Abstract
Cell-cell fusion in sexually reproducing organisms is a mechanism to merge gamete genomes, and in multicellular organisms, it is a strategy to sculpt organs such as muscles, bones, and placenta. Moreover, this mechanism has been implicated in pathological conditions such as infection and cancer. Study of genetic model organisms has uncovered a unifying principle: cell fusion is a genetically programmed process. This process can be divided in three stages: (i) competence: cell induction and differentiation, (ii) commitment: cell determination, migration and adhesion, and (iii) cell fusion: membrane merging and cytoplasmic mixing. Recent work has led to the discovery of fusogens, cell fusion proteins that are necessary and sufficient to fuse cell membranes. Two unrelated families of fusogens have been discovered, one in mouse placenta and one in Caenorhabditis elegans (Syncytins and F proteins, respectively). Current research aims to identify new fusogens and determine the mechanisms by which fusogens merge membranes.
Fusion proteins – the elusive key to cell-cell fusion
The emerging field of cell-cell fusion aims to understand where, when, how and why two or more cells merge to develop into a new organism (sexual fusion) or to generate diverse giant multinucleate cells that sculpt distinct organs (non-sexual cell fusion). Fused cells can undergo dramatic changes in signaling and behavior and acquire new developmental fates. During fertilization there is a barrier to further fusion events, but in somatic cell-cell fusion, the fused cells are often competent and sometimes committed for new rounds of fusion forming giant syncytia as occurs in muscles and epithelia.
Genetic model organisms have been used to characterize the process of cell-cell fusion at different levels, often by studying fusion-defective phenotypes, unraveling different stages of this process. Fusion competence is the first step of this process (Figure 1a), allowing cells to overcome mechanisms that prevent fusion. The next step, commitment, requires cell-cell interactions, induction, and activation of the fusion machinery (Fig. 1b). Finally, cells are fused through the merging of the main barriers that define cells. The fusion of the plasma membranes and mixing of the cytoplasms constitute two unique steps in the pathway (Fig. 1c).
Figure 1. Simplified vision of steps required for cell-cell fusion.
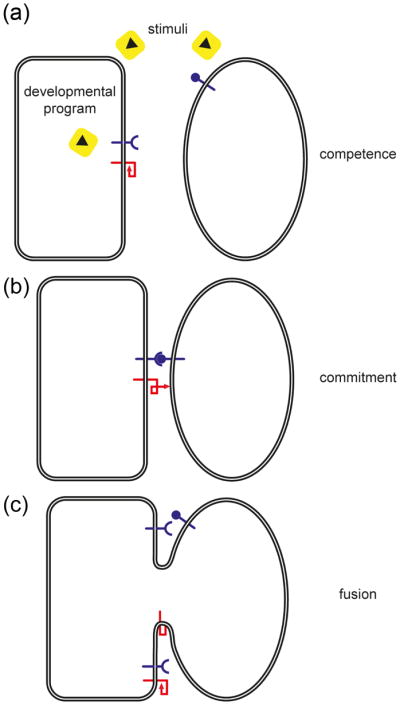
The representation of this multi-step pathway is purely schematic, and numerous cellular features are not incorporated- for example, signaling events and regulation of gene expression. A hypothetical three step scenario of heterotypic cell-cell fusion executed by a unilateral fusogen (red) is presented. (a) Competence. Differentiation into fusion-competent cells involves one or more of these complex processes: reception and response to extracellular signals, execution of developmental programs, cell polarization, cell migration, morphological changes, polarized secretion and ultimately surface display of key molecules required for the next step. (b) Commitment. This step involves cell-cell adhesion, continuous signaling and cell polarization, and consequential full exposition and/or activation of the fusogenic machinery. (c) Cell-Cell Fusion. Correct merging of plasma membranes connects both cytoplasms leading to further signaling and developmental changes. Unlike fertilization, somatic fused cells can still be competent and therefore ready for new rounds of fusion forming giant syncytia in some cases of cell-cell fusion in multicellular organisms.
A major conceptual framework in the field is the existence of specific proteins on the surface of cells that are essential and sufficient to mediate merging of the cell membranes by taking part directly in cell-cell fusion. These cell fusion proteins (fusogens) have been the holy grail of the field for decades, but recently two independent and unrelated families of fusogens have been identified and characterized. The two families are required for fusion of epithelial cells. One family (the Syncytins) contains diverse proteins that originated from endogenous retroviruses related to the HIV gp41 envelope glycoprotein and function during fusion of the placental trophoblasts that form the syncytial trophoblasts essential for mouse placentation. The second family (the F fusion proteins) is responsible for diverse and numerous cell fusions in the skin and digestive and reproductive organs of the nematode Caenorhabditis elegans.
The hunt is on now to identify the missing fusogens in other species. These may be members of the two known families of cell-cell fusogens, related to another family of viral membrane fusogens, or novel. Identifying these proteins may be complicated because based on the work on viral fusogens, it is the overall structure of these proteins rather than the primary sequence that is conserved [1–3].
This review covers emerging genetic mechanisms and cell biological principles that control and mediate the process of sexual and somatic cell-cell fusion. Although there are a number of intriguing reports of cell-cell fusion involving stem cells, there is little information on the genetic basis of stem cell-cell fusion mechanisms. Therefore, we focus here on recent findings about sexual and somatic cell fusion in a variety of genetic systems, highlighting the distinct contributions of each model system to the field. We discuss examples of sexual non-self fusion in diverse genetic model organisms followed by somatic self-fusion in fungi, flies, worms, mice, and humans. In particular, we focus on how hyphal cells fuse to form tubular networks in Neurospora, myoblasts fuse to form multinuclear myofibers in diverse genetic model animals, macrophages fuse to form osteoclasts and giant cells, epithelia fuse to give rise to syncytiotrophoblasts in the placenta, and one third of all somatic cells fuse to form diverse organs in worms.
Sexual cell-cell fusion (gamete fusion)
Despite the great diversity in size, shape, and behavior of gametes from different species, it still takes two to tango: sexual reproduction requires two gametes that fuse and merge their genomes. Here, we discuss recent advances in sexual cell fusion in some representative genetic model organisms including mice, fungi, plants, protists, and worms.
Egg CD9 and sperm IZUMO1 are required for fertilization in mice
In mice, only two key genes have been identified that are essential in sperm-egg fusion, Izumo1 in the spermatozoon and Cd9 in the oocyte. Loss of these factors leads to a sterile phenotype, but currently there is no demonstration that they act as fusogens. However, the fact that the requirement for these factors can be overcome by bypassing the fusion step via intracytoplasmic sperm injection suggest that they make very specific contributions to egg-sperm fusion [6–8].
