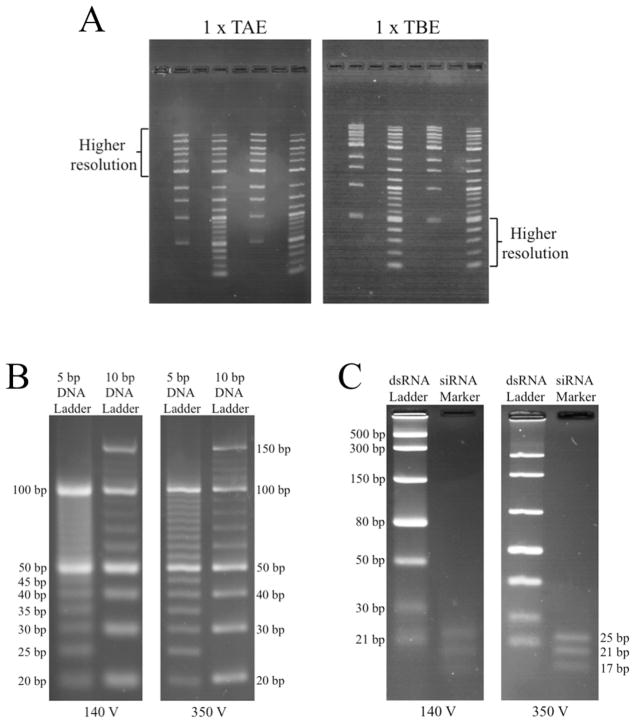
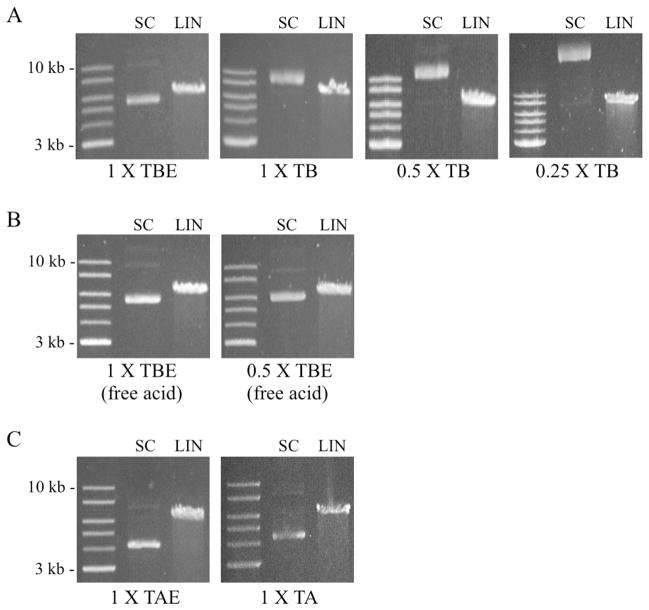
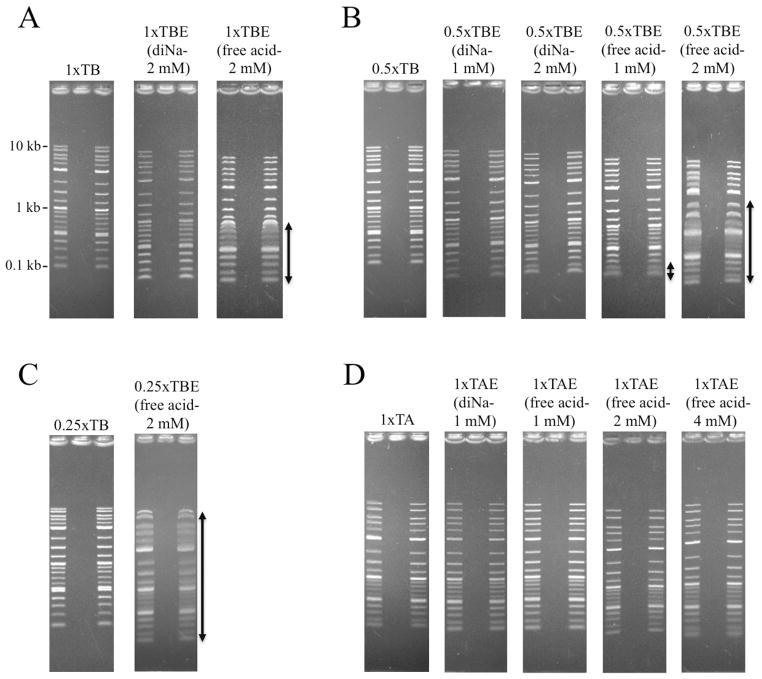
Abstract
Agarose gel electrophoresis of DNA and RNA is routinely performed using buffers containing either Tris, acetate and EDTA (TAE) or Tris, borate and EDTA (TBE). Gels are run at a low, constant voltage (~ 10 V/cm) to minimize current and asymmetric heating effects, which can induce band artifacts and poor resolution. In this study, alterations of gel structure and conductive media composition were analyzed to identify factors causing higher electrical currents during horizontal slab gel electrophoresis. Current was reduced when thinner gels and smaller chamber buffer volumes were used, but was not influenced by agarose concentration or the presence of ethidium bromide. Current was strongly dependent upon the amount and type of EDTA used and on the concentrations of the major acid-base components of each buffer. Interestingly, resolution and the mobilities of circular versus linear plasmid DNAs were also affected by the chemical form and amount of EDTA. With appropriate modifications to gel structure and buffer constituents, electrophoresis could be performed at high voltages (20–25 V/cm), reducing run times by up to 3-fold. The most striking improvements were observed with small DNAs and RNAs (10 – 100 bp): high voltages and short run times produced sharper bands and higher resolution.
