Abstract
Immunohistochemistry has recently been validated for the detection of the BRAFV600E mutation across a range of tumor types. In colorectal carcinoma, the presence of the BRAFV600E mutation can be used to virtually exclude Lynch syndrome in mismatch repair-deficient tumors. In mismatch repair-proficient tumors, BRAFV600E mutation assessed by molecular methods has been proposed as a poor prognostic factor. We investigated whether combined BRAFV600E and mismatch repair status assessment by immunohistochemistry alone can be used as a prognostic marker in the routine clinical setting. We performed immunohistochemistry for BRAFV600E, MLH1, PMS2, MSH2, and MSH6 on 1426 consecutive unselected colorectal carcinomas. Ninety-one (6.4%) carcinomas were mismatch repair-proficient and BRAFV600E mutant, and these tumors demonstrated a significantly worse 5-year survival of 49.7% compared with mismatch repair-proficient BRAF wild type (74.1% of tumors, 65.4% survival), mismatch repair-deficient BRAFV600E mutant (12.9% of tumors, 70.1% survival), and mismatch repair-deficient BRAF wild type (6.6% of tumors, 73.6% survival). The poor survival was confirmed by univariate analysis (P<0.01) but fell away in multivariate analysis (P=0.68) because of the strong effect of tumor stage and age on overall survival. We conclude that in addition to its utility in screening for Lynch syndrome, reflex BRAFV600E and mismatch repair assessment by immunohistochemistry can be used as a powerful predictor of all-cause survival.
Keywords: BRAFV600E, colorectal carcinoma, microsatellite instability, mismatch repair deficiency
To be adopted in the routine clinical setting, a biomarker must both demonstrate a clear additional benefit over standard clinical, radiological, and pathological examination (usually to predict outcome or response to treatment), and also be cost effective and readily available. Although literally thousands of biomarkers have been proposed, very few have entered routine clinical practice, owing to a combination of poor efficacy, expense, and lack of availability.1 For example, in colorectal carcinoma, the only biomarkers in widespread clinical use are KRAS mutation testing, which is used to predict response to anti-EGFR-targeted therapy in metastatic disease, and either microsatellite instability determination by molecular methods or immunohistochemistry for the DNA mismatch repair markers MLH1, PMS2, MSH2, and MSH6, which are essentially equally effective in triaging genetic testing for Lynch syndrome.2 There are currently no prognostic biomarkers for colorectal carcinoma in routine clinical use.
The presence of BRAF mutation in mismatch repair-deficient tumors is commonly used to virtually exclude Lynch syndrome, and many centers now perform routine BRAF testing in all microsatellite-unstable tumors.2 Recently, BRAF-mutant mismatch repair-proficient colorectal carcinomas have emerged as a poor prognostic phenotype3, 4 with unique features, including a poor or absent response to anti-EGFR therapy, despite being wild type for KRAS.5, 6, 7 Identification of these mismatch repair-proficient BRAF mutant tumors may be beneficial to predict poor outcome and to guide therapy.5, 6, 7 However, current testing, which is usually based on molecular techniques, has not been deployed universally because of the additional expense and because molecular testing is outside the routine workflow of many surgical pathology laboratories, which is based on morphology and immunohistochemistry.
Recently, four groups have demonstrated that mutation-specific immunohistochemistry using a novel commercially available mouse monoclonal antibody is a highly sensitive and specific technique to identify the BRAFV600E mutation in colorectal carcinomas.2, 8, 9, 10 To date, its main suggested utility has been to triage molecular testing for Lynch syndrome in mismatch repair-deficient tumors. It has been proposed that if BRAFV600E immunohistochemistry were performed universally in all tumors, it could also be used to detect the poor prognosis mismatch repair-proficient BRAFV600E mutant colorectal carcinomas.2 As the antibody is now commercially available and used in many diagnostic pathology laboratories, if validated as a prognostic marker BRAFV600E immunohistochemistry assessed in conjunction with mismatch repair status could become the first prognostic biomarker for colorectal cancer deployed into routine clinical use.
