SUMMARY
Ribosomes that stall before completing peptide synthesis must be recycled and returned to the cytoplasmic pool. The protein Dom34 and cofactors Hbs1 and Rli1 can dissociate stalled ribosomes in vitro, but the identity of targets in the cell is unknown. Here we extend ribosome profiling methodology to reveal a high-resolution molecular characterization of Dom34 function in vivo. Dom34 removes stalled ribosomes from truncated mRNAs, but, in contrast, does not generally dissociate ribosomes on coding sequences known to trigger stalling, such as polyproline. We also show that Dom34 targets arrested ribosomes near the ends of 3´ UTRs. These ribosomes appear to gain access to the 3 UTR via a mechanism that does not require decoding of the mRNA. These results suggest that ribosomes frequently enter downstream noncoding regions and that Dom34 carries out the important task of rescuing them.
INTRODUCTION
During translation, the ribosome faces a number of obstacles that have the potential to interrupt protein synthesis. Many such impediments originate within the mRNA, such as stable secondary structures, cleavages within the coding sequence (CDS), or encoded peptides that form stable interactions with the exit tunnel. Numerous other situations, such as limited availability of a particular tRNA, noncanonical base pairing between the mRNA and tRNA (i.e. wobble pairing), or damage to the ribosome itself may also impact the rate of peptide elongation. In principle, these disruptions arise either as a result of an anomalous perturbation, such as environmental stress, or as a programmed mechanism for regulating gene expression.
When ribosomes arrest with little or no chance of resuming translation, they must be either degraded or recycled for subsequent translation rounds. In addition, the associated mRNA, tRNAs, and unfinished protein must be removed from the ribosome for reuse or degradation. Failure to disassemble these arrested complexes would eventually lead to a critical shortage of translational machinery. However, such a rescue response is inappropriate for translational pauses that are only temporary. Such short-lived events provide time to allow folding of the nascent peptide (Zhang et al., 2009), change of reading frame (Dinman, 2012), or recruitment of regulatory factors, such as chaperones (Liu et al., 2013) or specialized elongation factors (Gutierrez et al., 2013; Ude et al., 2013). Pauses also allow time for the ribosome to localize to specific cellular loci, such as membranes (Yanagitani et al., 2011) or cytoplasmic projections (Chartrand et al., 2002). Other translational stalls are known to be indefinite; ribosomes remain poised to resume translation once an environmental change, such as starvation or exposure to toxins, triggers the ribosomes to resume translation (Wang and Sachs, 1997; Yanagitani et al., 2011). One of the challenges faced by the cell is to distinguish between those ribosomes that must be rescued to avoid a crisis and those that do not require such action.
In bacteria, the trans-translation system rescues many classes of stalled ribosomes by employing a specialized RNA called tmRNA (Moore and Sauer, 2007). In eukaryotes, no homologous system has been found but numerous proteins are known to carry out the individual steps of ribosome rescue, such as dissociation of ribosomal subunits and degradation of the mRNA and incomplete peptide. In a recent study, an assembly of factors termed the RQC complex was shown to associate with multiple classes of arrested ribosomes (Brandman et al., 2012). Other studies have speculated that the dissociation of stalled ribosomes detected by the RQC complex is facilitated by the protein Dom34 (PELO in H. sapiens) (Shao et al., 2013).
Dom34 is a homolog of release factor 1 (eRF1) (Graille et al., 2008) that has been shown in vitro to dissociate ribosome subunits when stimulated by the ATPase Rli1 (ABCE1 in H. sapiens) in a codon-independent fashion (Becker et al., 2012; Pisareva et al., 2011; Shoemaker and Green, 2011). Biochemical data suggest that Dom34 preferentially targets ribosomes where the 3´ tail of the mRNA is somewhat shortened, a preference that is conferred by the GTPase Hbs1 that gates this process (Shoemaker and Green, 2011). Structural data nicely rationalize this biochemical observation as the N-terminus of Hbs1 is positioned in the mRNA entrance channel where sterics could play an important role (Becker et al., 2011). Whether Hbs1 always functions as a gate for Dom34 activity in-vivo is not known.
Consistent with the biochemistry, in-vivo experiments with internally truncated synthetic reporter constructs indicated that Dom34 rescues ribosomes arrested at sites of truncation (Shao et al., 2013; Tsuboi et al., 2012). Truncated mRNAs could arise quite readily in the cell since many classes of stalled ribosomes in reporter systems appear to trigger upstream cleavage of the mRNA, including those that arrest at sites of mRNA damage (Gandhi et al., 2008), stable mRNA secondary structures (Doma and Parker, 2006), encoded polybasic peptides (Kuroha et al., 2010), poly(A) tails on genes targeted for “nonstop decay” (NSD) (Tsuboi et al., 2012) or a series of slow CGA codons (Chen et al., 2010). While it remains unknown whether Dom34 targets the primary ribosomes that stall on these motifs, it is generally agreed that Dom34 dissociates ribosomes that arrest at the resultant upstream cleavage sites. The mRNA degradation that follows as a result of these cleavage events is usually referred to as no-go decay (NGD) (Doma and Parker, 2006).
Clues about the importance of the ribosome rescue function of Dom34 to cellular fitness are hinted at by a number of studies characterizing the effects of DOM34 knockout. In yeast, reduction of small-subunit ribosomal protein levels by elimination of one of the duplicated ribosomal protein genes is poorly tolerated by the dom34Δ strain (Bhattacharya et al., 2010). Similarly, growth of the dom34Δ strain is particularly slow in situations where the supply of ribosomes is thought to be reduced, such as during transition out of stationary phase (Davis and Engebrecht, 1998) or when ribosomes are sequestered in an inactive state by overexpression of STM1 (Balagopal and Parker, 2011; Ben-Shem et al., 2011). One interpretation of these phenotypes is that Dom34 is important for maintaining a sufficient supply of ribosomes by rescuing those that become arrested during translation. Evidence that Dom34 is required for separation of free 80S subunits (van den Elzen et al., Embo J., in press), maturation of ribosomal particles (Strunk et al., 2011), and rescue of damaged ribosomes (Cole et al., 2009) are also consistent with this hypothesis. This rescue function may become more important when the cell faces challenging environmental conditions. It is known, for example, that the lag in logarithmic growth induced by the oxidizing agent diamide lasts ~10-fold longer in the absence of DOM34 (Fernandez-Ricaud et al., 2007). And, while DOM34 is not essential in S. cerevisiae, its absence is embryonic lethal in mammals and variants of an HBS1 ortholog in H. sapiens (HBS1L) lead to a variety of changes in peripheral blood cells. These data suggest that ribosome rescue is critical for key cellular processes (Adham et al., 2003; Menzel et al., 2007).
While Dom34 has been observed to act on some classes of stalled ribosomes in vitro, the full scope of Dom34 targeting in the cell remains unknown. To reveal the cellular targets of Dom34, we enhanced the recently-developed ribosome profiling method (Ingolia et al., 2009) to uncover detailed aspects of ribosome pausing across the transcriptome of S. cerevisiae. We found that Dom34 rescues ribosomes that stall at the 3´ end of mRNAs that are truncated, such as HAC1. Of particular interest, we also found that Dom34 rescues ribosomes in the 3´ untranslated region (3´ UTR) of many genes. Surprisingly, many ribosomes populate this noncoding region and even more unexpectedly, these ribosomes seem to move along the mRNA without recognizing codons, presumably not translating, and eventually arrest just short of reaching the poly(A) tail.
RESULTS
Ribosomes Protect Multiple Footprint Sizes
Ribosome profiling provides a snapshot of where ribosomes are bound throughout the transcriptome by using high-throughput sequencing to examine a transcriptome-wide ribosome footprinting experiment (Ingolia et al., 2012). The number of ribosome-protected footprints that map to the transcriptome (the “ribosome occupancy”) tends to be enriched at locations where ribosomes are known to pause (Ingolia et al., 2011), indicating that the relative length of time the ribosome spends at a given position is correlated with the site’s ribosome occupancy.
To quantify Dom34 activity, we performed ribosome profiling on wild-type and dom34Δ yeast strains with the expectation that ribosome occupancy would be augmented at Dom34 targets in the knockout strain. Ribosomes were stabilized prior to footprinting by adding the elongation inhibitor cycloheximide (CHX) to the lysis buffer. We avoided adding CHX to the cell culture as it has been shown that doing so can artifactually increase ribosome occupancy at start codons as initiation continues after elongation is inhibited (Ingolia et al., 2012). To rule out the possibility that addition of CHX at this later stage did not introduce artifacts or mask trends in the data, we tested alternative approaches for ribosome stabilization. In the presence of CHX, footprints over a length range of 25–34 nt were tightly distributed around a peak ~28 nt in length (Figure 1A), as observed in previous reports (Ingolia et al., 2009). Substituting CHX with GMP-PNP, a mixture of GMP-PNP and CHX, high (10 mM) MgCl2, or no extra additive led to only minor changes in the properties of the footprints obtained (Figure S1A) and negligible effects on the distribution of mapping across the transcriptome (Figure S1B). The most noticeable difference was a slight increase in the size of protected fragments in the presence of GMP-PNP. Given that differences between these datasets are minimal and that the broad trends described below were found in all samples, the five datasets were pooled for all comparative analysis (yielding a depth of 30–40 million mapped reads in both the wild-type and knockout datasets). Subsequent ribosome profiling experiments were stabilized only with CHX.
