Abstract
Antibodies have been extensively used as capture and detection reagents in diagnostic applications of proteomics-based technologies. Proteomic assays need high sensitivity and specificity, a wide dynamic range for detection, and accurate, reproducible quantification with small confidence values. However, several inherent limitations of monoclonal antibodies in meeting the emerging challenges of proteomics led to the development of a new class of oligonucleotide-based reagents. Natural and derivatized nucleic acid aptamers are emerging as promising alternatives to monoclonal antibodies. Aptamers can be effectively used to simultaneously detect thousands of proteins in multiplex discovery platforms, where antibodies often fail due to cross-reactivity problems. Through chemical modification, vast range of additional functional groups can be added at any desired position in the oligonucleotide sequence, therefore the best features of small molecule drugs, proteins and antibodies can be brought together into aptamers, making aptamers the most versatile reagent in proteomics. In this review we discuss the recent developments in aptamer technology, including new selection methods and the aptamers’ application in proteomics.
Keywords: aptamer-chips, bead-based selection, cancer biomarkers, X-aptamers, thioaptamers
1. Introduction
Selection of reagents that specifically recognize proteins and differentiate various natural protein variants, such as splice variants and post-translational modifications, is a major challenge in the growing field of proteomics [1] for both protein pull-down assays and sandwich assays. These protein capture and detection reagents should also have high sensitivity of detection (low detection limit), and the ability to detect proteins in a wide dynamic range of concentrations in a variety of biological fluids with often wildly different chemical environments. In order to have both high selectivity and high affinities, the reagents should recognize a broad surface area on the protein. Generally, small molecules do not have sufficient surface area to bind to a large enough surface of the protein to generate nanomolar (nM) binding affinities that can differentiate local modifications. Traditionally, these requirements have been met by antibodies, developed as the affinity ligands of choice for identifying proteins in research, diagnostics, biosensors, imaging, and therapeutics [2]. The impact of antibodies on biology and medicine has been profound and is an increasingly important component of the biotechnology industry [3, 4].
Although monoclonal antibodies (mAbs) provide highly selective, high binding affinity ligands and have been “game changers” over the past 20 years, they have significant limitations. Selection difficulties, selectivity problems, preparation difficulties, high costs of production, stability and cross reactivity issues are the major limitations in using monoclonal antibodies as detection reagents in the rapidly evolving and demanding needs of proteomics. In the vast majority of proteomics discovery platforms, mass spectrometry (MS) is used as the analytical method of choice to quantitate the target protein. In the majority of cases, particularly in the case of protein biomarkers such as the inflammatory cytokines, the target protein would be very low in concentration relative to that of the antibody, and the abundant signals from the antibody will interfere with the MS signal detection signals of the protein of interest, particularly after digestion of antigen/antibody complex. Therefore, non-protein based reagents that do not produce interfering peptide sequences are highly desired. Although these limitations have not significantly diminished the use of mAbs in biotechnology applications to date, future advances will demand better reagents that circumvent these limitations.
More recently, high-throughput screening of small molecule libraries, as well as combinatorial libraries such as nucleic acid aptamers, has yielded excellent protein-binding ligands. These aptamers have sufficient surface areas to recognize and bind their targets, much like mAb's and thus have, the ability to differentiate between isoforms and splice variants of a protein.
2. Nucleic acid aptamers
Nucleic acid aptamers are emerging as attractive alternatives to antibodies and small molecules in diagnostic, therapeutic, imaging and targeting applications [5-10]. Aptamers are small oligonucleotide molecules that have been shown to mimic antibodies, and exhibit high binding affinity, having dissociation constants typically in the nM and even picomolar (pM) range, and high selectivity towards their targets [11-15]. They are 1/10th of the molecular weight of antibodies and yet provide complex tertiary, folded structures with sufficient recognition surface areas to rival and even surpass the binding affinities of antibodies. In a proximity-dependent assay using two DNA aptamers, detection limit of zeptomole (10-21 mol) amounts was achieved [16]. The ability of single stranded nucleic acids to fold into unique and stable secondary structures allow them to form tertiary structures that not only recognize and bind specifically to protein targets, but also to discriminate between subtle molecular differences within the target [17,18]. The extremely high degree of ligand recognition and discrimination is demonstrated by several aptamers. For e.g., an RNA aptamer selected against bronchodilator theophylline binds to the target with a Kd of 0.1μM, but has a 10,000-fold lesser binding affinity to caffeine, which differs from theophylline by only a methyl group attached to nitrogen at N-7 position [19]. As such, aptamers exhibit the potential ability to differentiate between the splice variants and post transcriptional modifications of the same protein.
Aptamers’ promise was first demonstrated by their high-affinity binding to a target after in vitro combinatorial library screening, known as SELEX (Systemic Evolution of Ligands by EXponential enrichment) [11, 12]. Since then aptamers have been extensively sought and studied as protein-capture and detection reagents, targeted therapy, as well as for therapeutic, diagnostic and biosensor applications [6-10]. The first Food and Drug Administration (USA) approved aptamer-based drug, Macugen [20], is used to treat age-related macular degeneration, and several other aptamers are in various stages of clinical trails [6, 21, 22]. Numerous aptamers against a wide range of targets have already been selected and show affinities in the pM to low nM range. Aptamer targets range from small organic molecules [23, 24], ions [25, 26], small peptides [27, 28], large proteins [29-33] and even whole cells [34-38] and viruses [39-42]. Protein targets include viral proteins, cytokines, enzymes and transcription factors such as NF-κB [31, 32, 43-46].