Keywords: gel electrophoresis, agarose, nucleic acids, current, resolution
Gel electrophoresis is routinely used for detection and size analysis of proteins and nucleic acids [1–6]. Separation by this method utilizes an electrical current to propel charged biomolecules through a porous gel matrix at a rate that is a function of the charge, size and shape of the molecules. Proteins are frequently analyzed using vertically-oriented gels made of polyacrylamide, while DNA and RNA are most commonly analyzed using horizontal agarose slab gels. Agarose exists as a copolymer containing 1,3-linked B-D-galactose and 1,4-linked 3,6-anhydro-α-L-galactose. A small number of sulfate and other anionic groups are also present [3,4,7]. Boiling the carbohydrate in water and cooling leads to the formation of helical bundles of agarose strands held together by hydrogen bonds. The bundles associate with each other to create pores that permit molecular sieving of biomolecules.
Standard horizontal agarose slab gels allow separation of DNAs ranging in size from ~ 75 bp to 30,000 bp [3,4,8,9]. Large DNAs at the top of this range are studied using low percentage agarose gels (0.3–0.5%), while small DNAs are typically analyzed using higher percentages (2–4%) [8–10]. Highest resolution of single-stranded and double-stranded DNAs smaller than 75 nucleotides in length is achieved using polyacrylamide gels. This is because polyacrylamide’s strong, covalently bonded gel framework can produce smaller and more stable gel pores, allowing finer molecular sieving [2,3,6].
Electrophoresis of DNA in slab gels is primarily performed using one of two standard buffers, either TAE, composed of Tris (2-amino-2-[hydroxymethyl]-1,3-propanediol), acetic acid and EDTA (ethylenediaminetetraacetic acid) or TBE (Tris, boric acid and EDTA). These solutions contain a weak acid, acetic or boric acid, that can exist in neutral and anionic forms (e.g., COOH and COO− species) and a weak base, Tris, that exists in either neutral or cationic forms (Tris-NH2 and Tris-NH3+). These ions carry the electrical current, buffer the pH, and maintain a low conductivity medium. EDTA is not absolutely essential [4], but is added as a preventative because it chelates Mg2+ ions and therefore inactivates potential DNA nucleases that may be present.
For separation of DNA molecules ranging in size from several hundred to several thousand bp, both TAE and TBE provide good resolution of DNA fragments, with slightly improved separation of smaller fragments in TBE and of larger sizes in TAE. Despite their similar performance in most applications, some buffer-specific effects have been observed, especially in borate-containing buffers [11–15]. Over the last several years, other conductive media have also been developed for electrophoresis of nucleic acids. Examples include solutions containing L-histidine, sodium borate, sodium threonine, lithium borate, and mixtures of pKa-matched organic acids and bases [16–21].
DNA electrophoresis is usually performed at constant voltage. During an experiment, the current (measured in mA) increases and warming of the running buffer occurs. The voltage is kept low, ~ 10 V/cm, where cm refers to the length of the gel, to minimize heating effects. For a typical small- or medium-sized horizontal gel rig (10–15 cm length), power supplies are set to between 100 and 150 volts and gels are run for 50–90 min. Increasing the voltage above this range using standard 1 x TAE or TBE buffers increases the speed of movement of the DNAs, but leads to asymmetric heating of the gel and solution. Such heating is undesirable because it promotes broadening, slanting, and compression of the bands into each other, as well as faster movement of samples in the center than on the outside of the gel (“smiling”) and other lane anomalies [3,4,6,22].
Although TAE and TBE are almost universally employed in biochemistry laboratories, their compositions have never been rigorously optimized and therefore essentially arbitrary concentrations are still used today (89 mM Tris, 89 mM boric acid, and 2 mM EDTA in 1 x TBE and 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA in 1 x TAE) [4,8,9]. In the current study, we have systematically examined the impacts of acid, base and EDTA concentrations on electrical current and on resolution of linear and circular DNAs within agarose slab gels. In addition, the impact of altering gel structure, percent agarose and electrophoresis buffer volume on electrical current was investigated. Using optimized electrophoresis conditions arising from these experiments, we demonstrate that much higher voltages are achievable with traditional buffers, resulting in shorter run times and improved resolution of small DNA and RNA molecules.
Materials and Methods
Materials
Tris base was purchased from J. T. Baker and glacial acetic acid was from Mallinckrodt Chemicals. Ethylenediaminetetraacetic acid (EDTA)-free acid, EDTA-disodium salt and Omnipur agarose were obtained from EMD Chemicals, Inc. Boric acid was purchased from Sigma-Aldrich and ethidium bromide (EtBr) was from Shelton Scientific, Inc. The 2-Log DNA ladder, 1 Kb DNA ladder, dsRNA ladder and siRNA ladder standards were purchased from New England Biolabs.
Gel electrophoresis methods
Gel electrophoresis experiments were performed using 1.0% agarose gels and 6 cm × 10 cm mini-sub cell GT 8 gel rigs (Bio-Rad) with a default gel volume of 40 ml unless specified otherwise. Some experiments utilized 11 cm × 14 cm gels and Horizon rigs (LabRepco) along with 70 ml gel volumes. Agarose was dissolved by microwaving (for 25–30 s on high setting initially, then in 5–10 s increments) in the same electrophoresis buffer that was used to cover the gel in the rig. After pouring into a plastic tray with tape attached to two ends, gels were allowed to solidify at room temperature for at least 20 min. Allowing less time than this made gels prone to band artifacts, possibly because agarose near the wells had not solidified completely. EPS601 (Amersham Bioscience) and EC-105 (E-C Apparatus Corporation) power supplies were employed for all electrophoresis experiments. Electrophoresis was performed until the front edge of the bromophenol blue tracking dye had migrated to a point that was 70% down the gel. A line was placed on the sides of each gel tray to ensure that the tracking dye traveled the same distance in each gel. After electrophoresis was completed, gels were stained for 15–20 min with EtBr (~ 0.5 ug/ml), rinsed 3–4 times with deionized water, and images were captured using an Alpha Innotech RED gel imager or a Kodak Digital Science 440CF Imagestation instrument. For experiments testing the effects of EtBr on electrical currents, the chemical was added to the gel after microwaving and to the electrophoresis buffer covering the gel at concentrations ranging from 0.5 μg/ml to 2.0 μg/ml.