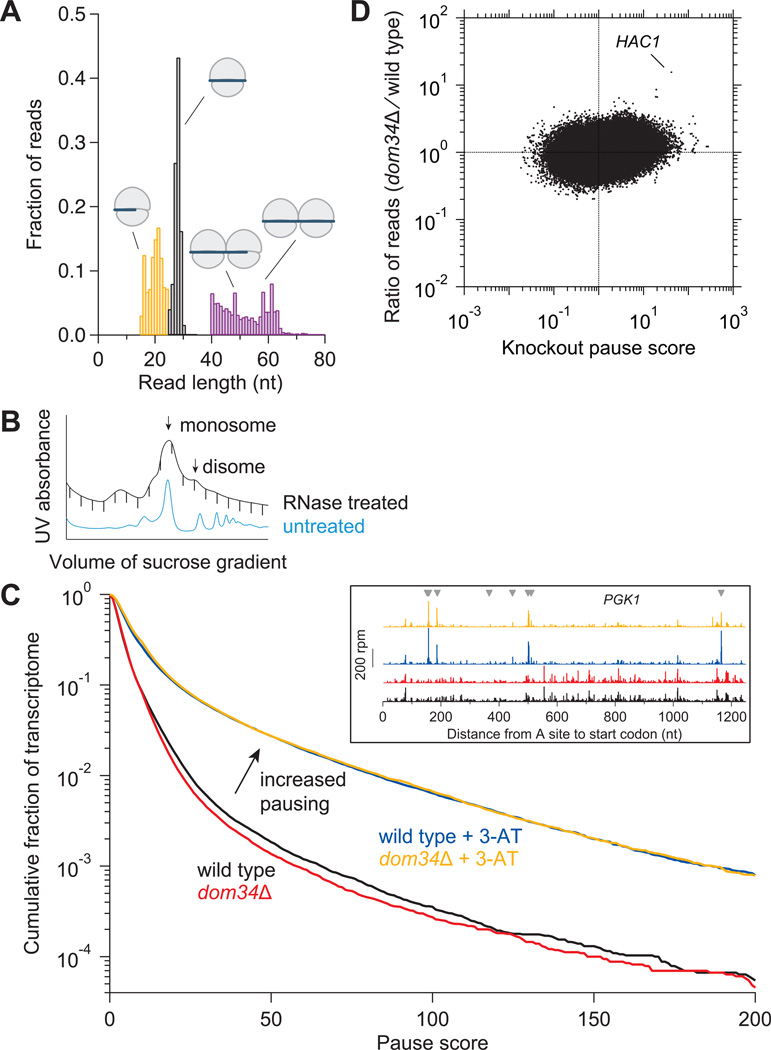
Figure 1. Analysis of Ribosome Footprint Size and Pausing.
(A) Size distributions of untrimmed ribosome footprints that mapped without mismatches to the genome or splice junctions. Short monosome-protected fragments (15–24 nt, orange), long monosome-protected fragments (25–34 nt, black), and disome-protected fragments (40–80 nt, purple). Each distribution came from a separate sample and thus sums to 1.0. Black and purple distributions obtained from wild-type cells and orange from ski2Δ cells.
(B) Sucrose gradient profile for dom34Δ yeast showing both RNase-treated and untreated control samples. Monosome and disome peaks are indicated.
(C) Effect of DOM34 knockout and 3-AT on ribosome pausing. Cumulative histogram of pause score values (smoothed reads at a position / median reads in gene) computed for all positions in the transcriptome with adequate read density. (Inset) Ribosome footprint reads that map to an example gene (PGK1). Histidine codons are marked with gray arrowheads.
(D) Enrichment ratio of reads mapped at single nucleotide positions in the dom34Δ strain compared to the wild-type strain shown as a function of knockout pause score (smoothed reads at a position / average reads in gene) for all positions in the transcriptome with adequate read density. Sites where pauses are amplified by knockout of DOM34, such as on HAC1, are located in the upper right quadrant.
See also Figures S1, S2, and S6.
Footprints of shorter lengths were also isolated since we suspected that the footprint of a ribosome stalled on a truncated mRNA would be smaller if the ribosome translated until the truncated end entered the A site. Since truncated transcripts are more vulnerable to 3´ to 5´ degradation by the exosome, we performed this experiment in a ski2Δ strain to disable this activity (Anderson and Parker, 1998). In the distribution of fragment lengths from 15–24 nt, we discovered peaks at approximately 16 and 21 nt (Figure 1A). In general, the 5´ ends of reads taken from a region around each peak started mapping ~13 nt upstream of the start codon, just like reads taken from around the 28-nt peak (Figure S1C). This alignment coincidence suggests that the 3´ end of the shorter reads is truncated rather than the 5´ end. The distribution correlation across the transcriptome between the 21-nt reads and the 28-nt reads was reasonably high whereas the correlation between the 16-nt reads and 28-nt reads was poorer (Figure S1D). An interpretation of these data that has been reported elsewhere (L. Lareau, personal communication) is that the 21-nt reads are derived from an alternative state in the peptide elongation cycle where the 3´ end of the mRNA is more vulnerable to RNase treatment. As we believe that the 16-nt reads represent truncated mRNAs, we do not expect them to be strongly correlated with 28-nt reads across the transcriptome.
We also looked for cases where ribosomes had collided to form stacked “disome” structures that would be expected to protect longer mRNA fragments (Wolin and Walter, 1988). We therefore examined 40–80 nt fragments from the disome peak (Figure 1B) and found that these reads protected a broad range of sizes under ~65 nt in length (Figure 1A). The peak in disome occupancy from ribosomes that were paused at the stop codon was shifted ~30 nt upstream from the peak generated by a single terminating “monosome,” a distance consistent with the presence of an extra ribosome in the disome (Figure S1E). Overall levels of ribosome coverage in the disome-protected reads were only somewhat correlated with 25–34 nt monosome reads, consistent with the expectation that disomes should be enriched upstream of sites where single ribosomes tend to pause (Figure S1D). Intriguingly, disome-protected reads were rare in the first ~25 nt of the open reading frame (ORF) (Figure S1F), suggesting that the ribosome does not start translation if a previously-initiated ribosome is blocking access to any of the first ~25 nt of the mRNA. One reason the initiating ribosome may require this extra space is to accommodate the size of initiation factors.
Most Paused Ribosomes are Not Targeted by Dom34
To broadly quantify pausing, we computed a pause score (Ingolia et al., 2011) at every nucleotide position in the transcriptome by taking the ratio of the ribosome occupancy at each position and the median occupancy of the gene. Yeast strains with a greater number of paused ribosomes should have a greater proportion of sites with high pause scores. To test this expectation, we treated yeast with the drug 3-amino-1,2,4-triazole (3-AT), an inhibitor of histidine biosynthesis (Klopotowski and Wiater, 1965), to induce widespread ribosome pausing. As expected, significant pauses appeared at His codons, leading to a dramatic shift in a cumulative histogram of pause scores (Figure 1C). If knocking out DOM34 also resulted in a widespread increase in pausing, we would expect a shift in the cumulative histogram for the knockout strain. Instead, the histogram was not shifted in the knockout strain, either in the presence or absence of 3-AT, suggesting that Dom34 is not essential for rescuing many classes of stalled ribosomes.
Similarly, other classes of known pauses, such as peptide tunnel stalls (Kurian et al., 2011; Wang and Sachs, 1997) or proline-rich motifs (Gutierrez et al., 2013; Ingolia et al., 2011; Woolstenhulme et al., 2013) did not exhibit Dom34-dependent differential ribosome occupancy (Figures S2A and S2B). To perform a more exhaustive search for enrichment of ribosome occupancy in the knockout strain, we computed the enrichment ratio of reads between the knockout and the wild-type strain for nucleotide positions with adequate read density across the transcriptome (Figure 1D). Analysis of these values revealed a site on the gene HAC1 with ~30-fold enrichment in the dom34Δ strain. In addition, a few other sites of enrichment emerged. Many correspond to known sites of premature polyadenylation that are subject to NSD, such as those within the ORF of the gene SIR1, and are known to be targets of Dom34 (Frischmeyer et al., 2002; Ozsolak et al., 2010; Tsuboi et al., 2012). This dearth of robust Dom34 targets was also evident when we instead assessed the change in gene-wide density (read occupancy per length of gene) of ribosome footprints when we knocked out DOM34 (Figure S2C). A similar analysis of mRNA-Seq density revealed no large changes in mRNA level for any gene in the knockout strain. While it is possible that Dom34 regulation is small in magnitude, it is also possible that it occurs abundantly but without specificity for particular sites or that it primarily affects lowabundance transcript isoforms that would only be revealed by deeper sequencing. Additionally, it is possible that redundant mechanisms exist that mask phenotypes associated with a DOM34 deletion (for example mRNA decay pathways).
