Abstract
FOXO transcription factors are conserved regulators of longevity downstream of insulin signaling. These transcription factors integrate signals emanating from nutrient deprivation and stress stimuli to coordinate programs of genes involved in cellular metabolism and resistance to oxidative stress. Here we discuss emerging evidence for a pivotal role of FOXO factors in promoting the expression of genes involved in autophagy and the ubiquitin-proteasome system – two cell clearance processes that are essential for maintaining organelle and protein homeostasis (proteostasis). The ability of FOXO to maintain cellular quality control appears to be critical in processes and pathologies where damaged proteins and organelles accumulate, including aging and neurodegenerative diseases.
The FOXO family of transcription factors
FOXO transcription factors are involved in a number of physiological and pathological processes, including aging, cancer, and neurological diseases [1, 2]. The conserved function of FOXO transcription factors in the extension of organismal lifespan [3] has fueled the search for the mechanisms underlying this pro-longevity function of FOXO. Mechanisms of cellular quality control have recently been found to be involved in longevity and could play an important role in mediating the ability of FOXO to extend lifespan and delay signs of age-related diseases.
The FOXO family is a subclass of Forkhead transcription factors characterized by a winged helix DNA binding domain known as a Forkhead box [4]. C. elegans and Drosophila each possess one FOXO factor, whereas the mammalian FOXO family comprises four members (FOXO1, FOXO3, FOXO4 and FOXO6) that mainly differ in their tissue-specific expression. FOXO transcription factors function mostly as transcriptional activators, and their activity is inhibited by insulin and growth factor signaling. In the presence of insulin and insulin-like growth factor (IGF), the PI3K-AKT signaling pathway is activated and protein kinases such as AKT and the related protein kinase SGK (serum and glucocorticoid-induced kinase) directly phosphorylate FOXO factors at three conserved residues, resulting in FOXO exclusion from the nucleus and repression of transcriptional activity [5–8]. In the absence of insulin or growth factor signaling, or during starvation, FOXOs translocate to the nucleus where they activate programs of gene expression. The first FOXO targets that were identified include stress resistance genes [7, 9, 10] and metabolism genes [11–13]. In addition to being phosphorylated by AKT and SGK, FOXOs can also be post-translationally modified at many other residues. FOXO post-translational modifications have been proposed to serve as a combinatorial ‘FOXO code’ which can be recognized by binding partners to regulate select programs of gene expression (metabolism versus stress resistance) in response to diverse external stimuli [14]. For example, FOXO factors can be phosphorylated at diverse residues by a number of different stress-responsive protein kinases, including AMPK [15], JNK [16, 17], MST1 [18], as well as ERK and p38 MAPK [19, 20]. FOXOs are also regulated by acetylation/deacetylation, ubiquitination, and arginine and lysine methylation in response to a variety of stimuli, including oxidative stress and changes in nutrient status [21–24]. The central position of FOXO transcription factors at the integration hub for many stimuli has sparked interest in identifying additional gene expression programs and cellular functions that could mediate the ability of these transcription factors to regulate longevity.
Emerging evidence from multiple systems indicate that FOXOs orchestrate the expression of genes involved in cellular quality control, and in particular the protein homeostasis (proteostasis) network [25]. Moreover, recent findings suggest that maintenance of proteostasis may in part underlie FOXO’s role as a pro-longevity factor. Here we discuss the role of FOXO factors in the regulation of two intracellular clearance mechanisms, autophagy and the ubiquitin-proteasome system, that function to rid the cell of damaged and aggregated proteins and could contribute to the role of FOXO factors in cellular homeostasis. In addition, we discuss the interplay between FOXOs and other regulatory mechanisms, in particular the mTOR pathway, a central regulator of autophagy and aging. Finally, we discuss the implications of FOXO-mediated cellular quality control for aging and age-related diseases.
FOXOs trigger autophagy in specific cell types
FOXOs have been implicated in autophagy in a variety of cell types in vertebrates and invertebrates [26–33]. Autophagy is an evolutionarily conserved process that allows cells to degrade and recycle cytoplasmic proteins and organelles in response to starvation. Autophagy is also responsible for the degradation of protein aggregates, which would otherwise accumulate and cause cytotoxicity. Three main types of autophagy exist: chaperone-mediated autophagy, microautophagy, and macroautophagy, all of which function to deliver intracellular contents to the lysosome. In chaperone-mediated autophagy, cytosolic proteins are selectively and individually transported into the lysosome for degradation. In microautophagy, a portion of the cytoplasm is directly engulfed by the lysosome. In macroautophagy, proteins and organelles are engulfed by a double-membraned autophagosome, which subsequently fuses with the lysosome. Macroautophagy (hereafter referred to as autophagy for simplicity) functions as a protective response that is stimulated under stress conditions including nutrient deprivation and starvation. Decreased autophagy is associated with hallmarks of premature aging [34–36] and age-related degenerative diseases [37–41].