Evaluation of parameters affecting temperature and pH during electrophoresis
For experiments involving measurement of gel buffer temperatures, two thermometers were placed into the electrophoresis buffer, one at each end of the rig, and the results were averaged. pH was determined using a Corning 220 pH meter after removing 2 ml of electrophoresis buffer from above the anode and cathode both before and immediately after completion of electrophoresis.
Analysis of the relative migration of supercoiled and linear plasmid DNAs
Experiments using plasmid DNAs employed the yeast shuttle vector p426URA3 [23]. Plasmids were purified using a miniprep kit (Qiagen) and digested with EcoRI (New England Biolabs) to generate linearized DNAs, which were precipitated with ethanol and resuspended in water prior to electrophoresis as described [9].
Results
The buffers 1 x TAE and 1 x TBE are almost universally used for agarose gel electrophoresis, but their tendency to build up electrical current and heat places limits on the voltages that can be applied, which are typically 100–150V for gels that are 10–15 cm long. To expand upon previous work [4] and identify factors that affect heating, we initially investigated the effects of altering several physical and chemical parameters on the generation of electrical current using 10 cm agarose gels.
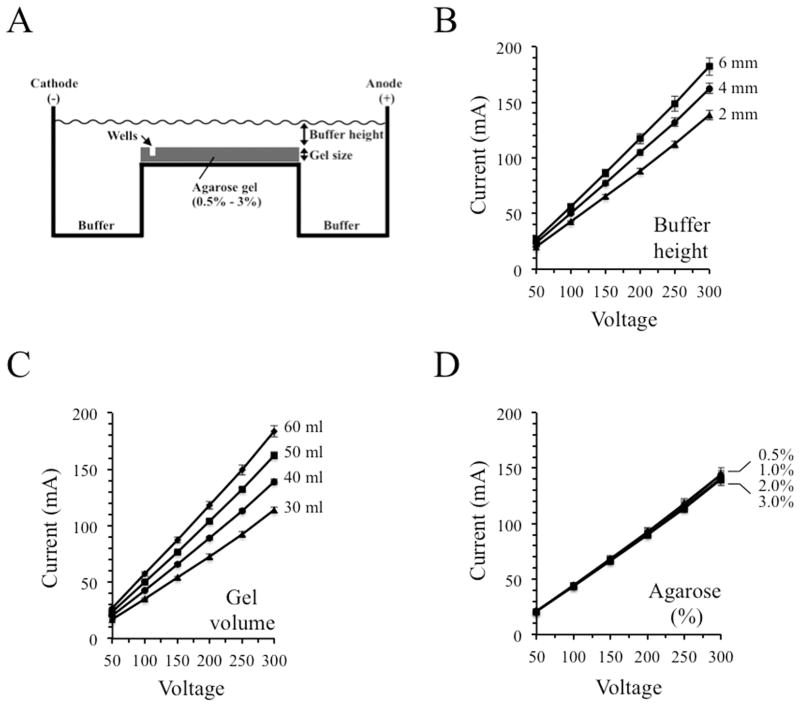
A typical horizontal slab gel electrophoresis system is depicted schematically in Figure 1A. The length and width of a gel in a commercially available rig are fixed, but other parameters such as percent agarose, total gel size, and electrophoresis buffer volume can be adjusted by the experimenter. Initial tests assessed the effects of changing the volume of buffer within the rig, determined by measuring the height of the buffer above the top surface of the gel (Figure 1B). The experiments employed a 1% agarose gel (6 cm × 10 cm) prepared in 40 ml of 1 x TBE buffer and electrical currents were measured over a range of 50 – 300 V. In this experiment and the others shown in Figure 1, measurements were taken in 4 different gel rigs and the resulting averages and standard deviations are shown. Increasing the height of the buffer above the gel from 2 mm to 6 mm produced a consistent increase in current. For example, adjusting the height from 2 mm to 6 mm increased the current at 150 V (15 V/cm) from 61 mA to 85 mA, an increase of 40% (Figure 1B).
Fig.1.
Altering gel size and buffer volume influences the electrical current generated during agarose gel electrophoresis. 10 cm gels were employed for each assay. (A) Schematic representation of a typical horizontal gel electrophoresis system. (B) Raising the height of the electrophoresis buffer above the surface of the gel increased the current. (C) Current was elevated when the height of the gel was increased while maintaining a constant 2 mm buffer height above its surface. (D) Currents were not affected when agarose gel concentrations were varied from 0.5% – 3.0%. Averages and standard deviations from measurements performed using four different gel rigs are shown.
Expanding the volume of the gel (producing changes in gel height because the length and width are fixed by the gel molding tray) while maintaining a constant 2 mm buffer height above the gel revealed that larger gels produce higher currents. At 150 V, the current increased by 64% when gel volume was raised from 30 ml to 60 ml (Figure 1C). Average heights for the gels analyzed in Figure 1C were determined to be 4.1, 6.8, 7.3 and 9.0 mm for the 30, 40, 50 and 60 ml gels, respectively.
Altering the percentage of agarose in a gel changes its strength and average pore size and determines the effective size fractionation range. Molecular biologists routinely adjust this percentage within the range of approximately 0.5% to 3.0%, depending on the sizes of the nucleic acids being analyzed [8,9,24]. Interestingly, electrical currents did not change when agarose concentrations were varied between 0.5% and 3.0% (Figure 1D). These results indicate that gel volume and buffer height, but not agarose concentration and average pore size, exert strong effects on current and the potential of a system to produce excess heating during electrophoresis.
EDTA chelates magnesium, an essential cofactor for many nucleases, and thereby protects DNA from degradation, but its necessity in electrophoresis has been questioned [4]. The major electrophoresis buffers 1 x TAE and 1 x TBE contain 1 mM and 2 mM EDTA, respectively. At the pH of these buffers (approximately 8.0), three of the four carboxylic acid groups of EDTA exist primarily in their negatively charged carboxylate ion forms and therefore can contribute to the conductivity of the solution.