Dom34 Dissociates Ribosomes that Stall at Sites of mRNA Truncation
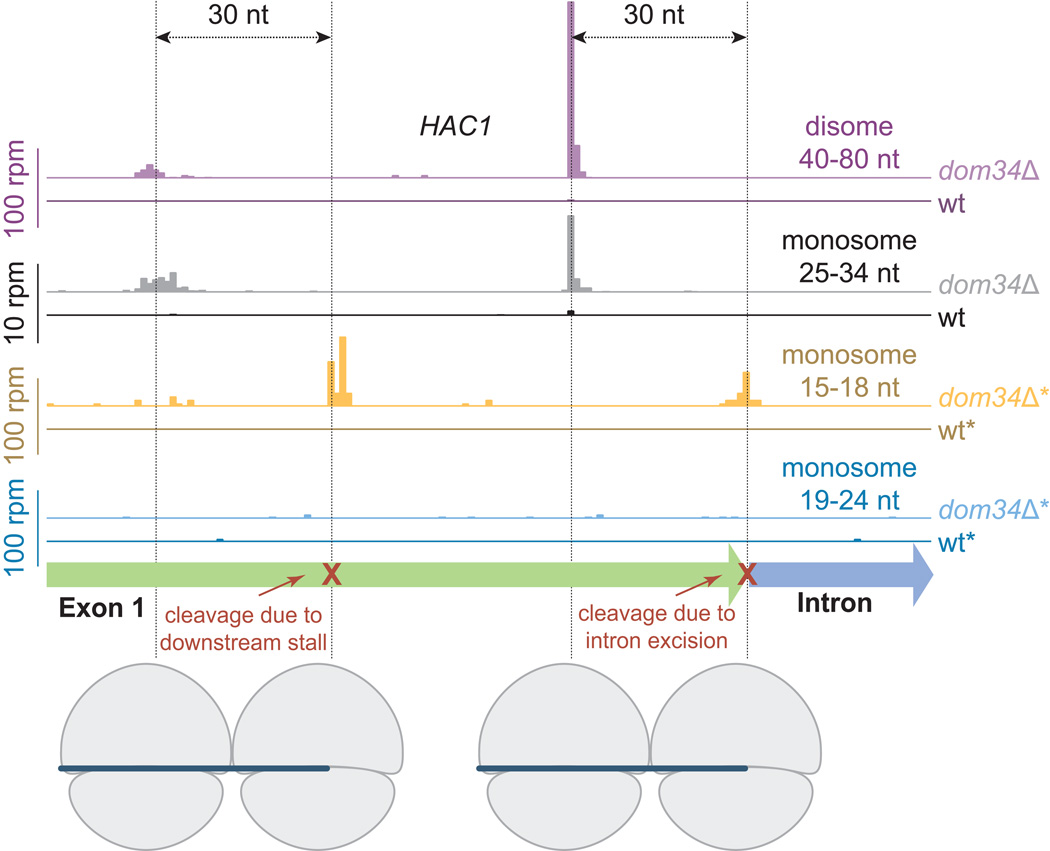
The increased density on HAC1 in the dom34Δ strain primarily results from higher ribosome occupancy at a distinct site in the 25–34 nt fragment sample from the monosome fraction (Figure 2). A similar increase in occupancy was apparent in the 40–80 nt disome-protected footprint sample and the magnitude of this peak is more than 10-fold greater (Figure 2, note adjusted vertical scale). This pattern is consistent with a model where ribosomes at this position are stacked against a stalled ribosome downstream. When we looked downstream for the primary stalling event, we found a peak in occupancy in the 15–18 nt reads taken from the monosome peak of the sucrose gradient. These reads exactly overlapped the very 3´ end of the first exon, suggesting that the exon had been severed from the intron but not ligated to the second exon. In such a scenario, the ribosomes would stall as they arrive at this truncation point (Figure 2) and then be rescued by Dom34. In the knockout strain, the ribosomes at the truncated end are not rescued and additional upstream ribosomes then collide with this stalled ribosome. In support of this model, the majority of these disome reads measured ~47 nt in length, the approximate size expected for a fragment composed of one ribosome protecting a full ribosome footprint (28–30 nt) and another protecting a smaller footprint at a site of mRNA truncation (16–17 nt).
Figure 2. Dom34 Targets Ribosomes on a Truncated mRNA.
Mapped footprints of varied sizes from wild-type (darker shades) and dom34Δ (lighter shades) strains on the gene HAC1. Data were collected from the disome and monosome fractions of a sucrose gradient. Note that scales vary according to sample and reads are plotted at approximate position of the ribosome A site. Ribosome drawings below the plots illustrate how the first set of ribosomes stall at the end of the exon and the second set of ribosomes stall at an upstream cleavage site, presumably caused by the stalled ribosomes downstream. Data for short (<25 nt) reads were taken in a ski2Δ background (indicated by *) to inhibit NSD.
See also Figures S3 and S4.
Why do there appear to be isolated copies of the first exon of HAC1 (XBP1 in higher eukaryotes) in the cytoplasm? In S. cerevisiae, the HAC1 transcript is exported from the nucleus with its intron retained and is then spliced only in the presence of protein folding stress (Aragon et al., 2009; Cox and Walter, 1996; Yanagitani et al., 2011). Translation of this mRNA is normally minimal because sequences in the intron base pair with the 5´UTR and block most ribosome initiation until endonucleolytic cleavage by the membrane-bound factor Ire1 excises the intron. In response to certain cellular stresses, the two exons are ligated together by tRNA ligase Rlg1 to complete splicing (Mori et al., 2010; Sidrauski et al., 1996). Importantly for our result, a recent study isolated a population of mRNA enriched in severed 5´ ends and found evidence for constitutive cleavage between the first exon and the intron in the absence of any cellular stress (Harigaya and Parker, 2012). The isolated first exon has also been directly observed by Northern blot though in relatively low abundance (Cox and Walter, 1996). These data support our interpretation of read assignment on HAC1. Initiation of translation is presumably more efficient on the excised first exon because the inhibitory intron is missing, allowing ribosomes to translate the mRNA and then stall at the site of cleavage, protecting a short (15–18 nt) footprint. Ribosome profiling enriches for ribosomes found on the rare isolated first exon in comparison with the essentially ribosome-free unspliced transcript.
Interestingly, we observed an enrichment on HAC1 in the 15–18 nt monosome-protected footprints at a position ~40 nt behind the stacked ribosomes in the knockout strain. These observations are consistent with the idea that the original stalling event (at the excised intron junction) induces a secondary endonucleolytic cleavage ~70 nt upstream from the 3´ end of the first exon, similar to observations of reporter constructs that were designed to induce NGD (Tsuboi et al., 2012). Stacked ribosomes were also apparent mapping ~30 nt further upstream of this second cleavage site in both the 25–34-nt monosome- and disome-protected reads. These data offer a high-resolution view of the NGD process where a stalled ribosome induces upstream mRNA cleavage events. We note that there was no enrichment in this region in the 19–24 nt reads obtained from the monosome-protected peak from the dom34Δ strain, suggesting that reads of this size are not created by ribosomes that are stalled on truncated messages nor by ribosomes that stall after colliding with a downstream ribosome.
We did observe a handful of other sites where a similar pattern of footprint mapping (a cluster of 45–50 nt disome-protected reads positioned ~30 nt upstream of a cluster of 15–18 nt monosome-protected reads) implies that the mRNA is truncated under normal conditions and the ribosomes are rescued by Dom34 (for example PAN6 in Figure S3). While we suspect that Dom34 generally plays such a role on all truncated mRNAs, many truncation sites will be stochastically generated by the cell, for example when translated mRNAs undergo 3´ to 5´ degradation, and therefore escape detection by a global approach like ribosome profiling.
To ask whether this Dom34 activity on HAC1 was important for the gene’s known role in the protein-misfolding response, we assessed the growth rate of the dom34Δ strain in the presence of protein misfolding agents such as tunicamycin and DTT, but found no substantial differences when compared to that of the wild-type strain (data not shown). We also performed ribosome profiling in the presence of DTT and found that HAC1 was spliced and efficiently translated in both strains (data not shown). It is conceivable that Dom34 may be critical to other putative functions of HAC1, for example, as suggested by the observed activation of HAC1 during late-stage meiosis (Brar et al., 2012).
