Abstract
Entamoeba histolytica is a eukaryotic intestinal parasite of humans, and is endemic in developing countries. We have characterized the E. histolytica putative low molecular weight protein tyrosine phosphatase (LMW-PTP). The structure for this amebic tyrosine phosphatase was solved, showing the ligand-induced conformational changes necessary for binding of substrate. In amebae, it was expressed at low but detectable levels as detected by immunoprecipitation followed by immunoblotting. A mutant LMW-PTP protein in which the catalytic cysteine in the active site was replaced with a serine lacked phosphatase activity, and was used to identify a number of trapped putative substrate proteins via mass spectrometry analysis. Seven of these putative substrate protein genes were cloned with an epitope tag and overexpressed in amebae. Five of these seven putative substrate proteins were demonstrated to interact specifically with the mutant LMW-PTP. This is the first biochemical study of a small tyrosine phosphatase in Entamoeba, and sets the stage for understanding its role in amebic biology and pathogenesis.
Keywords: Entamoeba histolytica, low molecular weight protein tyrosine phosphatase (LMW-PTP), LMW-PTP crystal structure, substrate-trapping, LMW-PTP putative substrate identification
1. Introduction
The unicellular protozoal human parasite Entamoeba histolytica has two stages in its life cycle: infective cysts and motile trophozoites [1]. E. histolytica infection can result in amebic colitis and liver abscesses; an estimated 50 million symptomatic clinical cases of amebiasis occur every year worldwide, resulting in 100,000 deaths [1, 2]. E. histolytica cysts are spread to human hosts via the fecal-oral route via contaminated food or water, and infection with this organism is endemic in many parts of the developing world [2]. Outbreaks in developed countries have occurred when drinking water has become contaminated with human fecal matter such as in the city of Tbilisi in the Republic of Georgia in 1998 [3], and in Chicago in 1933 during the World's Fair [4].
Phosphorylation and dephosphorylation of protein tyrosine residues play important roles in regulating cellular processes [5]. Low molecular weight protein tyrosine phosphatases (LMW-PTPs) are found in most organisms including Archaea, bacteria, and eukaryotes [6]. In general, an organism has one or two LMW-PTP genes: E. histolytica has two, the commensal species E. dispar and the reptile parasite E. invadens each have one, as does the green alga Chlamydomonas reinhardtii and the plants Arabidopsis thaliana and Oryza sativa [7]. The black cottonwood tree Populus trichocarpa has two [7], as does Drosophila [8]. All mammals, including humans [6], have a single gene yielding two active isoforms [9]. Mammalian LMW-PTPs have been observed to be overexpressed in certain tumors, and thus are considered oncogenes [10].
The active site, or P loop, of LMW-PTPs has the conserved sequence CLGNICR, conforming to the general PTP sequence CX5R [5, 11]. The cysteine residue performs the nucleophilic attack on the phosphorus atom of the substrate phosphate group, producing a covalent phosphoenzyme intermediate [12, 13]. Mutating the active site cysteine to a serine or alanine creates an enzyme lacking detectable catalytic activity [13]. Cysteine to serine (Cys to Ser) mutants bind substrates and substrate analogs with the same affinity as the wild-type PTP [12]. These mutants are used to isolate and identify PTP substrates by “substrate trapping” either in vivo or in vitro, since they bind substrate but do not carry out the catalytic reaction.
E. histolytica has 20 genes annotated as PTPs or putative PTPs [14, 15], far fewer than the 107 PTPs that the human genome contains [7, 16]. The two E. histolytica LMW-PTP proteins (GenBank: XP_656359, coded by GenBank: XM_651267, and GenBank: XP_653357, coded by GenBank: XM_648265), are identical except for a single conservative residue change at position 85 in the protein sequence: XP_656359 has an alanine and XP_653357 a valine. Both genes are expressed in cultured trophozoites, clinical isolates, and cysts [17, 18]. XM_651267, the gene encoding XP_656359, was cloned and expressed for this study, as was its Cys to Ser substrate-trapping mutant form. This LMW-PTP had never been studied before and determination of its structure could be a starting point for designing drugs targeting it. In mammalian cells, LMW-PTPs play roles in controlling cell proliferation, motility, and adhesion through dephosphorylation of such substrates as growth factor receptors and cytoskeleton-associated proteins [11, 16, 19, 20, 21]. Identifying E. histolytica LMW-PTP putative substrates by use of a substrate-trapping Cys to Ser mutant LMW-PTP is a start to elucidating cellular pathways regulated by the action of this LMW-PTP.
2. Materials and Methods
2.1. Alignment of LMW-PTP Protein Sequences
The wild-type E. histolytica LMW-PTP protein sequence (GenBank: XP_656359) was input into BLAST [22] to identify, select, and align LMW-PTP sequences from other representative species; LALIGN [23] was also used for alignments. The one exception was the E. invadens LMW-PTP, where the ameba database AmoebaDB [14] hosted by EuPathDB [24] was used for selection and alignment. Phylogenetic trees for the relative genetic distances between the LMW-PTPs were created with or without an outgroup using the phylogeny.fr web site [25] or mirror site [26], which was used in its “One-Click” mode, utilizing MUSCLE for alignment, PhyML for phylogeny, and TreeDyn for tree rendering [25, 26]. Gblocks (which can be used to eliminate divergent regions and poorly aligned positions after alignment is performed) was not used in this analysis. PTP1B was included as the outgroup for one tree. The sequence for human PTP1B was input into BLAST [22] to obtain the sequence for the E. histolytica PTP1B homologue, which in turn was input into BLAST [22] to identify sequences for PTP1B homologues in the same organisms for which the LMW-PTPs had been selected for comparison.
2.2. Cloning of Substrate-Trapping Mutant LMW-PTP
The substrate-trapping mutant Cys to Ser LMW-PTP gene was cloned using E. histolytica genomic DNA as a template. PCR was used for site-directed mutagenesis to alter the codon at residue number 7 in the XM_651267 sequence from TGT to AGT in the LMW-PTP active site, using the forward primer BamH1TyrPhosFmut (5′-ATG GGA TCC ATG AAG TTG TTG TTT GTA AGT TTA GGC AAC ATT TGT CGA TCT CCT-3′) and the reverse primer SalITyrPhosR (5′-GGC GTC GAC TTA ATT AAT AAG TTT TCC TTC TTC TAG TTT AAT GAT TTA ATT CTC ACA AGC ATC-3′). PCR products were digested with Bam H1 and Sal I and cloned into plasmid pBluescript II KS(+) (Agilent Technologies, Santa Clara, CA, USA). Mutant LMW-PTP protein was recombinantly expressed and purified at the Seattle Structural Genomics Center for Infectious Disease (SSGCID) as previously described [27, 28].
2.3. Recombinant Protein Crystallization and X-Ray Diffraction
Recombinant wild-type LMW-PTP protein was expressed and purified as previously described [27, 28]. Crystallization (26-31 mg/ml) and X-ray diffraction conditions were as follows for the wild-type LMW-PTP structures: 3ILY (apo structure): Molecular Dimensions ProPlex Screen condition A9, 0.1 M MES pH 6.0, 20% PEG MME 2000, 0.2 M NaCl, 20% glycerol as cryo-protectant; 3IDO (substrate analog HEPES bound): Molecular Dimensions ProPlex condition F1, 0.1 M HEPES pH 7.0, 20% PEG 8000, 25% glycerol as cryo-protectant; 3JS5 (substrate analog HEPES bound; higher resolution than 3IDO) Emerald Bio PACT screen condition E6, 20% PEG 3350, 0.2 M sodium formate; 3JVI (product analog sulfate bound) Emerald Bio Wizard II screen condition 41, 2.0 M ammonium sulfate, 0.1 M Tris-HCl pH 7.0, 0.2 M lithium sulfate. The expression, crystallization, and X-ray diffraction work was performed by the SSGCID protein crystallization team. The crystallographic X-ray data and refinement statistics are shown in Supplemental Table 1.
2.4. Comparison of LMW-PTP Solved Structures
To identify and compare LMW-PTPs with solved structures similar to the E. histolytica LMW-PTP, the structure PDB: 3JS5 (substrate mimic HEPES bound) was input into both the RSCB protein database [29] and NCBI's VAST+ database [30] in order to find structural homologues. VAST+ was then used to compare the three-dimensional structures of selected LMW-PTP homologues to the E. histolytica LMW-PTP using geometric criteria.
2.5. Testing Recombinant Wild-Type and Mutant LMW-PTPs for Phosphatase Activity
Phosphatase activity was tested with the SensoLyte pNPP Colorimetric Protein Phosphatase Assay Kit (AnaSpec, Fremont, CA, USA) using the supplied Assay Buffer in 100 μl volumes in a 96-well plate. 10-200 ng wild-type LMW-PTP was tested, and absorbance was measured at 405 nm (A405). HALT™ Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA) was used to test inhibition in one set of samples.
2.6. Culture of Trophozoites
E. histolytica strain HM1:IMSS trophozoites were grown axenically in TYI-S-33 (trypticase-yeast extract-iron-serum) medium supplemented with 1X Diamond's vitamins (SAFC Biosciences, Lenexa, KS, USA), 15% heat-inactivated bovine serum (Gemini Bio-Products, West Sacramento, CA), 100 U penicillin/ml and 100 mg streptomycin sulfate/ml (Gibco/Life Technologies, Grand Island, NY, USA), at 37°C in T-25 tissue culture flasks [31].
2.7. Transfection of Trophozoites
HM1:IMSS strain E. histolytica trophozoites were transfected as previously described using the lipofection technique [32], but with the following changes: 30 μg plasmid DNA (>1 μg/μl solution, or a dried pellet) was suspended in supplemented M199 medium in 200 μl volume in 2 ml sterile microcentrifuge tubes, and 30 μl Attractene transfection reagent (Qiagen, Valencia, CA, USA) was added per tube. After overnight incubation at 37°C, 6 μg/ml G418 (geneticin) (Gibco/Life Technologies, Grand Island, NY, USA) was added for selection [33]. Transfectant stocks were maintained with 6 μg/ml G418 (geneticin) (Gibco/Life Technologies, Grand Island, NY, USA).
2.8. Lysis Buffer
Lysis buffer consisted of 5X SigmaFast protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA), 150 mM NaCl, 50 mM Tris, 10% glycerol, 1% Nonidet P-40 or 1% Triton X-100, and phosphatase inhibitors: 100 μM NaF, 1 mM Na3VO4 or pervanadate, and 5 mM iodoacetamide.
