Abstract
No-go decay (NGD) is one of several mRNA surveillance systems dedicated to the removal of defective mRNAs from the available pool. Two interacting factors, Dom34 and Hbs1, are genetically implicated in NGD in yeast. Here we show, using a reconstituted yeast translation system, that Dom34:Hbs1 interacts with the ribosome to promote subunit dissociation and peptidyl-tRNA drop-off. Our data further indicate that these recycling activities are shared by the homologous translation termination factor complex, eRF1:eRF3, suggesting a common ancestral function. As Dom34:Hbs1 activity exhibits no dependence on either peptide length or A-site codon identity, we propose that this quality-control system functions broadly to recycle ribosomes throughout the translation cycle whenever stalls occur.
The quality of actively translated mRNA is monitored through multiple cellular mechanisms including: nonsense-mediated decay (NMD) (1), non-stop decay (NSD) (2, 3) and no-go decay (NGD) (4). NMD and NSD target messages containing premature stop codons or lacking stop codons, respectively. The in vivo targets of NGD are poorly characterized, but appear to include a broad range of stalled translation complexes containing, for example, mRNAs with inhibitory secondary structure (4), chemical damage (5), premature stop or rare codons (4) or ribosomal defects (a process more broadly referred to as nonfunctional ribosome decay (NRD)) (6, 7).
In Saccharomyces cerevisiae, two proteins, Dom34 (Pelota in higher eukaryotes) and Hbs1, were previously implicated in the endonucleolytic event characteristic of NGD (4), although neither factor directly catalyzes this cleavage (8). These proteins associate both in vivo (9) and in vitro (10), and while neither protein is essential in yeast, both deletion strains (DOM34Δ and HBS1D) exhibit synthetic growth defects with strains lacking a subset of 40S ribosomal subunit proteins (9), suggesting involvement in a common process. Dom34 is evolutionarily related to the eukaryotic translation termination factor eRF1, with two notable differences: the central domain of Dom34 lacks the conserved GGQ motif required for catalysis of peptide release and the N-terminal domain of Dom34 lacks the NIKS motif involved in codon recognition (Fig. S1) (10–12). Hbs1 exhibits significant sequence identity to eRF3-related GTPases, including eEF1a, which delivers aminoacylated-tRNA to the A site of the ribosome, and Ski7, a factor implicated in NSD in yeast (3, 11). These observations suggested that a Dom34:Hbs1 complex may directly engage the ribosome (similar to eRF1:eRF3) to initiate the events of no-go decay that ultimately result in mRNA degradation.
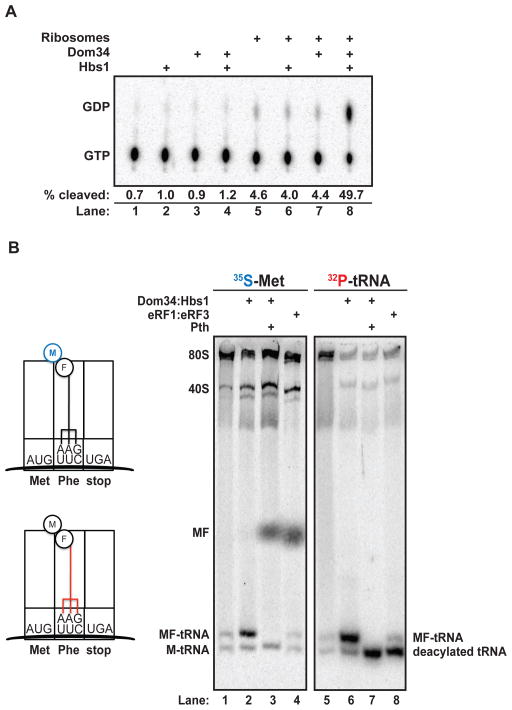
To ask whether Dom34:Hbs1 productively interacts with the ribosome, we measured the GTP hydrolysis activity of purified recombinant protein (Fig. S2) in the presence or absence of S. cerevisiae ribosomal subunits (13). Robust hydrolysis occurred only when both Dom34 and Hbs1 were incubated together with both ribosomal subunits (40S and 60S) (Fig. 1A). This activity parallels the stimulation observed with the homologous translation termination factors eRF1 and eRF3 (14). To further characterize the reaction catalyzed by Dom34:Hbs1 on eukaryotic ribosomes, we employed an in vitro reconstituted yeast translation system (13, 15). Programmed complexes were typically comprised of 80S ribosomes stalled on a defined mRNA with a peptidyl-tRNA in the P site, and either a sense or stop codon poised in the A site (for “elongation” and “termination” complexes, respectively). We followed the fate of various components by labeling the peptidyl-tRNA either on the peptide moiety (with 35S-methionine) or on the P-site tRNA itself (with 32P at the terminal adenosine) (Fig. 1B).