2.1 Advantages of aptamers over antibodies
Aptamers offer significant advantages over antibodies [8]. They are in general more stable than antibodies, and have a longer shelf life. Aptamers are produced through a simple and inexpensive process and the time required to generate aptamers is comparatively short. Unlike antibodies, aptamers do not need animals or an immune response for their production. Because aptamers are chemically synthesized, batch-to-batch variation can be greatly reduced allowing economical, high-accuracy large-scale production of aptamers for clinical applications. Furthermore, aptamer's affinity can be modulated by optimizing their recognition sequence and/or by manipulating binding reaction conditions. Once selected, the stability of the aptamers can be further increased by chemical modification of the nucleotides as well as by altering their secondary structures (for e.g., introducing additional base pairs). Because aptamers are chemically synthesized, chemical modifications can be introduced into them at any desired position in the nucleotide chain. Although antibodies can be chemically modified, site-specific modifications are extremely difficult [47, 48]. Furthermore, through established solid-phase chemical synthetic methods and site-directed chemistries, labels for detection and linkers for conjugation can be easily inserted at desired sites in the oligonucleotide sequence without compromising the binding affinity or selectivity [49]. The in vitro selection process allows aptamers to be generated against otherwise toxic compounds that would kill the animal in antibody production. Also, aptamers are more stable at high temperature and they can be regenerated easily after denaturation and can be repeatedly used. The chemical synthesis and the in vitro selection process can be completely automated [50, 51] allowing rapid, parallel production of multiple aptamers against complex target sets such as proteomes. Aptamers are smaller in size compared to antibodies, thus allowing improved transport and tissue penetration compared to antibodies.
2.2 Chemical modification of aptamers
Since native oligonucleotides are susceptible to digestion by cellular nucleases present in body fluids or cells, chemical modifications of the oligonucleotides are often required to increase resistance for degradation by nucleases. Several strategies have been developed to increase the stability of aptamers without compromising the binding affinity and specificity towards their targets. These strategies include chemical modification of the phosphate backbone [52], sugars and/or the bases [53-56], end-capping at the 3’ or 5’ termini [57] and locked nucleic acids [58, 59]. Among the chemical modifications reported for oligonucleotides, sulfur substitution of the phosphate backbone (for both DNA and RNA) and the modification of the 2’ position of the ribose sugar (for RNA) are the most common. The 2’-sugar modifications found to increase the resistance and the overall stability of the functional oligonucleotides are the O-Me and fluoro substitutions, and the locked nucleic acid. The 2’-amino and 2’-fluoro substitutions are shown to increase the half-life of RNA aptamers in human serum from 8s to 86 hours [60]. The LNA modification contains an intramolecular 2’-O to 4’-C methylene bridge, and has been reported to exhibit enhanced secondary structure stability [58, 59, 61].
2.3 Thioaptamers
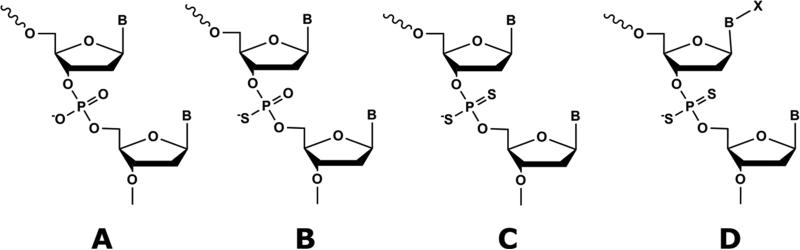
We have developed thio-substituted aptamers, called thioaptamers (TAs) [31, 32, 45, 46, 62-64], where one or both of the non-bridging phosphoryl oxygens in the oligonucleotide phosphate backbone are substituted with sulfur (Figure 1). TAs are attractive choices for aptamer development for several reasons: (a) The sulfur substitutions of the phosphodiester backbone render oligonucleotides more stable in cellular and plasma environments, mostly due to their enhanced nuclease resistance [66-68]; (b) TAs have higher affinities towards proteins than do unmodified ODNs [69]. Based on molecular dynamics and theoretical calculations, we and others have suggested that increased affinity of TA may be attributed to the decreased interaction of solvated cations with the sulfur atoms, which act as softer Lewis bases on the polyanionic backbone [70]; (c) Thioaptamers are easy to synthesize by chemical or enzymatic methods, and their sequences can be amplified and read out by PCR methods. The monothio aptamers can be PCR amplified, as the Taq DNA polymerase is capable of incorporating up to three different monothio dNTPs [71]. For RNA thioaptamers, T7 RNA polymerase is shown to be capable of incorporating (∞S)-rNTPS to produce monothio phosphate containing RNA molecules [72]. Both DNA and RNA dithioaptamers can be chemically synthesized by the standard solid-phase synthetic methods using commercially available thiophosphoramidite.
Figure 1.
Chemical structures of modified oligonucleotides. (A) normal, phosphodiester backbone (B). mono-thio modified thioaptamer (C) di-thio modified thioaptamers (D) di-thio modified X-aptamer.
The dithio-oligonucleotides contain an internucleotide phosphodiester group with sulfur substituted for both non-bridging phosphoryl oxygens (Figure 1), so they are both isosteric and isopolar with the normal phosphodiester bond, and are also highly nuclease-resistant. Importantly, the dithioate oligonucleotides are achiral about the dithiophosphate center (in contrast to the chemically synthesized monothiophosphate oligonucleotides), so problems associated with diastereomeric mixtures are completely avoided [73]. Although sulfur substitution often results in enhanced binding to the target proteins, complete substitution of the phosphate backbone might result in non-specific binding. To address this problem, we have developed a novel combinatorial selection method, where we can optimize the number as well as the position of the thio substitution in the TA [52]. Using this method, we have identified TAs against several viral and human protein targets [31, 32, 45, 46, 62-64].
2.4 The next generation aptamers: X-aptamers
Although aptamers are chemically synthesizable, and more easily selected than antibodies, they achieve their selectivity through a very limited repertoire of functional groups – the sugar phosphate backbone and four bases. In contrast, antibodies use 20 amino acids with a full range of chemical substituents including positively-charged, sulfhydryl, hydrophobic sidechains, etc. Aptamers are poly anions and it is often difficult to select an aptamer targeted to very acidic proteins because there are no cationic groups to neutralize anionic surfaces on the protein. These limitations are overcome by chemically adding various functional groups to the oligonucleotide bases. Using DNA SOMAmers (Slow Off-rate Modified Aptamers), DNA aptamers uniformly modified at the 5-position of dU residues, as capture reagents in a highly multiplexed assay platform, Kraemer et al., [74] demonstrated that the problems of cross-reactivity and non-specific adsorption to chip surfaces can be largely eliminated.