2.9. Immunoblotting
Trophozoites were harvested by tapping culture flasks on the benchtop or by icing for 20 min, contents were transferred to 50 ml conical tubes, centrifuged at 200 × g for 5 min, pelleted trophozoites were resuspended in ~1 ml supernatant and transferred to 2 ml microcentrifuge tubes, and were re-pelleted as above. Pellets were resuspended in lysis buffer; after lysis, 10 mM DTT was added. Whole lysate protein was quantified with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Samples were subjected to SDS-PAGE under reducing conditions on 13.5% or 15% Tris-glycine gels in running buffer (25 mM Tris, 192 mM glycine, 1% SDS, pH 8.3) and were transferred to Immobilon-P PVDF transfer membrane with pore size 0.45 μm (Millipore, Billerica, MA, USA). Membranes were blocked in either 5% nonfat dry milk or 3.5% BSA in TBST (43 mM Tris-HCl, 7 mM Tris base, 200 mM NaCl, 0.1% Tween 20, pH 7.35), and incubated with either anti-LMW-PTP rabbit sera or with commercial antibodies recognizing the E, S, or KT3 epitope tags (GeneTex, San Antonio, TX, USA and Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C. Secondary antibody incubation with goat anti-mouse Fc-specific peroxidase-conjugated or goat anti-rabbit whole molecule peroxidase-conjugated IgG (Sigma-Aldrich, St. Louis, MO, USA) was performed at room temperature. The ECL kit from Roche (Roche Applied Science, Indianapolis, IN, USA) was used to treat membranes, bands were visualized on film, and film images were electronically scanned.
2.10. Coupling Recombinant Mutant LMW-PTP and BSA to Affi-Gel 15 beads
250 or 500 μg recombinant mutant LMW-PTP or control BSA protein was diluted in 500 μl 100 mM HEPES (pH 7.5), and coupled overnight to 50 or 100 μl Affi-Gel 15 beads (Bio-Rad, Hercules, CA, USA) at 4°C in microcentrifuge tubes on a nutator overnight, and beads-alone (mock-coupled) samples were incubated with buffer alone. Coupled beads were blocked using 0.1 ml of 1 M ethanolamine-HCl (pH 8) per ml beads [34], and washed five times in cold 100 mM HEPES (pH 7.5) to remove unbound protein. If beads with coupled proteins were not to be immediately used, they were stored in microcentrifuge tubes at 4°C in 100 mM HEPES plus 0.2% sodium azide.
2.11. Pulldowns of Putative Substrate Proteins Using Mutant LMW-PTP and BSA Coupled to Affi-Gel 15 Beads
HM1:IMSS strain non-transfected E. histolytica trophozoites were pre-treated with 1 mM pervanadate by direct addition to the TYI medium for 30 min before harvesting to inhibit phosphatase activity. Sodium orthovanadate (Na3VO4) and pervanadate solutions were prepared as described [35, 36]. TYI was removed from sample flasks and 10 ml PBS/1 mM pervanadate was added. Trophozoites were harvested as described for immunoblotting. Pellets were resuspended in lysis buffer, and sonicated on ice using a Branson 150D sonifier set to 6 (Branson Ultrasonics Corporation, Danbury, CT, USA), pulsing twice for 10 seconds each with a 10 second rest between pulses. Lysates were incubated in microcentrifuge tubes on a nutator for 30 min at 4°C. DTT was added to 10 mM, and lysates were microcentrifuged at 10,000 × g for 5 min to remove insoluble material [37, 38]. Total lysate protein was quantified with the Bio-Rad Protein Assay Kit, and diluted to 1 μg/μl with cold lysis buffer. 1 ml lysate was added to each sample tube with prepared Affi-Gel 15 beads, and tubes were incubated on a nutator at 4°C overnight. Beads were washed five times with cold PBS, suspended in lysis buffer, and SDS-PAGE sample buffer was added. Samples were frozen in liquid N2 and stored at -80°C until subjected to SDS-PAGE and silver staining. Silver staining of samples was performed with the mass spectrometry-compatible SilverQuest Staining Kit (Invitrogen/Life Technologies, Grand Island, NY, USA). Silver-stained whole gels were submitted to the W.M. Keck Biomedical Mass Spectrometry Laboratory for trypsin digestion and mass spectrometry analysis of tryptic peptides of the selected bands.
2.12. Protein Band Sequencing and Analysis
Silver-stained bands were excised from gels and protein sequencing was performed at the W.M. Keck Biomedical Mass Spectrometry Laboratory as described previously [39] but with the following changes: the liquid chromatography (LC)-MS system consisted of a Thermo Electron Orbitrap Velos ETD mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed 8 cm × 75 μm id Phenomenex Jupiter 10 μm C18 reversed-phase capillary column. 7.5 μl of the extract was injected, and the peptides were eluted from the column by an acetonitrile/0.1 M acetic acid gradient at a flow rate of 0.5 μl/min over 30 min. The nanospray ion source was operated at 2.5 kV. This mode of analysis produces 10000 collision-activated dissociation (CAD) spectra of ions over several orders of magnitude. Data were analyzed by database searching using the Sequest search algorithm against E. histolytica.
2.13. Cloning and Epitope-Tagging of Putative LMW-PTP Substrate Protein Genes Identified by Mass Spectrometry Analysis
Each selected protein gene identified as a putative LMW-PTP substrate was cloned with epitope tags added to the 5′ or the 3′ gene terminus to generate an N-terminally (N-term) or C-terminally (C-term) epitope-tagged protein. Oligo pairs listed in Supplemental Data Set 1 were used to clone the selected genes via PCR. A combination tag of three epitope tags back-to-back was added to the 5′ or 3′ end of the cloned genes: the E epitope tag (GAPVPYPDPLEPR), the S epitope tag (KETAAAKFERQHMDS), and the KT3 epitope tag (KPPTPPPEPET). Tagged protein genes were cloned into pGIR209 (Supplemental Fig. 1).
2.14. Pulldowns of Epitope-Tagged Putative Substrate Proteins Using Mutant LMW-PTP and BSA Coupled to Affi-Gel 15 Beads
Transfected amebae overexpressing epitope-tagged putative substrate proteins identified by mass spectrometry were selected with 24 μg/ml G418 24 hours before harvesting as described above. Sample lysates were subjected to immunoblotting to verify expression and sizes of epitope-tagged proteins. Beads with mutant LMW-PTP, BSA, or mock protein-attached (beads alone) were incubated overnight at 4°C on a nutator with either 1 ml sample lysate, 1 ml (2.5 mg) BSA, or with lysis buffer alone. Beads were washed twice in cold PBS + 0.02% Tween 20, twice in PBS, resuspended in lysis buffer, and SDS-PAGE sample buffer was added. Samples were frozen in liquid N2 and stored at -80°C until subjected to SDS-PAGE. Immunoblots were probed with S tag antibody.
3. Results
3.1. The E. histolytica LMW-PTP protein sequences are conserved and are most closely related to those of Bacteroides or plants
Selected representative LMW-PTP protein sequences from other organisms were aligned with the E. histolytica LMW-PTP sequence (GenBank: XP_656359) to compare similarities and conservation of residues. Representative LMW-PTPs were included from both multicellular and unicellular organisms. Chosen LMW-PTPs were from the closely related species Entamoeba dispar, the less related species Entamoeba invadens, and from six species of other unicellular eukaryotes (Dictyostelium discoideum, Giardia lamblia, Giardia intestinalis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Kluyveromyces lactis), seven bacterial species (Pseudomonas aeruginosa, Bdellovibrio bacteriovorus, Bacteroides caccae, Bacteroides thetaiotaomicron, Bacteroides fragilis, Parabacteroides merdae) five species of cyanobacteria (Nostoc punctiforme, Lyngbya aestuarii, Arthrospira maxima, Prochlorococcus marinus, Cyanothece sp. PCC 7822), eight species of plants (sorghum, maize, rice, barley, grape, black cottonwood tree, castor bean, Arabidopsis thaliana) and seven species of multicellular animals (human, house mouse, domestic cow, chicken, African clawed frog, fruit fly, zebrafish) (Supplemental Fig. 2, Supplemental Data Set 2). Certain regions of the LMW-PTP sequences were highly conserved across kingdoms, especially the active site CLGNICRS, and the DPYY loop containing the catalytically critical aspartic acid residue which acts as a general acid and base during phosphate removal from the substrate, and the two back-to back tyrosines which in mammalian LMW-PTPs can be phosphorylated to regulate enzyme activity and recruit adapter proteins (Supplemental Fig. 2) [11, 19, 40, 41]. Average query coverage (the actual length of the compared region of the query protein, here the E. histolytica protein) and overall sequence conservation between the E. histolytica LMW-PTP and those from bacteria (95% query coverage and 48% identity), plants (92% query coverage and 51% identity) and cyanobacteria (94% query coverage and 50% identity) was higher than that of non-Entamoeba unicellular eukaryotes (93% query coverage and 35% identity) and multicellular animals (87% query coverage and 35% identity) (Supplemental Data Set 2).
To analyze the similarity and conservation between these selected LMW-PTPs graphically, two phylogenetic trees were constructed with online software [25, 26], one with and one without an outgroup. These trees show graphically, in terms of sequence identity and similarity, that the E. histolytica LMW-PTP proteins are most closely related to those of Bacteroides species, then those of plants, then cyanobacteria (Supplemental Fig. 3). Interestingly, the LMW-PTPs from the other non-Entamoeba unicellular eukaryotes are clustered by themselves, and are more closely related to LMW-PTPs from multicellular animals (Supplemental Fig. 3). PTP1B (PTPN1) was included as an outgroup in a second tree (Supplemental Fig. 4). A nonreceptor classical PTP, PTP1B was chosen as the outgroup because it has been extensively studied (homologues exist in many organisms); it was the first PTP to be identified and its structure solved [42, 43], and in human cells has some of the same substrates as the LMW-PTP, such as the PDGF receptor [44] and insulin receptor [45]. With the PTP1B outgroup included, all the eukaryotic PTP1B homologues are on one branch (with the exception of K. lactis), and the bacterial/cyanobacterial PTP1Bs are on a distant branch, more closely related to the LMW-PTPs from unicellular eukaryotes (including Entamoeba) and multicellular animals, and all of the plant and most of the bacterial and cyanobacterial LMW-PTPs form groups off of their own branch (Supplemental Figure 4). The Entamoeba LMW-PTPs are still quite closely related to some of the bacterial LMW-PTPs (although two bacterial PTP1B homologues are grouped with the Entamoeba LMW-PTPs), but are closer to the other unicellular eukaryotes and to multicellular animals. The reason for this could be that since the bacterial and cyanobacterial PTP1B homologues differ quite substantially from those of eukaryotes (and the size range for the PTP1B proteins is larger than that of the LMW-PTPs) the query coverage tends to be substantially less (Supplemental Data Set 2), including just those residues important in phosphatase function, therefore affecting the branch and distance calculations.
