Abstract
Ni(II) and Zn(II) complexes were synthesized from tridentate 3-formyl chromone Schiff bases such as 3-((2-hydroxyphenylimino)methyl)-4H-chromen-4-one (HL1), 2-((4-oxo-4H-chromen-3-yl)methylneamino)benzoic acid (HL2), 3-((3-hydroxypyridin-2-ylimino)methyl)-4H-chromen-4-one (HL3), and 3-((2-mercaptophenylimino)methyl)-4H-chromen-4-one (HL4). All the complexes were characterized in the light of elemental analysis, molar conductance, FTIR, UV-VIS, magnetic, thermal, powder XRD, and SEM studies. The conductance and spectroscopic data suggested that, the ligands act as neutral and monobasic tridentate ligands and form octahedral complexes with general formula [M(L1–4)2]·nH2O (M = Ni(II) and Zn(II)). Metal complexes exhibited pronounced activity against tested bacteria and fungi strains compared to the ligands. In addition metal complexes displayed good antioxidant and moderate nematicidal activities. The cytotoxicity of ligands and their metal complexes have been evaluated by MTT assay. The DNA cleavage activity of the metal complexes was performed using agarose gel electrophoresis in the presence and absence of oxidant H2O2. All metal complexes showed significant nuclease activity in the presence of H2O2.
1. Introduction
Metal complexes of O, S, and N containing Schiff bases have been the subject of current and growing interest because it has wide range of pharmacological activities [1]. In particular Schiff bases with 2-amino thiophenol, 2-amino phenol, 2-amino benzoic acid, and 2-amino 3-hydroxy pyridine exhibit various biological activities such as antimicrobial activity, protein tyrosine phosphatases inhibition, and nuclease activity [2, 3]. Several aromatic amine Schiff bases have been investigated but few works deal with chromone skeleton derivatives. Chromones are a group of naturally occurring compounds that are ubiquitous in nature especially in plants. Molecules containing the chromone skeleton have extensive biological applications including antimycobacterial, antifungal, anticancer, antioxidant, antihypertensive, and anti-inflammatory applications and tyrosinase and protein kinase C inhibitors [4–13].
DNA plays an important role in the life process since it contains all the genetic information for the cellular function. However, DNA is the primary intracellular target of anticancer drugs, damaged under various conditions such as interactions with some small molecules, which cause DNA damage in cancer cells blocking the division of cancer cells and resulting in cell death. Metal complexes such as cobalt, nickel, copper, and zinc with Schiff base ligands have shown excellent binding and cleavage activities [14–16]. There is substantial literature supporting the DNA binding studies of chromone Schiff bases and their metal complexes [17–19]. Compounds showing the properties of effective binding as well as cleaving double stranded DNA under physiological conditions are of great importance since these could be used as diagnostic agents in medicinal and genomic research.
Fluorescent transition metal centres are particularly attractive moieties because they often possess distinctive electrochemical or photophysical properties, thus enhancing the functionality of the binding agent [20].
In previous papers we presented synthesis, detailed characterization, and biological activity studies of 3-formyl chromone Schiff bases such as 3-((2-hydroxy phenylimino)methyl)-4H-chromen-4-one (HL1), 2-((4-oxo-4H-chromen-3-yl)methylneamino)benzoic acid (HL2), 3-((3-hydroxypyridin-2-ylimino)methyl)-4H-chromen-4-one (HL3), 3-((2-mercapto phenylimino)methyl)-4H-chromen-4-one (HL4), and their Cu(II), Co(II), and Pd(II) complexes [21–23]. The present paper deals with the synthesis, characterization of Ni(II) and Zn(II) complexes of 3-formyl chromone Schiff bases (HL1, HL2, HL3, and HL4), and various biological activity studies, that is, antimicrobial, antioxidant, and nematicidal activities and cytotoxicity and DNA cleavage.
2. Experimental
2.1. Materials and Physical Measurements
All the chemicals used were of analytical grade and procured from Spectrochem Pvt. Ltd., Mumbai, India. 3-Formyl chromone was synthesized according to the literature [24].