Using the ‘click chemistry’ [75-77] synthetic approach, additional functional groups can be easily added to the 5- position of pyrimidine residues at selected positions, allowing virtually unlimited possibilities for chemical modifications [77, 78]. Now, we can combine the features of high throughput small molecule library selection with high throughput bead-based aptamer combinatorial library selection. The resulting aptamers, which we call X-aptamers, could revolutionize the next generation of protein-targeted ligands. Through established synthetic chemistry methods, a vast range of additional functional moieties, from small organic molecules, to amino acid side chains to large macromolecules, including antibodies and large proteins can be conjugated to the aptamer sequence at any desired position. Therefore, the best features of small molecule drugs and antibodies can be brought together into the X-aptamers that will fold into unique tertiary structure scaffolds to present to the target protein.
As a proof-of-principle, we modified previously selected thioaptamers specific for the HA binding domain of CD44 [65], by introducing a chemical drug demonstrated to bind to the target protein. The drug was selected as a lead compound by in-silico screening, and was conjugated to the dU residue in the aptamer via either ‘click chemistry’ or amide coupling chemistry. The modified X-aptamers showed increased binding of up to 100 fold to the CD44-HABD (He, et al., submitted for publication).
3 Aptamer Selection Methods
Unlike antibodies, aptamers are selected by in-vitro methods, by screening a large library of oligonucleotides against the target to find the tightest binding candidates. The selection methods can be broadly divided into two categories. The SELEX-type method, originally reported by Gold Tuerk, Szostak and Ellington [11, 12], involves iterative cycles of screening and PCR amplification at each round. The other is the most recent, bead-based method where the oligonucleotide library is synthesized on non-cleavable beads and the high-affinity binders are identified in a single-step screening. Aptamers are also selected on microfludics chips [79, 80]. Like the bead-based selection method, this method would also significantly reduce the time required for aptamer selection. Platt et al described the CLADE technology for aptamer selection that combines the discovery, evolution and optimization in a single-step process [80]
3.1 The SELEX method
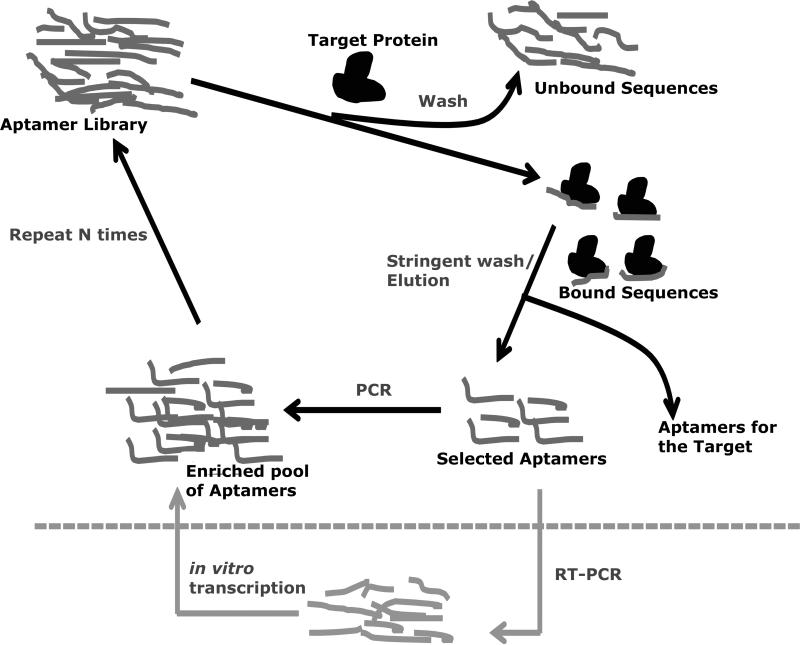
In a typical SELEX experiment, an initial oligonucleotide library containing ca.1014 unique but random sequences is designed and chemically synthesized. The sequence has a 30-40 nucleotide random region, flanked by PCR primer regions (18-22 residues each). The initial library is PCR amplified, and the single stranded DNA oligos are isolated following denaturation. This DNA pool is first heated at 95 °C and subsequently cooled in a binding buffer in order to fold into stable structures and subsequently incubated with the target. The bound sequences are partitioned from the unbound and weakly bound sequences by membrane filtering or other such separation methods. The bound aptamers are eluted from the targets, amplified by PCR and taken to the next cycle of the selection process. In the case of the RNA aptamers, the library sequence includes the T7 RNA polymerase promoter region to transcribe the initial DNA library into an RNA library and each iterative cycle also involves an RT-PCR step (Figure 2). At each iterative cycle, the stringency of the selection is progressively increased by increasing the relative ratio of aptamers to the target in order to increase the competition between the aptamers and the target. The iterative cycle of the selection process is continued until convergence towards a single or a few sequences with high affinity to the target is obtained. PCR of the aptamers pool is sub-cloned and plasmids with individual aptamers inserts are isolated and sequenced and analyzed. The selected aptamer sequences are synthesized and their binding affinities and specificities will be characterized. Most aptamers are selected by similar manual protocols. However, automation of the SELEX protocol is also described for several targets [47, 81, 82].
Figure 2.
Scheme showing the traditional SELEX procedure. The iterations are continued until the sequences converge and the tightly binding sequences are selected and identified. For the RNA oligonucleotides, two additional steps are needed for each iteration. The selected RNA sequences are converted and amplified into DNA sequences via RT-PCR, and then converted back into RNA sequences via in vitro transcription.
Aptamers can also be selected against whole cells. The process, called cell-SELEX, is used to select aptamers targeting specifically to a cell type of interest. This method is useful to select aptamers that binds to a diseased (e.g., cancer) cell line and the selected aptamers are used to identify the cell type and differentiate cancer cells from normal cells [83-88]. Cell-SELEX is the method of choice for aptamer selection in instances where a clear target is not known, or the target is hidden or shielded from the surface. (for e.g., by permeabilizing cell membranes or by chemically removing sugar units in glycoproteins [89]). In this method, aptamers are selected against the cell surface where the target protein is present in their native conformation and/or with post translational modifications retaining their biological function. The cell-SELEX method usually yields a pool of aptamers against many protein targets on the cell surface, generating a molecular signature to the cells that can be useful in disease diagnosis and treatment. In the selection process, an initial random library is incubated with target cells and control cells. The library is first screened against the control (normal) cells to filter out the sequences that bind to the control cells. The unbound sequences are separated and screened against the diseased cells. The selection cycles are continued similar to the SELEX procedure until the selected sequences show convergence.