In this study, we sought to validate combined BRAF and mismatch repair status as determined by immunohistochemistry as a prognostic biomarker in colorectal carcinoma.
Materials and methods
We searched the pathology database of the Royal North Shore Hospital, Sydney, Australia, for all colorectal carcinomas undergoing surgical resection during the calendar years 2004–2009. During this period, this center performed centralized pathological testing for two quaternary referral hospitals with dedicated colorectal surgery units and four community hospitals. Therefore, this patient cohort represents a true snapshot of colorectal carcinoma cases encountered in the community as a whole. Tissue microarrays containing duplicate 1 mm cores were created and immunohistochemistry for the four mismatch repair proteins (MLH1, MSH2, PMS2, and MSH6) and BRAFV600E (using clone VE1, SpringBioscience, Pleasonton, CA) were performed using previously described methods.2
For BRAFV600E mutation-specific immunohistochemistry, slides were stained using the Leica BondIII autostainer (Leica Microsystems, Mount Waverley, VIC, Australia) used according to the manufacturer's protocol. The slides were dewaxed in Bond Dewax solution (AR9222, Leica Microsystems) and hydrated in Bond Wash solution (AR9590, Leica Microsystems). Heat-induced epitope retrieval was performed for 60 min in the manufacturer's alkaline retrieval solution ER2 (VBS part no: AR9640, Leica Microsystems). Slides were then incubated with the primary antibody at a dilution of 1 in 80 for 30 min at room temperature. Antibody detection was performed using the biotin-free Bond Polymer Defined Detection System (DS9713, Leica Microsystems) according to the manufacturer's protocol. Slides were then counterstained with hematoxylin. Staining was interpreted by a single pathologist blinded to all other data. Slides were considered positive if >20% of neoplastic cells stained positively. If the tissue microarray staining was not clearly interpretable (eg, if there were no good internal controls for the mismatch repair markers), it was repeated on whole sections.
Overall survival was defined as the duration alive from time of definitive surgery. Follow-up was obtained by the examination of hospital medical records, the records from surgeons' private rooms, and archival public death notices and obituaries in the state of New South Wales, Australia. Patients were followed up until death or their last date of follow-up not >7 years after definitive surgery.
The survival for each of the four immunohistochemical phenotypes was determined as a function of cumulative survival over time (Kaplan–Meier method). Pairwise log rank test was used to determine significance in survival differences. Multivariate Cox regression was employed to explore the effect of mismatch repair/BRAF immunohistochemistry phenotype on survival, using a model that included gender, age at diagnosis, tumor anatomic location, tumor histologic grade, presence or absence of lymphovascular space invasion, peritumoral lymphocyte reaction status, and American Joint Committee on Cancer/TNM seventh edition tumor stage. A P-value<0.05 was taken to be significant. All analyses were performed using IBM SPSS Statistics version 21 on OSX.
This study was approved by the Northern Sydney Local Health District Human Research Ethics Committee under protocol 1201-035M.
Results
A total of 1426 colorectal carcinomas were assessed in the tissue microarray. The clinical and pathological features are presented in Table 1. Briefly, the mean age was 74 years (range 17–100 years) and 52.1% were females. Most patients (79.2%) presented with stage 2 or 3 disease. One thousand one hundred and forty-eight (80.5%) were mismatch repair-proficient, comprising 1057 (74.1%) mismatch repair-proficient BRAF wild type and 91 (6.4%) mismatch repair-proficient/BRAFV600E mutant. One hundred and eighty-four cases (12.9%) were mismatch repair-deficient/BRAFV600E mutant and 94 (6.6%) mismatch repair-deficient/BRAF wild type.
