equal amount of proteins were electrophoresed on a 12% SDS-PAGE, and the bands were transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories). The membrane was blocked with 5% (w/v) non-fat dry milk for 3 hours and incubated with Isl1 antibody (40.2D6; Developmental Studies Hybridoma Bank, Iowa City, IA, USA) and internal control Gapdh antibody (AM4300;= Ambion, Austin, TX, USA) overnight at 4°C. The PVDF membrane was then washed three times for 30 minutes in 0.1% Tween-20 in Tris-buffered saline (TBST) and incubated for 1 hour with horseradish peroxidase-conjugated goat anti-mouse IgG (Zhongshan, Beijing, China). After washing for 30 minutes with three changes of TBST, the membrane was treated with the pierce™ ECL 2 Western Blot Substrate (Thermo Scientific, Rockford, IL, USA).
Chromatin immunoprecipitation
Chromatin was prepared from the pyloric regions of C57BL/6 mouse embryos at E14.5 using the manufacturer’s instructions (17–371; Millipore, Darmstadt, Germany). Tissues were dissected in ice-cold PBS. Following a gentle digestion, cells were cross-linked with 1% formaldehyde (252549, Sigma) and chromatin was sheared by sonication to an average length of 500 bp. The antibody used for immunoprecipitation was the 39.4D5 Isl1 (Developmental Studies Hybridoma Bank). Reverse cross-linked immunoprecipitated chromatin was subjected to both RT-PCR and RT-qPCR using primers corresponding to the specific region (spanning the 10 putative Isl1 binding sites). Primers are listed in Additional file 2: Table S3. In all, we collected pylorus regions of more than 400 embryos and repeated ChIP reactions four times.
Plasmid construction
P1 and P6 regions of Gata3 and α-SMA[41] promoter gene were amplified from mouse genomic DNA by RT-PCR method using specific primers. Primers are listed in Additional file 2: Table S2. The forward primer contained a restriction enzyme site of SacI and the reverse primer contained a restriction enzyme site of HindIII. The PCR product was purified from agarose gel, digested, and cloned into SacI and HindIII sites of pGL3.0-basic luciferase reporter vector (E1910; Promega). Mutated Gate3-P1 and Gata3-P6 promoter regions were done using over-lap PCR, and ATTA/TAAT were mutated to CGGC/GCCG in each putative Isl1 binding site sequence. All of the constructs were verified by sequencing.
Transient transfection and luciferase assays
Human embryonic cells (293FT) were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Invitrogen) supplemented with 100 IU/ml penicillin and 100 IU/ml streptomycin. 293FT cells were plated at a density 5 × 104 cells per well in 24-well plates. After 24 hours in culture, cells were transfected with the Isl1 expression vector (Institute of Molecular and Cell Biology, Singapore) or pXJ40-Myc control vector, Gata3 or α-SMA luciferase reporter vectors, and pTK-Ranilla vector (E2241; Promega) at a ratio of 10:4:1 using the VigoFect transfection reagent (Vigorous Biotechnology). Cells were harvested 24 hours after transfection. Using the same method, the pcDNA-Gata3 expression vector (plasmid 1332; Addgene, Cambridge, MA, USA) and α-SMA luciferase reporter vector were co-transfected into 293FT cells. Luciferase activity was measured using a dual-luc assay kit (E1960; Promega) on a Modulus™ Microplate Luminometer (Turner Biosystems, Sunnyvale, CA, USA). Values shown by the fluc to rluc ratio were normalized to an empty luciferase reporter control. All transfection experiments were performed at least three times.
Hematoxylin and eosin staining
Hematoxylin and eosin staining was performed as previously described [42]. Briefly, sections were dewaxed, rehydrated, stained with hematoxylin, incubated in bluing solution, counterstained with eosin, dehydrated, and equilibrated with xylene. Glass coverslips were mounted with Permount Mounting Media (SP15-100; Fisher Scientific, Pittsburgh, PA, USA). Sections were photographed under bright-field microscope photograph system (Leica Microsystems, Buffalo Grove, IL, USA).
Immunofluorescence
Stomach samples or embryos were fixed in 4% paraformaldehyde in PBS and embedded in paraffin. Serial sections were dewaxed and rehydrated, and antigen retrieval was performed by microwaving the sections in 0.01 M sodium citrate buffer (pH 6.0). Sections were then blocked using 10% normal animal serum in PBS for 1 hour at room temperature, and incubated with primary antibodies overnight at 4°C. Subsequently, sections were washed and incubated with appropriate secondary antibodies for 2 hours at room temperature. For signal amplification, slides were washed and incubated with appropriate tertiary antibodies for 2 hours. Sections were counterstained with DAPI (10236276001; Roche Applied Science, Basel, Switzerland) for 10 minutes and mounted on plus-coated slides that were cover-slipped using Vectashield (H-1000; Vector Laboratories, Burlingame, CA, USA). Finally, sections were photographed under a fluorescence microscope photograph system (Leica Microsystems).