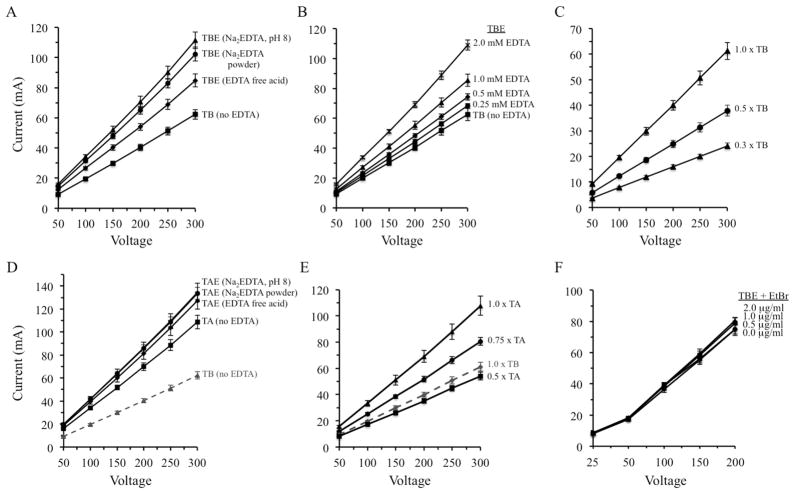
EDTA is poorly soluble below neutral pH and is commercially available in both free acid and sodium salt forms. The disodium form is more soluble than the free acid and is commonly used in concentrated TAE and TBE mixtures that are sold commercially or prepared in research labs. The impacts of the different forms of EDTA on electrical current were investigated in Figure 2. Standard 1 x TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA), prepared using either free acid or disodium EDTA, produced higher currents than 1 x TB buffer containing no EDTA at each voltage between 50 V and 250 V (Figure 2A). The disodium form produced higher currents than the free acid, presumably because of the conductivity contributed by the Na+ ions.
Fig.2.
Current levels are dependent upon the concentrations and chemical forms of electrophoresis buffer components. (A) The presence of 2 mM EDTA, added either as powders of the free acid and disodium forms or as the disodium form prepared as a stock solution set to pH 8.0, increases current when Tris-borate buffers are employed. (B) Current is dependent upon the concentration of disodium EDTA (added from powder during buffer preparation). (C) Reduction of Tris and borate concentrations decreases the current. (D) Current levels are only modestly affected by varying the form of EDTA used to prepare TAE gels. (E) Current is high in 1 x TA solutions but can be decreased by reduction of Tris and acetate concentrations. (F) Currents are not affected by inclusion of ethidium bromide in the gel and buffer during electrophoresis. Averages and standard deviations are shown from tests performed using four different gel rigs.
Some protocols describing the preparation of TBE recommend adding EDTA from a 50 mM or 0.5 M disodium EDTA stock solution that has previously been adjusted to pH 8.0 to allow the chemical to dissolve [8,9,24]. Because sodium hydroxide is used to set the pH, solutions prepared in this way have higher levels of Na+ ions and higher electrical conductivity (Figure 2A; listed as Na2EDTA, pH 8). Varying the concentration of the most commonly used form, disodium EDTA (using the powder, not a stock solution set to pH 8.0, to prepare the buffer), revealed that the concentration must be decreased to approximately 0.25 mM to reduce currents to the same level as a solution without EDTA (Figure 2B).
1 x TBE, containing 89 mM Tris and 89 mM boric acid, is the most common form of this buffer used, but some labs prepare and run gels using 0.5 x TBE [9,25,26]. Decreasing the concentration of the major electrolytes in TB buffer (Tris-NH3+ and B[OH]4−) by using 0.5 x TB substantially reduced the electrical current (Figure 2C). The current observed at 150 V in 1 x TB was reduced by 36% in 0.5 x TB and by 61% using 0.3 x TB buffer.
Currents produced using 1 x TAE as electrophoresis buffer were also influenced by the presence and the form of EDTA used, but the effects were not as strong as those seen with TBE. 1 x TAE prepared using disodium EDTA and free acid EDTA generated currents that were not statistically separable from each other (overlapping standard deviations) (Figure 2D). In addition, currents in TAE buffers containing EDTA were only modestly higher than those observed in TA buffer without EDTA. The more moderate impact of eliminating EDTA from the Tris-acetate buffer is consistent with the fact that the concentration of EDTA in 1 x TAE is only 1 mM whereas it is 2 mM in 1 x TBE. However, results obtained with 1 x TA and 1 x TB (each without EDTA) were also quite different, with currents using TA being approximately twice those observed with TB (Figure 2D). 1 x TA contains 40 mM Tris and 20 mM acetic acid. Reduction of the major charged species within this buffer (Tris-NH3+ and CH3COO−) using 0.5 x TA lowered the current modestly, to approximately the level seen with 1 x TB (Figure 2E). These experiments demonstrate that the concentrations of both EDTA and the major acid-base components can be modified to reduce the current produced using TBE and TAE buffers.
It is standard practice in many molecular biology labs to add ethidium bromide, a DNA-binding fluorophore, to the gel and to the conductive media covering the gel so that a post-electrophoresis DNA staining step is unnecessary (9). Although addition of this chemical introduces charged species into the system, electrical currents were not increased when it was incorporated into gels run in 1 x TBE at the standard concentration (0.5 μg/ml) or at concentrations up to 2.0 μg/ml (Figure 2F).
The experiments above indicated that 1 x TB has lower conductivity than 1 x TA and that currents are even lower in 0.5 x and 0.3 x TB buffers. Standard 1 x TBE buffer produces sharp bands and good resolution in gels run at typical low voltages of ~ 10 V/cm. However, such gels usually require run times of 50 – 90 min to achieve optimum separation of DNA fragments. We performed tests to determine if good resolution could be maintained when electrophoresis was performed at higher voltages, producing shorter run times, if appropriate modifications were made to decrease current and therefore reduce heating and potential DNA banding artifacts. A series of 1% agarose gels that were 10 cm long were prepared using 1 x TB, 0.5 x TB and 0.3 x TB. EDTA was left out of the buffers because it increases the current and therefore limits the voltage that can be applied. Electrophoresis was also performed using low buffer height (2 mm) and low gel volume (40 ml) to minimize the current.