Table 1. Clinical and pathological characteristics of 1426 consecutive colorectal cancer patients (2004–2009).
| Variable | Count (%) unless otherwise stated | Single variable P-valuea | Univariate analysis HR (95% CI), P-value | Multivariate analysis HR (95% CI), P-value |
|---|---|---|---|---|
| Gender | 0.13 | |||
| Female | 743 (52.1) | 1.00 | 1.00 | |
| Male | 683 (47.9) | 0.82 (0.67–1.01), 0.07 | 1.24 (0.94–1.65), 0.13 | |
| Age at diagnosis, median (range) | 74 (17–100) | 1.03 (1.02–1.04), <0.01 | 1.04 (1.03–1.05), <0.01 | |
| Anatomic location | <0.01 | |||
| Rectum | 363 (25.4) | 1.00 | 1.00 | |
| Cecum | 312 (21.9) | 1.36 (1.00–1.85), 0.05 | 1.08 (0.72–1.62), 0.71 | |
| Ascending colon | 219 (15.3) | 1.22 (0.85–1.75), 0.29 | 1.00 (0.62–1.62), 0.99 | |
| Transverse colon | 168 (11.8) | 1.95 (1.37–2.77), <0.01 | 1.17 (0.72–1.90), 0.54 | |
| Descending colon | 51 (3.6) | 1.65 (0.96–2.83), 0.07 | 0.88 (0.42–1.86), 0.74 | |
| Sigmoid colon | 302 (21.2) | 1.14 (0.83–1.58), 0.42 | 1.02 (0.67–1.55), 0.93 | |
| Histologic grade | <0.01 | |||
| Low | 826 (57.9) | 1.00 | 1.00 | |
| High | 214 (15.0) | 1.87 (1.42–2.47), <0.01 | 1.27 (0.87–1.84), 0.21 | |
| Lymphovascular space invasion | 0.02 | |||
| Absent | 538 (37.7) | 1.00 | 1.00 | |
| Present | 459 (32.2) | 2.26 (1.72–2.95), <0.01 | 1.57 (1.13–2.19), <0.01 | |
| Peritumoral lymphocyte reaction | <0.01 | |||
| Absent | 45 (3.2) | 1.00 | 1.00 | |
| Present | 999 (70.1) | 2.21 (0.91–5.37), 0.08 | 1.90 (0.74–4.89), 0.18 | |
| Overall stage AJCC/TNM 7th edn | <0.01 | |||
| I | 235 (16.5) | 1.00 | 1.00 | |
| IIA | 415 (29.1) | 2.00 (1.30–3.17), <0.01 | 2.21 (1.20–4.11), 0.01 | |
| IIB | 85 (6.0) | 2.44 (1.34–4.42), <0.01 | 2.66 (1.20–5.89), 0.02 | |
| IIC | 15 (1.1) | 9.66 (4.17–22.40), <0.01 | 11.93 (4.68–30.41), <0.01 | |
| IIIA | 64 (4.5) | 1.10 (0.50–2.43), 0.82 | 0.94 (0.27–3.33), 0.93 | |
| IIIB | 376 (26.3) | 3.05 (1.97–4.72), <0.01 | 2.75 (1.49–5.08), <0.01 | |
| IIIC | 174 (12.2) | 6.48 (4.12–10.17), <0.01 | 5.85 (3.11–11.02), <0.01 | |
| IVA | 32 (2.2) | 8.06 (4.24–15.31), <0.01 | 11.76 (4.88–28.32), <0.01 | |
| IVB | 30 (2.1) | 14.10 (7.47–26.64), <0.01 | 15.86 (6.71–37.48), <0.01 | |
| Mismatch repair IHC status | <0.01 | |||
| Proficient | 1148 (80.5) | 1.00 | ||
| Deficient | 278 (19.5) | 0.74 (0.55–0.99), 0.04 | ||
| BRAFV600E immunohistochemistry status | <0.01 | |||
| Wild type | 1151 (80.7) | 1.00 | ||
| Mutant | 275 (19.3) | 1.14 (0.88–1.49), 0.32 | ||
| Immunhoistochemistry phenotypes | <0.01 | |||
| Mismatch repair-proficient/BRAF wild type | 1057 (74.1) | 1.00 | 1.00 | |
| Mismatch repair-deficient/BRAFV600E mutant | 184 (12.9) | 0.84 (0.60–1.19), 0.32 | 0.57 (0.35–0.93), 0.03 | |
| Mismatch repair-deficient/BRAF wild type | 94 (6.6) | 0.66 (0.40–1.08), 0.10 | 0.65 (0.34–1.27), 0.21 | |
| Mismatch repair-proficient/BRAFV600E mutant | 91 (6.4) | 1.79 (1.24–2.60), <0.01 | 1.10 (0.69–1.76), 0.68 |
Reports on the significance of differences between two or more categories within each variable as a one sample non-parametric binomial or χ2-test.