IZUMO1, which was named after a Japanese marriage shrine, was initially identified by a sperm-egg fusion-inhibiting monoclonal antibody. Mice deficient in Izumo1 produce spermatozoa that appear morphologically normal, bind and penetrate the zona pellucida surrounding the egg, but are not capable of fusing with eggs [9]. IZUMO1 is initially hidden in the acrosomal organelle under the plasma membrane. After exocytic fusion between the acrosome and the plasma membrane, IZUMO1 relocates to the surface of the sperm head, suggesting that redistribution of IZUMO1 is essential for fusion [4].
CD9 (a tetraspanin) was shown to be required for fusion on the egg plasma membrane [6–8]. It was proposed that exosome-like CD9-containing vesicles are secreted from unfertilized eggs, thereby conferring fusion competence to the spermatozoon [10]. However, this experiment could not be reproduced in independent laboratories [11, 12]. In addition to the sterility phenotype, Cd9−/− mice have altered length, thickness, and density of eggs’ microvilli, suggesting that CD9 participates in microvilli formation and that microvilli are important for sperm-egg fusion [13]. There is no evidence indicating that IZUMO1 and CD9 directly interact during sperm-egg fusion. Even though IZUMO1 and CD9 are essential for sperm-egg fusion, it is still unclear how they participate in the process and if other proteins are also involved.
Fertilization in plants, protists, and invertebrates
Investigation of fertilization in other model organisms has uncovered a diverse set of genes required for this process. Recently, a male gamete-specific protein from lily pollen, Generative Cell Specific 1 (GCS1)/HAP2, was identified as a crucial factor in double fertilization in plants [14, 15]. Interestingly, GCS1/HAP2 orthologs have been found in various protists, including Chlamydomonas and Plasmodium, and in some cases, these orthologs have been shown to have a role in fertilization [14, 16, 17]. Despite this, GCS1/HAP2 and related proteins have not been reported to be sufficient for cell-cell fusion, thus they are not considered fusogens.
Self-fertilizing C. elegans hermaphrodites generate equal numbers of sperm and oocytes (~300) and all of them fuse as pairs. Similarly, high fusion efficiencies are found in sexual crosses where the number of gametes produced is higher (~1000). These and other characteristics make C. elegans a convenient genetic system to study sperm-egg fusion. Extensive genetic screens identified genes required for different stages of fertilization, but none have been shown to be sufficient to fuse gametes without the participation of other genes [18].
Although there are essential genes for fertilization in some species, the sufficiency for cell membrane fusion has not been reported in heterologous cells. The fact that no genes have been found in plants, protists or worms that are sufficient for fusion does not necessarily imply that no such genes exist in these species. However, it is not expected that these fusogens will be conserved between worms and mice, based on the diverse fusogens that have been found (see below; [2]).
Gamete fusion is regulated by PRM1 in yeast and filamentous fungi
In the Saccharomyces cerevisiae mating process, haploid cells differentiate into gametes, which fuse and form a diploid zygote (Figure 2a). PRM1 was identified as a candidate protein involved in cell fusion through a combination of in silico analysis and functional characterization of mating-specific integral membrane proteins [20]. PRM1 is a tetra-spanning integral membrane protein that forms disulfide-linked homodimers and localizes at the cell fusion zone [21, 22]. prm1 mating pairs show three alternative phenotypes: arrest with both plasma membranes apposed, successful fusion, or lysis [20, 23]. Lysis appears to be a byproduct of the fusogenic activity, suggesting that PRM1 has a regulatory role in this process. It has been hypothesized that in its absence, a misregulated fusogen compromises membrane integrity leading to lysis instead of fusion [23–25]. However, this model remains untested because there are no known fusogens in yeast. Several candidate fusogens have emerged from recent genetic screens, including genes involved in cell polarization or cell wall remodeling [19, 26–28], but none appear to act as a bona fide fusogen. PRM1 has a conserved function in the filamentous fungus Neurospora crassa both in sexual fusion and in vegetative cell merger (see below). Similar to yeast, approximately 50% of N. crassa prm1 fusion pairs arrest at the stage of plasma membrane merger [29]. It is currently unknown whether lysis occurs in N. crassa prm1 fusion events.
Figure 2. Non-self fusion in S. cerevisiae and self-fusion in N. crassa is a multi-step process involving MAP kinase cell signaling.
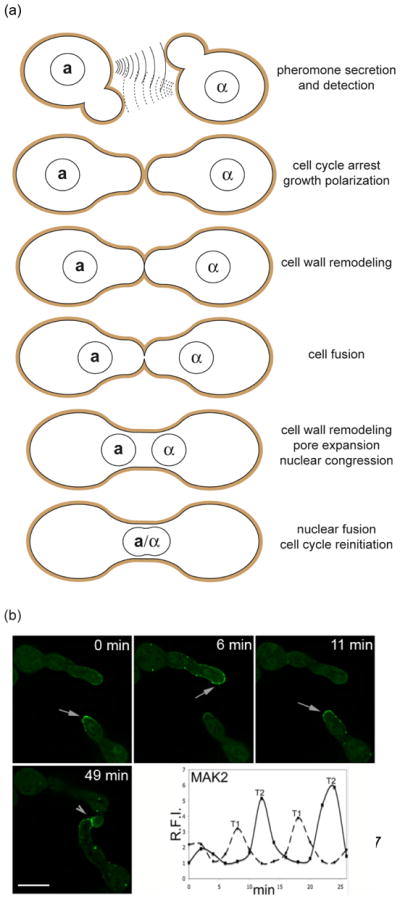
(a) Mating in S. cerevisiae involves the fusion of two haploid cells of opposite mating types (a andα) into an a/α diploid zygote. Haploid cells secrete peptide pheromones that can be detected by their cognate partners. Pheromone detection induces cell-cycle arrest, transcriptional induction of pheromone-specific genes, and polarization of growth towards the pheromone source (shmooing). Shmoo formation leads to cell-cell contact and cell wall (brown) merging. To avoid the risk of lysis caused by high internal osmotic pressure, the cell wall is degraded only at the zone of cell-cell contact. Plasma membrane fusion occurs within a few minutes after contact. Further cell wall removal allows pore expansion followed by congression and fusion of the nuclei. (b) Fusion germlings of N. crassa alternate between two physiological stages during chemotropic interaction. While two germ tubes approach each other, the cytoplasmic MAP kinase MAK-2 is recruited to the plasma membrane of the fusion tips (arrows) in an oscillating manner. Once the cells have established physical contact the kinase accumulates at the fusion point (arrowhead). Time points: time after observation started. Scale bar: 5 μm. The graph indicates fluorescence intensity of MAK2-GFP at two fusion cell tips. T1 = cell tip 1, T2 = cell tip 2 (adapted from [32]).