Primary antibodies used were goat polyclonal to Isl1 (AF1837; R&D, Minneapolis, MN, USA); mouse monoclonal to α-SMA (A2547; Sigma); mouse monoclonal to Gata3 (sc-268; Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit polyclonal to Pdx1 (ab47267; Abcam, Cambridge, UK); rabbit polyclonal to PGP9.5 (AB1761; Millipore); rabbit polyclonal to Sox9 (AB5535; Millipore); rabbit monoclonal to cleaved Caspase 3 (9664S; Cell Signaling), and mouse polyclonal to BrdU (G3G4; Developmental Studies Hybridoma Bank). Secondary antibodies used were biotinylated conjugated donkey anti-goat IgG (sc-2042; Santa Cruz Biotechnology), CY2-conjugated goat anti-mouse IgG (115-225-146; Jackson ImmunoResearch, West Grove, PA, USA), and 488 donkey anti-rabbit IgG (A21206; Life Technologies, Carlsbad, CA, USA). Tertiary antibodies used were TRITC-conjugated streptavidin (7100–03; SouthernBiotech, Birmingham, AL, USA). See Additional file 2: Table S4 for details of specific immunofluorescence protocols.
For BrdU immunofluorescence, DNA was denatured in 2 N HCl at 37°C for 30 minutes and BrdU-incorporated sites were exposed by 0.01% trypsin at 37°C for 12 minutes. After incubation with animal serum, other-step process described above.
Immunohistochemistry
Paraffin sections were processed as described above (see Immunofluorescence). Mouse monoclonal antibody to Cxd2 (AM392; BioGenex, San Ramon, CA, USA) and Isl1 antibody (40.2D6; Developmental Studies Hybridoma Bank, Iowa City, IA,USA) were incubated on sections overnight at 4°C. Sections were washed and incubated with a biotinylated goat anti-mouse IgG (115-065-146; Jackson ImmunoResearch) for 2 hours at room temperature. Slides were then washed and incubated for horseradish peroxidase-conjugated streptavidin (123-065-021; Jackson ImmunoResearch) for 2 hours at room temperature. Peroxidase activity was detected with the addition of diaminobenzidine (D4293; Sigma) and 0.1% H2O2. Sections were counterstained with hematoxylin, dehydrated, and covered with coverslips. Sections were photographed as described above (see Hematoxylin and eosin staining). See Additional file 2: Table S4 for details of specific immunohistochemistry protocols.
Measurement of pyloric sphincter constriction
A single section from at least six independent Isl1F/+and Isl1MCM/Del embryos was examined by immunofluorescence for α-SMA, as described above (see Immunofluorescence). The shortest distance between the smooth muscle layers on opposite sides of the pyloric lumen was measured with Image J (United States National Institutes of Health, Bethesda, MA, USA) [43].
BrdU labeling
BrdU was conducted by intraperitoneal injection of BrdU (50 mg/kg) into the pregnant female 2 hours before euthanasia by cervical dislocation. The embryos were removed and analyzed as described above.
Whole mount in situ hybridization
WISH was performed as previously described [44]. Tissues were fixed in 4% paraformaldehyde for 4 hours, dehydrated in methanol, and stored in 100% MeOH at -20°C until use. Samples were rehydrated, pretreated with proteinase K, and hybridized with DIG-labeled cRNA probes after washing with 2× SSC/50% formamide three times at 70°C. The signal was detected using an alkaline phosphatase-conjugated anti-DIG antibody (11093274910; Roche). Tissues were incubated in the BM Purple alkaline phosphatase substrate (11442074001; Roche) at 4°C for several hours until the signal developed to the desired extent. Probes for Gata3 564 nucleotide (1028 to 1591 bp), Nkx2.5 825 nucleotide (628 to 1452 bp), Gremlin 550 nucleotide (758 to 1307 bp), and Isl1 780 nucleotide (524 to 303 bp) were generated using DIG RNA Labeling Kit (11 175 025 910; Roche). Primers are provided in Additional file 2: Table S2.