3.2. Structures of the Entamoeba histolytica LMW-PTP
This E. histolytica LMW-PTP was expressed recombinantly, crystallized, and subjected to X-ray diffraction. Several structures of the LMW-PTP were solved representing different states along the catalytic pathway (Fig. 1). These structures include enzyme with no ligand bound (apo structure), enzyme with the substrate mimic HEPES bound, and enzyme with the product mimic sulfate bound. Structures were deposited in the RSCB Protein Data Bank [29]. Supplemental Table 1 shows the crystallographic X-ray data and refinement statistics.
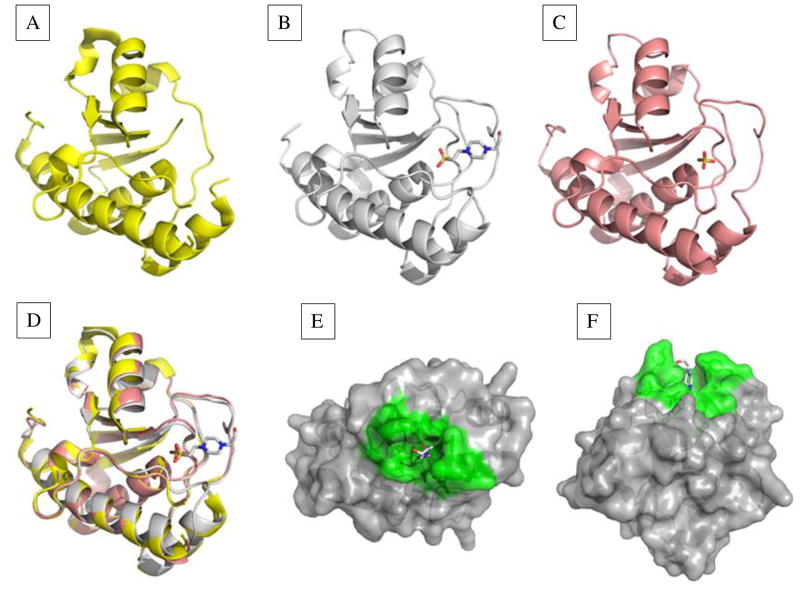
Fig. 1. Structures of the E. histolytica LMW-PTP with no bound ligand, substrate mimic bound, or product mimic bound.
Structures were solved for each given resolution. (A) The apoenzyme structure of 2.2 Å showed a disordered active site (P- loop) (PDB: 3ILY). (B) Two structures, of 2.2 Å and 1.95 Å (PDB: 3IDO; PDB: 3JS5) were obtained with substrate mimic HEPES bound. The P-loop and interacting residues were well-ordered. (C) Structure with the product mimic sulfate (PDB: 3JVI) bound in the active site (1.8 Å). This structure had a well-ordered P-loop, but additional residues on the surface of the active site were disordered. (D) A triple overlay image of structures in (A), (B), and (C) showed changes in enzyme conformation when no substrate, substrate, or product was bound. (E) side view and (F) top view of the LMW-PTP structure with HEPES bound in the active site. Active site and contributing residues are green, and other residues are gray.
The non-ligand-bound (apo) structure of the LMW-PTP showed a disordered P-loop (active site), presumably due to the absence of substrate (Fig. 1A). (PDB: 3ILY). Two nearly identical structures (PDB: 3IDO; PDB: 3JS5) were obtained from different crystallization conditions with HEPES as a substrate mimic bound in the P-loop (Fig. 1B, 1E). Here, the P-loop residues were well-ordered, as were the residues that interacted with the P-loop residues (Fig. 1B, 1E). An enzyme structure was obtained with the product mimic sulfate bound in the P-loop (PDB: 3JVI). The P-loop was well-ordered, but Y125 formed a different conformation than that observed in the substrate analog bound state, and Y126 and G127 were disordered, demonstrating mobility of the highly conserved DPYY region involved in substrate recognition and catalysis (Fig. 1C). An overlay of all three structures for comparison is depicted in Fig. 1D, demonstrating that aside from the P-loop and nearby DPYY residues the remaining enzyme structure remained constant throughout the catalytic cycle. The apo structure and the sulfate-bound structures were discontinuous, indicating more than one conformation of those regions in the enzyme. A space-filling model viewed from the top (Fig. 1E) and side (Fig. 1F) show how the E. histolytica LMW-PTP recognizes the substrate analog HEPES in the active site. The active site in PTPs forms a crevice where the substrate binds; this crevice is ~9 Å deep in all phosphotyrosine (pTyr)-specific phosphatases, allowing specificity for pTyr substrates [46, 47].
The E. histolytica LMW-PTP structure (Fig. 1) has a central twisted four-stranded β-sheet surrounded by five α-helices [48, 49]. The two β—α—β motifs in LMW-PTP structures form a Rossmann fold, with the P-loop located in the first β—α—β motif, at the C-terminus of β1 and the N-terminus of α1 [48, 49]. The P-loop is stabilized by a network of hydrogen bonding, and is the most rigid and ordered part of the enzyme when substrate, substrate analogs or inhibitors are bound [48]. The E. histolytica LMW-PTP active site residues were completely conserved, having the characteristic CLGNICR sequence (Fig. 2) [5, 11]. The DPYY loop was also conserved, containing the aspartic acid residue [19, 50] and two back-to-back tyrosines whose phosphorylation plays regulatory roles in the mammalian LMW-PTP [11, 51].
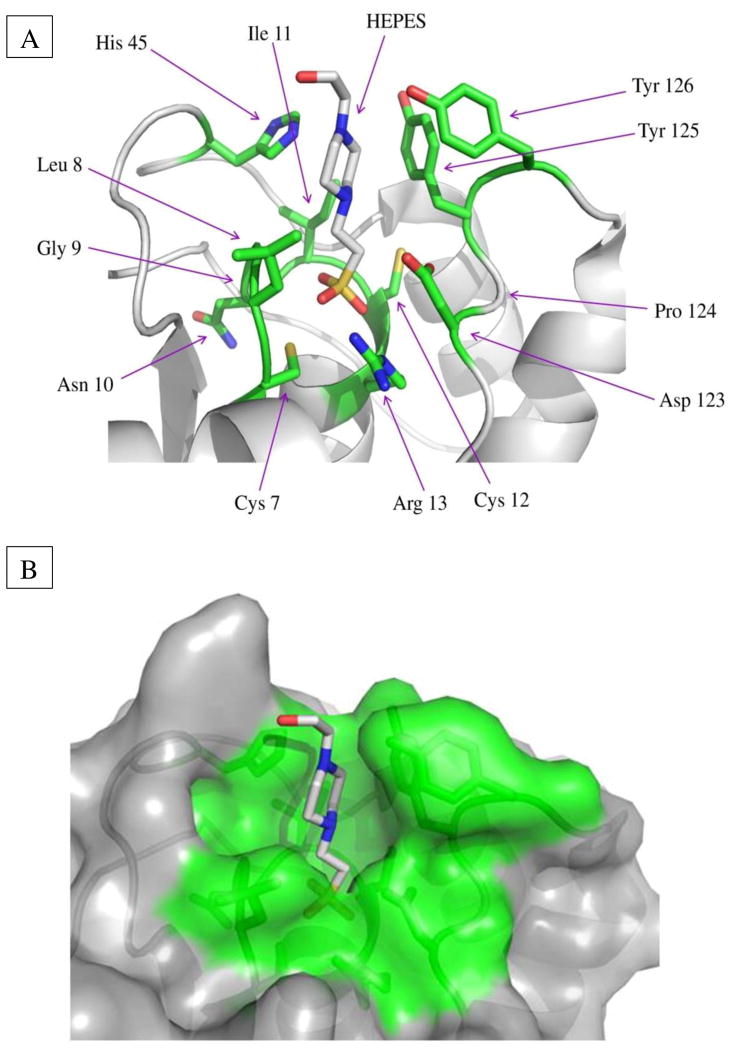
Fig. 2. Active site structure and residues of the E. histolytica LMW-PTP.
(A) In the ribbon model with the substrate HEPES bound, active site residues are green, sulfur atoms yellow, nitrogen atoms blue, and oxygen atoms red. Residues are numbered according to the E. histolytica LMW-PTP protein sequence. Arg, arginine; Asn, asparagine; Asp, aspartic acid; Cys, cysteine; Gly, glycine; His, histidine; Ile, isoleucine; Leu, leucine; Pro, proline; Tyr, tyrosine. Key residues apart from the Cys7 catalytic residue include Cys12, which can form a disulfide bond with Cys7 to prevent irreversible oxidation of Cys7 in the presence of reactive oxygen species, and Arg13, which stabilizes the transition state via hydrogen bonding. The surface loop contains the conserved Asp123, which functions as a general acid/base in catalysis, as well as Tyr125 and Tyr126 residues, which can be phosphorylated by Src family kinases in mammalian LMW-PTPs to regulate enzymatic activity. The His45 residue appears to function in substrate specificity recognition. (B) In the space-filling model, active site and contributing residues are green; residues not contributing to catalysis are gray.
The structure of the E. histolytica LMW-PTP was found to have a similar structure to other solved LMW-PTP structures. To identify LMW-PTPs with solved structures similar to the E. histolytica LMW-PTP, the structure PDB: 3JS5 (substrate mimic HEPES bound) was input into both the RCSB Protein Data Bank [29] and NCBI's VAST+ macromolecule structure database [30]. VAST+ was then used to identify how similar each selected LMW-PTP was to PDB: 3JS5. VAST+ compares three-dimensional structures of molecules by the use of geometric criteria, and thus can uncover structural homologs that are divergent in sequence. VAST+ optimally superimposes the structures, and calculates the root mean square deviation (RMSD), in angstroms (Å), between all aligned residues based on the position of equivalent C-alpha atoms of each amino acid in the sequence of the compared structures. The E. histolytica PDB: 3IDO structure is almost identical to PDB: 3JS5 (RMSD = 0.33Å), with the PDB: 3JCVI (product mimic sulfate bound), and PDB: 3ILY (apo structure) having very similar structures, as expected (RMSD = 0.64Å and 0.69Å) (Supplemental Data Set 3). The ten most similar solved LMW-PTP structures to the E. histolytica LMW-PTP outside of E. histolytica are three from Bos taurus (RMSD = 0.92Å-0.97Å), one from Thermus thermophilus (RMSD = 1.01Å), one from Homo sapiens (RMSD = 1.04Å), one from Mus musculus (RMSD = 1.06Å), three from Saccharomyces cerevisiae (RMSD = 1.09Å-1.22Å), and one from Thermoanaerobacter tengcongensis (RMSD = 1.22Å) (Supplemental Data Set 3). For comparison, a hydrogen atom is about 1 Å and a carbon atom about 2 Å in diameter [52] and a carbon-carbon bond has a length of 1.54 Å [53].