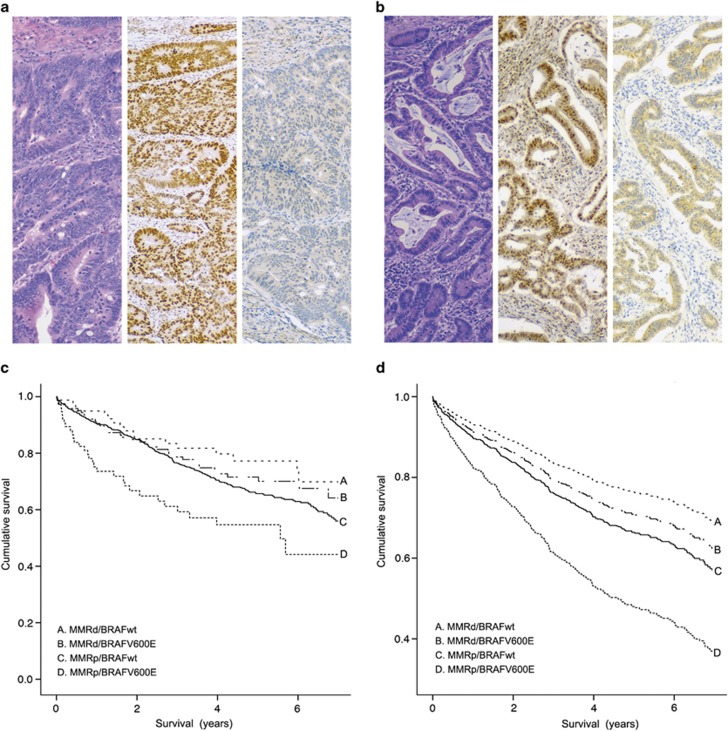
During follow-up, (mean 5.29 years, 75th centile 3.21 years), 353 patients died. Survival curves correlating immunohistochemical staining pattern and survival by both Kaplan–Meier and Cox regression methods are presented in Figure 1 along with photomicrographs of representative staining patterns. The 5-year survivals progressively deteriorated from mismatch repair-deficient/BRAF wild type (73.6%) to mismatch repair-deficient/BRAFV600E mutant (70.1%), to mismatch repair-proficient/BRAF wild type (65.4%) to mismatch repair-proficient/BRAFV600E mutant (49.7%). Pairwise comparisons with mismatch repair-proficient/BRAF wild-type colorectal carcinomas as the baseline group in univariate Cox regression modeling revealed overlapping 5-year survival figures for all tumor groups, except mismatch repair-proficient/BRAFV600E mutant tumors, which showed a statistically significant worse outcome—hazard ratio of death 1.79 (95% CI=1.24–2.60), P<0.01. In multivariate analysis, the poor prognosis of mismatch repair-proficient/BRAFV600E mutant tumors was negated (hazard ratio of 1.10 (95% CI=0.69–1.76), P=0.68) by the dominant effect of stage and age on overall survival. However, the better prognosis of mismatch repair-deficient/BRAFV600E mutant tumors became significant (hazard ratio of 0.57 (95% CI=0.35–093), P=0.03) when compared with the baseline group of mismatch repair-proficient/BRAF wild-type tumors (Table 1).