The lack of success in identifying fusogens suggests they may be essential, redundant, bilateral (required in both fusing cells), or multigenic. Thus, biochemical or bioinformatics approaches may be more helpful in finding fusogens. Furthermore, future studies in a range of organisms should identify other fusion-related proteins involved in gamete fusion, and it will be interesting to determine if there are common principles for gamete fusion, or if this process varies in different species. Sex determination evolves very rapidly, and it is possible that gamete fusion follows this trend.
Asexual cell-cell fusion (somatic cell fusion)
Asexual fusion, which is usually homotypic, is important for vegetative growth in fungi and sculpting organs in multicellular animals. However there are exceptions to these generalizations. During asexual development of multicellular organisms, numerous somatic cells fuse to create and sculpt organs and build reticular networks in a process called self-fusion.
The formation of tubular networks in N. crassa
N. crassa colonies consist of a syncytial network of highly polarized, growing, multinucleated hyphae. Its life cycle comprises at least four different experimentally tractable cell fusion events, allowing comparison of specific molecular factors in these different processes. During colony initiation, germinating spores mutually attract, grow towards each other, and fuse, forming a cellular network, which further develops into a mycelial colony. Genetic analysis identified numerous mutants affected in this asexual fusion. However, with the exception of Δprm-1, all mutants are defective in steps preceding the plasma membrane merger, such as competence, cell-cell signaling, or directed growth (Figure 1). Cell communication in germling fusion depends on components homologous to the highly conserved yeast pheromone response MAP kinase module [30, 31], which adopt, however, unusual subcellular dynamics in N. crassa. While two fusion germlings grow towards each other, MAK-2 (homologous to Fus3p) oscillates from the cytoplasm to the plasma membrane in the region of the growing cell tip (Figure 2b) [32]. This dynamic localization alternates with SO, a cytoplasmic protein of unknown molecular function. The anti-phase oscillation of MAK-2 and SO indicates that tightly controlled and coordinated protein concentration and localization ensures fusion occurs when and where it should. The two cells that will fuse alternate between signal sending and receiving in a type of cell-cell dialogue [32–34]. Mathematical modeling indicated this would allow efficient fusion partner signaling by a single signal-receptor system while preventing self-excitation [34]. This example illustrates that sophisticated mechanisms have evolved to insure that two fusion cells establish a stable and efficient interaction within a larger population of genetically and developmentally identical individuals.
Myoblast fusion: invasion by Podosome-Like Structures (PLS)
During mammalian muscle development thousands of myoblasts fuse to form the myofiber. Work in fly, zebrafish, and mouse has identified many genes required for myoblast fusion, which have been detailed in recent reviews [35–37]. One of the unifying features of these factors is their participation in Arp2/3-mediated actin polymerization [38]. In Drosophila, muscle fibers form via fusion between a founder cell (FC)/myotube and a fusion competent myoblast (FCM). The fusion interface between the FC/myotube and FCM is identified by an actin-rich structure [39–42] (Figure 3). The distribution of actin at the fusion site is asymmetric: the FC/myotube builds a thin sheath of actin [43], whereas the FCM provides a dense actin network termed the actin focus (Figure 3a) [43–45]. Consistent with cellular asymmetry of the fusion site, many actin regulatory genes including WASp [40–42, 46, 47], Verpolin/WIP/Solitary [41–43], Blown Fuse [48], and Mbc/Dock180 [44] are necessary only in the FCM, whereas SCAR/WAVE [39, 43, 46, 47] is necessary in both fusion partners (Figure 3c).
Figure 3. Podosome-like structures asymmetrically invade the myotube.
(a) Stage 14 Drosophila embryo showing FCMs attached to a developing myotube (FC/myotube). Phalloidin staining reveals F-actin at the cell cortex and prominent F-actin foci at the FC/myotube//FCM interfaces. Arrowheads show late stage F-actin foci of protruding invadosomes shortly before cell-cell fusion. Scale: 2.1 μM. Images taken on a Leica SP5 63x oil immersion NA 1.4 objective [39]. (b) Schematic of (a), detailing actin focus structure in each FCM. (c) Schematic of one actin focus from (b, boxed area), indicating FCM Arp2/3 dependent pathways required for actin dynamics for the PLS and fusion. Each actin focus is surrounded by FC/myotube/FCM specific adhesion proteins (not shown) that, upon engagement, signal to actin regulators [(MyoblastCity (Mbc) ->Rac ->SCAR ->Arp2/3; BlownFuse (Blow) ->Verprolin/WIP/Solitary (Vrp1) -> WASp -> Arp2/3]. Links between these pathways exist (e.g., Blow -> Kette). Please refer to recent reviews for details on the genetic and physical interactions required for actin during myoblast fusion [35–37].
3D reconstruction indicates that the actin focus is lined with the FC-FCM Immunoglobulin (Ig)-domain containing adhesive proteins [43, 45, 49]. Ultrastructural studies have further identified 3–4 protrusions from the FCM, which invade the apposing FC/myotube [43]. Based on their invasiveness, size, and actin core with a surrounding ring of adhesive proteins, these structures have been called “Podosome-Like Structures” (PLS) [43, 45, 49]. Similar structures have been identified in cancer cells (invadopodia), macrophages, osteoclasts, and tissue culture cells (podosomes), and they are associated with cell adhesion, migration, and invasion as well as ECM protease secretion— all functions that could facilitate cell fusion [43, 45, 49]. Unlike the other invasive structures, PLS are unique in that they form a single domain of 300 – 1,500 nm between the FCM and FC/myotube that is believed to participate in the generation of cytoplasmic continuity between fusing myoblasts [43, 50]. However, it is unclear how the initial fusion pores form and whether the PLS participates in pore formation and expansion. Although the existing invadopodia/podosome literature suggests new regulators of fusion [45, 51], for example, ECM proteases and cortactin are hallmarks of these structures [51], whether these function in myoblast fusion is unknown. Moreover, the links between actin, PLS, and membrane dynamics that lead to cytoplasmic continuity are still unclear.
Macrophage fusion: regenerative and pathologic
Macrophage fusion, both homotypic (macrophage-macrophage) and heterotypic (macrophage-somatic), occurs in the dynamic process of tissue remodeling/regeneration and in pathogenic states. Continued investigation into aspects of macrophage fusion promises to shed insight into the regenerative-pathogenic axis of this unique and important function.