FOXO transcription factors induce autophagy and mitophagy in muscle
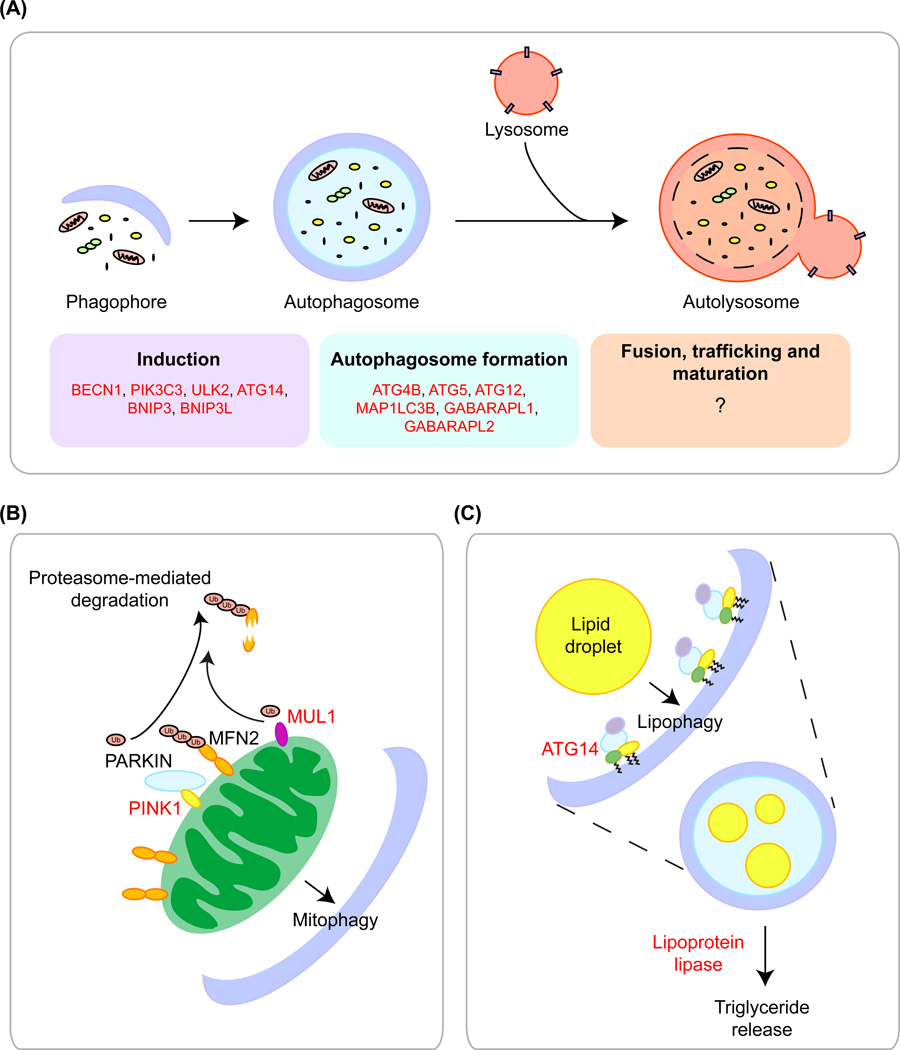
FOXO factors were initially found to regulate autophagy in muscle in response to starvation or denervation. Overexpression of FOXO3 in C2C12-derived myotubes, isolated mouse muscle, or myofibers increases a marker of autophagosome formation (LC3-GFP) [27, 32]. Conversely, knockdown of FOXO3 results in decreased autophagosome foci in adult skeletal muscle [27]. The process of autophagy is coordinated by a network of autophagy-related genes (atg genes), which encode proteins that are involved in the recruitment of cargo, formation of autophagosomes, fusion with the lysosome, and release of degradation products (amino acids, lipids, nucleic acids and carbohydrates) [42, 43]. FOXO3 induces the expression of a number of autophagy genes involved in various stages of the process, including LC3b (Map1lc3b), Gabarapl1, Pi3kIII, Ulk2, Atg12l, Beclin1, Atg4b and Bnip3 (Figure 1A). Chromatin immunoprecipitation (ChIP) experiments revealed that several of these genes are direct FOXO targets in muscle cells, including LC3b, Gabarapl1, Atg12l, Bnip3 and Bnip3l [27, 32]. Interestingly, fasting increases FOXO3 binding at the LC3b, Bnip3 and Bnip3l promoters in adult mouse skeletal muscle, and FOXO3 is required for fasting-induced autophagy in myotubes [27]. Thus, that FOXO-induced autophagy may be critical for adaptation to fasting and recycling of nutrients from the cell/tissue in the face of starvation.
Figure 1. FOXOs regulate the expression of genes involved in autophagy in mammalian cells.
(A) FOXO regulates induction of the autophagic pathway (phagophore development) and formation of the autophagosome. Genes regulated by FOXO in various cell types are listed in red. Whether FOXO also regulates genes that promote the fusion, trafficking and maturation of the autolysosome is not yet known. (B) FOXO factors regulate mitophagy via the E3 ubiquitin ligase MUL1 (muscle) and PINK1, which recruits the E3 ubiquitin ligase PARKIN (fibroblasts and MCF-7 cells). Both enzymes tag MFN2 for degradation, which targets mitochondria for mitophagy. (C) FOXOs activate the transcription of ATG14, a protein that localizes to the inner face of the phageophore, to promote lipophagy. FOXO1 can also promote transcription of lipoprotein lipase, an enzyme that promotes triglyceride release, but it not known if this regulation is direct. In all panels, genes highlighted in red have been shown to be FOXO targets.
Precisely how FOXO3 is activated in skeletal muscle in response to starvation and denervation is not fully understood. Inhibition of the insulin pathway is likely involved in the activation of FOXO3 in response to starvation. A recent study implicates the protein kinase MST1, a known regulator of FOXO activity, in the activation of FOXO3 in response to denervation [44]. MST1 is activated upon denervation and knock-out of the Mst1 gene results in decreased denervation-induced muscle wasting in adult mice, as well as reduced expression of LC3b (Map1lc3b) and Bnip3 seven days post-denervation [44]. The activation of MST1 and atg gene expression correlates with the phosphorylation and nuclear translocation of FOXO3 [44], though it remains to be determined whether FOXO3 is the main intermediary between MST1 and muscle atrophy, or if other factors contribute as well.
In muscle, FOXO transcription factors also promote a specialized form of autophagy – termed mitophagy – in which mitochondria are recognized and degraded by the autophagy-lysosomal pathway [45]. At the mechanistic level, mitophagy is induced upon degradation of Mitofusin 2 (MFN2), a large GTPase that normally promotes mitochondrial fusion [46]. In muscle, overexpression of FOXO1 or FOXO3 activates the expression of Mul1, a mitochondrial E3 ubiquitin ligase which colocalizes with autophagosomes during mitophagy and targets MFN2 for degradation (Figure 1B) [47]. MFN2 degradation in turn results in the induction of mitophagy and skeletal muscle atrophy [47].
The ability of FOXO transcription factors to trigger autophagy in muscle is conserved in Drosophila [26]. Muscle-specific overexpression of dFOXO activates autophagy in this tissue, which delays the accumulation of protein aggregates and mitigates the loss of muscle functionality during aging [26]. dFOXO activates transcription of 4E-BP, a known inhibitor of protein translation, and 4E-BP overexpression itself is sufficient to activate autophagy and increase performance in flight assays [26]. While the induction of autophagy by FOXOs in muscle is conserved from flies to mammals, it is unknown whether the mechanisms involved have been maintained throughout evolution. For example, it is currently unknown whether activation of 4E-BP by FOXO transcription factors in mammalian cells affects autophagy levels. Conversely, whether mammalian FOXO autophagy targets, regulation of autophagy by MST1, or FOXO-mediated mitophagy are conserved in Drosophila has yet to be addressed.