We also asked whether Hbs1, the GTPase binding partner of Dom34, contributes to ribosome rescue events in the cell and whether it plays a particular role in substrate selection. To test these ideas, we performed ribosome profiling on a hbs1Δ strain. Interestingly, the observed pattern of read occupancy on HAC1 mirrored that observed in the dom34Δ strain, though the trends were a bit less dramatic (Figure S4A). These data are consistent with models where these factors work together as a complex on the ribosome (Shoemaker et al., 2010; van den Elzen et al., 2010).
Many Ribosomes Are Found in 3´ UTRs and Are Rescued by Dom34
We next examined whether ribosomes were enriched on portions of the transcript other than the ORF in the dom34Δ strain. In sharp contrast to the canonical view that 3´ UTRs are generally unpopulated by ribosomes, the 3´ UTRs of many genes in the dom34Δ strain were heavily enriched in ribosome occupancy relative to those in the wild-type strain (Figure 3A). We estimate that ~11% of genes are more than 3-fold enriched in ribosome occupancy in the knockout strain. Closer examination of these 3´ UTR reads revealed that footprints were clustered at the very 3´ end of the UTR and often included part of the poly(A) tail (Figure 3B). As for the HAC1 data, the pattern of reads in the hbs1Δ strain mimic what was observed in the dom34Δ strain, though somewhat muted in intensity (Figure S4B). As with all the reported phenomena in the dom34Δ strain, use of CHX to stabilize ribosomes in the lysis buffer and sucrose gradients did not affect ribosome accumulation in 3´ UTRs (Figure S4C).
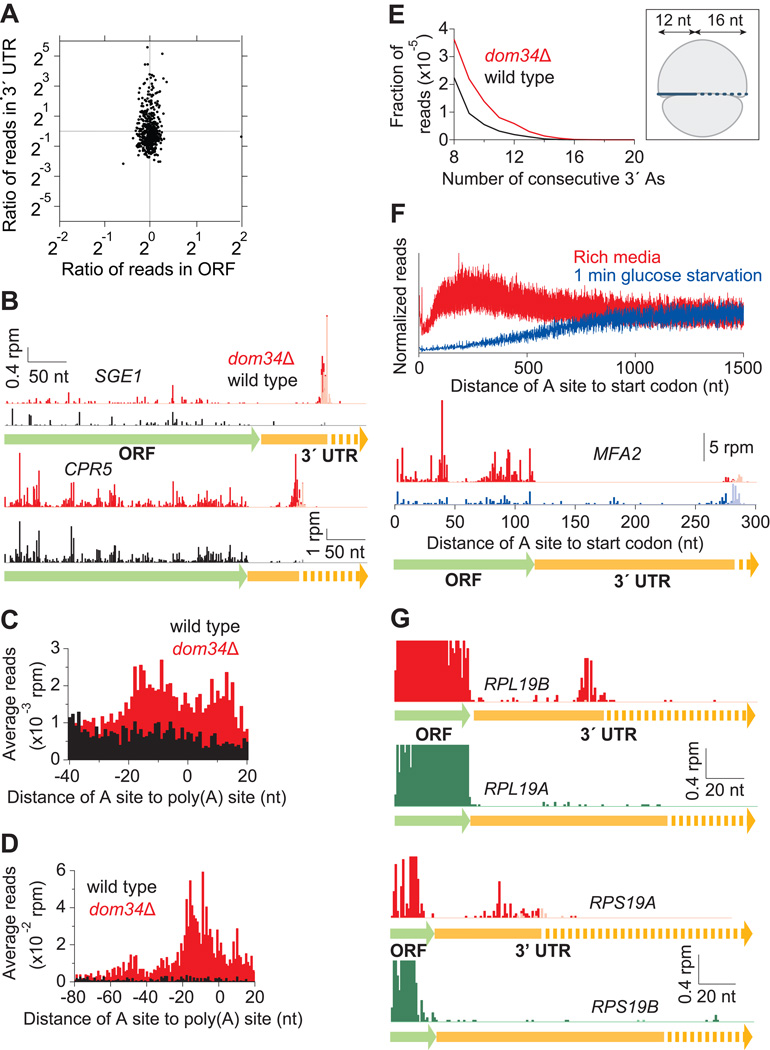
Figure 3. Dom34 Targets Arrested Ribosomes in the 3´ UTR.
(A) Enrichment ratio of reads on genes is plotted for 3´ UTRs as a function of ORFs between wild-type and dom34Δ strains. Vertical asymmetry of distribution indicates read enrichment in the dom34Δ 3´ UTRs. Genes were included with >10 rpkm in the ORF and >0.17 rpm (5–6 raw reads) in the 3´ UTR in both strains.
(B) Examples of mapped footprints on genes (SGE1 and CPR5) showing enrichment near the end of the 3´ UTR in the dom34Δ strain. Reads that mapped only after consecutive As were removed from the 3´ ends are shown in a lighter shade. Approximate region where polyadenylation takes place is indicated with a dotted arrow.
(C) Average ribosome occupancy from 3´ UTRs aligned at the annotated site of polyadenylation for wild-type and dom34Δ strains.
(D) Same as (C) except that only 99 genes with high 3´ UTR ribosome occupancy in the dom34Δ strain were included (>0.33 rpm and >3× as many reads as in the wild-type sample) and a broader 5´ window region was used to reveal ribosome stacking.
(E) Histogram of A count on the 3´ ends of footprint reads before alignment to the genome. (Inset) Cartoon of a ribosome after a 16-nt incursion into the poly(A) tail (dashed line indicates poly(A)).
(F) Normalized average ribosome occupancy from genes aligned at their stop codons for dom34Δ cells under normal or runoff conditions (top). Traces were normalized to the average value of ribosome density over the last 200 nt of data shown. Footprints mapping to the MFA2 gene in the dom34Δ strain (bottom) reveal that reads in the 3´ UTR are enriched relative to those in the ORF during starvation.
(G) Examples of mapped footprints on ribosomal protein genes with identical (RPL19) or nearly identical (RPS19) ORF amino-acid sequences and very different 3´ UTR sequences. Data show substantial differences in 3´ UTR ribosome occupancy in the dom34Δ strain. Reads that mapped only after consecutive As were removed from the 3´ ends are shown in a lighter shade. Note that ORF reads are truncated by the vertical axis scaling and are missing in some places because reads cannot be uniquely mapped. In each case, overall ORF ribosome density differs by a factor of 1.4 between the two copies of the gene.
See also Figures S4 and S5. See also Tables S1 and S2.
To determine the average pattern of ribosome occupancy across all 3´ UTRs, a “gene-average” plot for was created by aligning 3´ UTRs by their annotated sites of polyadenylation (Nagalakshmi et al., 2008) and then averaging the ribosome occupancy (Figure 3C). As expected, substantially more occupancy was present in the knockout strain, on average. We note that the peak in occupancy is broad, likely because the sites of polyadenylation are known to be heterogeneous, even among transcripts of the same gene (Ozsolak et al., 2010). Ribosomes that accumulate at the end of the 3´ UTR seem to originate upstream because a second peak, indicative of stacked ribosomes, was apparent when the gene average was limited to a set of genes with high ribosome occupancy in the 3´ UTR (Figure 3D). Consistent with this interpretation, reads from the disome peak of the sucrose gradient also mapped to this region of the 3´ UTR for some genes (Figure S4D).
To determine how far ribosomes advance into the poly(A) tail, we counted the number of consecutive 3´ As on all ribosome footprints prior to genomic alignment (Figure 3E). To our surprise, we found that ribosome footprints never protected more than ~15 consecutive As at their 3´ end. To rule out the possibility that the sequencing methodology could not capture tracts of polyA, we performed this analysis on our mRNA-Seq data and found that a significant fraction of those reads consisted of more than 30 As. These results show that, at most, the poly(A) tail only reaches the P site of the ribosome.
To ask whether ribosomes at the end of the 3´ UTR are truly arrested, we performed a ribosome runoff experiment by starving the yeast of glucose for 1 minute prior to harvesting (Ashe et al., 2000). As expected, based on the estimated rate of peptide bond formation of 6 aa/sec (Ingolia et al., 2011), ribosome occupancy at the 5´ end of ORFs was reduced for the first ~1000 nucleotides, on average (Figure 3F). When we looked at reads mapping to MFA2, a gene where the ORF and 3´ UTR are short enough that ribosome density should be reduced in both regions if ribosomes are progressing similarly along the mRNA, ribosome occupancy was strongly enriched in the 3´ UTR relative to the ORF after runoff (Figure 3F). These data suggest that the 3´ UTR-bound ribosomes are immobilized on the timescale of the experiment. Other genes with short CDSs, such as HUB1, showed a similar effect (data not shown).