Figure 1.
(A) Ribosome stimulation of GTP hydrolysis by Dom34:Hbs1 as visualized by thin layer chromatogram. (B) Native gel analysis of ribosomal termination complexes incubated with indicated components. Stylized diagrams of differentially labeled (35S-Met (blue) or 32P[CCA]-tRNA (red)) ribosome complexes are on the left. Pth, peptidyl-tRNA hydrolase.
Radiolabeled dipeptidyl 80S termination complexes were treated with either eRF1:eRF3:GTP or Dom34:Hbs1:GTP, and the products of the reaction were followed by native gel electrophoresis (16). eRF1:eRF3:GTP ternary complex promoted the release of two distinct species from 80S complexes: 35S-Met-Phe dipeptide (Fig. 1B, lane 4) and 32P-deacylated-tRNAPhe (Fig. 1B, lane 8). By contrast, Dom34:Hbs1:GTP ternary complex promoted the release of a distinct product that migrated at the same position under both labeling conditions (Fig. 1B, lanes 2 and 6) suggesting it represented intact peptidyl-tRNA. This was confirmed by treatment with recombinant peptidyl-tRNA hydrolase (Pth) (Fig. 1B, lanes 3 and 7), which produced the predicted shifts in product identity (17).
The kinetic parameters of the Dom34:Hbs1-promoted reaction were determined using a Pth-coupled reaction and an electrophoretic TLC system (Fig. S3) (18). Ribosome termination complexes (containing tripeptidyl-tRNA in the P site and a stop codon in the A site) were treated with various factors and the rate constants for the corresponding reactions were determined. The rate constant for peptide release by eRF1:eRF3 was about 3.7 min−1 (as previously reported in the yeast system (15)), while that of Dom34:Hbs1-promoted peptidyl-tRNA release was approximately 15-fold slower (Fig. 2A), in a reaction that was strictly dependent on the presence of both factors (Fig. S4). Importantly, the rate of peptidyl-tRNA hydrolysis by Pth substantially exceeds that of the Dom34:Hbs1-catalyzed peptidyl-tRNA release from the ribosome (Fig. S5). As previously observed for eRF1:eRF3 (19), catalysis by Dom34:Hbs1 was strongly inhibited in the presence of the non-hydrolyzable GTP analog, GDPNP. The observed inhibition of product formation by both GDPNP and heat inactivation of Dom34:Hbs1 (Fig. S6) excludes the uninteresting possibility that non-specific ribosome dissociation might account for the observed peptidyl-tRNA release.