The elemental analysis of carbon, hydrogen, nitrogen, and sulphur contents was performed using Perkin Elmer CHNS analyser. Molar conductance of the complexes was measured using a Digisun conductivity meter in DMF. The electronic absorption spectra of the complexes were recorded on JASCO V-670 Spectrophotometer in the wavelength region of 250–1400 nm in the solid state. The FTIR spectra of the complexes were recorded on Tensor 2 FTIR spectrophotometer in the region of 4000–400 cm−1 using KBr disc. The magnetic susceptibilities of Ni(II) complexes were measured with a Sherwood scientific balance. Diamagnetic corrections were calculated from Pascal's constants. The magnetic moment values were calculated using the relation μ eff = 2.83 (χ m T)1/2 B.M. Thermal studies of the complexes were carried out on a Perkin Elmer diamond TGA instrument at a heating rate of 10°C and nitrogen flow rate of 20 mL/min. The fluorescence spectra of the complexes were recorded on Fluorolog FL3-11 spectrofluorometer. The X-ray patterns of the complexes were recorded on Xpert-Pro X-ray diffractometer with Cu Kα radiation (λ = 1.5406 Å). The diffraction data are integrated by using the Nakamuta program. Scanning electron micrograph (SEM) of the complexes was obtained in a Hitachi S-520 electron microscope at an accelerated voltage of 15 kV.
2.2. Synthesis of Ligands
All the Schiff base ligands HL1, HL2, HL3, and HL4 (Figure 1) were prepared according to the literature methods [21, 25, 26].
Figure 1.
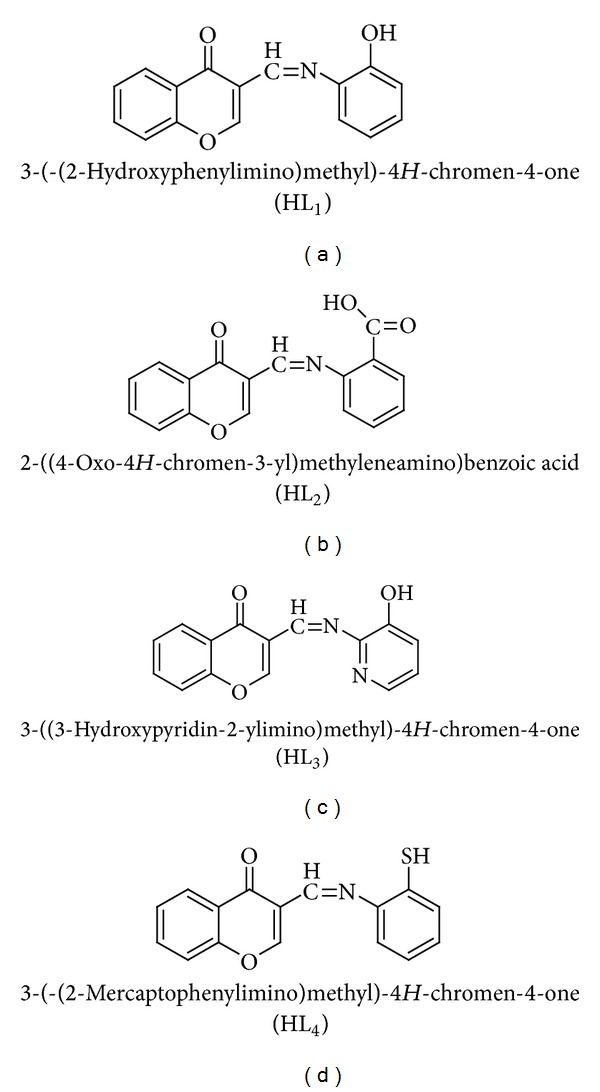
Structures of Schiff base ligands.
2.3. Synthesis of Metal Complexes
The ligands (2 mM) and metal acetates (Ni(CH3COOH)2·4H2O and Zn(CH3COOH)2·2H2O) (1 mM) were dissolved in methanol. The mixtures were stirred at room temperature for 2-3 h; various colour precipitates were separated from the solution by suction filtration, purified by washing several times with methanol, and dried for 12 h in vacuum.