The enrichment of the pool can be monitored with a fluorescently labeled DNA pool and the cells are monitored by flow cytometry or by fluorescent microscopy. The pool is then sub-cloned and the sequences of pool again identified to determine any convergence of the pool towards the target cells. Selected sequences from a converged pool are synthesized either chemically using the standard phosphoramidite chemistry or enzymatically by PCR from the plasmids with fluoresecence or dye labels at either the 5’ or 3’ end. Their binding affinities and Kd values are often determined by flow cytometry. Several high affinity aptamers have been identified successfully using the cell-SELEX method indicating that aptamers can be generated with complex targets such as tumor cells and tissues [34, 87]. The targets can also be identified with the aptamers in the tissue samples. A fluorophore labeled aptamer can also bind to tissue containing the target protein.
Over the last decade, several variations of the traditional SELEX method were reported, each designed to address specific requirements. These methods are discussed in detail in a recent review paper [90].
3.2 Bead-based selection
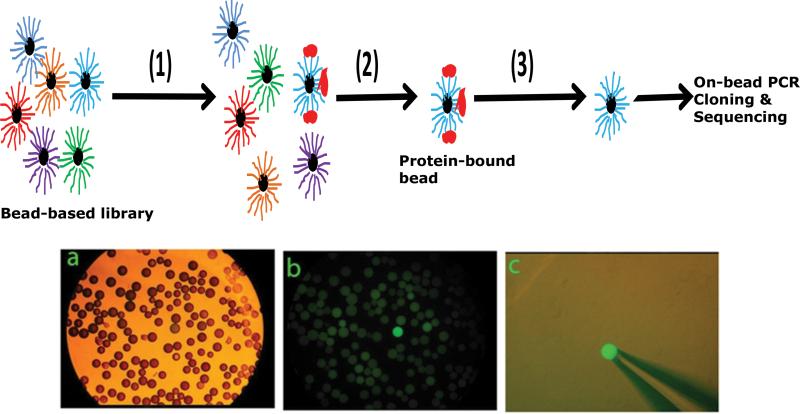
While a wide-range of chemical modifications can be made on the oligonucleotides to increase the diversity of the library, some of the modified oligonucleotides cannot be amplified by PCR methods, posing problems for aptamer selection by SELEX-type methods. We have developed a bead-based combinatorial library selection method to address this limitation (Figure 3). In the first step, a combinatorial oligonucleotide library is synthesized, via split/pool method on noncleavable beads and exposed to the target protein. We have developed a ‘split and pool’ synthesis method to create a combinatorial library of oligonucleotides, on micron-size beads, with any type of backbone modification [91]. In the second step, the bead-based library is incubated with the fluorescently tagged protein, and the protein-bound aptamer beads can be manually picked under the fluorescence microscope or are sorted by high-throughput flow-cytometry [92]. Alternatively, proteins can be labeled with biotin and the bound aptamer beads can be sorted by magnetic selection using streptavidin coated magnetic nanoparticles (Invitrogen). In the final step of the process, the sequences of the aptamers on the selected beads can then be read-out by PCR amplification [92, 93].
Figure 3.
Scheme showing the bead-based selection process. In the first step, the bead based library is incubated with fluorescently tagged protein. In the second step, the protein bound beads are separated. Third step shows the protein is stripped from the bead. The sequence on the bead is then PCR-amplified, cloned and sequenced. The bottom panel shows the bead-based aptamer library screen. (A) An aliquot of bead-based library is viewed under light microscope. (B). The same filed viewed under fluorescent microscope. The aptamer bead bound to the protein is easily identified among the many hundreds of non-reactive beads. (C). A single positive bead is retrieved using a hand-held micropipette under fluorescent microscope.
The bead-based selection has several significant advantages. Since the bead based method does not involve iterative selection and PCR amplification cycles as in SELEX procedure, selection of aptamer candidates can be achieved in much shorter time (hours). Several groups have reported that PCR can impose an “amplifiability bias” where certain sequences are preferentially amplified while other sequences are amplified with very low efficiencies during aptamer selection. This may significantly limit or even prevent recovery of desirable aptamers that amplify with low efficiency [94, 95]. The bead-based process allows diverse chemical modifications to be introduced into the oligonucleotides, and facilitates the development of the X-aptamer technology.
The bead-based selection method reverses the conventional SELEX approach to binding and separation. In traditional SELEX, a mixed library of oligonucleotides in solution is selected for binding to an immobilized target (e.g. a protein bound to a filter or chromatography resin). In this situation, a large number of weak-binding sequences are recovered along with the small number of tight binding aptamers. These weak binding aptamers are initially in much higher concentration than the small number of tight binding sequences and effectively compete with the tight binding ones. Thus, numerous rounds of selection are typically required to enrich for the tightest binding sequences since only about 10% of the library can be selected in a single iteration. In contrast, the bead based process incubates the soluble target with an immobilized aptamer library. Thus, there is no direct binding competition between tight and weak binders [92, 93, 96]. A single bead carrying a high-affinity aptamer will bind large amounts of target at low concentrations, while the majority of beads will show little or no binding.
4 Aptamers in proteomics applications
Detecting biomarkers is critically important for the early detection of diseases, particularly in complex diseases such as cancer and cardiovascular diseases, for the prognosis and treatment outcome. The majority of biomarkers are proteins and they are found in blood and other body fluids. Although single biomarker detections form the core of molecular diagnostics, sensitivity and specificity of diagnostics can be significantly enhanced by simultaneously detecting multiple biomarkers, both quantitatively and qualitatively. Initially, two-dimensional gel electrophoresis combined with MS/MS for protein identification was widely used for proteome screens [97]. Although it was relatively economical, it is a laborious method, and issues of sensitivity and reproducibility raise serious limitations. Antibody based techniques are more sensitive and reliable than 2D gel and mass spectrometry. Sandwich assays (e.g., ELISA), where two specific antibodies are required for each protein so any non-specific binding by the primary antibody is eliminated, are required to get increased specificity and sensitivity often required in clinical proteomic assays. For single-analyte tests, including sandwich assays, antibodies have proven to be very effective. However, in multiplex (multi-analyte) platforms using antibodies will have serious limitations due to cross reactivity of antibodies. Because of this cross-reactivity limitation, antibody-based detection systems have often failed when used for simultaneously detecting more than a few tens of proteins [98, 99].