3´ UTR Occupancy Is Modulated by Sequence
To test whether the sequence of the 3´ UTR is important for the accumulation of ribosomes in the knockout strain, we examined pairs of duplicate ribosomal protein genes where the amino-acid sequences are identical or nearly identical but the 3´ UTR sequences are considerably different. For example, data from the knockout strain showed accumulation of ribosome occupancy at the end of the 3´ UTR for RPL19B but not RPL19A, where ORF amino-acid sequences are identical (Figure 3G). We observed a similar trend between RPS19A and RPS19B (Figure 3G). These observations suggest that the 3´ UTR in some way specifies the accumulation of ribosomes in the dom34Δ strain. To further test this finding, we created a reporter plasmid that encodes CPR5 (Figure S5A), a gene where ribosomes accumulate in the 3´ UTR in the absence of DOM34 (Figure 3B), and found that changing the 3´ UTR sequence strongly diminished this accumulation. To gain insight about the sequence elements that might contribute to this phenomenon, we analyzed the nucleotide content of 201 3´ UTRs that showed significant ribosome enrichment in the dom34Δ strain (Table S1). We performed further analysis to consider properties of these 3´ UTRs such as sequence content (Figures S5B, S5C, and S5D), length (Figure S5E), gene ontology classification (Boyle et al., 2004), and choice of stop codon and its adjacent codon context, but found no significant trends. Overall, our finding that no one parameter serves as a robust identifier of 3´ UTRs that accumulate ribosomes in the dom34Δ strain suggests that the elements responsible may not be simply defined.
Many Ribosomes in the 3´ UTR Are Not Translating
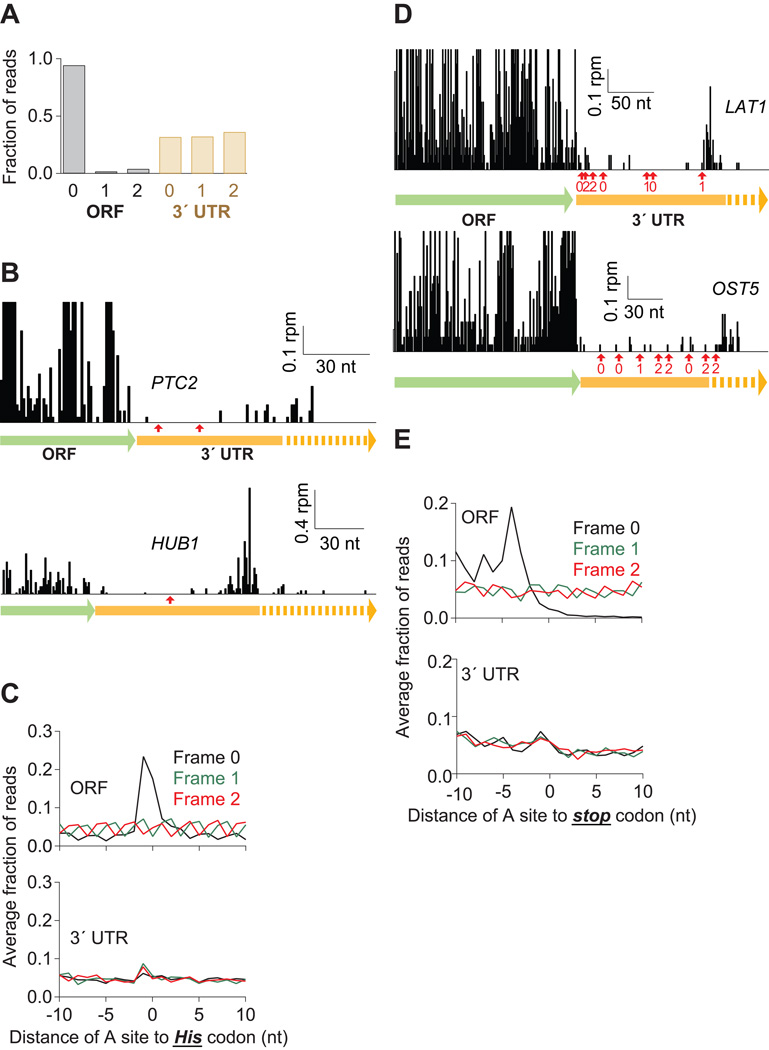
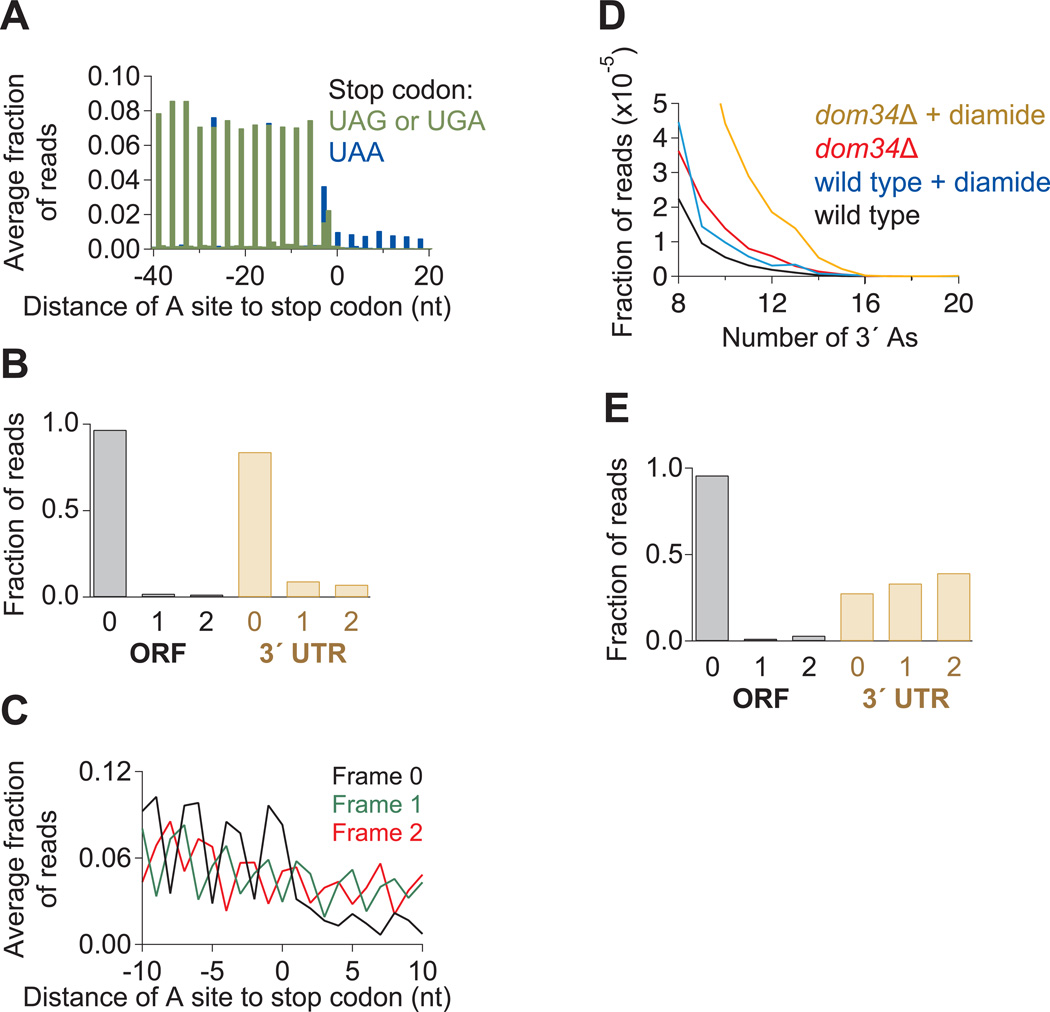
We next wished to determine the mechanism by which ribosomes might access the 3´ UTR. Normally, ribosomes should terminate translation and be recycled at stop codons, eliminating opportunities for reading into the 3´ UTR (Dever and Green, 2012). One possible explanation is that termination is inefficient and ribosomes mistakenly incorporate an amino acid at stop codons (“nonsense suppression”) and continue translating into the 3´ UTR. To rule out this hypothesis, we examined the reading frame to which the reads mapped and found that while 94% of 28-nt reads mapped to one frame of the ORF, no more than 36% of reads mapped to any one frame of the 3´ UTR (Figure 4A).
Figure 4. Many Ribosomes Do Not Translate the 3´ UTR.
(A) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for dom34Δ cells. Normalized data were averaged from ribosomes stabilized with CHX, 10 mM MgCl2, or no extra additive, but not GMP-PNP as frame was less well preserved.