2.4. Antimicrobial Activity
In vitro antimicrobial activity of the metal complexes towards the bacteria Proteus vulgaris, Klebsiella pnuemoniae, Staphylococcus aureus, and Bacillus subtili and fungi Candida albicans was carried out using disc diffusion method. The antibiotics kanamycin and clotrimazole are the standards for antibacterial and antifungal activity studies. Standard inoculum, 1-2 × 107 cfu/mL 0.5 Mc Farland standards [27], was introduced onto the surface of sterile nutrient agar plate and evenly spread by using a sterile glass spreader. Sterile antibiotic discs (6 mm in diameter, prepared using Whatmann number 1 paper) were placed over the nutrient agar medium. Each disc was spread by 100 μg of the compounds (initially dissolved in DMSO). The plates were incubated with bacterial cultures for 24 h at 37°C and fungal cultures at 25°C for 48 h. The activity of the compounds was determined by measuring the diameter of inhibition zone in “millimetres” and compared with standard antibiotics. DMSO (which has no activity) and standard antibiotics were used as negative and positive controls for antimicrobial activity studies. The activity results are calculated as a mean of triplicates.
Minimum inhibitory concentrations (MIC) of the complexes which showed antimicrobial activity were determined using the literature method [28]. All the compounds that were diluted within the range of 100–10 μg/mL were mixed in nutrient broth and 0.1 mL of active inoculums was added to each tube. The tubes were incubated aerobically at 37°C for bacteria and 25°C for fungi up to 24 h. The lowest concentration of the compound that completely inhibited bacterial growth (no turbidity) in comparison to control was regarded as MIC.
2.5. Nematicidal Activity
Root knot nematode, Meloidogyne incognita, is major plant parasitic nematodes affecting quantity and quality of the crop production in many annual and perennial crops. Meloidogyne nematode can develop galls and lesions in the roots, thereby causing stunted growth of the plants. Some of the chemicals can be used to control nematodes [29].
Nematicidal activity of the complexes was carried out on Meloidogyne incognita. Fresh egg masses of Meloidogyne incognita are collected from stock culture maintained on tomato (Lycopersicon esculentum) root tissues and kept in water for egg hatching. The eggs suspensions were poured on a cotton wool filter paper and incubated at 30°C to obtain freshly hatched juveniles (J2). Juveniles collected within 48 h were used for screening nematicidal activity of the compounds.
The compounds were initially dissolved in dimethyl sulfoxide (DMSO) and then in distilled water to make dilutions of 250, 150, and 50 μg/mL. Experiments were performed under laboratory conditions at 30°C. About 100 freshly hatched second stage juveniles were suspended in 5 mL of each diluted compound and incubated. Distilled water with nematode larvae was taken as control. The dead nematodes were observed under an inverted binocular microscope. After an incubation of 24 and 48 h, percentage of mortality was calculated. Nematodes were considered dead if they did not move when probed with a fine needle [30].
2.6. DPPH Radical Scavenging Activity
The free radical scavenging activities of the metal complexes were determined by using DPPH free radical scavenging method according to the literature [31]. DPPH is a stable free radical containing an odd electron in its structure and usually utilized for detection of the radical scavenging activity in chemical analysis. In the spectrophotometric assay the ability to scavenge the stable free radical DPPH is measured by decrease in the absorbance at 517 nm. Each compound was dissolved in methanol (10 mg/10 mL) and it was used as stock solution. From the stock solutions, 1 mL of each compound solution with different concentrations (0.25 μg–1.00 μg) was added to the 3 mL of methanolic DPPH (0.004%) solution. After 30 min, the absorbance of the test compounds was taken at 517 nm using UV-VIS spectrophotometer. BHT was used as standard, DPPH solution was used as control without the test compounds, and methanol was used as blank. The percentage of scavenging activity of DPPH free radical was measured by using the following formula:
| (1) |
where A o is the absorbance of the control and A i is the absorbance of the sample.