3.3. Anti-LMW-PTP rabbit immune sera immunoprecipitates native LMW-PTP
Rabbit antisera generated against the recombinant phosphatase recognized the native LMW-PTP in amebic lysate. Fig. 3 shows a representative immunoblot. A band of the expected size (17 kDa) was apparent after the immunoprecipitate was probed with immune serum. The two immune sera worked equally well in immunoprecipitation and immunoblotting. Unfortunately these immune sera did not function well in immunofluorescence staining, giving high background (data not shown).
Fig. 3. Anti-LMW-PTP rabbit sera immunoprecipitates native LMW-PTP from amebic lysate.
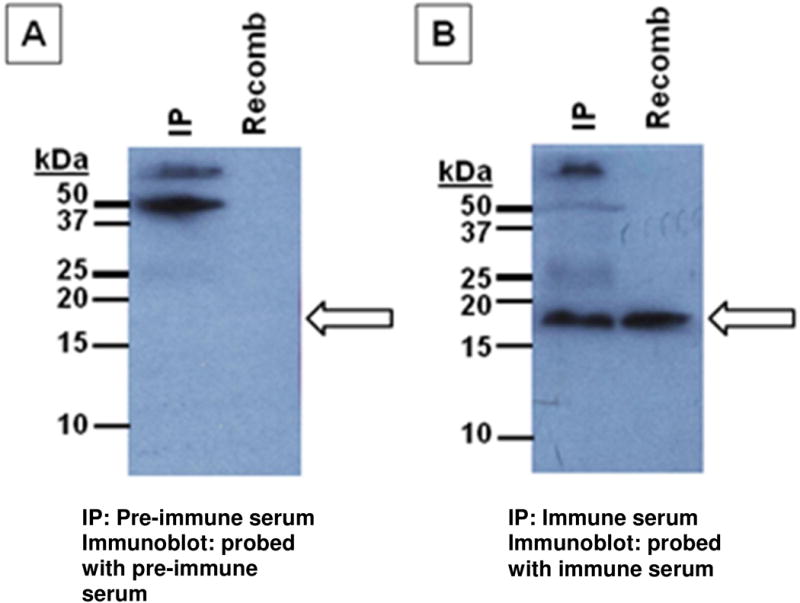
Two rabbits (UVA417 and UVA418) were immunized with wild-type recombinant LMW-PTP (Cocalico Biologicals, Inc., Reamstown, PA, USA). Lysate from 6.75x106 non-transfected trophozoites was immunoprecipitated with either pre-immune or immune rabbit serum using Dynabeads Protein A as per the manufacturer's instructions (Invitrogen/Life Technologies, Grand Island, NY, USA). Antibody (pre-immune or immune sera) was not crosslinked to the beads. Immunoblots were probed with either pre-immune or immune serum from the rabbit that was not used for immunoprecipitation of that sample. 100 ng purified recombinant wild-type LMW-PTP protein was loaded as a positive control. IP = LMW-PTP immunoprecipitated from lysate; Recomb = recombinant wild-type LMW-PTP protein. Arrow: expected size of the LMW-PTP (17 kDa). (A) Pre-immune serum from rabbit UVA417 was used to immunoprecipitate the native LMW-PTP from lysate, and the immunoblot was probed with pre-immune serum from rabbit UVA418. (B) Immune serum from rabbit UVA417 was used to immunoprecipitate the native LMW-PTP from lysate, and the immunoblot was probed with immune serum from rabbit UVA418.
3.4. The E. histolytica recombinant wild-type LMW-PTP possessed phosphatase activity, and the mutant lacked activity
The substrate-trapping mutant Cys to Ser LMW-PTP gene was generated via PCR to convert the catalytic cysteine to a serine in the expressed protein. Recombinant wild-type and mutant LMW-PTP proteins were tested for phosphatase activity using a colorimetric protein phosphatase assay kit. The wild-type recombinant LMW-PTP possessed phosphatase activity, and it was strongly inhibited by the addition of a commercial phosphatase inhibitor cocktail (Fig 4A). The phosphatase activity of recombinant mutant LMW-PTP protein was tested using 100 ng of mutant LMW-PTP protein, compared with 100 ng wild-type LMW-PTP protein as a positive control for phosphatase activity. The mutant recombinant LMW-PTP lacked phosphatase activity, as expected (Fig. 4B).
Fig. 4. Assays for phosphatase activity of recombinant wild-type and mutant LMW-PTP.
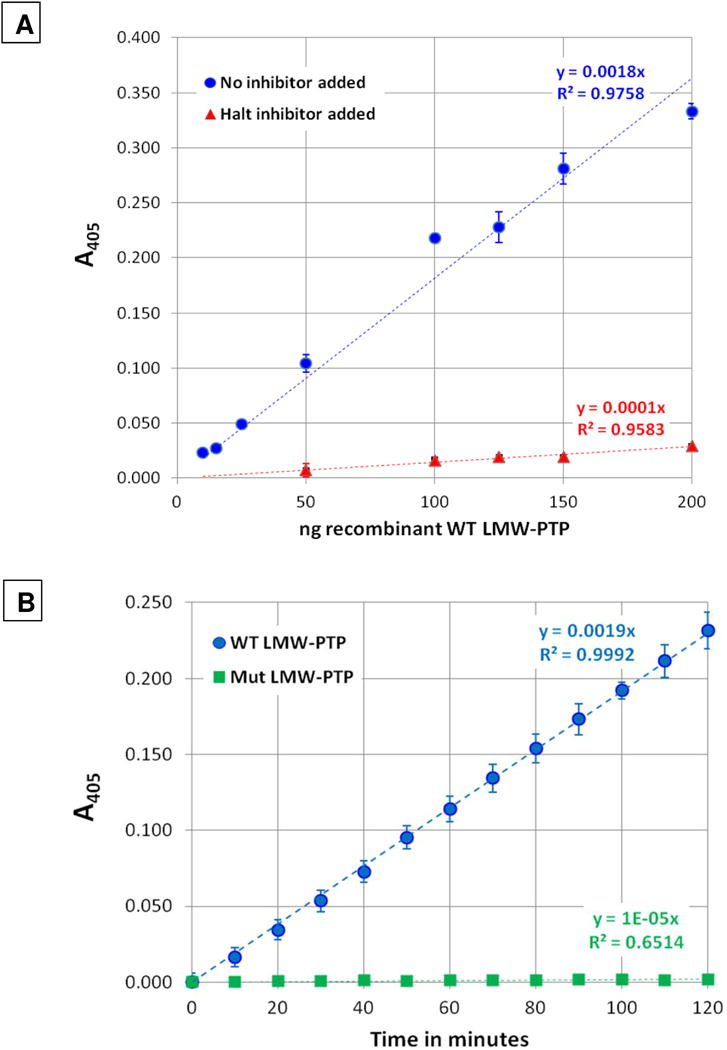
Recombinant wild-type and substrate-trapping mutant proteins were assayed using the SensoLyte pNPP Colorimetric Protein Phosphatase Assay Kit (AnaSpec, Fremont, CA, USA). (A). Phosphatase activity of recombinant wild-type LMW-PTP with or without phosphatase inhibitor added. 10 to 200 ng of recombinant wild-type LMW-PTP was assayed at the 60 minute time point. To test inhibition, HALT™ Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA) was added at 1X to reactions with 50 to 200 ng wild-type LMW-PTP. Values were plotted as ng of recombinant wild-type LMW-PTP protein vs absorbance at 405 nm using Microsoft Excel. The trendline slopes and the R2 values are also shown. Wild-type LMW-PTP phosphatase activity was inhibited by an average of 92.5 ± 1.0 % (± SD) for each sample. (B). Comparison of wild-type and mutant LMW-PTP phosphatase activity. 100 ng of either recombinant wild-type or substrate-trapping mutant LMW-PTP protein was assayed for 120 minutes, with A405 measured every five minutes. Data was plotted as A405 vs time (minutes) using Microsoft Excel. The slopes of the trendlines and the R2 values are also shown.
3.5. Substrate trapping of interacting proteins (putative substrates) using recombinant mutant LMW-PTP protein coupled to beads
To trap putative LMW-PTP substrates, pulldowns were performed with recombinant Cys to Ser mutant LMW-PTP protein or BSA protein coupled to Affi-Gel 15 beads or mock protein-coupled beads as negative controls. Beads were incubated with lysate from nontransfected HM1:IMSS strain amebae pre-treated with pervanadate to inhibit cellular PTPs and increase available tyrosine-phosphorylated substrates. Samples were subjected to SDS-PAGE, and gels were silver-stained to assay for unique bands found only in the mutant LMW-PTP protein-coupled samples incubated with lysate but not in the control samples.
Two unique bands of ~37 kDa and ~45 kDa were present in the silver-stained gels in mutant LMW-PTP + lysate samples (Fig. 5). As these bands were unique, it appeared that putative substrate (interacting proteins) had been recovered. Gels were submitted for mass spectrometry analysis of the two bands. 83 proteins total were identified in the ~37 and ~45 kDa bands from their tryptic peptides: 42 proteins were found in the ~37 kDa band, 49 total proteins were found in the ~45 kDa band, and eight proteins were present in both bands, including the LMW-PTP itself. No peptides with tyrosine phosphorylation were found. All proteins identified from peptides in both the ~37 kDa and ~45 kDa band are listed in Supplemental Data Set 4, as are their tryptic peptides.
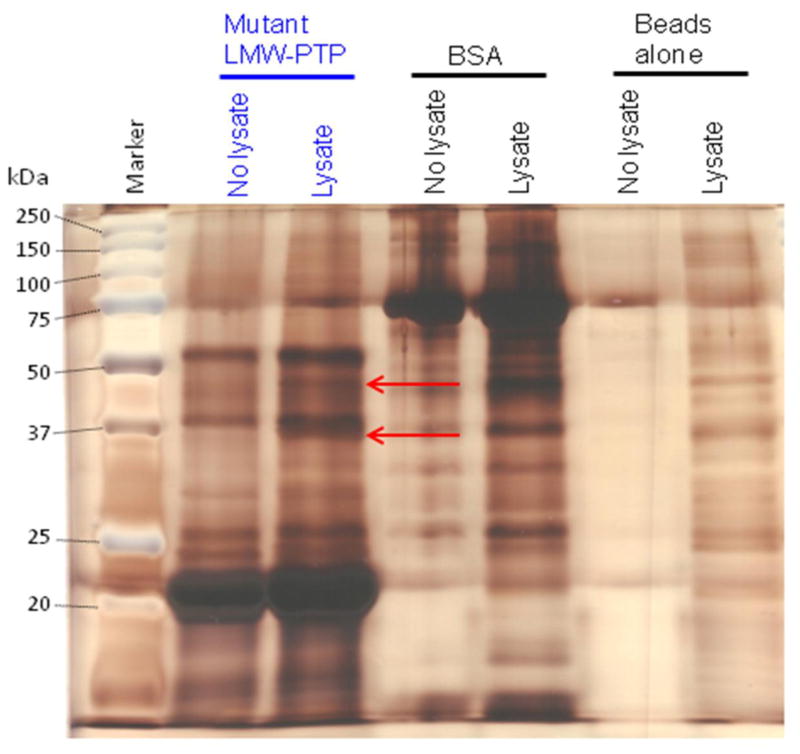
Fig. 5. A representative silver-stained gel of pulldowns using substrate-trapping mutant LMW-PTP or BSA protein coupled to Affi-Gel 15 beads and submitted for tryptic digest and mass spectrometry analysis.