Electrophoretic mobility shift assays
pcDNA3.1-Isl1 plasmid was used as a template for in vitro transcription and translation of Isl1 using the TNT Coupled Reticulocyte Lysate System (Promega; L4611) and pcDNA3.1 was used as control. 5′-biotin-labeled oligonucleotides were obtained from Sangon Biotech (Shanghai, China). Double-stranded DNA probes were generated by incubating complementary oligonucleotides at 90°C for 5 minutes, room temperature for 15 minutes, and 4°C for 5 minutes in a buffer containing 10 mM Tris, 1 mM EDTA and 100 mM NaCl (pH 8.0). pcDNA3.1-Isl1 was generated by cloning a fragment encoding C-terminal 216 amino acids of Isl1 into the pcDNA3.1/Hygro (+) vector. N-terminal 133 amino acids including Isl1 LIM domains have been shown previously to inhibit DNA binding in vitro[45]. Recombinant Isl1 protein was prepared by pcDNA3.1-Isl1 in vitro transcription and translation using the TNT Coupled Reticulocyte Lysate System (L4611; Promega) and pcDNA3.1 was used as control. DNA binding reactions (20 μl final volume) were proceeded at room temperature for 20 minutes in 1 × binding buffer (40 mM KCl, 15 mM HEPES (pH 7.9), 1 mM EDTA, 0.5 mM DTT, 5% glycerol and 50 ng/μl poly (dI•dC)) containing 2 μl of in vitro translated recombinant Isl1 or control reticulocyte lysate and 2 nM of 5′-biotin-labeled oligo probe. Oligonucleotide sequences were as follows: number 1 wild type: GTCCTCTTTCCCAATTACCCACTGTCAGTC, mutant: GTCCTCTTTCCCACGGCCCCACTGTCAGTC; number 2 wild type: GGACCGGCTGGGAATTACATGTTAAATACC, mutant: GGACCGGCTGGGACGGCCATGTTAAATACC; number 3 wild type: CCTGGAGGGGCCTATTAGATATTTTGTTTT, mutant: CCTGGAGGGGCCTCGGCGATATTTTGTTTT. Competition experiments were performed using 100-fold excess of unlabeled wild-type or mutant oligonucleotides pre-incubated with the Isl1 protein at room temperature for 10 minutes before adding the DNA probes. Antibody super-shift assays were performed using 1 μl of Isl1 antibody (40.2D6, 400 μg/mL) pre-incubated with Isl1 protein at room temperature for 20 minutes before adding the DNA probes. All DNA binding samples were electrophoresed on 6% non-denaturing polyacrylamide gels at 100 V for 45 minutes in 0.5 × tris-borate-EDTA buffer. Gels were transferred to a nylon membrane at 380 mA for 45 minutes in 0.5 × tris-borate-EDTA buffer. The biotin-labeled DNA was detected with a LightShift chemiluminescent EMSA kit (20148; Thermo Scientific).
Statistical analysis
Data are expressed as means ± SEM. Statistical analysis was performed with GraphPad Prism 6.0. Comparisons between two groups were analyzed by Student’s t-test. More than two groups were compared using a one-way factorial analysis of variance, followed by Student’s t-test. A value of P <0.05 was considered to be statistically significant.
Abbreviations
α-SMA: α-smooth muscle actin; bp: base pair; BrdU: bromodeoxyuridine; ChIP: chromatin immunoprecipitation; E: embryonic day; EMSA: electrophoretic mobility shift assays; Gata3: GATA binding protein 3; ICM: inner circular muscle; IgG: immunoglobulin G; Isl1: Insulin gene enhancer protein; Isl1F/F: Isl1flox/flox; Isl1MCM/Del: Isl1MCM/F-inducible knockout; LIM-HD: LIM homeodomain; mER: mutated estrogen receptor ligand-binding domain; OLM: outer longitudinal muscle; PBS: phosphate-buffered saline; Pdx1: Pancreatic and duodenal homeobox 1; PGP9.5: Protein gene protein 9.5; PVDF: polyvinylidene difluoride; RT-qPCR: real-time quantitative PCR; TBST: Tween-20 in Tris-buffered saline; WISH: whole mount in situ hybridization.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YSL and JRP were involved in experiment design, acquisition of data, analysis and interpretation of data, and drafting of the manuscript. CW, JC, YL, JLL, and XXZ performed experiments. SME was involved in critical revision of the manuscript for important intellectual content and provided Isl1F/F and Isl1MCM/+mice. YC was involved in study concept and design, critical revision of the manuscript for important intellectual content, and technical support, and provided human material. SC was involved in study concept and design, critical revision of the manuscript for important intellectual content, obtaining funding, and study supervision. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information. This file contains Figures S1 to S10.