Since the first report in 1996 of using oligonucleotides to detect proteins in “immuno” assays [100], significant progress has been made in utilizing nucleic acid aptamers in protein detection assays [101]. Aptamers have replaced antibodies in dot-blot [102] and western-blot applications [103, 104]. In the sandwich assay called ELONA (Enzyme Linked Oligonucleotide Assay), oligonucleotide-based aptamers replaced antibodies as capture and/ or detection reagents [105-108]. Recently, several groups have reported aptamer-based biomarker discovery platforms with multi-plexing capabilities. We have developed a protein chip using thioaptamers to identify unknown proteins from crude biological samples [109]. Proteins, captured by specific thioaptamers, are identified by ‘on-chip’ digestion followed by MS. Gold and co-workers have recently described biomarker discovery system that is capable of simultaneously measuring thousands of proteins from serum or plasma samples. Using their system, they discovered 58 potential biomarkers for chronic kidney disease [110]. The same group, using their patented aptamers, called SOMAmers [74, 111, 112], reported a large scale study of screening serum samples to discover biomarkers for non-small cell lung cancer. These two reports showcase the significant improvements in the development of highly sensitive, aptamer-based biomarker discovery platforms. Aptamer-facilitated biomarker discovery (AptaBiD) technology [113] was reported to detect biomarkers differently expressed on cell surfaces. Through multi-round selection of single-stranded DNA aptamers, biomarkers are isolated from cells and subsequently identified by mass spectrometric methods [114-117]. Interference from non-target proteins, some of them may be present in significantly higher concentrations in body fluids, is a serious limitation in proteomic analysis. In a thioaptamer-based integrated electrophoretic gel-shift detection platform [118], non-specific oligonucleotides were successfully used to suppress interference from non-target proteins present in body fluids.
Aptamer-based sensors have been developed in a variety of formats, including microarray slides that can be read in commercially available scanners designed for gene expression chips as well as flow cells that use fluorescence, quartz crystals micro-mechanical detection systems [119-121]. Some of these studies have demonstrated a unique feature of aptamers immobilized on biosensors: after measurement is made, aptamers may be chemically denatured (with 7M urea) to completely strip off the affinity bound analytes, and then refolded into their active conformations by washing in binding buffers. It is possible that the aptamers selected via the invitro processes may or may not function optimally when immobilized on a chip surface [122]. However, since aptamers are chemically synthesized, it is easier to covalently place additional linkers between the aptamer and the chip surface without altering the functional structure of the aptamer [123].
Since their discovery two decades ago, aptamers have attracted significant interest among scientists and clinicians. Significant progress has been made in the aptamer selection process and newer applications are being reported. The bead based selection process that significantly reduces the time required to select aptamers, advances in synthetic chemistry to incorporate diverse functional groups into the oligonucleotide chain and the next generation sequencing where hundreds of thousands of sequences can be read simultaneously, will revolutionize the aptamer field in the future. The enormous potential of aptamers in proteomics applications is revealed in the recent reports of aptamer based biomarker discovery platforms for simultaneous detection of thousands of proteins [111, 112]. Novel aptamer-based technologies will continue to evolve and there is little doubt that aptamers will provide enormous opportunities in the future for both biomarker discovery and clinical proteomics in general.
Acknowledgements
We thank Dr. David Volk for critical review of the manuscript. The authors’ research was supported by grants from the National Institutes of Heath (R01CA128797, AI27744, GM 084552, NCI U54CA151668, N01-HV-28184, HHSN 272200800048C), Department of Defense (W81XWH-09-1-0212) and the Welch Foundation (H-1296).
Abbreviations
- FDA
Food and Drug Administration
- nM
nano molar
- MS
mass spectrometry
- SELEX
systemic evolution of ligands by exponential enrichment
- TA
thioaptamer
Footnotes
Conflict of interest statement.
Dr. Gorenstein and the University of Texas Health Science Center at Houston have research related financial interests in AptaMed Inc. and AM Biotechnologies LLC, Houston, TX, USA.