FOXOs promote autophagy and mitophagy in neurons – implications for neurodegenerative diseases
Regulation of autophagy and mitophagy by FOXO transcription factors has also been observed in mammalian neurons [31, 48], which may have ramifications for neurodegenerative diseases. Similar to other cell types, several autophagy genes are activated in neurons in response to FOXO1 nuclear localization, including Bnip3, Beclin1 and Atg5 (Figure 1A) [31]. Recent evidence implicates FOXO3 in the clearance of defective mitochondria in dopaminergic neurons expressing α-synuclein, an aggregation-prone protein involved in familial Parkinson’s disease [48]. On the one hand, expression of a constitutively active form of FOXO3 induces degradation of α-synuclein and clearance of mitochondria [48]. On the other hand, dominant negative forms of FOXO3 protected against death of dopaminergic neurons [48]. Thus, while FOXO3 is able to prevent α-synuclein-associated toxicity in Parkinson’s disease models, it may also be responsible for neuronal death in diseased individuals [48]. Importantly, these conclusions were based on virally delivered mutants of FOXO3, and it will be necessary to investigate FOXO3’s role in α-synuclein-mediated toxicity using genetic models to determine the precise role of FOXO3 in mammalian models of Parkinson’s disease. Together, several lines of evidence indicate that in mammals, FOXO factors activate mitophagy at the transcriptional level by increasing expression of mitochondrial E3 ubiquitin ligases, which in turn target mitochondria to the phagophore for degradation, and this may be a protective mechanism in mammalian models of Parkinson’s disease.
In Drosophila, dFOXO expression protects dopaminergic neurons in models of Parkinson’s disease [49]. Global dFOXO overexpression ameliorates the mitochondrial defects and neuronal degeneration that are normally observed in Drosophila PINK1 mutants [49]. Whereas in mammalian cells, FOXO3 was shown to promote mitophagy by transcriptionally activating the gene encoding the serine/threonine kinase PINK1, which in turn recruits the ubiquitin ligase PARKIN (PARK2) to the mitochondrial membrane to promote MFN2 degradation (Figure 1B) [50], PINK1 appears to function upstream of FOXO in fly dopaminergic neurons [49]. It will be important to determine if PINK1 can also act upstream of FOXO in mammals in a feedback loop, and determine whether the ability of FOXO3 to promote mitophagy delays neurodegeneration in cellular or organismal models for Parkinson’s diseases in mammals. While FOXO plays a protective role in flies that are deficient for PARKIN (PARK2), it has also been reported to induce apoptosis in dopaminergic neurons under conditions of oxidative stress triggered by loss of DJ-1β (PARK7), another gene involved in Parkinson’s disease, in Drosophila [51]. Thus, FOXO may act as an oxidative stress rheostat in dopaminergic neurons, playing a protective role when oxidative stress levels are relatively low, but promoting cell death when the oxidative stress load is high.
Future studies will be necessary to tease apart the apparently paradoxical roles for FOXO transcription factors in survival and death of dopaminergic neurons. It is possible that cellular outcomes in Parkinson’s disease models depend on the level of FOXO activation or the context in which FOXO is activated, such as conditions of high oxidative stress. Alternatively, the differences observed may be due to disparities in experimental approaches. Thus far, experiments assessing the role of FOXOs in Parkinson’s disease have been performed in different cell types (fibroblasts, MCF-7 cells or dopaminergic neurons) [48, 50] or genetic conditions (PINK1 or PARK7 mutants) [49, 51]. In addition, some studies have used overexpression of wild type or mutant forms of exogenous FOXOs to determine function [48, 50]. The different approaches and conditions used make it difficult to determine the precise role of FOXO-mediated mitophagy in dopaminergic neurons. To begin to address these issues, future work should examine the role of FOXO factors in mammals in vivo by combining genetic knockouts of these transcription factors with models of Parkinson’s disease. Such experiments will help to reveal whether FOXOs are protective, detrimental, or both in the context of Parkinson’s disease.
FOXOs promote lipophagy, a specific form of autophagy, in the liver
In the liver, FOXO transcription factors regulate a specific form of autophagy known as lipophagy. Lipophagy is the subtype of autophagy in which cytoplasmic lipid droplets are broken down, resulting in the release of lipids for use as an energy source by the cell. Lipophagy occurs mainly in the liver, where it can be activated under conditions of nutrient deprivation to generate free fatty acids for the cell. During aging, reduction in lipophagy may result in increased liver steatosis and, in turn, metabolic syndrome [52]. In a recent study, liver-specific triple knockout of Foxo1, Foxo3, and Foxo4 revealed a role for FOXOs in both autophagy and lipid metabolism [30]. Either liver-specific triple knockout or Atg14 knockdown in liver resulted in increased hepatic serum triglyceride levels [30]. Consistent with this phenotype, mammalian FOXO1 is known to transcriptionally activate lipoprotein lipase, an enzyme responsible for the breakdown of triglycerides (Figure 1B) [53].
Role of FOXO in autophagy in other differentiated cell types
FOXO factors also regulate autophagy in many other differentiated cell types, such as cardiomyocytes [54] and mouse primary renal proximal tubular cells [33]. In cardiomyocytes, FOXO1 and FOXO3 activate the core autophagy genes LC3b, Gabarapl1 and Atg12 [54]. Bnip3 is also upregulated by FOXO3 in mouse primary renal proximal tubular cells cultured with serum from calorically restricted mice in combination with hypoxia, indicating that FOXOs may regulate similar targets in different cell types [33]. Thus, activation of autophagy by FOXO in differentiated cells appears to be a general phenomenon that is coordinated by a network of autophagy-related genes.