2.7. Cytotoxic Activity
The human breast carcinoma cell line (MCF-7), human colon carcinoma cell line (COLO 205), and murine microphage cell line (Raw 264.7) were obtained from the National Centre for Cell Science (NCCS), Pune, and grown in Dulbecco's Modified Eagles Medium (DMEM) containing 10% fetal bovine serum (FBS), amphotericin (3 μg/mL), gentamycin (400 μg/mL), streptomycin (250 μg/mL), and penicillin (250 units/mL) in a carbon dioxide incubator at 5% CO2. About 700 cells/well were seeded in 96-well plate using culture medium; the viability was tested using trypan blue dye with help of haemocytometer and 95% of viability was confirmed. After 24 h, the new medium with compounds in the concentration of 100, 10, and 1 μg/mL were added at respective wells and kept in incubation for 48 h. After incubation MTT assay was performed.
2.7.1. MTT Assay
After 48 h of the drug treatment the medium was changed again for all groups and 10 μL of MTT (5 mg/mL stock solution) was added and the plates were incubated for an additional 4 h. The medium was discarded and the formazan blue, which was formed in the cells, was dissolved with 50 μL of DMSO. The optical density was measured on microplate spectrophotometer at a wavelength of 570 nm. The percentage of cell inhibition was calculated by using the following formula [32]:
| (2) |
where A i is the absorbance of the sample and A o is the absorbance of the control. IC50 values were determined using Graph Pad Prism software.
2.8. DNA Cleavage Activity
The DNA cleavage activity of metal complexes was monitored by agarose gel electrophoresis. pUC19 plasmid was cultured, isolated, and used as DNA for the experiment. Test samples (1 mg/mL) were prepared in DMF. 25 μg of the test samples was added to the isolated plasmid and incubated for 2 h at 37°C. After incubation, 30 μL of plasmid DNA sample mixed with bromophenol blue dye (1 : 1) was loaded into the electrophoresis chamber wells along with the control DNA, 5 M FeSO4 (treated with DNA), and standard DNA marker containing TAE buffer (4.84 g Tris base, pH 8.0, 0.5 M EDTA/1 L). Finally, it was loaded on to an agarose gel and electrophoresed at 50 V constant voltage up to 30 min. After the run, gel was removed and stained with 10.01 μg/mL ethidium bromide and image was taken in Versadoc (Biorad) imaging system. The results were compared with standard DNA marker. The same procedure was followed in the presence of H2O2 also.
3. Results and Discussion
All the Ni(II) complexes were colored, stable, and nonhygroscopic in nature. The complexes are insoluble in common organic solvents but soluble in DMF and DMSO. The elemental analysis showed that the complexes have 1 : 2 stoichiometry of the type [M(L1–4)2]·nH2O, where L stands for singly deprotonated ligands. Molar conductance of the complexes was measured in DMF. The conductance values, which are presented in Table 1, indicate the nonelectrolytic nature of the complexes [33].
Table 1.
Elemental analysis and physical properties of metal complexes.