Figure 1.
(a,b) Representative photomicrographs serially stained for hematoxylin and eosin, PMS2 and BRAFV600E of (a) DNA mismatch repair-proficient/BRAF wild-type colorectal carcinoma and (b) DNA mismatch repair-proficient/BRAFV600E mutant tumor (original magnifications, × 400). (c) Kaplan–Meier survival functions of the four immunohistochemistry phenotypes. (d) Univariate Cox regression survival function of the four immunohistochemistry phenoytpes.
Discussion
The use of molecular markers to predict outcome in colorectal carcinoma, particularly mismatch repair deficiency/microsatellite instability and BRAF mutation, has been an area of active research. Briefly, most studies have indicated that mismatch repair deficiency is associated with a good outcome.3, 11, 12, 13, 14, 15, 16, 17 In contrast, the presence of BRAF mutation is usually found to be a marker of poor prognosis.3, 18, 19, 20, 21, 22, 23, 24 Although some studies have found that BRAF mutation does not predict outcome in an unselected population,25 this discrepancy appears to be because of the strong association between BRAF mutation (a poor prognostic factor) with mismatch repair deficiency (a good prognostic factor) through the somatic hypermethylation pathway.
Therefore, recently several groups have used the combination of MMR and BRAF mutation status as determined by molecular means to predict outcome in colorectal cancer. Using this approach, mismatch repair-deficient/BRAF wild-type colorectal carcinomas have been consistently found to have a good prognosis,4, 20, 26, 27 whereas mismatch repair-proficient/BRAFV600E mutant colorectal carcinomas have emerged as a poor prognostic group in most3, 4, 12, 27, 28 but not all studies.26
This combined mismatch repair/BRAF prognostic algorithm was tested recently by Lochhead et al29 who used molecular techniques to determine microsatellite instability and BRAF mutation status in 1253 colorectal carcinoma patients. Compared with the majority of tumors that were mismatch repair-proficient/BRAF wild type, mismatch repair-proficient/BRAF mutant colorectal carcinomas demonstrated a poor prognosis (hazard ratio of colon cancer-specific mortality 1.6 (95% CI=0.12–2.28)). Mismatch repair-deficient/BRAF mutant colorectal carcinomas demonstrated a good prognosis with a hazard ratio of 0.48 (95% CI=0.27–0.87) and mismatch repair-deficient/BRAF wild type demonstrated a very good prognosis with a hazard ratio of 0.25 (95% CI=0.12–0.52).
The results of our study, although based on all cause rather than cancer-specific survival, are essentially confirmatory of Lochhead et al,29 with the significant advantage that we did not use any molecular techniques, only immunohistochemistry—an approach that is readily available in virtually any diagnostic surgical pathology laboratory. In centers where universal Lynch syndrome screening is already undertaken with mismatch repair deficiency immunohistochemistry for MLH1, PMS2, MSH2, and MSH6, it would simply be a matter of performing immunohistochemistry for five markers rather than four, with estimated additional disposable costs of <US$10 and minimal extra labor costs.2
We note that the very poor prognosis of mismatch repair-proficient/BRAF mutant tumors found in univariate analysis (P<0.01) fell away in multivariate analysis because of the dominant effect of stage and age on overall survival. However, given the ease with which BRAF status can be determined in conjunction with mismatch repair deficiency in the routine clinical setting, and the established indication for combined mismatch repair deficiency and BRAF testing to triage formal genetic testing for Lynch syndrome,2 our study makes an argument for its potential use as a prognostic marker in all colorectal cancers.