References
- 1.Brody EN, Gold L, Lawn RM, Walker JJ, et al. High-content affinity-proteomics: unlocking protein biomarker discovery. Expert Rev. Mol. Diagn. 2010;10:1013–1022. doi: 10.1586/erm.10.89. [DOI] [PubMed] [Google Scholar]
- 2.Ritz J, Pesando JM, McConarty NJ, Clavell LA, et al. Use of monoclonal antibodies as diagnostic and therapeutic reagents in acute lymphoblastic leukemia. Cancer Res. 1981;41:4771–4775. [PubMed] [Google Scholar]
- 3.Borrebaeck CA. Antibodies in diagnostics- from immunoassays to protein chips. Immunol. Today. 2000;21:379–382. doi: 10.1016/s0167-5699(00)01683-2. [DOI] [PubMed] [Google Scholar]
- 4.Leong TY, Cooper K, Leong AS. Immunohistology: past present and futre. Adv. Anat. Pathol. 2010;17:404–418. doi: 10.1097/PAP.0b013e3181f8957c. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 6.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 7.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 8.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong H, Goel S, Zhang Y, Cai W. Molecular imaging with nucleic acid aptamers. Curr. Med. Chem. 2011;18:195–205. doi: 10.2174/092986711797189691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann AP, Bhavane RC, Somasunderam A, Montalvo-Oritz B, et al. Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget. 2011;2:298–304. doi: 10.18632/oncotarget.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Eaton BE, Gold L, Zichi DA. Lets get specific: the relationship between specificity and affinity. Chem Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 14.Bridonneau P, Chang YF, O'Connell D, Gill SC, et al. High-affinity aptamers selectively inhibit human nonpancreatic secretory phospholipase A2. J. Med. Chem. 1998;41:778–786. doi: 10.1021/jm970579k. [DOI] [PubMed] [Google Scholar]
- 15.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 16.Fredriksson S, Gullberg M, Jarvius J, Olsson C, et al. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 17.Conrad R, Ellington AD. Detecting immobilized protein kinase C isozymes with RNA aptamers. Anal Biochem. 1996;242:261–265. doi: 10.1006/abio.1996.0462. [DOI] [PubMed] [Google Scholar]
- 18.Conrad R, Keranen LM, Ellington AD, Newton AC. Isozyme-specific inhibition of protein kinase C by RNA aptamers. J Biol. Chem. 1994;269:32051–32054. [PubMed] [Google Scholar]
- 19.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 20.Ng EW, Adamis AP. Anti-VEGF aptamer therapy for ocular vascular diseases. Ann NY Acad. Sci. 2006;1082:151–171. doi: 10.1196/annals.1348.062. [DOI] [PubMed] [Google Scholar]
- 21.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 22.Dausse E, Gomes SDR, Toulme J-J. Aptamers: a new class of oligonucleotides in the drug discovery pipeline. Curr. Opinion Pharm. 2009;9:602–607. doi: 10.1016/j.coph.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Connell GJ, Illangesekare M, Yarus M. Three small ribonucleotides with specific arginine sites. Biochemistry. 1993;32:5497–5502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- 24.Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. RNA aptamers that bind l-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman HP, Limmer S, Hornung V, Sprinzl M. Ni2+ binding RNA motifs with an asymmetric purine-rich internal loop and a G:A base pair. RNA. 1997;3:1289–1300. [PMC free article] [PubMed] [Google Scholar]
- 26.Rajendran M, Ellington AD. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal. Chem. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 27.Nieuwlandt D, Wecker M, Gold L. In vitro selection of RNA ligands to substance P. Biochemistry. 1995;34:5651–5659. doi: 10.1021/bi00016a041. [DOI] [PubMed] [Google Scholar]
- 28.Williams KP, Liu XH, Schumacher TN, Lin HY, et al. Bioactive and nuclease-resistant l-DNA ligand of vasopressin. Proc. Natl. Acad. Sci. USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubik MF, Stephens AW, Schneider D, Marlar RA, Tasset D. High-affinity RNA ligands to human α-thrombin. Nucleic Acids Res. 1994;22:2619–2626. doi: 10.1093/nar/22.13.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somasunderam A, Ferguson MR, Rojo DR, Thiviyanathan V, et al. Combinatorial selection, inhibition, and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry. 2005;44:10388–10395. doi: 10.1021/bi0507074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang J, Lee MS, Watowich SJ, Gorenstein DG. Combinatorial selection of a RNA thioaptamer that binds to Venezuelan equine encephalitis virus capsid protein. FEBS Lett. 2007;581:2497–2502. doi: 10.1016/j.febslet.2007.04.072. [DOI] [PubMed] [Google Scholar]
- 33.Urvil PT, Kakiuchi N, Zhou DM, Shimotohno K, et al. Selection of RNA aptamers that bind specifically to the NS3 protease of hepatitis C virus. Eur. J. Biochem. 1997;248:130–138. doi: 10.1111/j.1432-1033.1997.t01-1-00130.x. [DOI] [PubMed] [Google Scholar]
- 34.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor micro vessels; selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 36.Hicke BJ, Marion C, Chang YF, Gould T, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 37.Sefah K, Phillips JA, Xiong X, Meng L, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shangguan D, Meng L, Cao ZC, Xiao Z, et al. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 39.Pan W, Craven RC, Qiu Q, Wilson CB, et al. Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl. Acad. Sci. USA. 1995;92:11509–11513. doi: 10.1073/pnas.92.25.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misono TS, Kumar PKR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface Plasmon resonance. Anal. Biochem. 2005;342:312–317. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Tang Z, Parekh P, Turner P, Moyer RW, Tan W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 43.Bielinska A, Shivdasani RA, Zhang LQ, Nabel GJ. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science. 1990;250:997–1000. doi: 10.1126/science.2237444. [DOI] [PubMed] [Google Scholar]
- 44.Lebruuska LL, Maher LJ. Selection and characterization of an RNA decoy for transcription factor NF-kappa B. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 45.Fennewald SM, Scott EP, Zhang L, Yang X, et al. Thioaptamer decoy targeting of AP-1 proteins influences cytokine expression and the outcome of arenavirus infections. J. Gen Virol. 2007;88:981–990. doi: 10.1099/vir.0.82499-0. [DOI] [PubMed] [Google Scholar]
- 46.King DJ, Bassett SE, Li X, Fennewald SA, et al. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappa B RelA (p65) and p50. Biochemistry. 2002;41:9696–9706. doi: 10.1021/bi020220k. [DOI] [PubMed] [Google Scholar]
- 47.Sharifi J, Khawli LA, Hornick JL, Epstein AL. Improving monoclonal antibody pharmacokinetics via chemical modification. Q. J. Nucl. Med. 1988;42:242–249. [PubMed] [Google Scholar]