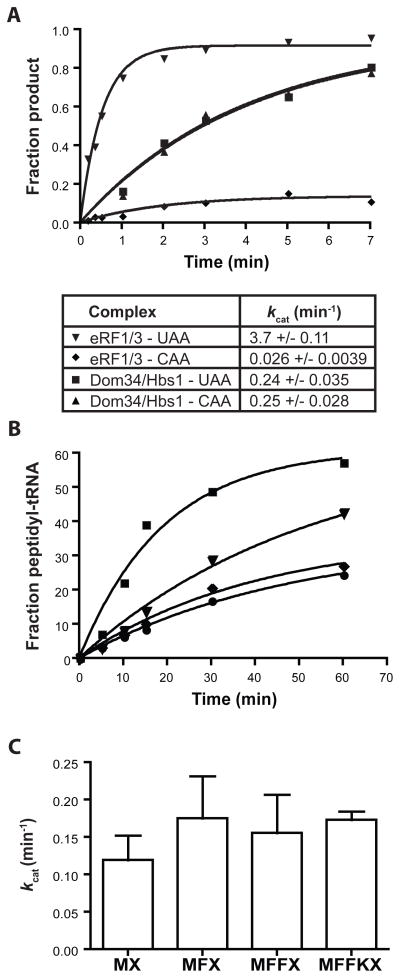
Figure 2.
Effects of codon identity and peptide length on Dom34:Hbs1-mediated peptidyl-tRNA release activity. (A) Kinetics of release reactions mediated by Dom34:Hbs1 and eRF1:eRF3 on UAA and CAA A-site-programmed ribosome complexes. kcat values are reported as mean +/− standard deviation (SD) (N=3). (B) Inhibition of Dom34:Hbs1-mediated reaction by a release-defective eRF1 variant (eRF1[AGQ]) on UAA-programmed ribosomes. Symbols: ■, Dom34/Hbs1 only; ▼, +5uM eRF1 (AGQ); ◆, +20uM eRF1(AGQ); ●, 20uM eRF1(AGQ) only. (C) Rate constants for Dom34:Hbs1-mediated peptidyl-tRNA release on programmed complexes containing Met-tRNAiMet, di-, tri- or tetra-peptidyl tRNA in the P site (N=4, +/− SD).
To test the specificity of these reactions, we compared the reactivity of Dom34:Hbs1 on complexes containing stop (UAA) or non-stop (CAA) codons in the A site. eRF1:eRF3 exhibits a high degree of specificity for stop codons relative to near-stop with rate constants for peptide release differing by about two orders of magnitude (Fig. 2A). By contrast, Dom34:Hbs1 exhibits no specificity for these complexes as the rate constants (and reaction endpoints) are indistinguishable (Fig. 2A). Given the codon-independent nature of the Dom34:Hbs1 reaction, we wanted to confirm that these factors engage the A site of the ribosome. This was accomplished by successfully competing Dom34:Hbs1 activity on termination complexes with increasing amounts of a catalytically inactive eRF1 variant, eRF1(AGQ) (20) (Fig. 2B).
Based on the known activities of other A-site-binding factors (e.g. eEF2), we were interested in whether Dom34:Hbs1 facilitates a translocation-like process on the ribosome, effectively pushing peptidyl-tRNAs out through the E site. However, because neither length of the peptide (Fig. 2C) nor E-site-bound inhibitors (Fig. S7) affected the release reaction, this model seemed unlikely.