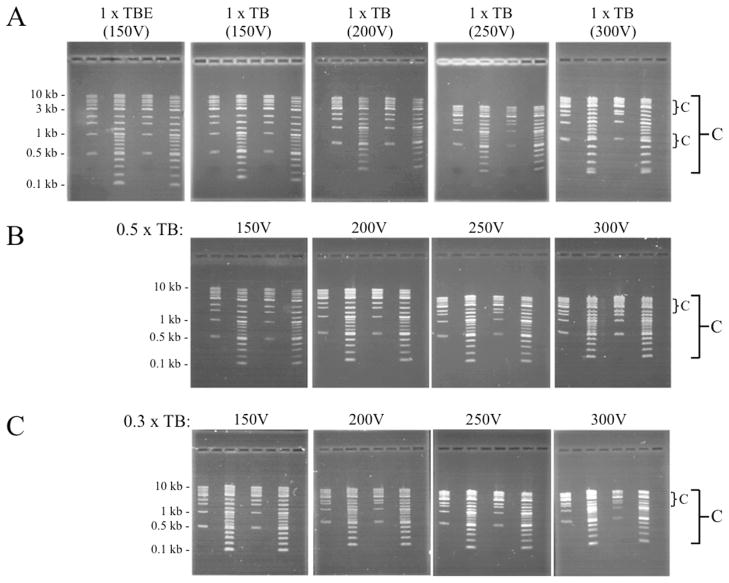
Electrophoresis of linear DNA fragments in two standard DNA ladders using 1 X TBE buffer at 150 V (15 V per cm) revealed good separation of bands (Figure 3A-upper left panel). Lanes 2 and 6 of the gel contain a collection of fragments between 500 bp and 10,000 bp in size and the DNAs in lanes 4 and 8 range from 100 bp to 10,000 bp. Gels were run until the bromophenol blue tracking dye migrated 7 cm down the 10 cm gel. The same ladders were subsequently run in 1 x TB buffer at 150, 200, 250 and 300 V (Figure 3A). Resolution remained high up to 250 V, but there was noticeable compression of bands at high voltages, whereby the distance separating the highest and lowest bands in the gel was reduced. This phenomenon is indicated by the large bracket and letter “C” on the right side of Figure 3A. In addition to the general compaction bringing all bands closer to each other, there was also localized compression of some bands into each other, seen primarily at 300 V (shown by the small brackets and small letter “C”). Use of 0.5 x TB also produced good separation of bands at voltages up to 250 V, though resolution was compromised at 300 V (Figure 3B). Gels prepared and run in 0.3 x TB exhibited low resolution and compression of bands toward each other at all voltages, with strongest compression and non-uniform band intensities apparent at 250 and 300 V (Figure 3C). Thus, good resolution was achieved using both 0.5 x TB and 1 x TB at voltages up to 250 V (25 V per cm), though all bands were closer to each other at the higher voltages.
Fig.3.
Resolution of linear DNA ladder fragments is maintained on 1% agarose gels run at elevated voltages. (A and B) Band resolution was similar in gels containing 1 x TB (without EDTA) and 0.5 x TB over a range of 150 – 250 V. (C) Band compression and other anomalies were present at all voltages when 0.3 x TB was employed. Gels used for the assays were 10 cm long and contained 40 ml gel volumes with buffer covering the surface by 2 mm. Large C indicates general compression of all bands; small c indicates a localized band compression. Each gel contained both 1 Kb and 2-Log DNA ladders.
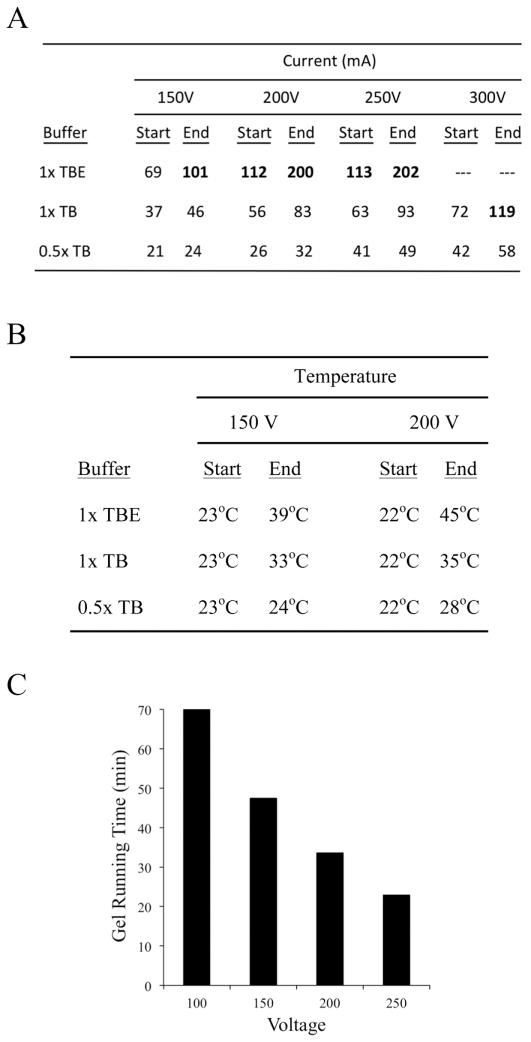
Electrical currents produced during electrophoresis of the gels shown in Figure 3A and 3B, plus control 1 x TBE gels, were measured at the beginning and at the end of runs performed using several different voltages. Starting and ending currents for gels run using 1 x TBE containing disodium EDTA were much higher than those observed when 1 x TB and 0.5 x TB were used (Figure 4A). Furthermore, the currents increased by larger percentages (nearly doubling) during the course of each run when 1 x TBE was used. When gels are run at constant voltage and the current increases with time, the temperature of the chamber buffer also tends to increase. Ten cm gels were prepared and run in 1 x TBE, 1 x TB and 0.5 x TB at 150 and 200 V as before, stopping each gel after the bromophenol blue tracking dye had migrated 7 cm. Average buffer reservoir temperatures were assessed by placing thermometers in the chambers at the beginning and immediately after the end of each run. At 150 V, temperature increased only 1 °C during the run in 0.5 x TB, but was up 10 °C in 1 x TB and 16 °C in 1 x TBE (Figure 4B). Even higher temperatures were observed at 200 V. These results confirm the postulate that reducing EDTA and buffer concentrations can have strong impacts on both the conductivity and the temperature produced during an electrophoresis experiment.