FOXO3 promotes autophagy in hematopoietic stem cells
Emerging evidence suggests that FOXOs not only promote autophagy in differentiated cells, but also elicit this pathway in stem cells. A recent report uncovered a critical role for FOXO3 in the maintenance of autophagy in hematopoietic stem cells (HSCs) [55]. Using an LC3-GFP reporter, the authors observed a two-fold reduction in autophagy in Foxo3-deficient HSCs relative to wild types. As in other cell types, FOXO3 regulates a program of genes important for the induction of autophagy, including Gabarapl2, Atg4b, and Bnip3. Interestingly, this study also revealed that autophagy was particularly important in HSCs during aging to maintain energy homeostasis and promote survival of the HSC pool [55].
In summary, FOXOs are key regulators of autophagy in various cell types, including stem cells. Specialized forms of autophagy, lipophagy and mitophagy, are also regulated by FOXOs, indicating that these transcription factors impinge on lipid and organelle homeostasis in addition to proteostasis. Intriguingly, depending on the cell type, upregulation of autophagy can be protective (HSCs) or detrimental (muscle) to the tissue. In some cases, whether autophagy is beneficial or harmful may be dependent on the cellular state (neurons). These cell type-specific differences raise the question of whether FOXO-mediated induction of autophagy is beneficial at the organismal level.
Organismal roles of FOXO in autophagy regulation
Studies in invertebrates have revealed that FOXO can induce autophagy in many tissues of the organism, potentially acting in a non-cell-autonomous manner. In C. elegans, FOXO/DAF-16 promotes autophagy in the context of pathogenic stimuli and heat stress. Global overexpression FOXO/DAF-16 induces autophagy in seam cells, and possibly other tissues, and increases lifespan in animals infected by the pathogen Salmonella [56]. Expression of the ATG genes bec-1 and lgg-1, the worm orthologs of Beclin-1 and LC3b, respectively, is required for the response to Salmonella infection [56]. Whether bec-1 and lgg-1 are directly activated by FOXO/DAF-16 in worms is unknown, but both genes are transcriptionally activated by FOXO in mammals. FOXO/DAF-16 also upregulates the expression of genes involved in the targeting of damaged proteins to the lysosome under heat stress conditions [57], suggesting that FOXO/DAF-16 may regulate autophagy under heat stress as well. In contrast, FOXO/DAF-16 is dispensable for the increased autophagy observed in response to reduction of insulin signaling [58], suggesting that other transcription factors (or transcription-independent mechanisms) may be required for autophagy in conditions of low insulin signaling. While autophagy is necessary for the lifespan extension of C. elegans insulin/IGF-1 receptor mutants (known as daf-2 mutants) [59], daf-2;daf-16 double mutants still show increased autophagy, but no longer exhibit lifespan extension. These studies show that activation of FOXO in worms can increase autophagic flux in some contexts (pathogenic stress), but not all (decreased insulin signaling). It will be important to gain a mechanistic understanding of how activation of FOXO/DAF-16 induces autophagy globally in some contexts but not others.
FOXO can also regulate autophagy at the organismal level in Drosophila. Interestingly, overexpression of dFOXO or 4E-BP only in muscle is sufficient to extend lifespan and to reduce protein aggregation in other tissues (brain, adipose tissue and the retina) [26]. This non-cell autonomous mechanism of lifespan extension by dFOXO or 4E-BP overexpression is likely to involve dietary restriction since the flies overexpressing FOXO in the muscle consume less food and have decreased hemolymph glucose levels [26]. Whether FOXOs can act non-cell autonomously in mammals to affect aggregate formation or lifespan has not been tested. One way in which FOXO-induced autophagy may promote organismal longevity in both invertebrates and mammals is by clearing protein aggregates in the tissue of action, and by regulating metabolism in a non-cell autonomous manner.
Interplay between FOXO and other autophagy regulatory mechanisms
Intersection between FOXOs and the mTOR pathway in autophagy
The mechanistic target of rapamycin (mTOR) pathway functions as a central regulator of cellular growth and metabolism. mTOR participates in two distinct multiprotein complexes, mTORC1 and mTORC2 [60]. While mTORC1 and mTORC2 both coordinate metabolism and growth, mTORC1 appears to be the mTOR-containing complex that is most pivotal in the regulation of autophagy [60]. In nutrient rich or high growth factor conditions, activated mTORC1 phosphorylates and inhibits ULK1 (ATG1 in yeast) to block autophagosome formation (Figure 2). Nutrient deprivation or high stress conditions cause mTORC1 to dissociate from ULK1, triggering the autophagic response [61, 62]. The mTORC1 complex plays a central role in the regulation of aging, in a manner that requires, at least in part, autophagy. Inhibition of mTORC1 with rapamycin or genetic interventions extends lifespan in various species, including mice [63–67]. In Drosophila, reduction of Atg5 eliminates rapamycin-induced lifespan extension [63]. Similarly, in C. elegans, BEC-1, a protein required for the initiation of autophagy, is required for the lifespan extension of mutants heterozygous for the TORC1 subunit daf-15/raptor [58]. The observation that mTORC1 inhibition extends lifespan in a manner that depends in part on autophagy raises the question of whether and how FOXOs and TORC1 interact.
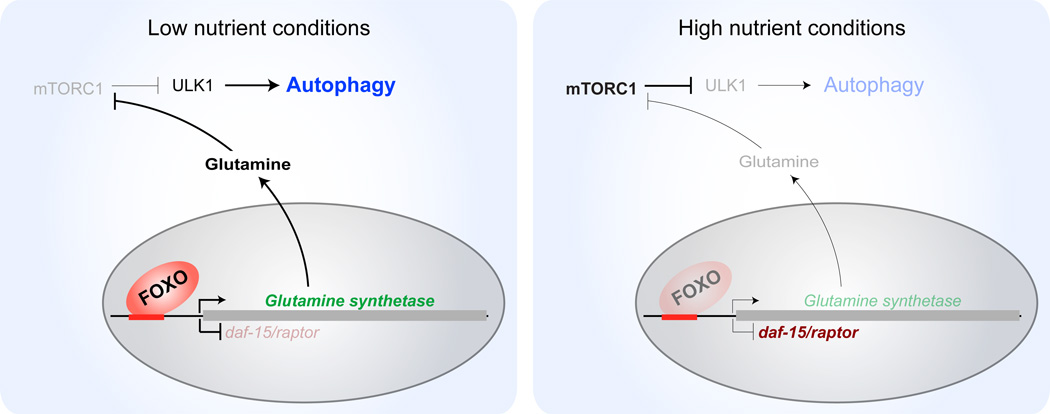
Figure 2. Antagonistic interaction between FOXOs and the mTORC1 pathway in autophagy.