| Molecular formula | Colour (% yield) | % found (cald.) | Molar conductivity (Ohm−1 cm2 mol−1) | ||||
|---|---|---|---|---|---|---|---|
| C | H | N | S | M (Ni/Zn) |
|||
| [Ni(L1)2] | Brick red | 64.49 | 3.21 | 4.23 | 9.91 | 10 | |
| [Ni(C16H10O3N)2] | (71) | (65.45) | (3.40) | (4.77) | (10.00) | ||
| [Ni(L2)2]·H2O | Light green | 61.59 | 3.02 | 4.21 | 8.79 | 12 | |
| [Ni(C17H10O4N)2]·H2O | (62) | (61.75) | (3.32) | (4.24) | (8.88) | ||
| [Ni(L3)2]·H2O | Brick red | 61.07 | 3.23 | 9.23 | 9.43 | 15 | |
| [Ni(C15H9O3N2)2]·H2O | (85) | (61.15) | (3.39) | (9.51) | (9.97) | ||
| [Ni(L4)2] | Black | 62.21 | 3.09 | 4.61 | 9.30 | 9.21 | 14 |
| [Ni(C16H10O2NS)2] | (82) | (62.06) | (3.23) | (4.52) | (10.34) | (9.49) | |
| [Zn(L1)2]·H2O | Orange | 62.91 | 3.52 | 4.62 | 10.58 | 15 | |
| [Zn(C16H10O3N)2]·H2O | (88) | (62.80) | (3.60) | (4.58) | (10.70) | ||
| [Zn(L2)2] | Yellow | 62.79 | 3.15 | 4.25 | 9.95 | 11 | |
| [Zn(C17H10O4N)2] | (61) | (62.83) | (3.08) | (4.31) | (10.07) | ||
| [Zn(L3)2] | Brick red | 60.21 | 3.25 | 9.35 | 10.51 | 18 | |
| [Zn(C15H9O3N2)2] | (87) | (60.46) | (3.02) | (9.40) | (10.98) | ||
| [Zn(L4)2]·H2O | Yellow | 60.22 | 3.35 | 4.21 | 10.01 | 10.31 | 20 |
| [Zn(C16H10O2NS)2]·H2O | (73) | (59.68) | (3.41) | (4.35) | (9.93) | (10.46) | |
3.1. Determination of the Metal Content of the Complexes
Known amount (0.150 g) of complexes was decomposed with concentrated nitric acid. This process was repeated till the organic part of the complexes got completely lost. The excess nitric acid was expelled by evaporation with concentrated sulphuric acid. The Ni(II) and Zn(II) contents of the complexes were determined as per the procedure available in the literature [34].
3.2. FTIR Spectra
The FTIR spectra of the complexes are compared with those of the ligands in order to determine the coordination sites that may involve in chelation. The position and/or the intensities of bands are expected to be changed while the coordination. The most important IR bands of the metal complexes with probable assignments are given in Table 2. The Schiff base ligands showing a band around 1650–1620 cm−1 is assigned to the (C=O) group of the chromone system. Upon complexation the ν(C=O) group is shifted to 20–35 cm−1 lower wavenumber region [35]. The ligands show the most characteristic ν(C=N) bands in the region 1605–1563 cm−1. In the spectra of their corresponding metal complexes, this band appears at 25–45 cm−1 to lower wavenumber region indicating the coordination of the azomethine nitrogen atom to the metal ion [36]. A broad band appeared in HL1 and HL3 at 3241 and 3246 cm−1, respectively, and is attributed to the ν(O–H) group. The absence of the band in the spectra of their corresponding metal complexes is due to the involvement of oxygen atom of (OH) group coordination to the metal ion. In HL2 ligand, a strong band appeared at 1365 cm−1 and is due to the ν(C–O) of carboxylic group. In its metal complexes it is shifted to 18–47 cm−1 lower wavenumber region [37]. Since SH stretching frequency band is very weak, the peak corresponding to SH is not clearly observed in the case of HL4 ligand [38]. Presence of band around 3400 cm−1 in all the complexes is the indication of water molecules present in the complexes. The presence of oxygen and nitrogen in the coordination sphere is further confirmed by the presence of ν(M–N) and ν(M–O) bands at 400–600 cm−1 region. The FTIR results show that all Schiff base ligands act as tridentate chelating ligands.
Table 2.
IR spectral data of ligands and their metal complexes (cm−1).