Recombinant Cys to Ser mutant LMW-PTP or BSA were coupled to Affi-Gel 15 beads, or beads alone (mock-attached) were incubated with buffer only. Beads were incubated with lysate from HM1:IMSS amebae pre-treated with pervanadate (Lysate) or with lysis buffer alone (No lysate). Samples were subjected to SDS-PAGE and silver-stained. There are two unique bands (red arrows) of ~37 and ~45 kDa in the Affi-Gel 15 beads with coupled mutant LMW-PTP incubated with amebic lysate. These bands were submitted for tryptic digest and mass spectrometry analysis. Some of the mutant LMW-PTP was released from the Affi-Gel 15 beads when samples were boiled before loading, and ran at ~22 kDa rather than at 17 kDa. This is likely due to the large amount of protein that had been attached to the Affi-Gel 15 beads. The BSA also ran at a larger apparent size, about ~75 kDa, rather than its expected ~66 kDa for likely the same reason.
3.6. Putative substrate proteins of the LMW-PTP selected for further examination
Seven of the putative substrate proteins were selected for further examination based on: (i) the number of unique peptides identified (Table 1); (ii) the correct size for being present in that band (± 5 kDa) (Table 1); (iii) multiple peptide coverage (Supplemental Fig. 5); and (iv) possessing potential sites of phosphorylation (Supplemental Fig. 6). Protein motifs, domains, function, and homology information were obtained using databases including the Protein Database at the NCBI website [54] for these selected proteins. The selected proteins were: hypothetical protein 328.t00002, a type A flavoprotein [55, 56], a putative serine/threonine protein kinase [57], conserved hypothetical protein gi56465028 [58], a putative Arp2/3 complex 34 kDa subunit [59], hypothetical protein 503.t00001 [60, 61], and a putative ribose-phosphate pyrophosphokinase [62, 63] (Table 1).
Table 1. Putative substrate proteins identified by mass spectrometry selected for cloning and epitope-tagging, overexpression, and further characterization.
Candidates from the ~37 kDa and ~45 kDa bands were chosen if they were the correct size (± 5 kDa), found exclusively in one band, had a number of unique peptides identified, good peptide coverage of the protein (Supplemental Fig. 5), and were predicted to be phosphorylated (Supplemental Fig. 6). Protein motifs, domains, functions, or homologies are indicated, and were obtained using the NCBI Protein Database at www.ncbi.nlm.nih.gov/protein/. The first column indicates the order in which the protein appeared in the mass spectrometry analysis (lower numbers = larger number of peptides identified). Supplemental Fig. 4 shows peptide locations and coverage. Supplemental Data Set 4 shows all proteins identified by mass spectrometry analysis in the substrate-trapping mutant LMW-PTP pulldown.
| # | Identified protein | GenBank accession number(s) | Molecular mass | Location (37 or 45 kDa band) | Number of unique peptides | Percent coverage of protein | Gene size (bp) | Motifs, domains, function, or protein homology |
|---|---|---|---|---|---|---|---|---|
| 5 | Hypothetical protein 328.t00002 | XM_644144, XP_649236 | 34 kDa | 37 kDa band | 12 | 36% | 915 | No conserved domains, matches only proteins in E. histolytica and the related ameba E. dispar. is found in the phagosome and appears to have a role in the regulation of erythrophagocytosis. |
| 6 | Type A flavoprotein | XM_651854, XP_656946 | 46 kDa | 45 kDa band | 15 | 44% | 1221 | Contains a lactamase B domain and a flavodoxin domain. May be involved in detoxifying oxygen and/or nitric oxide. |
| 9 | Ser/Thr protein kinase, putative | XM_645070, XP_650162 | 40 kDa | 45 kDa band | 9 | 31% | 1065 | Is ~50% identical to mammalian STK3/MST2, STK4/MST1, and yeast STE20. MST1 acts as a MAP kinase kinase kinase kinase. STK3/MST2 and STK4/MST1 are involved in caspase activation, apoptosis, and cell proliferation and differentiation. |
| 10 | Conserved hypothetical protein gi56465028 | XM_001914300, XP_001914335 | 43 kDa | 45 kDa band | 7 | 21% | 1041 | Belongs to the TLD superfamily (pfam 07534) containing an enzymatic domain, but matches only E. histolytica and E. dispar proteins. |
| 11 | ARP2/3 complex 34 kDa subunit, putative | DS571335, EAL49360 | 34 kDa | 37 kDa band | 7 | 27% | 882 | In mammalian cells, this protein is involved in control of actin polymerization (nucleates new actin strands). Involved in motility and formation of phagocytic cups. |
| 12 | Hypothetical protein 503.t00001 | XM_650927, XP_656019 | 43 kDa | 45 kDa band | 7 | 19% | 1080 | Contains a BAR and SH3 domain. BAR domain = involved in dimerization and lipid binding; SH3 domain = binds to proline-rich ligands, expecially PXXP-containing; involved in intramolecular interactions. |
| 18 | Ribose-phosphate pyrophospho-kinase, putative | XM_651049, XP_656141 | 37 kDa | 37 kDa band | 4 | 13% | 999 | Catalyzes the synthesis of phosphoribosyl-pyrophosphate, an intermediate in histidine and tryptophan biosynthesis, and in the de novo biosynthesis of purines and pyrimidines; essential for nucleotide synthesis. |
3.7. Overexpression in amebae of selected putative substrate proteins with added epitope tags
Oligo pairs were used to clone the selected putative substrate protein genes via PCR and to generate epitope tags (Supplemental Data Set 1). Proteins were overexpressed in amebae from pGIR209 (Supplemental Fig. 1). As a control for transfection, expression, and protein interaction, the calcium-responsive transcription factor URE3-BP [64] was cloned with an epitope tag and overexpressed. Lysates from N- or C-terminal epitope-tagged protein overexpressers of the same putative substrate protein were pooled for incubation with Affi-Gel 15 beads for pulldown experiments, and lysates were immunoblotted to verify expression of epitope-tagged protein immediately before incubation with sample beads. A representative immunoblot is shown in Fig. 6A.
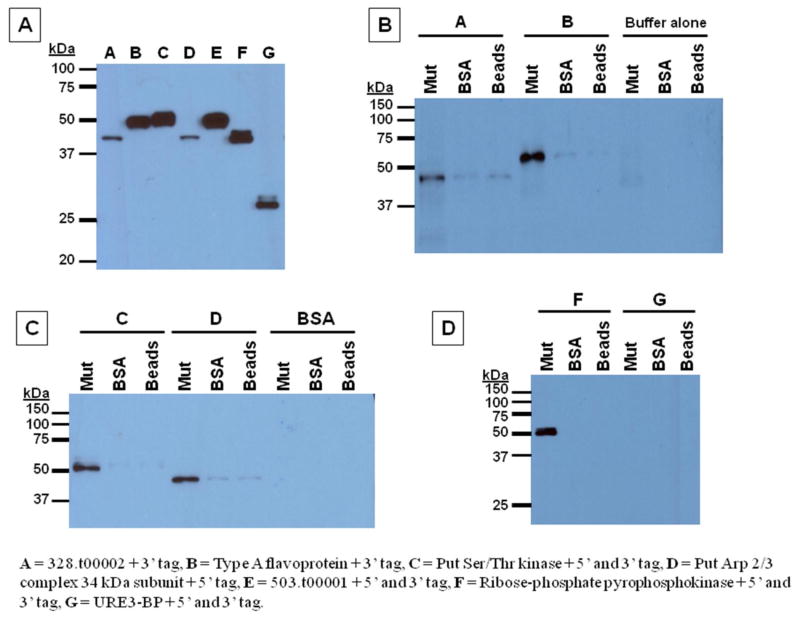
Fig. 6. Representative immunoblots of pulldowns of epitope-tagged putative substrate proteins using recombinant mutant LMW-PTP or BSA coupled to Affi-Gel 15 beads.
Equal amounts of mutant LMW-PTP or BSA protein, or buffer alone (mock-attached) were coupled to beads and incubated with lysate from pervanadate-treated transfected amebae overexpressing epitope-tagged putative substrate proteins or control URE3-BP protein, with BSA, or with lysis buffer only. Samples were subjected to SDS-PAGE, and immunoblots probed with S tag antibody. Overexpresser lysates: A = 328.t00002 + C-term tag, B = Type A flavoprotein + C-term tag, C = Putative Ser/Thr kinase + N- and C-term tag, D = Putative Arp 2/3 complex 34 kDa subunit + N-term tag, E = 503.t00001 + N- and C-term tag, F = Ribose-phosphate pyrophosphokinase + N- and C-term tag, G = URE3-BP + N- and C-term tag. (A). Sample lysates alone (5 μg/lane) were immunoblotted to test for epitope-tagged protein expression. (B), (C), (D). Substrate-trapping mutant LMW-PTP (Mut), BSA (BSA), or mock-attached (Beads) Affi-Gel 15 beads were incubated as indicated with lysate from putative substrate overexpressers, control URE3-BP overexpressers, BSA in lysis buffer, or lysis buffer alone.
3.8. Confirmation of LMW-PTP-putative substrate protein interaction via pulldowns of epitope-tagged putative substrate proteins
Mutant LMW-PTP or BSA coupled to Affi-Gel 15 beads or mock-coupled beads were incubated with either lysate from epitope-tagged putative substrate protein overexpresser transfectants, BSA in lysis buffer, or with lysis buffer alone. Samples were subjected to SDS-PAGE, and immunoblots were probed with antibody against the S epitope tag. Five proteins bound to mutant Cys to Ser substrate-trapping LMW-PTP coupled to beads, but not to BSA coupled to beads or to mock-coupled beads. These proteins are the hypothetical protein 328.t00002, the type A flavoprotein, the putative Ser/Thr kinase, the putative Arp 2/3 complex 34 kDa subunit, and the ribose-phosphate pyrophosphokinase (Fig. 6). The epitope-tagged control URE3-BP protein [64] did not bind to the coupled mutant LMW-PTP, to the BSA, or the Affi-Gel 15 beads alone (Fig. 6).