Homotypic macrophage fusion: Osteoclasts and multinucleated giant cells
Homotypic fusion of macrophages in mice (and men) occurs in bone and tissues affected by chronic inflammation. In bone, multinucleated macrophages (osteoclasts) mediate bone resorption essential for skeletal stability. In other circumstances (generally called multinucleated giant cells), the function of fused macrophages is unknown [52].
Macrophage fusion can generate osteoclasts through cytokine induction with macrophage colony stimulating factor (M-CSF) and Receptor Activator of NF-κB Ligand (RANKL) or giant cells through interleukin-4 (IL-4) and/or granulocyte macrophage colony stimulating factor (GM-CSF) [52]. Despite distinct pathways, the molecular machinery involved appears to be at least partially shared [52]. The function of the molecules upregulated during macrophage fusion supports the importance of proteolysis, cytoskeletal rearrangements, chemotaxis, migration, lipid recognition, and cell-cell attachment in the process [52–62] (Figure 4). Intriguingly, several molecules shown to be involved in macrophage fusion are also important in phagocytosis. Phagocytosis involves several membrane fusion events, and fusion may represent an alternative process to the engulfment of one macrophage by another.
Figure 4. Macrophage fusion is a highly-regulated, induced process.
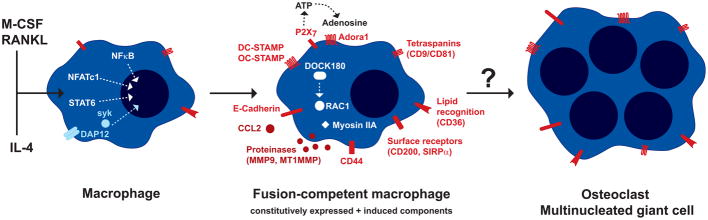
In order to fuse, macrophages acquire a fusion-competent phenotype through the integration of different signals: cytokines (e.g., RANKL + M-CSF or IL-4), cell-cell interaction (e.g., via TREM-2/DAP12 [53]), and the respective intracellular signaling pathways (NF-κB, NFATc1, STAT6, syk [52, 54, 55]). Genes essential for and upregulated during fusion include the chemokine CCL2, the putative multiple transmembrane receptors DC-STAMP and OC-STAMP, the cell adhesion molecule E-Cadherin, and the matrix metalloproteinase MMP9 [52, 55–57]. Cytoskeletal rearrangements required for fusion are mediated by RAC1 and DOCK180 [52]. Proteinases may influence fusion by cleavage/activation/degradation of other proteins such as CD44 or Myosin IIA [58, 59]. In contrast, the matrix metalloproteinase MT1MMP seems to regulate RAC1 activity during fusion independently of proteolytic function [60]. Another crucial factor for macrophage fusion is the release of ATP via the P2X7 receptor and recognition of adenosine by the receptor Adora1 [61, 62]. In addition, exposure of phosphatidylserine and lipid recognition by CD36 as well as surface receptors recognizing macrophage-expressed ligands such as CD200 and SIRPα were shown to be involved [52]. Tetraspanins (CD9/CD81) act as molecular membrane organizers and appear to play an inhibitory role in macrophage fusion [52]. The mechanistic basis of the actual membrane fusion step has not been elucidated so far.
For IL-4 induced macrophage fusion, it was shown that all fusion partners must receive the IL-4 stimulus [63], but that macrophages lacking selected molecules or expressing only part of the fusion machinery can still fuse with a fully fusion-competent macrophage [63, 64]. This shows that macrophage fusion between ‘donor’ and ‘acceptor’ macrophage is possible. As described below, a fusogenic ‘donor’ macrophage may even fuse with other cells such as tumor cells. Based on the current data from homotypic macrophage fusion events, we assume that these heterotypic fusion partners (e.g., cancer cells) display discrete molecular properties not found in other cells.
Heterotypic macrophage fusion in tumor pathogenesis
The concept that cell fusion plays a role in cancer pathogenesis spans a century, stemming from the observation of spontaneous cancer-somatic cell fusion in culture [65]. The combination of two genomes into a single cell presents an alluring basic mechanism for gain of chromosomal instability, aberrant gene expression, and acquisition of novel cellular behaviors that can potentiate tumor aggressiveness and metastatic disease. However, there is little mechanistic evidence and even less physiologic validation in human tumors substantiating the hypothetical link between cancer and cell fusion.
Recently, in vivo demonstration of robust fusion between cancer cells and macrophages was reported in mouse models that employed either bone marrow transplantation or parabiosis to facilitate introduction of marked bone marrow-derived cells into tumor-bearing mice [66, 67]. Although functional transfer of the macrophage cell behavior to the cancer cell remains to be demonstrated, transcriptome analysis of isolated cell fusion hybrids revealed retention of macrophage gene expression within the cell fusion hybrid cell [66]. This finding supports the possibility that macrophage fusion with cancer cells could provide a mechanistic link for the acquisition of metastatic properties encompassing those native to the macrophage (e.g., intravasation, extravasation, tissue colonization).
The evidence for cancer-blood cell fusion in humans is weaker, although there are a number of case reports from gender mis-matched bone marrow transplant patients that later went on to acquire a solid tumor malignancy (e.g., [68]). Further, evidence of cancer cell fusion has been frequently documented in multiple myeloma patients by tracking gene translocations [69]. However, the clear demonstration of a physiologic and biologic impact of cancer cell fusion in human disease remains elusive.
Epithelia fusion in placenta
The placenta is a transitory organ that connects the fetus to the uterine wall allowing nutrient uptake, waste elimination, and gas exchange to ensure embryonic development [70]. The syncytiotrophoblast (STB), formed by fused trophoblast cells, is a critical domain of the placenta required for nutrient exchange and secretion among other functions [70, 71]. In humans, trophectoderm cells in contact with the embryoblast and the maternal endometrium generate the primary STB by intercellular fusion during peri-implantation. The secondary STB, which is formed 1–2 weeks after implantation and throughout pregnancy, is a single, multinucleated and highly dynamic syncytium maintained by the continuous fusion of cytotrophoblast cells (CTBs) with the neighboring STB [70] (Figure 5a). The murine STB resides in the placental labyrinth and is a two-layered structure – SynT-I and SynT-II, which never fuse [72] (Figure 5b). Hormones, growth factors, cytokines, protein kinases, transcriptional factors, and membrane proteins regulate the complex anatomy and physiology of the placenta [70, 71].
Figure 5. Comparative anatomy of the human and mouse placental syncytiotrophoblasts and the localization of Syncytins.