Under low nutrient conditions, the serine/threonine kinase ULK1 initiates activation of autophagy. Under these conditions, FOXO activates transcription of glutamine synthetase, which increases intracellular glutamine levels, thereby blocking mTOR activity. In C. elegans, and possibly vertebrates, FOXO/DAF-16 represses expression of daf-15/raptor, a critical member of the TORC1 complex. In high nutrient conditions, low FOXO activity relieves the repression of daf-15/raptor. In addition, reduced glutamine synthetase levels increase mTOR activity and autophagy is impeded.
An antagonistic relationship between FOXOs and mTOR has been observed in both mammalian cells and invertebrates. Unlike FOXO3, which serves to preserve mammalian HSC pools [68, 69], mTORC1 activation leads to the premature depletion of these HSC pools [70, 71]. Experiments in C. elegans have also revealed opposing effects of FOXO and the mTOR pathway on autophagy [56, 58, 72], lifespan [72], and other physiological processes [73]. Whereas FOXO/DAF-16 promotes autophagy and longevity in worms [56, 74], mTORC1 inhibits these processes [58, 72]. The opposing effects of FOXO and mTORC1 on autophagy raises the question of whether these two ‘hubs’ intersect.
Recent evidence has highlighted crosstalk between FOXOs and mTOR in autophagy in mammals and invertebrates. Overexpression of FOXO3 in cultured mammalian cells leads to a four-fold increase in glutamine synthetase activity and subsequent increase in glutamine levels [29]. Interestingly, induction of autophagy by FOXO3 in a human colon carcinoma cell line requires glutamine synthetase activity, and glutamine treatment is sufficient to induce autophagy (Figure 2) [29]. Glutamine synthetase activity is sufficient to inhibit mTORC1’s localization to the lysosome, suggesting that high glutamine levels induce autophagy at least in part by blocking the TOR pathway [29]. Intersection between FOXOs and the mTORC1 complex also occurs in invertebrates. In C. elegans, FOXO/DAF-16 negatively regulates the expression of daf-15/raptor (Figure 2) [66], which encodes a component of the TORC1 complex, and this daf-15 downregulation may underlie part of the antagonistic relationship between FOXO and the mTOR pathway in longevity and possibly autophagy. Reciprocally, mTOR can inhibit FOXO/DAF-16 in C. elegans [75]. However, in this context, FOXO/DAF-16 is not directly required for general autophagy. Instead, FOXO/DAF-16 induces expression of lipl-4, a triacylglycerol lipase, which is important for lipophagy [75, 76].
Interplay between FOXOs and other regulatory pathways involved in autophagy
In addition to regulation by mTOR signaling, autophagy can be activated directly by the BECN1/VPS34/ATG14 complex [77]. This complex promotes formation the phagophore (a precursor of the autophagosome) and is itself regulated by a network of factors, including the protein kinase AKT. AKT directly phosphorylates and inhibits BECN1, resulting in the inhibition of autophagy in an mTOR-independent manner [78]. When activated by insulin or IGF-1, AKT also inhibits FOXO transcription factors [5], thereby inhibiting transcriptional activation of autophagy targets by FOXO factors. Thus, once activated, AKT has a dual mechanism for inhibiting autophagy: directly through the inhibition of BCN1 and indirectly by inhibiting FOXO-mediated transcriptional activation of autophagy genes.
FOXO transcription factors upregulate genes involved in the ubiquitin-proteasome system
In addition to autophagy, FOXO factors also regulate another clearance mechanism, the ubiquitin-proteasome system. Whereas autophagy is thought to be relatively specific to long-lived proteins and degradation under chronic starvation conditions [79], the proteasome system is responsible for the degradation of most short-lived and regulatory cytosolic proteins [80]. The proteasome is also critical for the removal of misfolded and damaged proteins [81, 82]. Decreased proteasomal activity during aging has been observed in several tissues, including muscle, liver, heart and skin, accounting for the observed accumulation of damaged proteins and inclusion bodies [83–87]. Moreover, an intact ubiquitin-proteasome pathway is essential to prevent neurodegenerative disease, including Parkinson’s, Alzheimer’s and Huntington’s diseases as well as amyloidosis [25, 88]. By contrast, excessive activity of the ubiquitin-proteasome system may lead to muscle wasting and sarcopenia [89].
FOXOs upregulate E3 ubiquitin ligases
Studies on skeletal muscle atrophy have highlighted the role of mammalian FOXOs in the upregulation of ubiquitin ligases. FOXO3 is a potent and direct transcriptional regulator of the muscle-specific E3 ubiquitin ligases atrogin-1 and Murf-1 (also known as Fbxo32 and Trim63, respectively) (Figure 3) [90–92]. Ubiquitin ligases are major effectors of protein degradation in skeletal muscle where they tag lysine residues of proteins with poly-ubiquitin chains, thereby targeting them to the proteasome for proteolysis in response to catabolic stress. Both ubiquitin ligases are highly upregulated by FOXO3 during muscle atrophy and required for normal rates of atrophy in response to denervation [93]. Since FOXO factors also upregulate autophagy genes in conditions of atrophy or denervation, this finding raises the question of whether there is crosstalk or redundancy between proteasome-dependent degradation and autophagy. In future studies, it will be important to determine the relative contribution of each pathway to muscle atrophy to better understand how to reverse this condition.