Supplementary Information. This file contains Tables S1 to S4.
Contributor Information
Yushan Li, Email: limanandliman@163.com.
Jirong Pan, Email: panjirong@163.com.
Chao Wei, Email: emotianshi1987@163.com.
Juan Chen, Email: Chenjuan09@163.com.
Ying Liu, Email: liuying1529@163.com.
Jiali Liu, Email: liujiali@cau.edu.cn.
Xiaoxin Zhang, Email: irise1006@163.com.
Sylvia M Evans, Email: syevans@ucsd.edu.
Yan Cui, Email: dryancui@yahoo.com.cn.
Sheng Cui, Email: cuisheng@cau.edu.cn.
Acknowledgements
The pXJ40-Myc-Isl1 plasmid was a kind gift from Dr Xinmin Cao from Institute of Molecular and Cell Biology, Singapore. This work was supported by the Natural Science Foundation of China (No. 31172287) and the National Basic Research Program of China (No. 2012CB124702).
References
- Roberts DJ. Molecular mechanisms of development of the gastrointestinal tract. Dev Dyn. 2000;12:109–120. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1047>3.3.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Smith DM, Grasty RC, Theodosiou NA, Tabin CJ, Nascone-Yoder NM. Evolutionary relationships between the amphibian, avian, and mammalian stomachs. Evol Dev. 2000;12:348–359. doi: 10.1046/j.1525-142x.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- Smith DM, Nielsen C, Tabin CJ, Roberts DJ. Roles of BMP signaling and Nkx2.5 in patterning at the chick midgut-foregut boundary. Development. 2000;12:3671–3681. doi: 10.1242/dev.127.17.3671. [DOI] [PubMed] [Google Scholar]
- de Santa BP, van den Brink GR, Roberts DJ. Molecular etiology of gut malformations and diseases. Am J Med Genet. 2002;12:221–230. doi: 10.1002/ajmg.10978. [DOI] [PubMed] [Google Scholar]
- Moniot B, Biau S, Faure S, Nielsen CM, Berta P, Roberts DJ, de Santa Barbara P. SOX9 specifies the pyloric sphincter epithelium through mesenchymal-epithelial signals. Development. 2004;12:3795–3804. doi: 10.1242/dev.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufderheide E, Ekblom P. Tenascin during gut development: appearance in the mesenchyme, shift in molecular forms, and dependence on epithelial-mesenchymal interactions. J Cell Biol. 1988;12:2341–2349. doi: 10.1083/jcb.107.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedinger M, Simon-Assmann PM, Lacroix B, Marxer A, Hauri HP, Haffen K. Fetal gut mesenchyme induces differentiation of cultured intestinal endodermal and crypt cells. Dev Biol. 1986;12:474–483. doi: 10.1016/0012-1606(86)90183-1. [DOI] [PubMed] [Google Scholar]
- Duluc I, Freund JN, Leberquier C, Kedinger M. Fetal endoderm primarily holds the temporal and positional information required for mammalian intestinal development. J Cell Biol. 1994;12:211–221. doi: 10.1083/jcb.126.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self M, Geng X, Oliver G. Six2 activity is required for the formation of the mammalian pyloric sphincter. Dev Biol. 2009;12:409–417. doi: 10.1016/j.ydbio.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa A, Narita T, Kameda T, Saitoh K, Nohno T, Iba H, Yasugi S, Fukuda K. The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development. 2000;12:1971–1980. doi: 10.1242/dev.127.9.1971. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Imanaka T, Takano T. Spatial pattern of smooth muscle differentiation is specified by the epithelium in the stomach of mouse embryo. Dev Dyn. 1998;12:448–460. doi: 10.1002/(SICI)1097-0177(199807)212:3<448::AID-AJA12>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vaezi MF, Singh S, Richter JE. Role of acid and duodenogastric reflux in esophageal mucosal injury: a review of animal and human studies. Gastroenterology. 1995;12:1897–1907. doi: 10.1016/0016-5085(95)90156-6. [DOI] [PubMed] [Google Scholar]
- Vaezi MF, Richter JE. Role of acid and duodenogastroesophageal reflux in gastroesophageal reflux disease. Gastroenterology. 1996;12:1192–1199. doi: 10.1053/gast.1996.v111.pm8898632. [DOI] [PubMed] [Google Scholar]
- Hermans D, Sokal EM, Collard JM, Romagnoli R, Buts JP. Primary duodenogastric reflux in children and adolescents. Eur J Pediatr. 2003;12:598–602. doi: 10.1007/s00431-003-1259-y. [DOI] [PubMed] [Google Scholar]