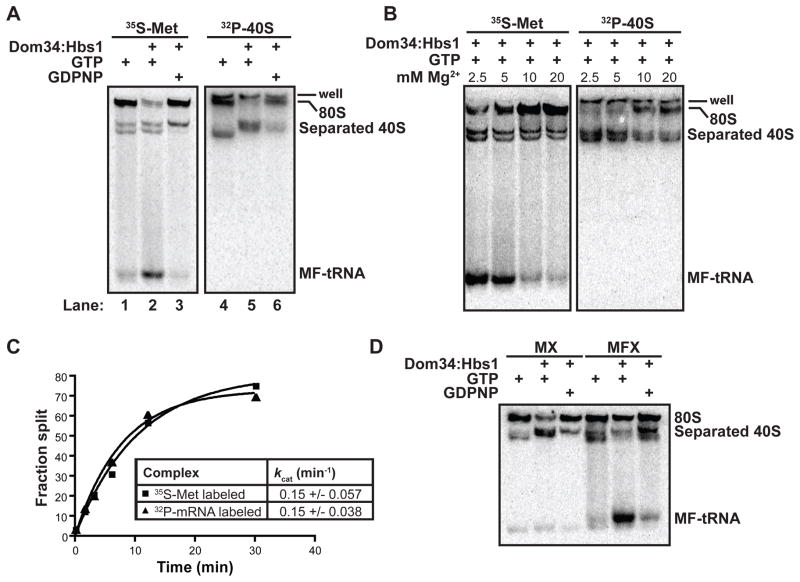
A subunit dissociation model was next considered based on the lack of influence of peptide length on the peptidyl-tRNA release reaction, and the likely difficulty of directly releasing such topologically constrained species from intact 80S particles. Subunit dissociation was followed with ribosome termination complexes containing either 32P-labeled 40S subunits or 35S-Met labeled peptidyl-tRNA. eIF6, a yeast protein known to bind to the interface region of the 60S subunit, served as an anti-association factor that “trapped” dissociation events (21, 22). Indeed, subunit dissociation was promoted by Dom34:Hbs1 (Fig. 3A, lane 5), and correlated nicely with peptidyl-tRNA release activity (Fig. 3A, lane 2). Both reactions were inhibited by the addition of GDPNP (Fig. 3A, lanes 3 and 6) and exhibited similar magnesium dependencies (Fig. 3B). We also followed Dom34:Hbs1-mediated subunit dissociation using complexes formed with 32P-labeled mRNA and saw that the mRNA segregated predominantly with 40S subunits (Fig. S8), consistent with the previously reported dependence of mRNA dissociation on additional initiation factors (23). Additionally, no cleavage of mRNA was observed during Dom34:Hbs1-mediated processes (Fig. S9), consistent with an earlier report (8).
Figure 3.
Dom34:Hbs1 promotes ribosome subunit dissociation. (A) Native gel analysis demonstrating GTP-dependence of Dom34:Hbs1-mediated products on differentially labeled ribosome complexes (B) Native gel analysis indicating similar magnesium sensitivities for Dom34:Hbs1-mediated formation of peptidyl-tRNA and separation of subunits. (C) Rates of peptidyl-tRNA formation and subunit separation are similar. kcat values are reported as mean +/− SD (N=3). (D) Native gel analysis reveals distinct products generated from Dom34:Hbs1 treatment of initiation (MX) and elongation (MFX) complexes. Met-tRNAiMet partitions with 40S subunits, while Met-Phe-peptidyl-tRNA is fully liberated from the ribosome.
As Dom34:Hbs1-driven subunit dissociation and peptidyl-tRNA release are correlated events (Fig. 3A,B), we sought to determine their order by measuring the rate constants of the two reactions using ribosome complexes containing either 32P-labeled mRNA or 35S-Met labeled peptidyl-tRNA (Fig. 3C). The observed rate constants for the disappearance of 80S ribosomes in a native gel were closely matched (~0.15 min−1) when either the labeled mRNA or peptidyl-tRNA was monitored. These data argue that subunit dissociation and peptidyl-tRNA formation by Dom34:Hbs1 are tightly coupled to one another, with one likely serving as the rate limiting step for the other (e.g. (24)).