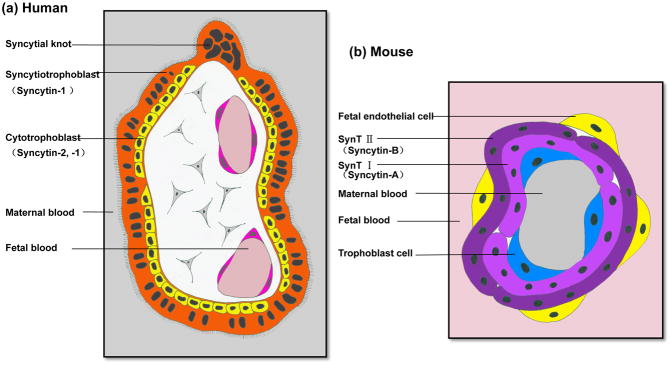
(a) A simplified cross-section through a human chorionic villus from the first trimester placenta. It is a two-layered structure composed of a layer of mononucleated cytotrophoblast cells (yellow) and a layer of multinucleated syncytiotrophoblast (orange), which is in contact with the maternal blood. Note Syncytin-1 is expressed in both layers whereas Syncytin-2 only localizes in the cytotrophoblast cells. (b) Schematic representation of the fetal-maternal interface in the mouse placental labyrinth. The mouse placental labyrinth contains maternal and fetal blood spaces separated by three layers of trophoblast cells and a layer of fetal endothelial cells. The three layers of trophoblast cells are: a single layer of trophoblast giant cells lining the maternal blood sinusoids and two layers of syncytiotrophoblast, SynT-I and –II. SynT-II is in contact with fetal endothelial cells. Note Syncytin-A is specifically expressed in SynT-I, and Syncytin-B is only detected in SynT-II.
Endogenous retroviral Syncytins are required and sufficient to fuse cells
One unique aspect of trophoblast syncytialization is the involvement of Syncytin-1 and -2 ENV genes of human endogenous retroviruses [71]. Syncytin-1 appears to be predominantly expressed in the STB whereas Syncytin-2 is only detected in the villous CTBs [70] (Figure 5a). Both Syncytins are sufficient to induce cell-cell fusion in different cell lines in a receptor-dependent manner, suggesting they are bona fide fusogens [70]. The disulfide bridge-forming CX2C and CX7C motifs of Syncytins are essential for their fusogenic activities [73].
Syncytin-A and Syncytin-B are murine endogenous retroviral ENV genes homologous to the primate Syncytin genes, and they localize respectively to SynT-I and SynT-II [74] (Figure 5b). Deletion of either Syncytin gene leads to impaired formation of the corresponding STB layer [75, 76], suggesting distinct yet essential roles during trophoblast fusion for these Syncytins. Syncytin-related genes have also been identified in Leporidae and Canidae [77, 78]. Because none of these Syncytins are orthologous [78], it has been suggested that they were captured by their hosts independently between 12–85 million years ago and might account for the different structures of placentas among different species [78]. However, it is still not clear why some mammals utilize fusogens of viral origin to execute the fusion process in the placenta, and although Syncytins are described as placenta-specific genes, the fusogenic activity of human Syncytin-1 has also been implicated in the fusion events of non-trophoblast cells, including cancer cells [79] and osteoclasts [80].
The observation that syncytiotrophoblast is not completely absent in Syncytin knockout mice [75] suggests the existence of yet unknown fusogens. The mechanism of trophoblast fusion supports the working hypothesis that viral-like fusogens will also merge myoblasts, macrophages, and gametes, but this has not yet been shown.
Programmed cell fusion is essential to sculpt organs in C. elegans
Over 300 somatic cells (out of ~1000) fuse to form syncytial muscles in the pharynx and epithelia in the skin, vulva, hymen, uterus, pharynx, excretory system, and glands in C. elegans [81–83]. Genetic screens have identified two genes, eff-1 and aff-1, that fuse and sculpt cells in C. elegans [1, 81–83]. Whereas fusion failure results in deformities, ectopic migration of unfused epithelial cells, behavioral defects, and low fertility, abnormal expression of EFF-1 or AFF-1 causes excess cell fusion and embryonic lethality [81–83].
EFF-1 and AFF-1 proteins are essential and sufficient to fuse cells
eff-1 is required for the initiation and expansion of fusion pores [81, 82, 84, 85]. EFF-1 and AFF-1 are type I membrane glycoproteins that belong to a family of homotypic cell-cell fusogens (F family) essential for somatic cell fusion. F proteins can fuse heterologous insect and mammalian cells via a hemifusion intermediate [83, 86, 87]. Moreover, enveloped viruses with F proteins substituted for their native fusogens are capable of infecting mammalian cells, which provides further evidence that they are bona fide fusogens (Figure 6) [87].
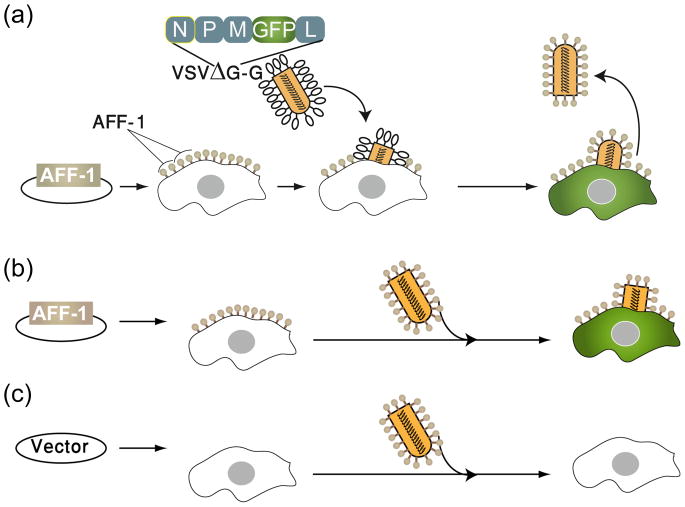
Figure 6. AFF-1 can substitute for the native fusogen glycoprotein G from Vesicular Stomatitis Virus (VSV) and fuse viral membranes to cells.
(a) AFF-1 from C. elegans can complement the generation of recombinant single round infective VSVΔG-AFF-1 virus-like particles in vitro. Baby Hamster Kidney cells (BHK) expressing AFF-1 protein on the cell surface can be infected with the G-complemented VSVΔG recombinant virus (VSVΔG-G). The viral genome encodes GFP in place of the fusogenic glycoprotein G. Infection results in viral induced expression of GFP by target cells (green cytoplasm). VSVΔG-AFF-1 virus-like particles are harvested from the supernatant. (b) BHK cells can be transfected with aff-1 and infected with virus-like particles obtained in (a). The infective virus-like particles express AFF-1 on their surface instead of VSV-G-glycoprotein. (c) Cells transfected with empty vector and infected with VSVΔG-AFF-1 do not result in green cells and serve as negative controls. These experimental paradigm shows that AFF-1, in contrast to VSV-G, is necessary in both membranes to mediate virus-cell fusion [87].