- Keynes WM. Simple and complicated hypertrophic pyloric stenosis in the adult. Gut. 1965;12:240–252. doi: 10.1136/gut.6.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe A, O'Leary D, Johnson AG. Duodenogastric reflux after gastric surgery and in gastric ulcer disease: continuous measurement with a sodium ion selective electrode. Gut. 1993;12:1047–1050. doi: 10.1136/gut.34.8.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou NA, Tabin CJ. Sox9 and Nkx2.5 determine the pyloric sphincter epithelium under the control of BMP signaling. Dev Biol. 2005;12:481–490. doi: 10.1016/j.ydbio.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Stanfel MN, Moses KA, Kim B-M, Zhang Y, Schwartz RJ, Shivdasani RA, Zimmer WE. Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development. Gastroenterology. 2009;12:1701–1710. doi: 10.1053/j.gastro.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Udager AM, Hu C, Qiao XT, Richards N, Gumucio DL. Dynamic patterning at the pylorus: formation of an epithelial intestine-stomach boundary in late fetal life. Dev Dyn. 2009;12:3205–3217. doi: 10.1002/dvdy.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udager AM, Prakash A, Saenz DA, Schinke M, Moriguchi T, Jay PY, Lim KC, Engel JD, Gumucio DL. Proper development of the outer longitudinal smooth muscle of the mouse pylorus requires Nkx2-5 and Gata3. Gastroenterology. 2014;12:157–165. doi: 10.1053/j.gastro.2013.10.008. e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;12:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Heilig JS. DeLIMiting development. Bioessays. 1998;12:58–69. doi: 10.1002/(SICI)1521-1878(199801)20:1<58::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;12:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;12:309–320. doi: 10.1016/S0092-8674(00)80985-X. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;12:877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H. Multistep control of pituitary organogenesis. Science. 1997;12:1809–1812. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Li E, Oh SP, Schrewe H, Harmon EB, Lee JS, Melton DA. Activin receptor patterning of foregut organogenesis. Genes Dev. 2000;12:1866–1871. [PMC free article] [PubMed] [Google Scholar]
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;12:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Das P, May CL. Expression analysis of the Islet-1 gene in the developing and adult gastrointestinal tract. Gene Expr Patterns. 2011;12:244–254. doi: 10.1016/j.gep.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;12:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;12:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;12:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;12:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Braunstein EM, Qiao XT, Madison B, Pinson K, Dunbar L, Gumucio DL. Villin: A marker for development of the epithelial pyloric border. Dev Dyn. 2002;12:90–102. doi: 10.1002/dvdy.10091. [DOI] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;12:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Kiefer SM, Robbins L, Rauchman M. Conditional expression of wnt9b in six2-positive cells disrupts stomach and kidney function. PLoS One. 2012;12:e43098. doi: 10.1371/journal.pone.0043098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieuw KH, Li G, Zhou Y, Grosveld F, Engel JD. Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev Biol. 1997;12:1–16. doi: 10.1006/dbio.1997.8575. [DOI] [PubMed] [Google Scholar]
- Kim BM, Miletich I, Mao J, McMahon AP, Sharpe PA, Shivdasani RA. Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen. Development. 2007;12:3603–3613. doi: 10.1242/dev.009308. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai C-L, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;12:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;12:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Makarenkova HP, Gonzalez KN, Kiosses WB, Meech R. Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle alpha-actin gene. J Biol Chem. 2009;12:14866–14874. doi: 10.1074/jbc.M807208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Y, Wang X, Zheng X, Cui S. Developmental changes of Islet-1 and its co-localization with pituitary hormones in the pituitary gland of chick embryo by immunohistochemistry. Cell Tissue Res. 2005;12:279–287. doi: 10.1007/s00441-005-0021-3. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;12:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. RNA detection using non-radioactive in situ hybridization. Curr Opin Biotechnol. 1995;12:20–23. doi: 10.1016/0958-1669(95)80004-2. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Osada H, Forster A, Rabbitts TH. The cysteine-rich LIM domains inhibit DNA binding by the associated homeodomain in Isl-1. EMBO J. 1993;12:4243–4250. doi: 10.1002/j.1460-2075.1993.tb06108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information. This file contains Figures S1 to S10.
Supplementary Information. This file contains Tables S1 to S4.