If subunit dissociation occurs first, it seemed possible that an intermediate product might be generated in which subunits have dissociated but peptidyl-tRNA has not yet departed (i.e. peptidyl-tRNA:40S subunit conjugates). In a comparison of Dom34:Hbs1 activity on an initiation-like ribosome complex (carrying initiator Met-tRNAiMet in the P site) and on an elongated ribosome complex (carrying Met-Phe-tRNAPhe in the P site), free Met-Phe-tRNAPhe was the predominant product from the Dom34:Hbs1-treated elongated ribosome complex, while Met-tRNAiMet-bound 40S subunit was the predominant product from the Dom34:Hbs1-treated initiation complexes (Fig. 3D, Fig. S10A). The appearance of a novel Met-tRNAiMet-bound 40S complex suggests that subunit separation can take place independently of peptidyl-tRNA release. To confirm that the observed Met-tRNAiMet-bound 40S complex represented an authentic stable product of the Dom34:Hbs1-catalyzed reaction, rather than re-association of Met-tRNAiMet with ribosomes following initial dissociation, we repeated the Dom34:Hbs1-catalyzed reaction in the presence of a large excess of unlabeled Met-tRNAiMet and saw that the chase had no effect on the reaction products (Fig. S10B). These data are broadly consistent with the previously reported high affinity of 40S subunits for Met-tRNAiMet (25) and argue that the Dom34:Hbs1 complex initially promotes subunit dissociation and that peptidyl-tRNA dissociation typically follows.
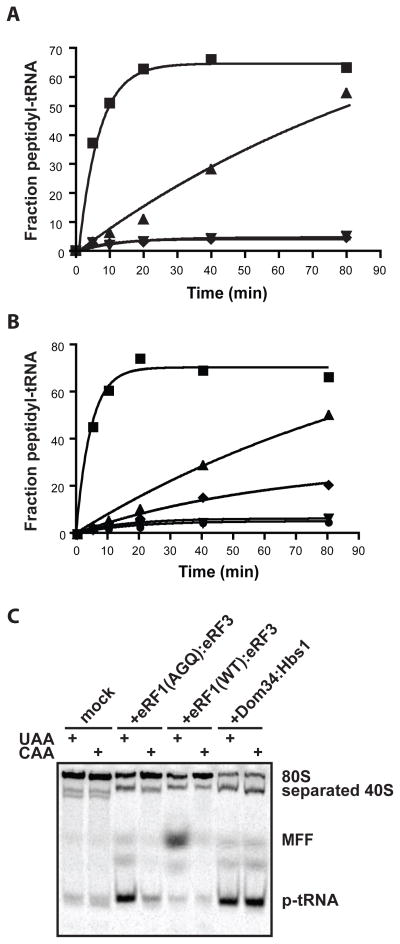
In light of the structural similarities between Dom34:Hbs1 and eRF1:eRF3, we wondered whether the canonical eukaryotic release factors might also promote subunit separation, independent of peptide release, and thereby contribute to ribosome recycling during termination. Treatment of termination complexes (with 35S-Met-labeled peptidyl-tRNA) with catalytically inactive eRF1(AGQ):eRF3 led to the formation of free peptidyl-tRNA in a reaction inhibited by GDPNP. This activity is distinguished from that promoted by Dom34:Hbs1 only by its slower rate (0.012 min−1 vs. 0.21 min−1) (Fig. 4A). Like the Dom34:Hbs1-catalyzed reaction, the eRF1(AGQ):eRF3 reaction depends on both protein components for full activity (Fig. 4B). Lastly, we find that eRF1(AGQ):eRF3-mediated peptidyl-tRNA release exhibits robust codon specificity, taking place only when a stop codon is presented in the A site (Fig. 4C).
Figure 4.
Intrinsic subunit separation activity associated with eRF1:eRF3. (A) Kinetics of peptidyl-tRNA release stimulated by eRF1(AGQ):eRF3. Symbols: ■ Dom34:Hbs1; ▲, eRF1(AGQ)/eRF3, ▼, mock; ◆, eRF1(AGQ)/eRF3 (GDPNP). (B) Stimulation of subunit separation capacity of eRF1(AGQ) by eRF3. Symbols: ■, Dom34/Hbs1; ▲, eRF1(AGQ)/eRF3; ◆, eRF1(AGQ); ▼, eRF3; ●, mock. (C) Native gel analysis of the codon dependence (UAA vs. CAA) of eRF1(AGQ):eRF3 recycling activity.