To date, four of the five independent studies investigating BRAFV600E mutation-specific immunohistochemistry in colorectal carcinoma have determined that it is highly sensitive and specific for the presence of the BRAFV600E mutation with only one study suggesting limited utility.2, 8, 9, 10, 30 It would be reasonable to conclude that local factors such as tissue processing techniques and staining methods can affect the performance of the antibody, but most laboratories including our own2 have found it to be a robust and reliable marker. However, we do caution that introduction of BRAF immunohistochemistry should only be performed with the appropriate quality control measures, including the use of controls and the validation of the accuracy of the testing in individual laboratories.
Previous studies of BRAFV600E mutation-specific immunohistochemistry in colorectal carcinoma have concentrated on its utility in triaging formal genetic testing for Lynch syndrome in microsatellite-unstable tumors.2, 8, 9, 10 Other studies have concentrated on the prognostic predictive power of combined BRAF and mismatch repair deficiency assessment when determined by molecular means.3, 4, 20, 26, 27, 28, 29 This is the first study to demonstrate that BRAF determination by immunohistochemistry alone, when interpreted in conjunction with mismatch repair deficiency status, also identifies subgroups of colorectal carcinomas with different overall survivals—most significantly the poor prognostic group of mismatch repair-proficient/BRAFV600E mutant tumors.
In summary, our results suggest that the addition of BRAFV600E immunohistochemistry to mismatch repair deficiency immunohistochemistry identifies distinct prognostic subgroups of colorectal carcinomas, including the poor prognostic group of mismatch repair-proficient/BRAFV600E mutant tumors. If our results are confirmed in other large independent cohorts, a strong argument could be made to perform routine BRAFV600E immunohistochemistry at the same time as mismatch repair deficiency assessment on all colorectal carcinomas, not just to facilitate universal screening for Lynch syndrome in mismatch repair-deficient tumors but also to identify the poor prognosis mismatch repair-proficient/BRAFV600E mutant group.
Acknowledgments
We thank Ms Nicole Watson for her assistance with the initial stages of the data assembly. This study was approved by the Northern Sydney Local Health District (NSLHD) Human Research Ethics Committee (HREC) under protocol 1201-035M.
The authors declare no conflict of interest.
References
- Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- Toon CW, Walsh MD, Chou A, et al. BRAFV600E Immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 2013;37:1592–1602. doi: 10.1097/PAS.0b013e31828f233d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai RK, Jayachandran P, Koong AC, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744–752. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCN Clinical practice guidelines in oncology. Colon Cancer. Version 3. 2013. Available at http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed: 1 August 2013.
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Capper D, Voigt A, Bozukova G, et al. BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer. 2013;133:1624–1630. doi: 10.1002/ijc.28183. [DOI] [PubMed] [Google Scholar]
- Sinicrope FA, Smyrk TC, Tougeron D, et al. Mutation-specific antibody detects mutant BRAF(V600E) protein expression in human colon carcinomas. Cancer. 2013;119:2765–2770. doi: 10.1002/cncr.28133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A, Heinonen M, Kantonen J, et al. BRAF mutation in sporadic colorectal cancer and Lynch syndrome Virchow's Arch(Epub ahead of Print)2013 [DOI] [PubMed]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinicrope FA, Foster NR, Thibodeau SN, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertagnolli MM, Redston M, Compton CC, et al. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer—a study of CALGB 9581 and 89803. J Clin Oncol. 2011;29:3153–3162. doi: 10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AD, Delorenzi M, Tejpar S, et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst. 2012;104:1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- Eklöf V, Wikberg ML, Edin S, et al. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108:2153–2163. doi: 10.1038/bjc.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21:1792–1798. doi: 10.1158/1055-9965.EPI-12-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- Kalady MF, Dejulius KL, Sanchez JA, et al. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55:128–133. doi: 10.1097/DCR.0b013e31823c08b3. [DOI] [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- French AJ, Sargent DJ, Burgart LJ, et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlobec I, Kovac M, Erzberger P, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. 2010;127:2569–2575. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adackapara CA, Sholl LM, Barletta JA, et al. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology. 2013;63:187–193. doi: 10.1111/his.12154. [DOI] [PubMed] [Google Scholar]