4. Discussion
There are many important similarities between the E. histolytica LMW-PTP and other LMW-PTPs. The apoenzyme, substrate mimic HEPES bound, and product mimic sulfate bound structures have been solved for the E. histolytica LMW-PTP, allowing for comparison to those of other LMW-PTPs. The E. histolytica LMW-PTP has the same overall structure as LMW-PTPs from other organisms; all have a central twisted four-stranded β-sheet surrounded by five α-helices. The E. histolytica LMW-PTP protein sequence has all the features of known LMW-PTPs: a conserved active site with a catalytic cysteine and stabilizing arginine, a second cysteine in the active site which can form a disulfide bond with the catalytic cysteine to prevent irreversible oxidation, and the conserved DPYY loop containing the general acid/base asparagine and the two tyrosines that can be phosphorylated to regulate phosphatase activity or allow binding of adapter proteins. The E. histolytica LMW-PTP protein sequence is most similar to selected plant or bacterial LMW-PTPs, with ~50% sequence identity, and is the least similar to those from other unicellular eukaryotes or multicellular animals (33%-40% identity). The structure of the E. histolytica LMW-PTP is also very similar to some of the other solved LMW-PTP structures when their protein structures are superimposed, and of these other structures, it is most similar to that of Bos taurus (Supplemental Data Set 3). It should be noted that there are many more LMW-PTP protein sequences available than there are solved structures; for example, searching the RSCB Protein Data Bank [29] using the Pfam Accession number PF01451 (LMW-PTPs) yields a total of 44 hits, including ten for Staphylococcus aureus, eight for Bacillus subtilis, seven for Bos taurus, four for Entamoeba histolytica (which are described herein), three for Saccharomyces cerevisiae, three for Mycobacterium tuberculosis, three for Synechocystis sp. PCC 6803, three for Homo sapiens, and one for Mus musculus; 34 bacterial structures, 19 eukaryotic structures, and one from Archaea have been deposited.
There are important differences between the E. histolytica LMW-PTPs and the better-characterized mammalian LMW-PTPs, which may indicate dissimilar substrate specificity, function, and regulation. For example, the His45 residue (Fig. 2) in the E. histolytica LMW-PTP is conserved in all the compared plant, bacterial, and unicellular eukaryotic LMW-PTPs (except for Giardia lamblia and Giardia intestinalis), but is lacking in LMW-PTPs from multicellular organisms except for Drosophila melanogaster (Supplemental Fig. 2). This His residue is in a variable loop forming a wall of the active site pocket [50]. These loop residues affect the pattern of the charge distribution and conformation of the active site, and thus affect substrate specificity [65]. Therefore, the LMW-PTP or its substrates may be dissimilar between mammalian and non-mammalian LMW-PTPs, and could be targeted for drug development.
Another difference is that mammalian LMW-PTPs are known to be phosphorylated by cytoplasmic Src family protein kinases Src, Lck, and Fyn at residues Tyr131 and Tyr132 in the DPYY loop [40, 41, 51]. Tyr131 phosphorylation by Src induces a 25-fold increase in LMW-PTP activity, while Tyr132 phosphorylation recruits Grb2 adapter protein to bind via its SH2 domain [11, 40, 51]. Tyr132 is close to the active site, so binding of Grb2 could limit substrate access, leading to selection of substrates by size, or even functional enzyme inactivation, if the active site is occluded [40, 51]. Lck and Fyn phosphorylate the LMW-PTP to a lesser extent than Src [11, 41, 51]. These tyrosines are completely conserved among all compared LMW-PTPs except in Saccharomyces cerevisiae (Supplemental Fig. 2), so they appear to be generally functionally necessary to regulate LMW-PTP activity via their phosphorylation/dephosphorylation. However, Src family analogs in E. histolytica contain transmembrane regions and are thus unlikely to be functionally analogous [66], so phosphorylation of DPYY loop tyrosines in the E. histolytica LMW-PTP is likely performed by a different, not-yet-identified kinase. This is the case in Mycobacterium tuberculosis, where a novel protein tyrosine kinase, the Rv2232 gene product PtkA, phosphorylates its LMW-PTP at these residues [67, 68].
A third difference is that mammalian LMW-PTPs have the sequence GND following the Tyr132 residue in the DPYY loop, conforming to the XNX consensus motif for pTyr recognition by the SH2 domain of Grb2 [11, 69]. The corresponding sequence in E. histolytica is GGE. This sequence, GGX, is conserved among all the selected compared plant and bacterial LMW-PTPs except for that of Parabacteroides merdae (Supplemental Fig. 2). Therefore, non-mammalian LMW-PTPs likely do not interact with Grb2. However, there may be a different adapter protein that recognizes this GGX consensus motif in the context of a pTyr if these LMW-PTPs are regulated in the same manner as mammalian LMW-PTPs. The kinases and adapter proteins that regulate LMW-PTP function may be quite divergent between the mammalian LMW-PTPs and those of Entamoeba, plants, and bacteria.
A recombinant substrate-trapping mutant LMW-PTP was used to trap putative LMW-PTP substrates, which were identified by mass spectrometry analysis of their tryptic peptides. Five of the seven top hits from the mass spectrometry analysis were overexpressed in amebae with an epitope tag, were found to specifically interact with the mutant recombinant LMW-PTP, and thus appeared to be actual substrates of the LMW-PTP. These five proteins have functions which likely play important roles in E. histolytica virulence: one in erythrophagocytosis, one in motility and erythrophagocytosis, two in metabolism, and one in signal transduction. The hypothetical protein 328.t00002 [70, 71] and the Arp2/3 complex 34 kDa subunit [70] have been found in the early phagosome during erythrophagocytosis. Amebic phagocytic ability is highly correlated with virulence [70, 72]: the presence of trophozoites with ingested red blood cells in dysenteric stool is strongly correlated with E. histolytica infection and invasive disease [1], The Arp2/3 complex is also required to nucleate actin to allow cellular motility [73, 74]. Motility appears to be necessary for virulence in E. histolytica, as only motile trophozoites can cause disease in the hamster liver abscess model [75]. Two of these proteins appear to play a metabolic role. The type A flavoprotein is thought to detoxify nitric oxide and oxygen [76, 77], which could allow E. histolytica to better survive exposure to reactive oxygen (ROS) and nitrogen species (RNS) produced by a host's immune system [76, 78]. The type A flavoprotein could conceivably also help maintain or recover the activity of the LMW-PTP itself, as LMW-PTPs can be inactivated by nitric oxide [79]. The ribose-phosphate pyrophosphokinase synthesizes phosphoribosylpyrophosphate, which is essential for the de novo biosynthesis of purines and pyrimidines, and is an intermediate in histidine and tryptophan biosynthesis [62], so thus feeds into both DNA/RNA and protein synthesis pathways. The putative serine/threonine protein kinase likely functions in signal transduction as a MAP kinase kinase kinase kinase, based on its similarity to mammalian STK4/MST1 and STK3/MST2, and may play a role in cell proliferation, differentiation, caspase activation, and/or apoptosis [57, 80].
The mass spectrometry analysis of the E. histolytica LMW-PTP putative substrate proteins did not include any phosphorylated peptides. Assuming the substrate-trapping mutant is trapping phosphorylated interacting proteins, the phosphate group of the phosphotyrosine is relatively labile, and during fragmentation of sample during mass spectrometry it can often be released from its peptide and is thus lost [81, 82]. E. histolytica LMW-PTP putative substrates identified by mass spectrometry analysis did not include receptor kinases. Many mammalian LMW-PTP substrates are receptor protein tyrosine kinases such as the PDGF receptor [11, 21], insulin receptor [11, 20], fibroblast growth factor receptor [11, 16], ephrin A2 receptor [11, 83] and ephrin B1 receptor [11, 19, 84]. E. histolytica has a large family of transmembrane kinases which appear to be receptor protein kinases [78], but their ligands or regulatory kinases and phosphatases have not yet been identified [85]. These may be regulated by a different phosphatase and not be LMW-PTP substrates. As the E. histolytica LMW-PTP is more closely related to bacterial and plant LMW-PTPs, it may be more functionally similar to those than to the better-characterized mammalian LMW-PTPs. Far fewer LMW-PTP substrates have been identified in non-mammalian cells, and none as of yet have been receptor kinases [86]. The study of many non-mammalian LMW-PTPs is still at the phosphatase-activity testing stage with pNPP or other substrate analogs by using wild-type and substrate-trapping mutants, for example in Staphylococcus aureus [87], Mycobacterium tuberculosis [50], and Campylobacter jejuni [88]. Alternately, a possible reason why no receptor kinases were identified as putative substrates could be that they were insufficiently solubilized, as integral membrane proteins may require harsher than standard conditions to solubilize [89].
There are still a number of experiments that could be performed in order to answer questions still remaining and to lay the groundwork for future studies on this LMW-PTP and its substrates. To address if phosphorylation of the DPYY loop tyrosines of the LMW-PTP affects its activity, for example, mutant forms of the LMW-PTP with substituted non-phosphorylatable residues at those positions could be expressed in vivo or in vitro and then assayed for phosphorylation (by immunoprecipitation with anti-phosphotyrosine antibody) and phosphatase activity; in vitro testing of these mutants alongside the recombinant wild-type LMW-PTP for phosphatase activity on pNPP after phosphorylation with Src kinase could be performed. Other follow-up experiments are to show that these interacting proteins identified using the substrate-trapping mutant are actual rather than putative substrates, by assaying the LMW-PTP for its ability to dephosphorylate the phosphorylated forms of the interacting putative substrate proteins (either in vivo or in vitro), or if it can dephosphorylate tyrosine-phosphorylated peptides containing the predicted tyrosine phosphorylation sites from these putative substrate proteins. Future work could also focus on how identified substrates are themselves regulated by phosphorylation, and how this affects the phenotype of E. histolytica. Since there are two E. histolytica LMW-PTP genes which are both expressed [17, 18] and code for proteins that differ by a single residue, these could be individually cloned and tagged, perhaps expressed by the use of an inducible vector, and assayed for their subcellular location via immunofluorescence staining. There may be differences in their roles, functions, or regulation or represent different pools targeting certain cellular functions, such as the separate cytoplasmic or cytoskeletal-associated pools with distinct substrates found for the human LMW-PTP [90].
In conclusion, we have partially characterized the E. histolytica LMW-PTP, annotated as a putative phosphatase, which has never been previously studied. Its active site and regulatory regions are completely conserved as compared with LMW-PTPs from other organisms, and it possesses phosphatase activity. The structures for this LMW-PTP without bound substrate, with the substrate mimic HEPES bound, and the product mimic sulfate bound have been solved, and the three-dimensional structures are conserved when compared with other solved LMW-PTP structures. The conformational changes the LMW-PTP structure undergoes in its reaction cycle can be used as a starting point for drug design, as the E. histolytica LMW-PTP appears to mediate activity of a number of virulence-related proteins. Five of the seven putative substrate proteins that interacted specifically with the mutant LMW-PTP appeared to play important roles in E. histolytica virulence: one in erythrophagocytosis, one in both motility and erythrophagocytosis, two in metabolism, and one in signal transduction. This is a promising beginning in the study of this LMW-PTP, and there remains much to be uncovered in how it regulates virulence-associated proteins in this organism, and how it itself is regulated.