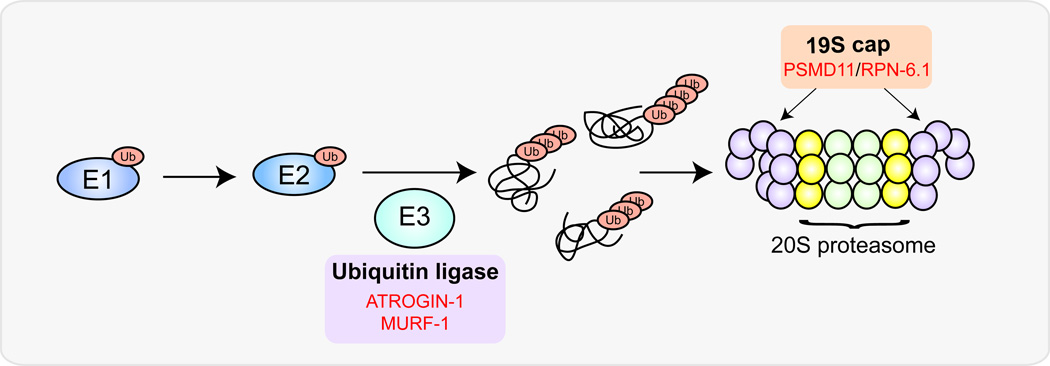
Figure 3. FOXOs coordinate the activation of the ubiquitin-proteasome pathway in mammalian cells.
FOXO directly regulates transcription of the E3 ubiquitin ligases Atrogin-1 and Murf1, which target substrates for proteasomal degradation. In hESC, FOXO4 activates transcription of the proteasomal regulatory subunit Psmd11 (rpn-6.1 in C. elegans) thereby maintaining high proteasomal activity in these cells.
Regulation of proteasomal machinery
In addition to acting upstream of ubiquitination, recent evidence indicates that FOXOs regulate proteolysis more directly by modulating the composition of the proteasome. In hESCs and induced pluripotent stem cells (iPS) cells, FOXO4 is both necessary and sufficient for expression of the proteasomal component PSMD11 (Figure 3), a subunit of the 19S regulatory cap and limiting constituent of the proteasome [94]. PSMD11 and total proteasome levels are high in hESCs where they are thought to prevent hESCs from accumulating damaged and aggregated proteins. During differentiation, proteasome activity declines due to a decrease in PSMD11 expression, which is no longer under FOXO4 regulation, and levels of poly-ubiquitinated proteins increase. In the absence of FOXO4, hESCs lose the potential to differentiate into neurons [95]. This effect is specific to FOXO4 since overexpression of FOXO1 or FOXO3 does not affect proteasome activity in hESCs. Interestingly, the role of FOXO in regulating activity of the 19S proteasomal subunit is conserved across species. In C. elegans, FOXO/DAF-16 regulates expression of the PSMD-11 ortholog, rpn-6.1, in the long-lived mutant glp-1 (Figure 3) [96]. In worms, rpn-6.1 also encodes a subunit of the 19S proteasomal cap that is a key regulator of proteasomal activity under proteotoxic stress [94, 97]. RPN-6.1 overexpressing worms are resistant to oxidative stress and poly-glutamine aggregation, and have extended lifespans under heat stress. FOXO/DAF-16 confers resistance to both oxidative [98] and heat stress [57] under low-insulin signaling conditions and daf-16 mutants have accelerated formation of poly-glutamine aggregates in C. elegans [99]. Together, these data suggest a model in which FOXO/DAF-16 promotes various types of stress resistance via the activation of rpn-6.1, although it remains to be determined whether RPN-6.1 itself is required downstream of FOXO/DAF-16 to mediate these effects.
FOXO-dependent cellular quality control in the regulation of aging and neuropathologies
Proteostasis pathways have a significant impact on aging and aging-related diseases. Generally, proteostasis mechanisms decrease with age, resulting in the accumulation of damaged, misfolded and aggregated proteins [100]. Such species are cytotoxic to the cell and contribute to tissue degeneration during aging. Defects in proteostasis are also associated with a number of aging-related diseases, including neurodegeneration and cancer [25, 88, 101–103]. Many unanswered questions remain as to whether and how FOXO’s regulation of proteostasis impacts aging or age-related diseases at the tissue or organismal level.
FOXO-mediated proteostasis in longevity
Whether FOXO-mediated proteostasis impacts aging in mammals has not been directly addressed, but several lines of evidence suggest that it may. First, work in Drosophila implicates dFOXO-induced autophagy in the non-cell autonomous regulation of longevity [26]. In addition, the function of FOXOs in selective forms of autophagy, lipophagy and mitophagy, may counter aging in specific tissues [30, 47, 48, 50]. For example, FOXO-dependent lipophagy may be critical for the maintenance of lipid metabolism with age, which may delay age-dependent changes in metabolism and the appearance of the metabolic syndrome. Similarly, removal of damaged mitochondria by FOXO-induced mitophagy could be particularly helpful during aging and age-related diseases to protect the cell from intracellular reactive oxygen species that are generated as byproducts of dysfunctional mitochondria.
It will be important to test whether FOXO-mediated autophagy and proteasomal function may influence mammalian lifespan, in particular human lifespan. In humans, single nucleotide polymorphisms (SNPs) in intronic regions of the FOXO3 locus have been associated with exceptional longevity in human centenarians, and better prognosis in disease states [104–110]. FOXO1 has also been linked with human longevity in one of these studies [106]. In the future, it will be interesting to test whether these FOXO variants correlate with different levels of proteostasis in human samples. It will also be crucial to dissect the respective contribution of FOXO transcription factors in mammalian longevity using mouse models and examine their ability to promote proteostasis in mammalian tissues.
Importantly, work from C. elegans challenges the view that increased proteostasis is always beneficial for longevity. In nematodes, increased autophagy is not sufficient to extend lifespan in the absence of FOXO/DAF-16 [58]. In contrast, increased autophagy is essential for lifespan extension in conditions of dietary restriction or TOR inhibition in C. elegans [58]. In addition, increased autophagy in mice, by the ubiquitous overexpression of Atg5, is sufficient to extend median lifespan 17.5% [111]. Why increased autophagy is not sufficient to extend lifespan in the absence of FOXO/DAF-16 in C. elegans remains unknown, but possibly because FOXO/DAF-16 is also needed to regulate other critical protective proteostasis mechanisms, such as the ubiquitin-proteasome system.