(B) Examples of mapped footprints from dom34Δ cells treated with 3-AT on genes (PTC2 and HUB1) with in-frame His codons (indicated with red arrows) in the 3´ UTR. Note that the vertical axis scaling truncates the upper range of ORF reads for PTC2.
(C) Average fraction of ribosome occupancy within a 20-nt window around His codons in a given frame. Data are shown for all three reading frames in ORFs and 3´ UTRs from dom34Δ cells treated with 3-AT.
(D) Examples of mapped footprints from dom34Δ cells on genes (LAT1 and OST5) with stop codons in all three reading frames of the 3´ UTR. Stop codons and their respective reading frame before the poly(A) tail are indicated (red arrows and values 0–2). Note that the vertical axis scaling truncates the upper range of ORF reads.
(E) Average fraction of ribosome occupancy within a 20-nt window around the first occurrence of a stop codon in a given frame. Data are shown for all three reading frames in ORFs and 3´ UTRs in dom34Δ cells.
Several alternative explanations could account for the abundance of ribosomes in the 3´ UTR and in all three reading frames. One possibility is that ribosomes may translate the 3´ UTR in all three reading frames if the ribosome also regularly shifts frame at stop codons, though we know of no robust mechanism for such a hypothesis. Alternatively, it is possible that ribosome termination is efficient and that partial recycling occurs, but that the 40S subunit (which is not isolated in our sucrose gradient preparation) remains associated with the 3´ UTR and translation reinitiates at downstream AUG codons. Another possibility is that ribosomes in the 3´ UTR are produced by a total recycling failure. The 80S ribosomes generated by such a failure might “scan” downstream and either reinitiate translation or continue to the end of the 3´ UTR in a scanning mode. In these situations where recycling is incomplete, we should see evidence for a population of non-translating ribosomes.
To more carefully assess whether the ribosomes in the 3´ UTR carry out translation, we looked for ribosome pausing at His codons in the data obtained from dom34Δ yeast treated with 3-AT. On several genes that were enriched in 3´ UTR ribosome occupancy, we failed to observe a substantial rise in ribosome occupancy at His codons in any frame (Figure 4B). Moreover, when we performed an average gene analysis to ask the same question, we found that in ORFs, average ribosome occupancy on His codons was substantial while in the 3´ UTR, ribosome occupancy was scarcely above background (Figure 4C). These data suggest that many ribosomes in the 3´ UTR are not translating.
To further evaluate the evidence for 3´-UTR translation in any frame, we looked at a number of 3´ UTRs where stop codons were present in all 3 reading frames. In each case, we noticed that ribosomes still accumulated at the end of the 3´ UTR in the dom34Δ strain (Figure 4D), implying that the stop codons were not recognized by the ribosome. Moreover, unlike termination codons at the end of ORFs, where average ribosome occupancy decreased by about 3 orders of magnitude following the stop codon, we found that the average occupancy was similar before and after stop codons in the 3´ UTR (Figure 4E). Given that this hallmark for ribosome translation (Guttman et al., 2013) was not met, these data again imply that a substantial population of ribosomes found in the 3´ UTR are not translating.
The alternative (trivial) explanation that reads mapping to the 3´ UTR are not ribosome footprints is unlikely because several hallmarks of ribosome protection are met, including a fragment size distribution that very closely mirrors the peaks found in both the monosome- and disome-protected samples (Figures S1A, S6A, and S6B). Additionally, we found that the size of the footprints in the 3´ UTR changed with addition of GMP-PNP, much like those in the ORF (Figure S1A).
Nonsense Suppression Reveals that 3´ UTRs Can Be Efficiently Translated
Since existing models for translation do not address the status of ribosomes in the 3´ UTR, we assessed the general capacity of ribosomes to translate 3´ UTR sequence by performing ribosome profiling on a strain that was transformed with an ochre suppressor tRNA, SUP4-o, so that 3´ UTRs of genes ending with a UAA stop codon would be translated (Pierce et al., 1987). We found that ribosome occupancy in the 3´ UTR was enriched, on average, by about 2 orders of magnitude for genes with UAA stop codons relative to those with UGA and UAG stop codons (Figure 5A), indicating that the suppressor tRNA is specific and reasonably efficient.
Figure 5. Ribosome Occupancy in 3´ UTR is Modulated by a Suppressor tRNA or Addition of Diamide.
(A) Average fraction of ribosome occupancy in a region around the stop codon of annotated ORFs. Data taken from ski2Δ cells transformed with the SUP4-o tRNA. Only reads that measured 28 nt in length and mapped without mismatches are plotted. Readthrough is increased only when the stop codon is UAA.
(B) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for ski2Δ cells transformed with the SUP4-o tRNA.
(C) Average fraction of ribosome occupancy within a 20-nt window around the first occurrence of a stop codon in a given frame. Data are shown only in 3´ UTRs that follow ORFs terminating with a UAA stop codon. Data from ski2Δ cells transformed with the SUP4-o tRNA.
(D) Histogram of A count on the 3´ ends of footprint reads before alignment to the genome. Data for wild-type and dom34Δ yeast in the presence and absence of diamide are shown.
(E) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for dom34Δ cells treated with diamide.
Reads that appeared in the 3´ UTR in the presence of the suppressor tRNA mapped to a single reading frame nearly as well as those in the ORF (Figure 5B), consistent with these ribosomes being engaged in active translation. These ribosomes also seemed to efficiently terminate once they reached downstream stop codons in the 3´ UTR since ribosome density decreased, on average, after the first in-frame stop codon (Frame 0) in the 3´ UTR (Figure 5C). We note that the observed drop in ribosome occupancy fell short of the 3 orders of magnitude change observed for normal ORF stop codons (Figure 4E), likely from dilution of signal by the ubiquitous level of ribosomes that enter the UTR via a non-translating mechanism as well as the readthrough that results from the expressed suppressor tRNA (UAA-specific). These results argue that the 3´ UTR, like the ORF, is readily translated by the ribosome.
Dom34-targeted 3´ UTR Ribosomes Increase in the Presence of Diamide
In a final set of experiments, we asked whether conditions known to exacerbate the phenotype of a DOM34 deletion strain also augmented the molecular signatures that we identified by ribosome profiling. We therefore performed ribosome profiling on yeast strains treated with the oxidizing agent diamide and found that the proportion of reads that were positioned near the poly(A) tail increased in both the wild-type and dom34Δ strain (Figure 5D). This effect is unlikely due to the suppression of stop codons since the proportion of reads mapping to the primary reading frame did not increase (Figure 5E). These results suggest that diamide augments the number of non-translating ribosomes in the 3´ UTR that must eventually be rescued by Dom34, a result that correlates well with the known exacerbation by diamide of the dom34Δ-delayed growth phenotype (Fernandez-Ricaud et al., 2007).
DISCUSSION
We have introduced a number of technical innovations to the ribosome profiling method that improve its capability to reveal mechanistic details about the function of ribosomes engaged throughout the transcriptome. In addition to the previously studied 28-nt ribosome footprints (Ingolia et al., 2009), and more recently 21-nt footprints (L. Lareau, personal communication), we found that single ribosomes also protect mRNA fragments of 15–18 nt in length at a known site of mRNA truncation. We also found that stacked ribosome footprints (40–65 nt in size) from disome fractions are a clear signal of ribosome stalling. Finally, we show that pauses induced by histidine starvation or the drop in ribosome occupancy following stop codons serve as signals for establishing whether ribosomes are engaged in active translation. We anticipate that these tools will be particularly useful for future studies defining ribosome pausing and ribosome interaction with noncoding regions.
Here we took advantage of these advances to reveal the in-vivo substrates of Dom34 and to address a number of mechanistic questions about the nature of ribosomes that engage the 3´ UTR. To our surprise, Dom34 does not target numerous known classes of paused ribosomes, including those induced by histidine starvation, implying that target selection is highly specific. A key example of a Dom34 target in the cell is a shortened isoform of the gene HAC1 (Figure 2). Our high-resolution data indicate that ribosomes do engage this rare isoform of HAC1 under normal conditions, and that Dom34 rescues these ribosomes. Moreover, it shows that no-go decay takes place on HAC1 and clearly illustrates the pattern of upstream stalling and mRNA cleavage that is apparently characteristic of the process. Given this evidence for NGD on an endogenous substrate, we anticipate that Dom34 broadly rescues ribosomes stalled at truncation sites, entrusting it with an important cellular role that was hinted at by previous evidence that Dom34 targets ribosomes stalled on truncated synthetic reporter mRNAs (Pisareva et al., 2011; Shao et al., 2013; Shoemaker and Green, 2011; Tsuboi et al., 2012). For example, we anticipate that ribosomes encountering the randomly-located mRNA truncations generated by exosome activity in the cell will be routinely rescued by Dom34 (Garneau et al., 2007).