Supplementary Material
Highlights.
Structures for the E. histolytica putative LMW-PTP have been solved.
The structures show the conformational changes necessary for binding of substrate.
This putative LMW-PTP has phosphatase activity on a generic phosphatase substrate.
A substrate-trapping mutant LMW-PTP was used to capture protein substrates.
Five proteins were demonstrated to interact specifically with the mutant LMW-PTP.
Acknowledgments
We would like to thank the SSGCID protein crystallization team for their work with the wild-type LMW-PTP recombinant protein expression and crystallization and the mutant LMW-PTP recombinant protein expression. We thank Carol Gilchrist for helpful advice and discussions, and Ellyn Moore, Christina Bousquet, Carrie Cowardin, and Debra Fisher for technical support.
Funding Sources: This work was supported by NIH grant AI 26649 to William A. Petri, Jr. This project was also funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract Nos.: HHSN272200700057C and HHSN272201200025C. The funding sources had no involvement in the study design, analysis and interpretation of data, in the writing of the article, or in the decision to submit the article for publication.
Footnotes
Conflict of Interest Statement: The authors of this paper declare there are no conflicts of interest or competing interests with respect to funding or any other issues.
Appendix B. Supplemental Data Sets: Supplemental Data Sets 1-4 (Excel Tables)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.World Health Organization. Amoebiasis. Weekly Epidemiol Record. 1997;72(14):97–100. No authors listed. [Google Scholar]
- 2.Petri WA, Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol. 2002;56:39–64. doi: 10.1146/annurev.micro.56.012302.160959. [DOI] [PubMed] [Google Scholar]
- 3.Barwick RS, Uzicanin A, Lareau S, Malakmadze N, Imnadze P, Iosava M, et al. Outbreak of amebiasis in Tbilisi, Republic of Georgia, 1998. Amer J Trop Med Hyg. 2002;67(6):623–31. doi: 10.4269/ajtmh.2002.67.623. [DOI] [PubMed] [Google Scholar]
- 4.Amebic dysentery in Chicago. Am J Public Health Nations Health. 1934;24(7):756–8. doi: 10.2105/ajph.24.7.756. No authors listed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramponi G, Stefani M. Structure and function of the low Mr phosphotyrosine phosphatases. Biochim Biophys Acta. 1997;1341:137–56. doi: 10.1016/s0167-4838(97)00087-3. [DOI] [PubMed] [Google Scholar]
- 6.Mustelin T. A brief introduction to the protein phosphatase families. Methods Mol Biol. 2007;365:9–22. doi: 10.1385/1-59745-267-X:9. [DOI] [PubMed] [Google Scholar]
- 7.Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146(2):351–67. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DT, Read R, Rusconi J, Cagan RL. The Drosophila primo locus encodes two low-molecular-weight tyrosine phosphatases. Gene. 2000;243(1-2):1–9. doi: 10.1016/s0378-1119(99)00553-3. [DOI] [PubMed] [Google Scholar]
- 9.Modesti A, Marzocchini R, Raugei G, Chiti F, Sereni A, Magherini F, et al. Cloning, expression and characterisation of a new human low Mr phosphotyrosine protein phosphatase originating by alternative splicing. FEBS Lett. 1998;431(1):111–5. doi: 10.1016/s0014-5793(98)00732-7. [DOI] [PubMed] [Google Scholar]
- 10.Malentacchi F, Marzocchini R, Gelmini S, Orlando C, Serio M, Ramponi G, et al. Up-regulated expression of low molecular weight protein tyrosine phosphatases in different human cancers. Biochem Biophys Res Commun. 2005;334(3):875–83. doi: 10.1016/j.bbrc.2005.06.176. [DOI] [PubMed] [Google Scholar]
- 11.Raugei G, Ramponi G, Chiarugi P. Low molecular weight protein tyrosine phosphatases: small but smart. Cell Mol Life Sci. 2002;59:941–9. doi: 10.1007/s00018-002-8481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang ZY. Mechanistic studies on protein tyrosine phosphatases. Prog Nucleic Acid Res Mol Biol. 2003;73:171–220. doi: 10.1016/s0079-6603(03)01006-7. [DOI] [PubMed] [Google Scholar]
- 13.Davis JP, Zhou MM, Van Etten RL. Kinetic and site-directed mutagenesis studies of the cysteine residues of bovine low molecular weight phosphotyrosyl protein phosphatase. J Biol Chem. 1994;269(12):8734–40. [PubMed] [Google Scholar]
- 14.http://amoebadb.org/amoeba/. AmoebaDB: Amoeba Genomics Resource. A EuPathDB project. Source publication: Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res 2010; 38(Database issue): D415-9.
- 15.Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Caler EV, Fischer S, et al. AmoebaDB and MicrosporidiaDB: functional genomic resources for Amoebozoa and Microsporida species. Nucl Acids Res. 2011;39:D612–9. doi: 10.1093/nar/gkq1006. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9(6):1426–44. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 18.Hon CC, Weber C, Sismeiro O, Proux C, Koutero M, Deloger M, et al. Quantification of stochastic noise of splicing and polyadenylation in Entamoeba histolytica. Nucleic Acids Res. 2013;41(3):1936–52. doi: 10.1093/nar/gks1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samet JM, Tal TL. Toxicological disruption of signaling homeostasis: tyrosine phosphatases as targets. Annu Rev Pharmacol Toxicol. 2010;50:215–35. doi: 10.1146/annurev.pharmtox.010909.105841. 2010. [DOI] [PubMed] [Google Scholar]
- 20.Chiarugi P, Cirri P, Marra F, Raugei G, Camici G, Manao G, et al. LMW-PTP is a negative regulator of insulin-mediated mitotic and metabolic signaling. Biochem Biophys Res Commun. 1997;238(2):676–82. doi: 10.1006/bbrc.1997.7355. [DOI] [PubMed] [Google Scholar]
- 21.Chiarugi P, Cirri P, Raugei G, Camici G, Dolfi F, Berti A, et al. PDGF receptor as a specific in vivo target for low Mr phosphotyrosine protein phosphatase. FEBS Lett. 1995;372:49–53. doi: 10.1016/0014-5793(95)00947-8. [DOI] [PubMed] [Google Scholar]
- 22.http://blast.ncbi.nlm.nih.gov/Blast.cgi. The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. Source publication: Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10.
- 23.http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi?rm=lalign. LALIGN/PLALIGN finds internal duplications by calculating non-intersecting local alignments of protein or DNA sequences.
- 24.http://eupathdb.org/eupathdb/. EuPathDB: Eukaryotic Pathogen Database Resources. EuPathDB Bioinformatics Resource Center for Biodefense and Emerging/Re-emerging Infectious Diseases is a portal for accessing genomic-scale datasets associated with…eukaryotic pathogens. Source publication: Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res 2010; 38(Database issue):D415-9.
- 25.http://www.phylogeny.fr/. Robust phylogenetic analysis for the non-specialist…a free, simple to use web service dedicated to reconstructing and analyzing phylogenetic relationships between molecular sequences. Source publication: Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36 (Web Server Issue):W465-9.
- 26.http://phylogeny.lirmm.fr/phylo_cgi/index.cgi. Robust phylogenetic analysis for the non-specialist…a free, simple to use web service dedicated to reconstructing and analyzing phylogenetic relationships between molecular sequences. Source publication: Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36 (Web Server Issue):W465-9.
- 27.Bryan CM, Bhandari J, Napuli AJ, Leibly DJ, Choi R, Kelley A, et al. High-throughput protein production and purification at the Seattle Structural Genomics Center for Infectious Disease. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 9):1010–4. doi: 10.1107/S1744309111018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myler PJ, Stacy R, Stewart L, Staker BL, Van Voorhis WC, Varani G, et al. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) Infect Disord Drug Targets. 2009;9(5):493–506. doi: 10.2174/187152609789105687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.http://www.rcsb.org/pdb/home/home.do. RCSB PDB Protein Data Bank: an information portal to biological macromolecular structures: biological macromolecular resource. Source publication: Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig IN, et al. The Protein Data Bank. Nucleic Acids Res 2000; 28:235-42.
- 30.http://www.ncbi.nlm.nih.gov/Structure/vastplus/vastplus.cgi. NCBI; VAST+ similar structures. 3D structural similarities among biological assemblies. Source publications: Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol 1996; 6(3): 377-85;; Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucl Acids Res. 2014;42(1):D297–303. doi: 10.1093/nar/gkt1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc of Trop Med Hyg. 1978;72(4):431–2. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 32.Linford AS, Moreno H, Good KR, Zhang H, Singh U, Petri WA., Jr Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 2009;9:38. doi: 10.1186/1471-2180-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramakrishnan G, Vines RR, Mann BJ, Petri WA., Jr A tetracycline-inducible gene expression system in Entamoeba histolytica. Mol Biochem Parasitol. 1997;84(1):93–100. doi: 10.1016/s0166-6851(96)02784-3. [DOI] [PubMed] [Google Scholar]
- 34.www.bio-rad.com/LifeScience/pdf/Bulletin_1085.pdf. Chromatography Tech Note 1085: Affi-Gel and 15 activated affinity media.
- 35.Gordon JA. Use of vanadate as protein-phosphotyrosine phophatase inhibitor. Methods Enzymol. 1991;201:477–82. doi: 10.1016/0076-6879(91)01043-2. [DOI] [PubMed] [Google Scholar]
- 36.www.scienceboard.net/resources/protocols.asp?action=article&protocol_id=584. The Science Advisory Board: Protocols: Sodium orthovanadate activation.