It remains unknown whether changes in FOXO expression or activity impact cellular quality control during aging. Since autophagy decreases with age in most tissues, it is possible that lower FOXO levels or activation underlie these changes. Interestingly, unlike the differentiated cell types that have been examined, HSCs appear to upregulate autophagy with age, which is important for energy homeostasis and cell survival. A portion of the autophagy program is under FOXO3 control in adult HSCs [55]. These findings, together with earlier reports of HSC dysfunction in Foxo3-deficient mice [68, 69], suggest that FOXOs contribute to the maintenance of the HSC reservoir during aging by limiting damage from reactive oxygen species, preserving metabolic homeostasis and promoting autophagic “flux”, or flow through the autophagic pathway. It will be important to directly test FOXO’s role in autophagy with age in HSCs, other stem cells, and differentiated cells to determine whether aging-related changes are due to FOXO activity and, in the long term, whether increases in FOXO activity could ameliorate aging-related cellular and tissue damage by upregulating proteostasis networks. Generally, understanding how the FOXO-induced proteostasis network changes with age and disease progression will provide key insights into the timing and tissue of action of this network.
FOXO-dependent proteostasis in neurodegeneration
FOXO transcription factors have been implicated in protection against neurodegenerative conditions in a number of animal models. In C. elegans, FOXO/DAF-16 activity is required for reduced toxicity of polyglutamine [99, 112, 113], Aβ [114] and SOD1 [115] aggregates in low insulin signaling conditions (daf-2 mutants), suggesting a protective role in Huntington’s disease, Alzheimer’s disease and amyotrophic lateral sclerosis (ALS). While there is some evidence, including work in mammals [48], that FOXO activity is protective in Parkinson’s disease, more work is required to understand how this is balanced with a potential pro-apoptotic role for FOXO3 in dopaminergic neurons. While several studies support a role for FOXO factors in combating aggregation-associated toxicity, it is not yet entirely clear whether this function is only dependent on FOXO-mediated proteostasis and clearance of aggregates. For example, protection against Aβ toxicity in mice with low IGF-1 signaling is associated with the formation of dense, low toxicity Aβ oligomers [116]. Whether FOXOs are involved in this protection against Aβ toxicity has not yet been tested; nevertheless, this study raises the possibility that FOXO factors may also trigger additional genes and pathways to alleviate the action of toxic aggregates in neurodegenerative diseases. It will be interesting to tease out the role of FOXO factors in clearing aggregates and regulating the survival of neurons, as some of these functions may be mediated by different, and independently regulatable, arms of the FOXO network.
FOXO-dependent proteostasis in cancer
In cancer, autophagy appears to have a dual role: autophagy can promote tumor suppression, but also supports tumor growth under low nutrient conditions. In mouse models, mutations in Atg genes are associated with tumorigenesis, and genes that are commonly mutated in cancer, such as p53, Pten, Tsc1 and Tsc2, positively regulate autophagy [117]. FOXO activity is associated with reduced tumor risks in humans [118] and indeed FOXOs can act as tumor suppressors in mice [119]. Whether FOXOs prevent tumor progression by promoting autophagy is not yet known, and will be important to test. Proteostasis via the ubiquitin-proteasome system significantly augments tumor development by degrading cell cycle regulators, and proteasome activity is commonly targeted in anti-cancer therapies [103]. Whether the specific E3 ubiquitin ligases downstream of FOXOs protect against cellular transformation has not been tested, and it remains possible that FOXOs induce additional modulators of the ubiquitin-proteasome pathway that prevent cancer development. It will be important to determine if FOXO factors’ ability to upregulate members of the proteasome pathway play an important role in their ability to act as tumor suppressors, since the antitumor efficacy of proteasome inhibitors could be diminished. It should also be noted that FOXO factors behave as oncogenes under some circumstances [120]. It will be interesting to understand the exact nature of the FOXO-proteostasis network in the balance of cancer formation versus tumor suppression in different cancer types.
Concluding Remarks
The relative role of FOXO-dependent proteostasis in overall FOXO functions is unknown. Given that FOXOs upregulate not only target genes involved in proteostasis, but also targets involved in cell cycle, oxidative stress, and metabolism, it will be important to dissect the extent to which various FOXO targets contribute to longevity and various diseases. Identification of mutant forms of FOXO transcription factors that specifically affect proteostasis, but not other aspects of FOXO function, as has been done for p53 [121], would be useful in elucidating the contribution of the FOXO proteostasis to organismal responses. It is possible that FOXO-mediated proteostasis is absolutely essential for longevity, but also conceivable that this activity of FOXO is not protective in aging in the absence of FOXO’s other functions. FOXOs may coordinate multiple mechanisms of quality control in the same cell to prevent damage accumulation.
While proteostasis is a cytoprotective mechanism that can increase organismal longevity and protect against aggregation-prone diseases and some cancers, excess autophagy or proteasomal activity can have detrimental consequences such as muscle wasting or cachexia [27, 32, 122]. Why does FOXO-induced proteostasis promote survival in some cases (neurons) but degeneration in others (skeletal muscle cells)? Some FOXO family members may be more potent at inducing proteostasis than others. As the expression of different FOXO family members vary in different tissues – for example FOXO3 and FOXO6 are highly enriched in the brain, FOXO1 in liver and adipose tissue, and FOXO4 in muscle – cell type-specific binding of FOXO to specific proteostasis targets may explain some differences, but the full story is likely to be more complex. It is possible that the protective effects of FOXO-induced proteostasis precedes in time the detrimental effects, but some cell types may resist for longer periods of time. Other cell type-specific parameters such as chromatin landscapes, binding of co-factors, and metabolic states, may contribute to the protective or degenerative consequences of proteostasis by FOXO factors. High throughput genomic approaches to identify transcription factor binding sites genome-wide are likely to shed new light on FOXO’s functions in autophagy and proteostasis. Genome-wide binding data are available for FOXO3 in neural stem cells and macrophages and for FOXO1 in B cells, liver and regulatory T cells [123–127], and these data indicate that FOXOs bind to proteostasis genes in all these cells. Future work comparing the kinetics of induction and the composition of the FOXO programs in tissues with different metabolic potential (liver, brain, muscle, fat, cardiac tissues) or in different cell types (differentiated vs. stem cells), particularly under low nutrient or high stress conditions, will expand our understanding of how FOXOs function to affect proteostasis. Identifying mechanisms to safely modulate these clearance mechanisms in vivo, without triggering negative effects will likely be of therapeutic benefit for diseases of age, in particular those linked to protein aggregates such as neurodegenerative diseases.