In addition to these targets, we found a new class of Dom34 substrate that comprises ribosomes immobilized near the junction between the poly(A) tail and the 3´ UTR (Figure 3). Intriguingly, we found that the majority of such ribosomes arrive at this noncoding location by a mechanism other than translation of the entire 3´ UTR (for example, as might be permitted by nonsense suppression or a frameshift event at the stop codon). In this regime, we find that the ribosome does not maintain a 3-nt reading frame, akin to the proposed scanning process that the small (40S) ribosomal subunit is thought to carry out in the 5´ UTR while it searches for the start codon during the initiation phase of translation (Hinnebusch and Lorsch, 2012). Evidence for “80S scanning” has been reported in bacteria when ribosome recycling factor (RRF) is mutated (Janosi et al., 1998), in yeast during the nonsense mediated decay response at premature termination codons (Amrani et al., 2004), or more generally when certain mammalian post-termination complexes are not recycled (Skabkin et al., 2013). Taken together, our data are consistent with a majority of the 3´ UTR bound ribosomes moving along the mRNA in an unconventional, non-translating mode for at least some distance (Figure 4).
Our data are also consistent with a fraction of ribosomes in the 3´ UTR eventually carrying out translation (Figures 4C and 4E) since it is conceivable that some scanning ribosomes may reinitiate translation of the mRNA, as has been observed for scanning bacterial ribosomes when RRF is mutated (Janosi et al., 1998). This reinitiation model could accommodate a scenario where the scanning phase is carried out by 40S or 80S ribosomes. In general, however, exclusive reinitiation models are unappealing because ~21% of the top 3´ UTRs targeted by Dom34 lack a start codon that would allow ribosomes to read to the observed arrest point for ribosomes without terminating. A reasonable explanation might include a combination of scanning ribosomes (including 80S or 40S) with reinitiation by a subset of these. This sort of mechanism could explain reports indicating that ribosomes bound to lincRNAs seem to bypass stop codons (Guttman et al., 2013).
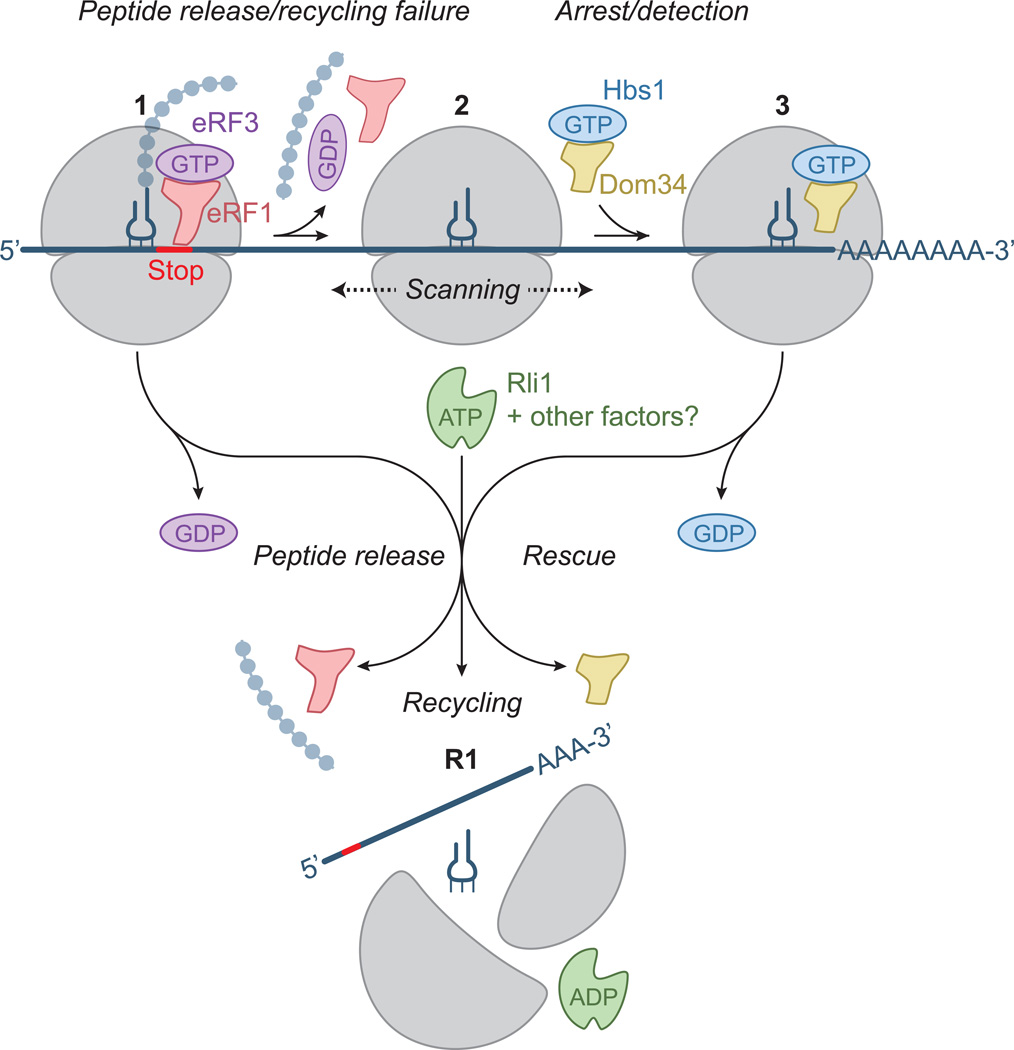
A plausible explanation for the origin of the unexpected ribosomes that populate many 3´ UTRs (and are then rescued by Dom34) is that the canonical termination and recycling process are not always efficient (Figure 6). In these situations, the subunit dissociation steps may not reach completion due to a defect in or inadequate levels of the recycling factor Rli1 or any potential cofactors that stimulate this step. As a result, the 80S or 40S ribosome could remain associated with the mRNA after peptide release and, unable to immediately resume translation, would scan along the mRNA. In principle, this process could be bidirectional, however, the downstream flow of translating ribosomes would tend to push the scanning ribosomes toward the 3´ UTR. This model also offers a potential explanation for the diamide-induced increase in the number of non-translating ribosomes in the 3´ UTR (Figures 5D and 5E). Because diamide is known to inactivate FeS proteins (Philpott et al., 1993), such as the recycling factor Rli1 (Barthelme et al., 2011), it is plausible that diamide exposure leads to Rli1 inactivation and a failure of ribosome recycling, and thus ultimately to an increase in 3´ UTR ribosome occupancy.
Figure 6. Model for Dom34-mediated Rescue of 3´ UTR-located Ribosomes.
When the ribosome reaches the 3´ end of an ORF, release factors (eRF1 and eRF3) recognize the stop codon in the A site and bind the ribosome (1). After peptide release, Rli1 typically mediates ribosome recycling (perhaps with other unknown factors) making the individual subunits available for reuse (R1). If the recycling step fails, the 80S ribosome (2) “scans” along the 3´ UTR. The ribosome then arrests upon reaching the poly(A) tail and rescue factors (Dom34 and Hbs1) bind the ribosome (3). These factors target the ribosome for recycling by Rli1, making the subunits available for reuse (R1). In some cases, the ribosome may reinitiate translation in the 3´ UTR prior to rescue.
It is puzzling that these ribosomes become arrested at the 3´ end of the UTR, sometimes after partially engaging the poly(A) tail. One mechanism to explain why these ribosomes do not fully enter the poly(A) region is that poly(A) binding protein (Pab1) or another 3´ UTR associated protein simply blocks movement of the ribosome once it reaches the end of the UTR. Alternatively, the poly(A) sequence itself may have affinity for the mRNA entry channel that prevents the ribosome from moving once a small series of As is stably bound (a “trap”).
Given the number of transcripts (~11%, Figure 3A) where termination and/or recycling appear to be somewhat inefficient, it is worth speculating whether non-translating ribosomes in the 3´ UTR serve some useful purpose. It has been proposed, for example, that translational readthrough resulting from a deficiency in release factor 3 (eRF3) activity (i.e. through activation of the [PSI+] prion in S. cerevisiae) may facilitate adaptation during periods of stress (Serio and Lindquist, 1999). Similarly, scanning readthrough may also be a signature of a larger system that regulates gene expression. For instance, 3´ UTR-binding proteins could regulate the efficiency of proper termination/recycling against an alternative pathway that releases the nascent peptide (perhaps to be degraded) but leaves the ribosome intact and capable of scanning into the 3´ UTR (and somehow affecting gene regulation). The importance of 3´ UTR sequence in determining the number of ribosomes that commence scanning after encountering the stop codon may derive from the sequence specificity of regulatory factors that, for example, impact recycling. Another hypothesis is that scanning ribosomes in the 3´ UTR may be able to directly funnel useful components of the translational machinery from the 3´ to the 5´ end of the mRNA for more efficient translation (Hinnebusch and Lorsch, 2012).