Fig.4.
Beneficial effects of reducing EDTA and Tris-borate concentrations on current, temperature and running time. (A and B) Initial and final currents and temperatures are lowest in gels prepared with 0.5 x TB. (C) Higher voltages lead to shorter run times. For each assay, bromophenol blue tracking dye was run until it had travelled 7 cm down a 10 cm gel.
Performing electrophoresis at higher voltages using modified TB media produced another advantage: large reductions in gel run times. For example, 0.5 x TB gels run at 100 V (10 V/cm) required 70 min for the bromophenol blue dye to travel 7 cm down the gel (Figure 4C). In contrast, gels run at 200 V and 250 V were completed in only 32 and 20 min, respectively, a time reduction of up to 3-fold.
Agarose is not commonly used for separation of nucleic acids that are approximately 75 nucleotides long or smaller [8,9,24]. Nucleic acids of this size are separated better and produce thinner bands in polyacrylamide gels. Part of the problem with the use of agarose is that the weak, hydrogen-bonded scaffold forming the pores of the gel permits greater diffusion of small molecules in all directions, resulting in broad, diffuse-looking bands. Since diffusion is a time-dependent process, we reasoned that use of electrophoresis conditions and buffers that permit higher voltages and much shorter run times should improve resolution of small molecules. Gels prepared with TBE produce better separation of smaller fragments than TAE gels (Figure 5A) and also generate less current. Therefore Tris-borate media was used to test the hypothesis.
Fig.5.
Use of optimized gel parameters and 0.5 x TB at higher voltages strongly enhances resolution of small nucleic acids. (A) Separation of small DNA fragments is better in TBE gels than in TAE gels. (B) Diffusion is reduced and resolution of small linear DNAs is improved when 4% agarose gels are run in 0.5 x TB at 25 V/cm (350 V) rather than the more conventional 10 V/cm (140 V). (C) Resolution of small RNAs is also improved under conditions allowing high voltages and short run times.
Agarose gels (4.0%) that were 14 cm long were prepared in 0.5 x TB and resolution of low molecular weight DNA ladders was compared at 140 V (10 V/cm) and 350 V (25 V/cm; equivalent to 250 V on a 10 cm gel). Bands were strikingly less diffuse at the higher voltage, with thin bands and good separation of fragments as small as 20 bp (Figure 5B). A similar experiment was performed using two different double-stranded RNA ladders containing fragments ranging in size from 17 bp to 500 bp. Band sharpness was greatly improved at 350 V, especially for the siRNA ladder containing 17, 21 and 25 bp fragments (Figure 5C).
The experiments described above established that 0.5 x TB, without EDTA, can be used at higher voltages with good resolution of large linear DNAs and with strongly improved resolution of small linear fragments. Since molecular biologists routinely analyze circular plasmid DNAs and their linear and nicked open circular derivatives on gels, migration of these molecules in the modified buffers was also investigated. Two forms of the yeast-E. coli shuttle vector p426TEF (6,352 bp) [23] were analyzed, representing both supercoiled (uncut) and linear (restriction enzyme-cut) conformations. Electrophoresis on a 1% gel in conventional 1 x TBE containing disodium EDTA produced migration patterns typically seen in TAE and TBE buffers, with supercoiled (SC) DNA migrating faster than linear DNA (LIN) (Figure 6A - left panel) [1,2,27]. The faint band in the SC lane co-migrating with the 10 kb linear DNA standard corresponds to nicked open circular (NOC) DNA, which migrated to a position above both the linear and SC forms as expected.
Fig.6.
Reduction of EDTA or Tris-borate concentrations alters the migration rates of supercoiled and linear plasmid DNAs. (A) The mobility of supercoiled plasmid DNA (SC) relative to linear DNA (LIN) seen in a conventional 1 x TBE gel (left panel) is progressively altered when 1 x TB, 0.5 x TB and 0.25 x TB are used. (B) Substitution of free acid EDTA for disodium EDTA in 1 x TBE and 0.5 x TBE buffers results in normal migration of the SC and LIN forms. (C) In contrast to Tris-borate buffers, removal of EDTA from 1 x TAE does not alter the relative migration rates of SC and LIN DNAs. Each gel contained 1% agarose and was run at 10 V/cm.
Surprisingly, use of 1 x TB buffer without EDTA strongly reduced the relative mobility of the SC DNA, causing it to run slower (higher) than the linear form. When ionic strength was lowered further to 0.5 x TB and 0.25 x TB, migration of the SC form was even slower (Figure 6A). 1 x TBE and 0.5 x TBE prepared using free acid EDTA restored the pattern to that seen with buffers prepared using disodium EDTA, with SC DNA migrating faster than linear DNA (Figure 6B). The migration rates of plasmids in Tris-acetate (TAE and TA) buffers were also tested. Unlike TBE buffers, removal of EDTA from 1 x TAE did not change the relative mobilities of the SC and LIN forms (Figure 6C).
During the performance of the experiments shown in Figure 6A and 6B, we observed that several lower bands in the 2-log DNA ladder appeared highly diffuse when TBE was prepared with free acid EDTA. To explore this further, several additional 1% agarose gels were run at 10 V/cm (Figure 7). Gels containing 1 x TB or conventional 1 x TBE (2 mM disodium EDTA) produced typical, thin ladder bands. However, in gels run with 1 x TBE containing free acid EDTA at the standard concentration (2 mM), all bands below 1 kb were broad and diffuse (Figure 7A-see double arrow). Similarly, 0.5 x TB and 0.5 x TBE gels containing disodium EDTA (1 mM or 2 mM) gave sharp bands (Figure 7B-left panels), but 0.5 x TBE gels containing 1 mM or 2 mM free acid EDTA exhibited diffusion that was greatest at the higher EDTA concentration (Figure 7B-double arrows). Interestingly, the number of bands affected was even stronger when 0.25 x TBE gels were used (Figure 7C). For completeness, band quality in Tris-acetate gels was also tested (Figure 7D). In contrast to the results with TBE, bands in 1 x TAE gels containing 1, 2 or 4 mM free acid EDTA did not differ substantially from those observed in standard 1 x TAE gels prepared with 1 mM disodium EDTA.