- 48.Constantinou A, Chen C, Deonarain MP. Modulating the pharmacokinetics of therapeutic antibodies. Biotechnol. Lett. 2010;32:609–622. doi: 10.1007/s10529-010-0214-z. [DOI] [PubMed] [Google Scholar]
- 49.Stoltenburg R, Reinemann C, Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 50.Cox JC, Rudolph P, Ellington AD. Automated RNA selection. Biotechnol. Prog. 1998;14:845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 51.Cox JC, Ellington AD. Automated selection of anti-protein aptamers. Bioorg Med Chem. 2001;9:2525–2531. doi: 10.1016/s0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 52.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 53.Vaught JD, Dewey T, Eaton BE. T7 RNA polymerase transcription with 5-position modified UTP derivatives. J. Am. Chem. Soc. 2004;126:11231–11237. doi: 10.1021/ja049009h. [DOI] [PubMed] [Google Scholar]
- 54.Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, et al. Expanding the chemistry of DNA for in vitro studies. J. Am. Chem. Soc. 2010:132. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 55.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J Clin Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faria M, Ulrich H. Sugar boost: When ribose modifications improve oligonucleotide performance. Curr. Opin. Mol Ther. 2008;10:168–175. [PubMed] [Google Scholar]
- 57.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 58.Crinelli R, Bianchi M, Gentilini L, Magnani M. Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002;30:2435–2443. doi: 10.1093/nar/30.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darfeuille F, Hansen JB, Orum H, Di Primo C, Toulme JJ. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–3107. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulrich H, Martins AH, Pesquero JB. RNA and DNA aptamers in cytomics analysis. Cytometry Part A. 2004;59A:220–231. doi: 10.1002/cyto.a.20056. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiviyanathan V, Somasunderam A, Gorenstein DG. Combinatorial selection and delivery of thioaptamers. Biochem Soc Trans. 2007;35:50–52. doi: 10.1042/BST0350050. [DOI] [PubMed] [Google Scholar]
- 63.Kang J, Lee MS, Copland JA, 3rd, Luxon BA, Gorenstein DG. Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-beta1 protein. Bioorg Med Chem Lett. 2008;18:1835–1839. doi: 10.1016/j.bmcl.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mann AP, Somasunderam A, Nieves-Alicea R, Li X, et al. Identification of thioaptamer ligand against E-selectin: potential application for inflamed vasculature targeting. PLoS One. 2010;2010;5:e13050. doi: 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somasunderam A, Thiviyanathan V, Tanaka T, Li X, et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry. 2010;2010;49:9106–9112. doi: 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andreola ML, Calmels C, Michel J, Toulme JJ, Litvak S. Towards the selection of phosphorothioate aptamers optimizing in vitro selection steps with phosphorothioate nucleotides. Eur. J Biochem. 2000;267:5032–5040. doi: 10.1046/j.1432-1327.2000.01557.x. [DOI] [PubMed] [Google Scholar]
- 67.Jhaveri S, Olwin B, Ellington AD. In vitro selection of phosphorothiolated aptamers. Bioorg Med Chem Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- 68.King DJ, Safar JG, Legname G, Prusiner SB. Thioaptamer interactions with prion proteins: sequence-specific and non-specific binding sites. J Mol Biol. 2007;369:1001–1014. doi: 10.1016/j.jmb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, Gorenstein DG. Progress in thioaptamers development. Curr. Drug Targets. 2004;5:705–715. doi: 10.2174/1389450043345074. [DOI] [PubMed] [Google Scholar]
- 70.Volk DE, Power TD, Gorenstein DG, Luxon BA. An ab initio study of phosphorothioate and phosphorodithioate interactions with sodium cation. Tet. Lett. 2002;43:4443–4447. [Google Scholar]
- 71.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 72.Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- 73.Lebedev AV, Wickstrom E. The chirality problem in P-substituted oligonucleotides. Pres Drug Discov Design. 1996;4:17–40. [Google Scholar]
- 74.Kraemer S, Vaught JD, Bock C, Gold L, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications; a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demko Z, Sharpless BA. A click chemistry approach to tetrazoles by Huisgen 1,3-dipolar cycloaddition: Synthesis of 5-Acyltetrazoles from azides and acyl cyanides. Angew. Chem. Int. Ed. 2002;41:2113. [PubMed] [Google Scholar]
- 76.Binder WH, Sachsenhofer R. Click chemistry in polymer and material science. Macromol Rapid Commun. 2007;28:15–54. [Google Scholar]
- 77.Gramlich PME, Wirges CT, Manetto A, Carell T. Post synthetic DNA modification through the copper-catalyzed azide-alkyne cycloaddition reaction. Angew. Chem. Int. Ed. 2008;47:8350–8358. doi: 10.1002/anie.200802077. [DOI] [PubMed] [Google Scholar]
- 78.Vaught JD, Bock C, Carter J, Fitzwater T, et al. Expanding the chemistry for in vitro selection. J. Am. Chem. Soc. 2010;132:4141–4145. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 79.Collett JR, Cho EJ, Ellington AD. Production and processing of aptamer microarrays. Methods. 2005;37:4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 80.Platt M, Rowe W, Wedge DC, Kell DB, et al. Aptamer evolution for array-based diagnostics. Anal. Biochem. 2009;390:203–205. doi: 10.1016/j.ab.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 81.Eulberg D, Buchner K, Maasch C, Klussmann S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res. 2005;33:e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blank M, Blind M. Aptamers as tools for target validation. Curr. Opin. Chem. Biol. 2005;9:336–342. doi: 10.1016/j.cbpa.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 83.Shangguan D, Li Y, Tang Z, Cao ZC, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cerchia L, Duconge F, Pestourie C, Boulay J, et al. Neutralizing Aptamers from Whole-Cell SELEX Inhibit the RET Receptor Tyrosine Kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li N, Ebright JN, Stovall GM, et al. Technical and biological issues relevant to cell typing with aptamers. J. Proteome Res. 2009;8:2438–2448. doi: 10.1021/pr801048z. [DOI] [PubMed] [Google Scholar]