F proteins execute self- and auto-fusion of cells in development
F proteins also mediate auto-fusion whereby specific cellular domains of pharyngeal muscles, epithelial excretory duct cell, glia, and neurons fuse with themselves in C. elegans [88–93]. For example, both pm8 and vpi1 auto-fuse to become two mononucleated donut-shaped cells [89]. Auto-fusion may have been undetected in other biological systems such as in phagocytosis, immunity, generation of small capillaries, and neurons. The difficulty of identifying an auto-fusion event is that the outcome of auto-fusion is the formation of a cell with a different shape. Because of the almost invariant cellular development of C. elegans, however, a number of auto-fusion events were characterized. But this was possible only after mutants that specifically failed to fuse were identified. In other genetic systems it will be necessary to first find the unidentified fusogens and then discover an intermediate that would reveal that the cell fused to itself to generate a new shape.
Diverse signaling pathways, transcription factors, and the vacuolar ATPase control the activities of eff-1 and aff-1 [83, 94–100]. For example, Notch signaling simultaneously induces AFF-1 and represses EFF-1 expression in pm8 muscle cells [89]. Thus, precise spatial and temporal regulation of F activity is essential to determine and maintain a stereotypic pattern of cell-cell and auto-fusions.
Although somatic fusogens have been characterized, the germ cell fusogens are as yet unknown (see sexual fusions above). F proteins have been identified in forty-seven species, comprising mostly nematodes, but also ctenophores, arthropods, one chordate, and one protist. These fusion proteins are believed to evolve rapidly and the conservation is probably structural [87]. To identify structurally related F proteins acting in sexual and asexual reproduction, muscle development, bone development, placenta formation, immunity, stem cell, and cancer cell fusion it will be necessary to identify the precise structural and functional signatures essential for cell fusion.
Concluding remarks
Different genetic model systems have recently contributed a distinct mechanistic understanding of the process of cell fusion. Although no universal mechanism may exist, the field is actively searching for unified concepts and unidentified fusogens. At the same time we are exploring the specific roles of the actin cytoskeleton in myoblast fusion, cytokine induction of macrophage fusion, the links between fusion and lysis in yeast, reciprocal, alternating signaling in Neurospora, the connections between cancer and cell fusion, the specific functions of different Syncytins in the generation of diverse placental giant multinucleated cells, and the biophysical mechanisms of F protein-mediated cell-cell fusion. Thus, cell fusion mechanistic research is still an embryonic field in a very exciting stage of development.
Three structural classes of authentic fusogenic membrane glycoproteins have been discovered in enveloped viruses, and recent research has uncovered two unrelated families of cellular fusogens, suggesting that dedicated fusogens specific for each cell fusion process are waiting to be discovered and characterized. Some researchers in the field believe that “spontaneous cell fusion” occurs in biology in the absence of fusogens or dedicated machinery. However, the F family of fusogens in nematodes and other species and the endogenous retroviral family of mammalian Syncytins in the placenta demonstrate that overcoming the barriers that prevent spontaneous cell-cell fusion in nature requires a dedicated machinery of fusion proteins. Genuine fusogens are thus expected to be found in all organisms. We strongly believe that a combination of genetic, biochemical, and biophysical approaches will eventually identify and characterize these elusive proteins required for fertilization and organ formation.
Acknowledgments
We apologize to our colleagues for the omission of important contributions to the field, and their references, owing to space constraints. We are grateful to I. Bothe, E. Folker, J. Grimm, X. Lu, and S. Yu for helpful comments on the manuscript. I. Bothe and S. Yu for Fig. 3., R. Wang for Fig. 5., O. Avinoam for Fig. 6. Work in our labs was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (21687018 to N. I), the National Institutes of Health (NIH GM078318 to M.B.), European Research Council (ERC Advanced grant 268843 to B.P.), the Israel Science Foundation (ISF grants 1542/07 and 826/08 to B.P.), International Centre for Genetic Engineering and Biotechnology (ICGEB grant CRP/URU11-01 to P.S.A.), Agencia Nacional de Investigación e Innovación (ANII-INNOVA grant -DCI-ALA/2007/19.040 URU-UE to P.S.A), the German Research Foundation (DFG, HE 5190/3-1 to L.H. and FL 706/1-1 to A.F.), the Ministry of Science and Technology of the People’s Republic of China (973 Program 2011CB944403 to H.W.), the National Natural Science Foundation of China Grant (30971088 to H.W.), the National Institute of Health/National Cancer Institute (CA118235 to M.W.).