Fig.7.
Free acid EDTA induces band broadening in gels containing TBE. (A) Lower bands appear diffuse (double arrow) in 1 x TBE gels containing free acid EDTA. (B and C) Band distortion/diffusion is increased in 0.5 x TBE and 0.25 x TBE gels containing 2 mM free acid EDTA (indicated by double arrows). (D) Use of free acid EDTA in TAE buffers, even at concentrations as high as 4 mM, does not diminish resolution.
In an attempt to understand this effect of the free acid, 10 cm gels containing 0.5 x TBE prepared with disodium or free acid EDTA were run as before except that aliquots were removed from above the positive and negative electrodes before and immediately after each run for pH measurements. The starting and final pH values measured at the cathode were similar for both types of TBE; however, the pH of the reservoir containing the anode was reduced by 0.5–0.6 pH units at the end of the run when the free acid was used. This result implies a breakdown in buffering capacity and, because the anode is closest to the bottom of each gel, is also consistent with the gel pictures in Figure 7 showing that the strongest effects were observed in the bands at the bottom of each gel.
Discussion
The findings presented here demonstrate that specific modifications of the physical setup and the major buffers, especially TBE, can permit gels to be run at higher voltages, without excessive current or heating, while retaining good band resolution. The tests revealed that use of thinner gels and lower chamber buffer volumes (minimizing the level of liquid above the gel surface) produced the lowest currents. In contrast, altering gel strength and average pore size by varying the agarose concentration from 0.5% to 3% had no effect. Inclusion of ethidium bromide in the gel and electrophoresis buffer during an experiment in amounts that were up to four times the standard concentration also had no effect on electrical current.
EDTA is not essential for electrophoresis and is added as a preventative to inhibit DNA nucleases. The pKa values of the four carboxylic acid groups in EDTA are 1.99, 2.67, 6.16 and 10.26 [28]. At the pH of TAE and TBE buffers (~ 8.0), three of these groups exist predominantly in their negatively charged carboxylate ion forms. In addition, one of the amine groups within EDTA may be protonated and carry a positive charge [28], though each molecule will have a net negative charge. As previously noted by Brody and Kern [4], we observed that the presence of EDTA in electrophoresis buffers influences conductivity. Media prepared using sodium-EDTA chemical powders or sodium-EDTA stock solutions that had previously been set to pH 8.0 using sodium hydroxide, as recommended by some molecular biology resources, generated higher electrical currents than solutions containing the free acid form. These results are logical, since the presence of Na+ ions should increase the conductivity of the solutions.
Strongest reductions in current were observed when the concentrations of Tris and acetate or borate were decreased, e.g., using 0.5 x instead of 1 x buffers. Since the conductivities of TB solutions (without EDTA) were lower than those of TA solutions, this buffer was chosen for additional tests. Analysis of gels containing DNA fragments ranging in size from 100 bp to 10,000 bp revealed that resolution remained high up to 20–25 V/cm (200–250 V on 10 cm gels) using both 1 x TB and 0.5 x TB. In contrast, strong compression of all bands was detected when 0.3 x TB was used, even at low voltages, indicating limitations to the reduction of Tris-borate concentration.
Analysis of the final current levels and final temperatures reached in gels run at 15 – 30 V/cm revealed that best results were achieved with 0.5 x TB compared to 1 x TB or 1 x TBE. Importantly, the increased voltages resulted in shorter run times. Run times of samples electrophoresed at 200V and 250 V were approximately one-half and one-third of those run at the more conventional 100 V.
Although supercoiled plasmid DNA migrates faster than the linear form in conventional 1 x TBE media, we observed that use of 1 x TB without EDTA caused the supercoiled form to migrate more slowly. Furthermore, supercoiled DNA mobility was reduced even further in 0.5 x TB and 0.25 x TB than in 1 x TB solutions. Removing EDTA and reducing TB concentrations decrease the overall ionic strength inside the gels. The results therefore appear to be consistent with previous work showing that changing the ionic strength affects the mobilities of supercoiled, nicked open circular and linear DNAs in different ways, with greatest impacts on supercoiled (form I) DNA [1,2].
Although TBE buffers made with free acid EDTA generated lower currents than buffers prepared with the disodium form, and did not alter migration of supercoiled vs. linear DNAs, use of this form of EDTA led to unusually strong diffusion and broadening of some bands. The precise mechanism causing this phenomenon is unclear, but appears to be linked to reduced buffering capacity and a change of pH in the reservoir containing the anode. Thus, although substitution of free acid EDTA for disodium EDTA in TBE buffers has the advantage of reducing electrical currents, the results presented in Figure 7 indicate that such a change would be detrimental to resolution.
Nucleic acids less than approximately 75 nt in length are poorly resolved on agarose gels, even when high agarose concentrations (e.g., 3–4%) are employed. This deficiency is caused primarily by the rapid diffusion of the small molecules in the weak agarose matrix, which leads to strong broadening as the DNAs migrate down the gel. We reasoned that higher voltages and shorter run times should improve band sharpness and separation of small molecules, primarily because there is less time for diffusion to occur. 4% agarose gels prepared in 0.5 x TB without EDTA and run at 25 V/cm exhibited thin bands and excellent resolution of linear DNAs and RNAs that were as small as 17 bp. Although polyacrylamide is usually used to resolve such small DNAs, these results demonstrate that agarose gels run using 0.5 x TB at high voltages can be substituted for many applications, potentially including analysis of PCR primers, modified DNA oligonucleotides, and short-interfering RNAs (siRNAs). This finding is important because preparation of agarose gels is faster and easier and agarose gel apparatuses are generally simpler than those used for polyacrylamide gels. In addition, most molecular biology laboratories have agarose gel rigs and power supplies that can achieve the voltages described here (200 – 300 V), and therefore experiments can be performed using existing instrumentation.