- 37.Garton AJ, Flint AJ, Tonks NK. Identification of substrates for protein-tyrosine phosphatases. In: Hardie DG, editor. Protein phosphorylation: a practical approach. 2nd. Oxford University Press Inc.; New York NY, USA: 1999. pp. 183–98. [Google Scholar]
- 38.Blanchetot C, Chagnon M, Dubé N, Hallé M, Tremblay ML. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35(1):44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Moreno H, Linford AS, Gilchrist CA, Petri WA., Jr Phospholipid-binding protein EhC2A mediates calcium-dependent translocation of transcription factor URE3-BP to the plasma membrane of Entamoeba histolytica. Eukaryot Cell. 2010;9(5):695–704. doi: 10.1128/EC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bucciantini M, Chiarugi P, Cirri P, Taddei L, Stefani M, Raugei G, et al. The low Mr phosphotyrosine protein phosphatase behaves differently when phosphorylated at Tyr131 or Tyr132 by Src kinase. FEBS Lett. 1999;456(1):73–8. doi: 10.1016/s0014-5793(99)00828-5. [DOI] [PubMed] [Google Scholar]
- 41.Tailor P, Gilman J, Williams S, Couture C, Mustelin T. Regulation of the low molecular weight phosphotyrosine phosphatase by phosphorylation at tyrosines 131 and 132. J Biol Chem. 1997;272(9):5371–4. doi: 10.1074/jbc.272.9.5371. [DOI] [PubMed] [Google Scholar]
- 42.Tonks NK, Diltz CD, Fischer EH. Purification of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem. 1988;263(14):6722–30. [PubMed] [Google Scholar]
- 43.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B (1994) Science. 1994;263(5152):1397–404. [PubMed] [Google Scholar]
- 44.Bourdeau A, Dubé N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17(2):203–9. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Seely BL, Staubs PA, Reichart DR, Berhanu P, Milarski KL, Saltiel AR, et al. Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes. 1996;45(10):1379–85. doi: 10.2337/diab.45.10.1379. [DOI] [PubMed] [Google Scholar]
- 46.Moorhead GBG, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–9. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 47.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268(5218):1754–8. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 48.Wang S, Tabernero L, Zhang M, Harms E, Van Etten RL, Stauffacher CV. Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate. Biochemistry. 2000;39:1903–14. doi: 10.1021/bi991348d. [DOI] [PubMed] [Google Scholar]
- 49.Madhurantakam C, Rajakumara E, Mazumdar PA, Saha B, Mitra D, Wiker HG, et al. Crystal structure of low-molecular-weight protein tyrosine phosphatase from Mycobacterium tuberculosis at 1.9-Å resolution. J Bacteriol. 2005;187(6):2175–81. doi: 10.1128/JB.187.6.2175-2181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madhurantakam C, Chavali VR, Das AK. Analyzing the catalytic mechanism of MPtpA: a low molecular weight protein tyrosine phosphatase from Mycobacterium tuberculosis through site-directed mutagenesis. Proteins. 2008;71(2):706–14. doi: 10.1002/prot.21816. [DOI] [PubMed] [Google Scholar]
- 51.Souza ACS, Azoubel S, Queiroz KC, Peppelenbosch MP, Ferreira CV. From immune response to cancer: a spot on the low molecular weight protein tyrosine phosphatase. Cell Mol Life Sci. 2009;66(7):1140–53. doi: 10.1007/s00018-008-8501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.http://www.uic.edu/classes/phys/phys450/MARKO/N003.html. Basic physical scales relevant to cells and molecules; Relevant length scales.
- 53.http://www.ncbi.nlm.nih.gov/books/NBK22567/. NCBI Bookshelf Biochemistry 5th edition. Berg JM, Tymoczko JL, Stryer L. New York: W.H. Freeman & Company, 2002. Section 1.3. Chemical Bonds in Biochemistry.
- 54.www.ncbi.nlm.nih.gov/protein/. The Protein database is a collection of sequences from several sources. Source publication: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (USA); 2002. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21101/.
- 55.http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=197917. NCBI Conserved Domains. smart00849: lactamase B. Metallo-beta-lactamase superfamily. Source publication for [55] through [61]: Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013; 41(Database issue):D348-52.
- 56.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam03358&islf=1. NCBI Conserved Domains. pfam03358: FMN_red. NADPH-dependent FMN reductase.
- 57.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=cd06612. NCBI Conserved Domains. cd06612: STKc_MST1_2. Catalytic domain of the protein serine/threonine kinases, mammalian Ste20-like protein kinase 1 and 2.
- 58.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=cl02144. NCBI Conserved Domains. cl02144: TLD superfamily. TLD; this domain is predicted to be an enzyme and is often found associated with pfam01476.
- 59.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=202863. NCBI Conserved Domains. pfam04045: P34-Arc. Arp2/3 complex, 34 kD subunit p34-Arc.
- 60.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=153271. NCBI Conserved Domains. cd07307: BAR. The Bin/Amphiphysin/Rvs (BAR) domain, a dimerization module that binds membranes and detects membrane curvature.
- 61.www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=212690. NCBI Conserved Domains. cd00174: SH3. Src homology 3 domain superfamily.
- 62.www.answers.com/topic/ribose-phosphate-pyrophosphokinase. AnswersTM. Ribose-phosphate pyrosphosphokinase. Oxford Dictionary of Biochemistry. Ribose-phosphate pyrosphosphokinase.
- 63.www.uniprot.org/uniprot/P60891. UniProt>UniProtKB. P60891 (PRPS1_HUMAN) reviewed, UniProtKB/Swiss-Prot. Recommended name: ribose-phosphate pyrophosphokinase 1.
- 64.Gilchrist CA, Moore ES, Zhang Y, Bousquet CB, Lannigan JA, Mann BJ, et al. Regulation of virulence of Entamoeba histolytica by the URE3-BP transcription factor. mBio. 2010;1(1):e00057–10. doi: 10.1128/mBio.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H, Xia B, Jin C. Solution structure of a low-molecular-weight protein tyrosine phosphatase from Bacillus subtilis. J Bacteriol. 2006;188(4):1509–17. doi: 10.1128/JB.188.4.1509-1517.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anamika K, Bhattacharya A, Srinivasan N. Analysis of the protein kinome of Entamoeba histolytica. Proteins. 2008;71:995–1006. doi: 10.1002/prot.21790. [DOI] [PubMed] [Google Scholar]
- 67.Stehle T, Sreeramulu S, Löhr F, Richter C, Saxena K, Jonker HRA, et al. The apo-structure of the low molecular weight protein-tyrosine phosphatase A (MptpA) from Mycobacterium tuberculosis allows for better target-specific drug development. J Biol Chem. 2012;287(41):34569–82. doi: 10.1074/jbc.M112.399261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bach H, Wong D, Av-Gay Y. Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem J. 2009;420:155–60. doi: 10.1042/BJ20090478. [DOI] [PubMed] [Google Scholar]
- 69.Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14(4):277–85. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, Sherman NE, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 2008;4(1):e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.King AV, Welter BH, Koushik AB, Gordon LN, Temesvari LA. A genome-wide over-expression screen identifies genes involved in phagocytosis in the human protozoan parasite, Entamoeba histolytica. PLoS ONE. 2012;7(8):e43025. doi: 10.1371/journal.pone.0043025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huston CD, Boettner DR, Miller-Sims V, Petri WA., Jr Apoptotic killing and phagocytosis by the parasite Entamoeba histolytica. Infect Immun. 2003;71(2):964–72. doi: 10.1128/IAI.71.2.964-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LeClaire LL, 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp 2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182(4):647–54. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 75.Coudrier E, Amblard F, Zimmer C, Roux P, Olivo-Marin JC, Rigothier MC, et al. Myosin II and the Gal-GalNAc lectin play a crucial role in tissue invasion by Entamoeba histolytica. Cell Microbiol. 2005;7(1):19–27. doi: 10.1111/j.1462-5822.2004.00426.x. [DOI] [PubMed] [Google Scholar]
- 76.Loftus B, Anderson I, Davies R, Alsmark UCM, Samuelson J, Amedeo P, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–8. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 77.Andersson JO, Hirt RP, Foster PG, Roger AJ. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol Biol. 2006;6:27. doi: 10.1186/1471-2148-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beck DL, Boettner DR, Dragulev B, Ready K, Nozaki T, Petri WA., Jr Identification and gene expression analysis of a large family of transmembrane kinases related to the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2005;4(4):722–32. doi: 10.1128/EC.4.4.722-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caselli A, Chiarugi P, Camici G, Manao G, Ramponi G. In vivo inactivation of phosphotyrosine protein phosphatases by nitric oxide. FEBS Lett. 1995;374:249–52. doi: 10.1016/0014-5793(95)01120-4. [DOI] [PubMed] [Google Scholar]
- 80.Radu M, Chernoff J. The DeMSTification of Mammalian Ste20 Kinases. Curr Biol. 2009;19(10):R421–5. doi: 10.1016/j.cub.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 81.Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell. 2013;24(5):535–42. doi: 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu HY, Liao PC. Analysis of protein phosphorylation using mass spectrometry. Chang Gung Med J. 2008;31(3):217–27. [PubMed] [Google Scholar]
- 83.Kikawa KD, Vidale DR, Van Etten RL, Kinch MS. Regulation of the EphA2 kinase by the low molecular weight tyrosine phosphatase induces transformation. J Biol Chem. 2002;277:39274–9. doi: 10.1074/jbc.M207127200. [DOI] [PubMed] [Google Scholar]
- 84.Stein E, Lane AA, Cerretti DP, Schoecklmann HO, Schroff AD, Van Etten RL, et al. Eph receptors discriminate specific ligand oligomers to determine alternative signaling. Genes Dev. 1998;12(5):667–78. doi: 10.1101/gad.12.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buss SN, Hamano S, Vidrich A, Evans C, Zhang Y, Crasta OR, et al. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int J Parasitol. 2010;40(7):833–43. doi: 10.1016/j.ijpara.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mukhopadhyay A, Kennelly PJ. A low molecular weight protein tyrosine phosphatase from Synechocystis sp strain PCC 6803: enzymatic characterization and identification of its potential substrates. J Biochem. 2011;149(5):551–62. doi: 10.1093/jb/mvr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherjee S, Dhar R, Das AK. Analyzing the catalytic mechanism of protein tyrosine phosphatase PtpB from Staphylococcus aureus through site-directed mutagenesis. Int J Biol Macromol. 2009;45(5):463–9. doi: 10.1016/j.ijbiomac.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 88.Tolkatchev D, Shavkhutdinov R, Xu P, Plamondon J, Watson DC, Young NM, et al. Three-dimensional structure and ligand interactions of the low molecular weight protein tyrosine phosphatase from Campylobacter jejuni. Protein Sci. 2006;15(10):2381–94. doi: 10.1110/ps.062279806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coats SR, Pledger WJ, Awazu M, Daniel TO. Detergent solubility defines an alternative itinerary for a subpopulation of PDGF beta receptors. J Cell Physiol. 1996;168(2):412–23. doi: 10.1002/(SICI)1097-4652(199608)168:2<412::AID-JCP20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 90.Cirri P, Chiarugi P, Taddei L, Raugei G, Camici G, Manao G, et al. Low molecular weight protein-tyrosine phosphatase tyrosine phosphorylation by c-Src during platelet-derived growth factor-induced mitogenesis correlates with its subcellular targeting. J Biol Chem. 1998;273(49):32522–7. doi: 10.1074/jbc.273.49.32522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.