Glossary
- Anchor cell Fusion Failure-1 (AFF-1)
protein essential for the fusion of the anchor cell with eight gonadal cells and additional epithelial and myoepithelial cell fusions in C. elegans. It was the second member of the F family of fusogens identified
- CD9
integral membrane protein required for fertilization in mice and member of the tetraspanin family. It is associated with integrins and other membrane proteins
- Cytotrophoblast cells (CTBs)
mononucleated progenitor cells of the placenta which can differentiate into extravillous trophoblast cells or fuse to form or increase the size of the multinucleated syncytiotrophoblast
- Epithelial Fusion Failure-1 (EFF-1)
integral membrane protein essential and sufficient for epithelial and myoepithelial fusions in C. elegans. EFF-1 was the first F family fusogen discovered
- Fusion Competent Myoblast (FCM)
naive myoblasts capable of fusing to founder cells during Drosophila muscle formation. These cells account for the bulk of the myoblasts that form the muscles in Drosophila
- Fusogen
molecule (usually a protein) that fuses biological membranes
- Founder Cell (FC)
Muscle precursor or pioneer cells that establish a muscle pre-pattern in Drosophila
- Generative Cell Specific 1 (GCS1)
a membrane-associated factor critical to gamete fusion in various eukaryotes
- Germ tube
polar growing hyphae emerging from a fungal spore
- Hypha
thread-like multinucleated growth form of many fungi
- IZUMO1
immunoglobulin superfamily type I transmembrane protein with one extracellular Ig-like domain. Spermatozoa from Izumo1-deficient mice fail to fuse with eggs
- Parabiosis
The surgical or natural union of anatomical parts of two organisms
- Podosome-Like Structures (PLS)
structures containing an actin core with a surrounding ring of adhesive proteins which forms a single domain 300 – 1,500 nm between the FCM and FC/myotube that provides cytoplasmic continuity
- PRM1
pheromone-regulated membrane protein 1
- Redundant
term used here in the context of yeast gamete fusion. A gene is redundant if additional loss of function of one or more genes in the same mating partner is needed to observe a phenotype
- Syncytiotrophoblast (STB)
multinucleated, terminally differentiated syncytium in the placenta formed by fusion of CTBs
- Syncytin
the ENV protein of an endogenous retrovirus
- Syncytium
multinucleated cell often resulting from cell fusion
- Unilateral
term used here in the context of yeast gamete fusion. A gene is unilateral if loss of function in both mating partners is needed to observe a phenotype. The viral fusion machinery and Syncytins are unilateral because the fusogen is only present in the virus or in one of the fusing cells. By contrast, EFF-1 and AFF-1 are required in both fusing cells, therefore the fusion machinery is not unilateral but homotypic or bilateral
- Vesicular Stomatitis Virus (VSV)
an enveloped virus related to rabies used as a model system to study membrane fusion and intracellular traffic
- Zona pellucida
glycoprotein shell that surrounds the oocyte
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Avinoam O, Podbilewicz B. Eukaryotic cell-cell fusion families. Curr Top Membr. 2011;68:209–234. doi: 10.1016/B978-0-12-385891-7.00009-X. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. Viral membrane fusion. Nature structural & molecular biology. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White JM, et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaney MC, Rey FA. Class II enveloped viruses. Cell Microbiol. 2011;13:1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- 5.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Advances in experimental medicine and biology. 2011;714:91–101. doi: 10.1007/978-94-007-0782-5_3. [DOI] [PubMed] [Google Scholar]
- 6.Miyado K, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- 7.Le Naour F, et al. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 8.Kaji K, et al. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 9.Inoue N, et al. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 10.Miyado K, et al. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci U S A. 2008;105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S, et al. Can the presence of wild-type oocytes during insemination rescue the fusion defect of CD9 null oocytes? Mol Reprod Dev. 2009;76:602. doi: 10.1002/mrd.21040. [DOI] [PubMed] [Google Scholar]
- 12.Barraud-Lange V, et al. Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilizing ability of Cd9-deleted oocytes. Reproduction. 2012 doi: 10.1530/REP-12-0040. [DOI] [PubMed] [Google Scholar]
- 13.Runge KE, et al. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Developmental biology. 2007;304:317–325. doi: 10.1016/j.ydbio.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Mori T, et al. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nature cell biology. 2006;8:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- 15.von Besser K, et al. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. doi: 10.1242/dev.02683. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirai M, et al. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol. 2008;18:607–613. doi: 10.1016/j.cub.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 18.Singson A, et al. Genes required for the common miracle of fertilization in Caenorhabditis elegans. Int J Dev Biol. 2008;52:647–656. doi: 10.1387/ijdb.072512as. [DOI] [PubMed] [Google Scholar]
- 19.Ydenberg CA, Rose MD. Yeast mating: a model system for studying cell and nuclear fusion. Methods in molecular biology. 2008;475:3–20. doi: 10.1007/978-1-59745-250-2_1. [DOI] [PubMed] [Google Scholar]
- 20.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engel A, et al. The yeast cell fusion protein Prm1p requires covalent dimerization to promote membrane fusion. PLoS One. 2010;5:e10593. doi: 10.1371/journal.pone.0010593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olmo VN, Grote E. Prm1 functions as a disulfide-linked complex in yeast mating. The Journal of biological chemistry. 2010;285:2274–2283. doi: 10.1074/jbc.M109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin H, et al. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar PS, et al. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol Biol Cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel A, Walter P. Membrane lysis during biological membrane fusion: collateral damage by misregulated fusion machines. The Journal of cell biology. 2008;183:181–186. doi: 10.1083/jcb.200805182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, et al. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. The Journal of cell biology. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heiman MG, et al. The Golgi-resident protease Kex2 acts in conjunction with Prm1 to facilitate cell fusion during yeast mating. J Cell Biol. 2007;176:209–222. doi: 10.1083/jcb.200609182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilar PS, et al. Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4170–4175. doi: 10.1073/pnas.0914094107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleissner A, et al. The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics. 2009;181:497–510. doi: 10.1534/genetics.108.096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey A, et al. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot Cell. 2004;3:348–358. doi: 10.1128/EC.3.2.348-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maerz S, et al. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics. 2008;179:1313–1325. doi: 10.1534/genetics.108.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleissner A, et al. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci U S A. 2009;106:19387–19392. doi: 10.1073/pnas.0907039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read ND, et al. Self-signalling and self-fusion in filamentous fungi. Curr Opin Microbiol. 2009;12:608–615. doi: 10.1016/j.mib.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Goryachev AB, et al. Excitable behavior can explain the “ping-pong” mode of communication between cells using the same chemoattractant. BioEssays: news and reviews in molecular, cellular and developmental biology. 2012;34:259–266. doi: 10.1002/bies.201100135. [DOI] [PubMed] [Google Scholar]
- 35.Haralalka S, et al. Recent advances in imaging embryonic myoblast fusion in Drosophila. Methods. 2012;56:55–62. doi: 10.1016/j.ymeth.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onel S, et al. Role of the actin cytoskeleton with FuRMAS during Drosophila myoblast fusion and first functionally conserved factors in vertebrates. In. In: Larsson LI, editor. Cell Fusions. Springer; 2011. pp. 139–170. [Google Scholar]
- 38.Gruenbaum-Cohen Y, et al. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11211–11216. doi: 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson BE, et al. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafer G, et al. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Massarwa R, et al. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Kim S, et al. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12:571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Sens KL, et al. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haralalka S, et al. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 2011;138:1551–1562. doi: 10.1242/dev.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onel SF, Renkawitz-Pohl R. FuRMAS: triggering myoblast fusion in Drosophila. Dev Dyn. 2009;238:1513–1525. doi: 10.1002/dvdy.21961. [DOI] [PubMed] [Google Scholar]