Canonical recycling, which occurs after termination, involves subunit dissociation, and mRNA and tRNA release, thus allowing for subsequent re-initiation of translation. In bacteria, a specialized ribosome recycling factor, RRF, is central to this GTP-dependent process (26). However, in eukaryotes, no RRF has been identified. Our results indicate that eRF1:eRF3 and Dom34:Hbs1 directly destabilize the subunit interface to promote recycling. Although additional factors likely accelerate various aspects of recycling in vivo (23, 27, 28), our observations could explain why no true RRF homologue is present in eukaryotes. We acknowledge that additional, and possibly redundant, mechanisms of recycling may exist in yeast (29). However, we argue that Dom34:Hbs1 acts in a specialized manner on malfunctioning ribosome complexes that, for example, do not appropriately engage the next factor in the translation cycle or are inherently defective and thus unable to properly elongate (i.e. NRD) (6). Subsequent to Dom34:Hbs1-mediated recycling, kinetic competition between translation re-initiation and mRNA decay (or rRNA decay in the case of NRD) will determine the partitioning of defective RNAs, with the opportunities for degradation accumulating with each passage through the quality control pathway.
Supplementary Material
Summary.
Biochemical analysis reveals a conserved mechanism for no-go decay (NGD), nonfunctional ribosome decay (NRD) and ribosome recycling.
Acknowledgments
We would like to thank J. Lorsch and his lab members, R. Parker and M. Moore for valuable discussions, H. Zaher and N. Guydosh for thoughtful comments on the manuscript, and B. Rogers for cloning and purification of Pth and eIF6.
References
- 1.Peltz SW, Brown AH, Jacobson A. Genes Dev. 1993;7:1737. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 2.Frischmeyer PA, et al. Science. 2002;295:2258. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 3.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Science. 2002;295:2262. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 4.Doma MK, Parker R. Nature. 2006;440:561. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi R, Manzoor M, Hudak KA. J Biol Chem. 2008;283:32218. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- 6.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. Mol Cell. 2009;34:440. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. EMBO J. 2009;29:80. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passos DO, et al. Mol Biol Cell. 2009;20:3025. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Mol Cell Biol. 2002;22:2564. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graille M, Chaillet M, van Tilbeurgh H. J Biol Chem. 2008;283:7145. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson GC, Baldauf SL, Hauryliuk V. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HH, et al. Mol Cell. 2007;27:938. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Methods Enzymol. 2007;430:111. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 14.Frolova L, et al. RNA. 1996;2:334. [PMC free article] [PubMed] [Google Scholar]
- 15.Saini P, Eyler DE, Green R, Dever TE. Nature. 2009;459:118. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Algire MA, et al. RNA. 2002;8:382. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel Z, Vogel T, Zamir A, Elson D. Eur J Biochem. 1971;21:582. doi: 10.1111/j.1432-1033.1971.tb01504.x. [DOI] [PubMed] [Google Scholar]
- 18.Youngman EM, Brunelle JL, Kochaniak AB, Green R. Cell. 2004;117:589. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 19.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. Cell. 2006;125:1125. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 20.Frolova LY, et al. RNA. 1999;5:1014. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartmann M, et al. J Biol Chem. 2010;285:14848. doi: 10.1074/jbc.C109.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si K, Maitra U. Mol Cell Biol. 1999 Feb;19:1416. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pisarev AV, Hellen CU, Pestova TV. Cell. 2007;131:286. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savelsbergh A, et al. Mol Cell. 2003;11:1517. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 25.Kolitz SE, Takacs JE, Lorsch JR. RNA. 2009;15:138. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavialov AV, Hauryliuk VV, Ehrenberg M. Mol Cell. 2005;18:675. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Pisarev AV, et al. Mol Cell. 2010;37:196. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoshnevis S, et al. EMBO Rep. 2010;11:214. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurata S, et al. PNAS. 2010;107:10854. doi: 10.1073/pnas.1006247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.