Acknowledgments
The authors wish to thank Crystal Flowers and Taylor Carter for their expert assistance during the initiation of this project. This work was supported in part by a grant from the National Institutes of Health (grant number 1R15GM09904901) to LKL and a departmental grant from the Welch Foundation.
Abbreviations
- nt
nucleotides
- bp
base-pairs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson PH, Grossman LI. Electrophoresis of DNA in agarose gels: optimizing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977;16:4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- 2.Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- 3.Serwer P. Agarose gels: properties and use for electrophoresis. Electrophoresis. 1983;4:375–382. [Google Scholar]
- 4.Brody JR, Kern SE. History and principles of conductive media for standard DNA electrophoresis. Anal Biochem. 2004;333:1–13. doi: 10.1016/j.ab.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 5.Kozulic B. Models of gel electrophoresis. Anal Biochem. 1995;231:1–12. doi: 10.1006/abio.1995.1495. [DOI] [PubMed] [Google Scholar]
- 6.Stellwagen NC. Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis. 2009;30:S188–195. doi: 10.1002/elps.200900052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P. New insight into agarose gel mechanical properties. Biomacromolecules. 2000;1:730–738. doi: 10.1021/bm005583j. [DOI] [PubMed] [Google Scholar]
- 8.Voytas D. Agarose gel electrophoresis. Curr Protoc Mol Biol. 2001;51:2.5A.1–2.5A.9. doi: 10.1002/0471142727.mb0205as51. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell SW, editors. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 10.Burland V, Curtis FP, Kusukawa N. Agarose gel analysis of 15–40-kb PCR amplimers. Biotechniques. 1996;21:142–144. doi: 10.2144/96211pf01. [DOI] [PubMed] [Google Scholar]
- 11.Maaloum M, Pernodet N, Tinland B. Agarose gel structure using atomic force microscopy: gel concentration and ionic strength effects. Electrophoresis. 1998;19:1606–1610. doi: 10.1002/elps.1150191015. [DOI] [PubMed] [Google Scholar]
- 12.Schwinefus JJ, Bloomfield VA. The greater negative charge density of DNA in Tris-borate buffers does not enhance DNA condensation by multivalent cations. Biopolymers. 2000;54:572–577. doi: 10.1002/1097-0282(200012)54:7<572::AID-BIP90>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 13.Stellwagon NC, Gelfi C, Righetti PG. DNA and buffers: the hidden danger of complex formation. Biopolymers. 2000;54:137–142. doi: 10.1002/1097-0282(200008)54:2<137::AID-BIP6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Biyani M, Nishigaki K. Sequence-specific and nonspecific mobilities of single-stranded oligonucleotides observed by changing the borate buffer concentration. Electrophoresis. 2003;24:628–633. doi: 10.1002/elps.200390073. [DOI] [PubMed] [Google Scholar]
- 15.Buchmueller KL, Weeks KM. Tris-borate is a poor counterion for RNA: a cautionary tale for RNA folding studies. Nucleic Acids Res. 2004;32:e184. doi: 10.1093/nar/gnh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandecki W, Hayden M. High-resolution polyacrylamide gel electrophoresis of oligonucleotides using L-histidine buffer. DNA. 1988;7:57–62. doi: 10.1089/dna.1988.7.57. [DOI] [PubMed] [Google Scholar]
- 17.Brody JR, Kern SE. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques. 2004;36:214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
- 18.Ishido T, Ishikawa M, Hirano K. Analysis of supercoiled DNA by agarose gel electrophoresis using low-conducting sodium threonine medium. Anal Biochem. 2010;400:148–150. doi: 10.1016/j.ab.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Singhal H, Ren YR, Kern SE. Improved DNA electrophoresis in conditions favoring polyborates and lewis acid complexation. PLoS One. 2010;5:e11318. doi: 10.1371/journal.pone.0011318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Li X, Sommer SS. pK-matched running buffers for gel electrophoresis. Anal Biochem. 1999;270:112–122. doi: 10.1006/abio.1999.4064. [DOI] [PubMed] [Google Scholar]
- 21.Brody JR, Calhoun ES, Gallmeier E, Creavalle TD, Kern SE. Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. Biotechniques. 2004;37:598–602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- 22.Kozulić B. Looking at bands from another side. Anal Biochem. 1994;216:253–261. doi: 10.1006/abio.1994.1039. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 24.Ogden RC, Adams DA. Electrophoresis in agarose and acrylamide gels. Methods Enzymol. 1987;152:61–87. doi: 10.1016/0076-6879(87)52011-0. [DOI] [PubMed] [Google Scholar]
- 25.Chalhoub B, Belcram H, Caboche M. Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnol J. 2004;2:181–188. doi: 10.1111/j.1467-7652.2004.00065.x. [DOI] [PubMed] [Google Scholar]
- 26.Furumoto K, Inoue E, Hiyama E, Miwa N. Age-dependent telomere shortening is slowed by enrichment of intracellular vitamin C. Life Sci. 1998;63:935–948. doi: 10.1016/s0024-3205(98)00351-8. [DOI] [PubMed] [Google Scholar]
- 27.Leloup C, Garty G, Assaf G, Cristovão A, Breskin A, Chechik R, Shchemelinin S, Paz-Elizur T, Livneh Z, Schulte RW, Bashkirov V, Milligan JR, Grosswendt B. Evaluation of lesion clustering in irradiated plasmid DNA. Int J Radiat Biol. 2005;81:41–54. doi: 10.1080/09553000400017895. [DOI] [PubMed] [Google Scholar]
- 28.Burgot J-L. Ionic Equilibria in Analytical Chemistry. Springer; New York, NY: 2012. [Google Scholar]