- 87.Guo KT, Ziemer G, Paul A, Wendel HP. CELL-SELEX: Novel perspectives of aptamer-based therapeutics. Int. J. Mol. Sci. 2008;9:668–678. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Francica JR, Varela-Rohena A, Medvec A, Plesa G, Riley JL, et al. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathogens. 2010;6:10.1371. doi: 10.1371/journal.ppat.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang X, Li N, Gorenstein DG. Strategies for the discovery of therapeutic aptamers. Expert Opin. Drug Discov. 2001;6:75–87. doi: 10.1517/17460441.2011.537321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang X, Bassett SE, Li X, Luxon BA, et al. Construction and selection of bead-bound combinatorial oligonucleotide phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang X, Li X, Prow TW, Reece LM, et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang X, Wang H, Beasley DW, Volk DE, et al. Selection of thioaptamers for diagnostics and therapeutics. Ann. NY Acad. Sci. 2006;1082:116–119. doi: 10.1196/annals.1348.065. [DOI] [PubMed] [Google Scholar]
- 94.Meyers LA, Lee J,F, Cowperthwaite M, Ellington AD. The robustness of naturally and artificially selected nucleic acid secondary structures. J Mol. Evol. 2004;58:681–691. doi: 10.1007/s00239-004-2590-2. [DOI] [PubMed] [Google Scholar]
- 95.Tsuji S, Hirabayashi N, Kato S, Akitomi J, et al. Effective isolation of RNA aptamer through suppression of PCR bias. Biochem Biophys Res Commun. 2009;386:223–226. doi: 10.1016/j.bbrc.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 96.Porschewski P, Grattinger M, Klenzke K, Erpenback A, et al. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screening. 2006;11:773–781. doi: 10.1177/1087057106292138. [DOI] [PubMed] [Google Scholar]
- 97.O'Farrell PH. The pre-omics era: the early days of two-dimensional gels. Proteomics. 2008;8:4842–4852. doi: 10.1002/pmic.200800719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fredriksson S, Dixon W, Ji H, Koon AC, et al. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 99.Schweitzer B, Roberts S, Grimwade B, Shao W, et al. Multiplexed protein profiling on micoarrays by rolling-cycle amplification. Nat. Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drolet DW, Moon-McDermott L, Romig TS. An enzyme-linked oligonucleotide assay. Nat Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 101.Agnew HD, Rohde RD, Millward SW, Nag A, et al. Iterative in situ click chemistry creates antibody-like protein-capture agents. Angew Chem Int. Ed. Engl. 2009;2009;48:4944–4948. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu J, Li T, Hu J, Wang E. A novel dot-blot DNAzyme-linked aptamer assay for protein detection. Anal Bioanal Chem. 2010;397:2923–2927. doi: 10.1007/s00216-010-3802-9. [DOI] [PubMed] [Google Scholar]
- 103.Tombelli S, Mascini M. Aptamers as molecular tools for bioanalytical methods. Curr. Opin. Mol. Ther. 2009;11:179–188. [PubMed] [Google Scholar]
- 104.Shin S, Kim I-H, Kang W, Yang JK, Hah SS. An alternative to Western blot analysis using RNA aptamer-functionalized quantum dots. Bioorg. Med. Chem. Lett. 2010;20:3322–3325. doi: 10.1016/j.bmcl.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 105.Joshi R, Janagama H, Dwivedi HP, Senthilkumar TMA, et al. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes. 2009;23:20–28. doi: 10.1016/j.mcp.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 106.Proschewski P, Grattinger M, Klenzke K, Erpenbach A, et al. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screen. 2006;11:773–781. doi: 10.1177/1087057106292138. [DOI] [PubMed] [Google Scholar]
- 107.Rye PD, Nustad K. Immunomagnetic DNA aptamer assay. Biotechniques. 2001;30:290–292. doi: 10.2144/01302st01. [DOI] [PubMed] [Google Scholar]
- 108.Baldrich E, Acero JL, Reekmans G, Laureyn W, O'Sullivan CK. Displacement enzyme linked aptamer assay. Anal. Chem. 2005;77:4774–4784. doi: 10.1021/ac0502450. [DOI] [PubMed] [Google Scholar]
- 109.Wang H, Yang X, Bowick GC, Herzog NK, et al. Identification of proteins bound to a thioaptamers probe on a proteomics array. Biochem. Biophys. Res. Commun. 2006;347:586–593. doi: 10.1016/j.bbrc.2006.06.132. [DOI] [PubMed] [Google Scholar]
- 110.Gold L, Ayers D, Bertino J, Bock C, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ostroff RM, Bigbee WL, Franklin W, Gold L, et al. Unlocking biomarker discovery: Large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One. 2010;5:e15003. doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kraemer S, Vaught JD, Bock C, Gold L, et al. From aptamer-based biomarker discovery to diagnostic and clinical applications: an aptamer based, streamlined multiplex proteomic assay. Nat. Preceding. 2010:4642.1. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-facilitated biomarker discovery (AptaBiD). J. Am. Chem. Soc. 2008;2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 114.Ahn JY, Lee SW, Kang HS, Jo M, et al. Aptamer Microarray Mediated Capture and Mass Spectrometry Identification of Biomarker in Serum Samples. J Proteome Res. 2010;9:5568–5573. doi: 10.1021/pr100300t. [DOI] [PubMed] [Google Scholar]
- 115.Cole JR, Dick LW, Jr., Morgan EJ, Linda B, McGown LB. Affinity Capture and Detection of Immunoglobulin E in Human Serum Using an Aptamer-Modified Surface in Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chem. 2007;79:273–279. doi: 10.1021/ac061256b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keller KM, Breeden MM, Zhang J, Ellington AD, Brodbelt JS. Electrospray ionization of nucleic acid aptamer/small molecule complexes for screening aptamer selectivity. J. Mass Spectrom. 2005;40:1327–1337. doi: 10.1002/jms.915. [DOI] [PubMed] [Google Scholar]
- 117.Zhao Y, Widen SG, Jamaluddin M, Tian B, et al. Quantification of activated NF-κB/RelA complexes using ss DNA aptamer affinity – Stable isotope dilution selected reaction monitoring—mass spectrometry. Mol Cell Proteomics. 2011;10:M111.008771. doi: 10.1074/mcp.M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hecht AH, Sommer GJ, Durland RH, Yang X, et al. Aptamers as affinity reagents in an integrated electrophoretic lab-on-a-Chip platform. Anal. Chem. 2010;82:8813–8820. doi: 10.1021/ac101106m. [DOI] [PubMed] [Google Scholar]
- 119.Bock C, Coleman M, Collins B, Davis J, et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4:609–18. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 120.Kirby R, Cho EJ, Gehrke B, Bayer T, et al. Aptamer-based sensor arrays for the detection and quantification of proteins. Anal. Chem. 2004;76:4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 121.Brody EN, Willis MC, Smith JD, Jayasena S, et al. The use of aptamers in large arrays for molecular diagnostics. Mol. Diagn. 1999;4:381–388. doi: 10.1016/s1084-8592(99)80014-9. [DOI] [PubMed] [Google Scholar]
- 122.Collett JR, Cho EJ, Ellington AD. Production and processing of aptamer microarrays. Methods. 2005;37:4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 123.McCauley TG, Hamaguchi N, Stanton M. Aptamer based biosensor arrays for detection and quantification of biological macromolecules. Anal. Biochem. 2003;319:244–250. doi: 10.1016/s0003-2697(03)00297-5. [DOI] [PubMed] [Google Scholar]