Highlights.
-
-
FOXO transcription factors are key regulators of autophagy and the ubiquitin-proteasome system
-
-
FOXOs have been implicated in protection against proteotoxicity
-
-
Reduced proteotoxicity is associated with protection against neurodegeneration
-
-
FOXO and mTOR reciprocally regulate proteostasis, depending on nutrient availability
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maiese K, et al. OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins. Trends Mol Med. 2008;14:219–227. doi: 10.1016/j.molmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008;192:19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 3.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 4.Kaestner KH, et al. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 5.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 6.Brunet A, et al. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 8.Biggs WH, 3rd, et al. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 10.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 11.Altomonte J, et al. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am J Physiol Endocrinol Metab. 2003;285:E718–E728. doi: 10.1152/ajpendo.00156.2003. [DOI] [PubMed] [Google Scholar]
- 12.Ren H, et al. FoxO1 target Gpr17 activates AgRP neurons to regulate food intake. Cell. 2012;149:1314–1326. doi: 10.1016/j.cell.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo H, et al. FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 2013;32:2589–2602. doi: 10.1038/emboj.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 16.Essers MA, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh SW, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehtinen MK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 19.Yang JY, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asada S, et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal. 2007;19:519–527. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 22.van der Horst A, et al. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 23.Calnan DR, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY) 2012;4:462–479. doi: 10.18632/aging.100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Kikis EA, et al. Protein homeostasis in models of aging and age-related conformational disease. Adv Exp Med Biol. 2010;694:138–159. doi: 10.1007/978-1-4419-7002-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Mammucari C, et al. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4:524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- 29.van der Vos KE, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 30.Xiong X, et al. The Autophagy-related Gene 14 (Atg14) Is Regulated by Forkhead Box O Transcription Factors and Circadian Rhythms and Plays a Critical Role in Hepatic Autophagy and Lipid Metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu P, et al. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Kume S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 37.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 39.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 40.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Codogno P, et al. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, et al. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 44.Wei B, et al. MST1, a key player, in enhancing fast skeletal muscle atrophy. BMC Biol. 2013;11:12. doi: 10.1186/1741-7007-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lokireddy S, et al. The ubiquitin ligase mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Pino E, et al. FOXO3 determines the accumulation of alpha-synuclein and controls the fate of dopaminergic neurons in the substantia nigra. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt530. [DOI] [PubMed] [Google Scholar]
- 49.Koh H, et al. Silent information regulator 2 (Sir2) and Forkhead box O (FOXO) complement mitochondrial dysfunction and dopaminergic neuron loss in Drosophila PTEN-induced kinase 1 (PINK1) null mutant. J Biol Chem. 2012;287:12750–12758. doi: 10.1074/jbc.M111.337907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mei Y, et al. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc Natl Acad Sci U S A. 2009;106:5153–5158. doi: 10.1073/pnas.0901104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang S, et al. Drosophila DJ-1 decreases neural sensitivity to stress by negatively regulating Daxx-like protein through dFOXO. PLoS Genet. 2013;9:e1003412. doi: 10.1371/journal.pgen.1003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamei Y, et al. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003;536:232–236. doi: 10.1016/s0014-5793(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 54.Sengupta A, et al. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warr MR, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia K, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McColl G, et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 60.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 66.Jia K, et al. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 67.Powers RW, 3rd, et al. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C, et al. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vellai T, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 73.Meissner B, et al. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J Biol Chem. 2004;279:36739–36745. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- 74.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 75.Lapierre LR, et al. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang MC, et al. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu LL, et al. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 78.Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 80.Low P. The role of ubiquitin-proteasome system in ageing. Gen Comp Endocrinol. 2011;172:39–43. doi: 10.1016/j.ygcen.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 81.Gidalevitz T, et al. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 82.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shibatani T, et al. Alteration of rat liver 20S proteasome activities by age and food restriction. J Gerontol A Biol Sci Med Sci. 1996;51:B316–B322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- 84.Conconi M, et al. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch Biochem Biophys. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- 85.Petropoulos I, et al. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J Gerontol A Biol Sci Med Sci. 2000;55:B220–B227. doi: 10.1093/gerona/55.5.b220. [DOI] [PubMed] [Google Scholar]
- 86.Bulteau AL, et al. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 87.Husom AD, et al. Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 88.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 89.Gumucio JP, Mendias CL. Atrogin-1, MuRF-1, and sarcopenia. Endocrine. 2012 doi: 10.1007/s12020-012-9751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 92.Sandri M, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 94.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vilchez D, et al. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell. 2013 doi: 10.1111/acel.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 97.Santamaria PG, et al. Rpn6p, a proteasome subunit from Saccharomyces cerevisiae, is essential for the assembly and activity of the 26 S proteasome. J Biol Chem. 2003;278:6687–6695. doi: 10.1074/jbc.M209420200. [DOI] [PubMed] [Google Scholar]
- 98.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 99.Hsu AL, et al. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 100.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Son JH, et al. Neuronal autophagy and neurodegenerative diseases. Exp Mol Med. 2012;44:89–98. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lecker SH, et al. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 103.Micel LN, et al. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31:1231–1238. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anselmi CV, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 105.Flachsbart F, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pawlikowska L, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Willcox BJ, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JC, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Donlon TA, et al. FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci. 2012;67:1132–1139. doi: 10.1093/gerona/gls067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pyo JO, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morley JF, et al. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 114.Cohen E, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 115.Boccitto M, et al. Daf-2 signaling modifies mutant SOD1 toxicity in C. elegans. PLoS One. 2012;7:e33494. doi: 10.1371/journal.pone.0033494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cohen E, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bhutia SK, et al. Autophagy: cancer's friend or foe? Adv Cancer Res. 2013;118:61–95. doi: 10.1016/B978-0-12-407173-5.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu MC, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 119.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dobrowolny G, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 123.Shin DJ, et al. Genome-wide analysis of FoxO1 binding in hepatic chromatin: Potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Litvak V, et al. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012;490:421–425. doi: 10.1038/nature11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Webb AE, et al. FOXO3 shares common targets with ASCL1 genomewide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]