In general, our data are consistent with the view that Dom34 is critical for regulating the overall level of ribosomes in the cell by liberating specific classes of stalled ribosomes; as such, Dom34 contributes to determining the amount of ribosomes available for the initiation of translation. This role in homeostasis of ribosome components reasonably accounts for why the dom34Δ strain is so poorly tolerant of situations that limit the supply of ribosomes. Because ribosome availability closely tunes the rate of cell growth, further studies of Dom34 may be useful for understanding various phenomena, such as cancer, where growth rates become uncontrolled. In addition, our unexpected finding that ribosomes populate the noncoding 3´ UTR of the cell raises a number of questions about the regulation of efficient termination and recycling at the end of open reading frames and the significance of ribosome occupancy on noncoding RNAs in the cell. We anticipate that future ribosome profiling studies will unveil a number of mechanistic insights to address these puzzles.
EXPERIMENTAL PROCEDURES
Preparation of Ribosome Footprint Libraries
Ribosome footprints were prepared essentially as described (Ingolia et al., 2012). Briefly, yeast cells were rapidly extracted from the media via vacuum filtration and frozen in liquid nitrogen. Cells were lysed in a freezer mill with lysis buffer (20 mM Tris pH 8, 140 mM KCl, 1.5 mM MgCl2, 1% Triton X-100) that sometimes contained an added stabilizing agent: 0.1 mg/ml CHX, 5 mM GMP-PNP (plus equimolar MgCl2), both CHX and GMP-PNP, or 10 mM MgCl2. Monosomes and disomes of RNase treated samples were separated by sucrose gradient and purified mRNA footprints were ligated to a universal linker. Following reverse transcription, circularization, rRNA subtraction, and PCR amplification, cDNA fragments were sequenced on an Illumina HiSeq2000 or HiSeq2500 machine at facilities at UC Riverside or the Johns Hopkins Institute of Genetic Medicine.
Growth Conditions
All cells were grown in YPD (BD Biosciences) media with the following exceptions: those expressing SUP4-o (CSM-His), the CPR5 reporter (CSM-Ura), or those that were 3-AT treated (CSM-His) (All CSM media was from Sunrise Science Products). All media was sterile filtered and cultures were grown at 30°C. Cultures were usually harvested at an OD of ~0.6 after ~5 doubling times. For the 3-AT experiments, cells were grown for ~2.5 doubling times in 100 mM 3-AT prior to harvest. For experiments with diamide, cultures at an OD of ~0.3 were treated with 2 mM diamide and 300 µM CaCl2 for 2 hours before harvest. For glucose starvation experiments, cells at OD ~0.6 were pelleted from YPD media, resuspended in YP, and shaken for 1 min prior to harvest.
Basic Read Preparation and Sequence Alignment
The R64-1-1 S288C reference genome assembly (sacCer3) was used for all analysis (Saccharomyces Genome Database Project). De-multiplexed sequences were first processed to remove low quality reads (any position with Phred score <20 or assigned N). Following a search for the linker (footprints) or truncation at 35 nt (mRNA-Seq), contaminating gel ladder oligos were removed and alignment to the RNA gene database (http://downloads.yeastgenome.org/sequence/S288C_reference/rna/archive/rna_coding_R64-1-1_20110203.fasta.gz) was performed to remove noncoding contaminants. Remaining reads were then aligned to the genome and those that did not align were aligned to a database of annotated splice junctions. In some cases, footprint reads left over at this stage were then trimmed of consecutive 3´ As and realigned to the genome and splice junction database. Matches in these various alignment rounds were pooled for calculation of ribosome occupancy. All reads that aligned to multiple sequences were discarded. Read occupancy was set by giving one count per read 5´ end and then shifted to correlate with various active sites in the ribosome. Read counts were then normalized to units of reads per million (rpm) (by dividing by the total number of million mapped reads in a sample). Alignments were performed with Bowtie 0.12.7 (Langmead et al., 2009)
Supplementary Material
ONLINE HIGHLIGHTS.
Varied footprint sizes (15–65 nt) distinguish classes of stalled ribosomes.
Dom34 dissociates ribosomes on truncated mRNAs.
Non-translating ribosomes are found in the 3´ UTR.
Dom34 dissociates ribosomes that become arrested in the 3´ UTR.
ACKNOWLEDGEMENTS
We thank Allen Buskirk and Kristin Smith-Koutmou for comments on the manuscript; Nick Ingolia, Gloria Brar, and Jonathan Weissman for protocols and helpful discussions about ribosome profiling; Toshi Inada for thoughtful discussions; Roy Parker for wild-type, dom34Δ and hbs1Δ yeast strains; Peter Walter for informative discussions about HAC1; Mick Tuite for advice on creating the suppressor tRNA strain; Alan Hinnebusch for advice on use of 3-AT; Pavan Vaidyanathan for advice on performing the runoff experiment; Grace Yeo for preliminary calculations of reading frame and help with choosing rRNA subtraction oligos; and Carla Connelly for assistance with tetrad dissection. The work was supported by the NIH and HHMI (R.G.) and the Damon Runyon Cancer Research Foundation (N.R.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Sequencing data (debarcoded fastq files and wig files) were deposited in the NCBI GEO database with series accession number GSE52968.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures, 3 tables, and Supplemental Experimental Procedures. It can be found with this article online at X.
REFERENCES
- Adham IM, Sallam MA, Steding G, Korabiowska M, Brinck U, Hoyer-Fender S, Oh C, Engel W. Disruption of the pelota gene causes early embryonic lethality and defects in cell cycle progression. Molecular and cellular biology. 2003;23:1470–1476. doi: 10.1128/MCB.23.4.1470-1476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. The EMBO journal. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Molecular biology of the cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. Rna. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Molecular and cellular biology. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Molecular cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nature structural & molecular biology. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Molecular cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Davis L, Engebrecht J. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics. 1998;149:45–56. doi: 10.1093/genetics/149.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harbor perspectives in biology. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley interdisciplinary reviews RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ricaud L, Warringer J, Ericson E, Glaab K, Davidsson P, Nilsson F, Kemp GJ, Nerman O, Blomberg A. PROPHECY--a yeast phenome database, update, 2006. Nucleic acids research. 2007;35:D463–D467. doi: 10.1093/nar/gkl1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Manzoor M, Hudak KA. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. The Journal of biological chemistry. 2008;283:32218–32228. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nature reviews Molecular cell biology. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. The Journal of biological chemistry. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A Promotes Translation of Polyproline Motifs. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Parker R. Global analysis of mRNA decay intermediates in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11764–11769. doi: 10.1073/pnas.1119741109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L, Mottagui-Tabar S, Isaksson LA, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. The EMBO journal. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopotowski T, Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Archives of biochemistry and biophysics. 1965;112:562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Kurian L, Palanimurugan R, Godderz D, Dohmen RJ. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature. 2011;477:490–494. doi: 10.1038/nature10393. [DOI] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO reports. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Molecular cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel S, Jiang J, Silver N, Gallagher J, Cunningham J, Surdulescu G, Lathrop M, Farrall M, Spector TD, Thein SL. The HBS1L-MYB intergenic region on chromosome 6q23.3 influences erythrocyte, platelet, and monocyte counts in humans. Blood. 2007;110:3624–3626. doi: 10.1182/blood-2007-05-093419. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annual review of biochemistry. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Mori T, Ogasawara C, Inada T, Englert M, Beier H, Takezawa M, Endo T, Yoshihisa T. Dual functions of yeast tRNA ligase in the unfolded protein response: unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Molecular biology of the cell. 2010;21:3722–3734. doi: 10.1091/mbc.E10-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Haile D, Rouault TA, Klausner RD. Modification of a free Fe-S cluster cysteine residue in the active iron-responsive element-binding protein prevents RNA binding. The Journal of biological chemistry. 1993;268:17655–17658. [PubMed] [Google Scholar]
- Pierce MK, Giroux CN, Kunz BA. Development of a yeast system to assay mutational specificity. Mutation research. 1987;182:65–74. doi: 10.1016/0165-1161(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO journal. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Lindquist SL. [PSI+]: an epigenetic modulator of translation termination efficiency. Annual review of cell and developmental biology. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Molecular cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and Other Unconventional Posttermination Events during Eukaryotic Translation. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3' end of aberrant mRNA. Molecular cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Seraphin B, Graille M. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nature structural & molecular biology. 2010;17:1446–1452. doi: 10.1038/nsmb.1963. [DOI] [PubMed] [Google Scholar]
- Wang Z, Sachs MS. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Molecular and cellular biology. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO journal. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nature structural & molecular biology. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.