- 46.Mukherjee P, et al. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 2011;138:2347–2357. doi: 10.1242/dev.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gildor B, et al. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO reports. 2009;10:1043–1050. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin P, et al. Competition between Blown fuse and WASP for WIP binding regulates the dynamics of WASP-dependent actin polymerization in vivo. Developmental cell. 2011;20:623–638. doi: 10.1016/j.devcel.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kesper DA, et al. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS) Dev Dyn. 2007;236:404–415. doi: 10.1002/dvdy.21035. [DOI] [PubMed] [Google Scholar]
- 50.Duan R, et al. Group I PAKs function downstream of Rac to promote podosome invasion during myoblast fusion in vivo. The Journal of cell biology. 2012;199:169–185. doi: 10.1083/jcb.201204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Helming L, et al. Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci Signal. 2008;1:ra11. doi: 10.1126/scisignal.1159665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K, et al. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP) Mol Endocrinol. 2008;22:176–185. doi: 10.1210/me.2007-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu M, et al. NF-kappaB signaling participates in both RANKL- and IL-4-induced macrophage fusion: receptor cross-talk leads to alterations in NF-kappaB pathways. J Immunol. 2011;187:1797–1806. doi: 10.4049/jimmunol.1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang M, et al. Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J Cell Physiol. 2008;215:497–505. doi: 10.1002/jcp.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skokos EA, et al. Lack of TNF-alpha-induced MMP-9 production and abnormal E-cadherin redistribution associated with compromised fusion in MCP-1-null macrophages. Am J Pathol. 2011;178:2311–2321. doi: 10.1016/j.ajpath.2011.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMichael BK, et al. Regulated proteolysis of nonmuscle myosin IIA stimulates osteoclast fusion. J Biol Chem. 2009;284:12266–12275. doi: 10.1074/jbc.M808621200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui W, et al. The intracellular domain of CD44 promotes the fusion of macrophages. Blood. 2006;107:796–805. doi: 10.1182/blood-2005-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalo P, et al. MT1-MMP Is Required for Myeloid Cell Fusion via Regulation of Rac1 Signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemaire I, et al. The P2X7 receptor and Pannexin-1 are both required for the promotion of multinucleated macrophages by the inflammatory cytokine GM-CSF. J Immunol. 2011;187:3878–3887. doi: 10.4049/jimmunol.1002780. [DOI] [PubMed] [Google Scholar]
- 62.Kara FM, et al. Adenosine A1 receptors (A1Rs) play a critical role in osteoclast formation and function. FASEB J. 2010;24:2325–2333. doi: 10.1096/fj.09-147447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helming L, Gordon S. Macrophage fusion induced by IL-4 alternative activation is a multistage process involving multiple target molecules. Eur J Immunol. 2007;37:33–42. doi: 10.1002/eji.200636788. [DOI] [PubMed] [Google Scholar]
- 64.Yagi M, et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 66.Powell AE, et al. Fusion between Intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rizvi AZ, et al. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci U S A. 2006;103:6321–6325. doi: 10.1073/pnas.0508593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakraborty A, et al. Donor DNA in a renal cell carcinoma metastasis from a bone marrow transplant recipient. Bone Marrow Transplant. 2004;34:183–186. doi: 10.1038/sj.bmt.1704547. [DOI] [PubMed] [Google Scholar]
- 69.Andersen TL, et al. Osteoclast nuclei of myeloma patients show chromosome translocations specific for the myeloma cell clone: a new type of cancer-host partnership? J Pathol. 2007;211:10–17. doi: 10.1002/path.2078. [DOI] [PubMed] [Google Scholar]
- 70.Huppertz B, Gauster M. Trophoblast fusion. Advances in experimental medicine and biology. 2011;713:81–95. doi: 10.1007/978-94-007-0763-4_6. [DOI] [PubMed] [Google Scholar]
- 71.Handwerger S. New insights into the regulation of human cytotrophoblast cell differentiation. Molecular and cellular endocrinology. 2010;323:94–104. doi: 10.1016/j.mce.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simmons DG, et al. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development. 2008;135:2083–2091. doi: 10.1242/dev.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen CP, et al. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biology of reproduction. 2008;79:815–823. doi: 10.1095/biolreprod.108.069765. [DOI] [PubMed] [Google Scholar]
- 74.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1164–1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heidmann O, et al. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cornelis G, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E432–441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bjerregaard B, et al. Syncytin is involved in breast cancer-endothelial cell fusions. Cellular and molecular life sciences: CMLS. 2006;63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soe K, et al. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone. 2011;48:837–846. doi: 10.1016/j.bone.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 81.Shemer G, et al. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol. 2004;14:1587–1591. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 82.del Campo JJ, et al. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr Biol. 2005;15:413–423. doi: 10.1016/j.cub.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 83.Sapir A, et al. AFF-1, a FOS-1-Regulated Fusogen, Mediates Fusion of the Anchor Cell in C. elegans. Dev Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mohler WA, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion in C. elegans. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 85.Gattegno T, et al. Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol Biol Cell. 2007;18:1153–1166. doi: 10.1091/mbc.E06-09-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Podbilewicz B, et al. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 87.Avinoam O, et al. Conserved Eukaryotic Fusogens Can Fuse Viral Envelopes to Cells. Science. 2011;332:589–592. doi: 10.1126/science.1202333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oren-Suissa M, et al. The Fusogen EFF-1 Controls Sculpting of Mechanosensory Dendrites. Science. 2010;328:1285–1288. doi: 10.1126/science.1189095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rasmussen JP, et al. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Dev Cell. 2008;14:559–569. doi: 10.1016/j.devcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghosh-Roy A, et al. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J Neurosci. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Procko C, et al. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development. 2011;138:1371–1381. doi: 10.1242/dev.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stone CE, et al. Lipocalin signaling controls unicellular tube development in the Caenorhabditis elegans excretory system. Dev Biol. 2009;329:201–211. doi: 10.1016/j.ydbio.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neumann B, et al. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev Dyn. 2011;240:1365–1372. doi: 10.1002/dvdy.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kontani K, et al. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev Cell. 2005;8:787–794. doi: 10.1016/j.devcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 95.Brabin C, et al. The Caenorhabditis elegans GATA factor ELT-1 works through the cell proliferation regulator BRO-1 and the Fusogen EFF-1 to maintain the seam stem-like fate. PLoS genetics. 2011;7:e1002200. doi: 10.1371/journal.pgen.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pellegrino MW, et al. LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development. 2011;138:4649–4660. doi: 10.1242/dev.071951. [DOI] [PubMed] [Google Scholar]
- 97.Choi J, et al. N-ethylmaleimide sensitive factor is required for fusion of the C. elegans uterine anchor cell. Dev Biol. 2006;297:87–102. doi: 10.1016/j.ydbio.2006.04.471. [DOI] [PubMed] [Google Scholar]
- 98.Friedlander-Shani L, Podbilewicz B. Heterochronic control of AFF-1-mediated cell-to-cell fusion in C. elegans. Adv Exp Med Biol. 2011;713:5–11. doi: 10.1007/978-94-007-0763-4_2. [DOI] [PubMed] [Google Scholar]
- 99.Huang X, et al. The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev Biol. 2009;333:337–347. doi: 10.1016/j.ydbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 100.Mason DA, et al. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development. 2008;135:2373–2382. doi: 10.1242/dev.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]