| Compound | ν(OH) | ν(C=O) (γ pyrone) | ν(C=N) | ν(CO) (carboxylate) | ν(C–S) | ν(M–N) | ν(M–O) |
|---|---|---|---|---|---|---|---|
| HL1 | 3241 | 1643 | 1605 | ||||
| HL2 | 1651 | 1605 | 1365 | ||||
| HL3 | 3246 | 1647 | 1591 | ||||
| HL4 | 1622 | 1597 | 790 | ||||
| [Ni(L1)2] | 1619 | 1563 | 467 | 586 | |||
| [Ni(L2)2]·H2O | 1618 | 1570 | 1318 | 419 | 514 | ||
| [Ni(L3)2]·H2O | 1615 | 1563 | 480 | 500 | |||
| [Ni(L4)2] | 1598 | 1564 | 743 | 443 | 524 | ||
| [Zn(L1)2]·H2O | 1614 | 1574 | 460 | 599 | |||
| [Zn(L2)2] | 1615 | 1563 | 1347 | 480 | 500 | ||
| [Zn(L3)2] | 1615 | 1563 | 488 | 521 | |||
| [Zn(L4)2]·H2O | 1609 | 1581 | 751 | 441 | 505 |
3.3. Electronic Spectra and Magnetic Moments
The electronic absorption spectra of the Schiff base metal complexes in solid state were recorded at room temperature and the band positions of the absorption maxima, band assignments, ligand field parameters, and magnetic moment values are listed in Table 3. The electronic spectra of [Ni(L4)2]·H2O and [Zn(L4)2]·H2O complexes are depicted in Figure 2. The electronic spectra of Ni(II) complexes displayed three absorption bands in the range 8000–9000, 14000–16000, and 20000–24000 cm−1. Thus, these bands may be assigned to the three spin allowed transitions 3A2g (F) →3T2g (F) (ν 1), 3A2g (F) →3T1g (F) (ν 2), and 3A2g (F) →3T1g (P) (ν 3), respectively, characteristic of octahedral geometry. The values of transition ratio ν 2/ν 1 and β lie in the range of 1.70–1.80 and 0.89–0.95, respectively, providing further evidence for octahedral geometry of Ni(II) complexes [39]. The B values for the complexes are lower than the free ion value, thereby indicating orbital overlap and delocalisation of d-orbitals. The β-values obtained are less than unity suggesting the covalent character of the metal-ligand bonds. All Ni(II) complexes are paramagnetic and the magnetic movement values at room temperature are in the range of 2.91–3.25 B.M which is well agreed with the reported octahedral Ni(II) complexes [40]. All Zn(II) complexes showed two bands around 25000 and 30000 cm−1 and are attributed to the n → π* and π → π* transitions, respectively. Zn(II) complexes that are in d10 configuration are diamagnetic and do not show any d-d transitions.
Table 3.
Electronic, magnetic, and ligand field parameters of Ni(II) complexes.
| Compound | Absorption maxima (cm−1) |
Tentative assignments | Magnetic moment (B.M) |
ν 2/ν 1 | 10 Dq (cm−1) |
B (cm−1) |
β | LFSE (kJ·mol−1) |
|---|---|---|---|---|---|---|---|---|
| [Ni(L1)2] | 8532 15200 24900 |
3A2g(F) → 3T2g(F) (ν
1) 3A2g(F) → 3T1g(F) (ν 2) 3A2g(F) → 3T1g(P) (ν 3) |
3.12 | 1.78 | 8532 | 966 | 0.92 | 122.50 |
|
| ||||||||
| [Ni( L2)2]·H2O | 8787 16000 24390 |
3A2g(F) → 3T2g(F) (ν
1) 3A2g(F) → 3T1g(F) (ν 2) 3A2g(F) → 3T1g(P) (ν 3) |
3.18 | 1.82 | 8787 | 935 | 0.89 | 126.19 |
|
| ||||||||
| [Ni(L3)2]·H2O | 8313 14705 20833 24390 |
3A2g(F) → 3T2g(F) (ν
1) 3A2g(F) → 3T1g(F) (ν 2) 3A2g(F) → 3T1g(P) (ν 3) |
3.25 | 1.76 | 8313 | 943 | 0.90 | 119.40 |
|
| ||||||||
| [Ni(L4)2] | 8628 16313 24570 |
3A2g(F) → 3T2g(F) (ν
1) 3A2g(F) → 3T1g(F) (ν 2) 3A2g(F) → 3T1g(P) (ν 3) |
2.91 | 1.89 | 8628 | 999 | 0.95 | 123.92 |
Figure 2.
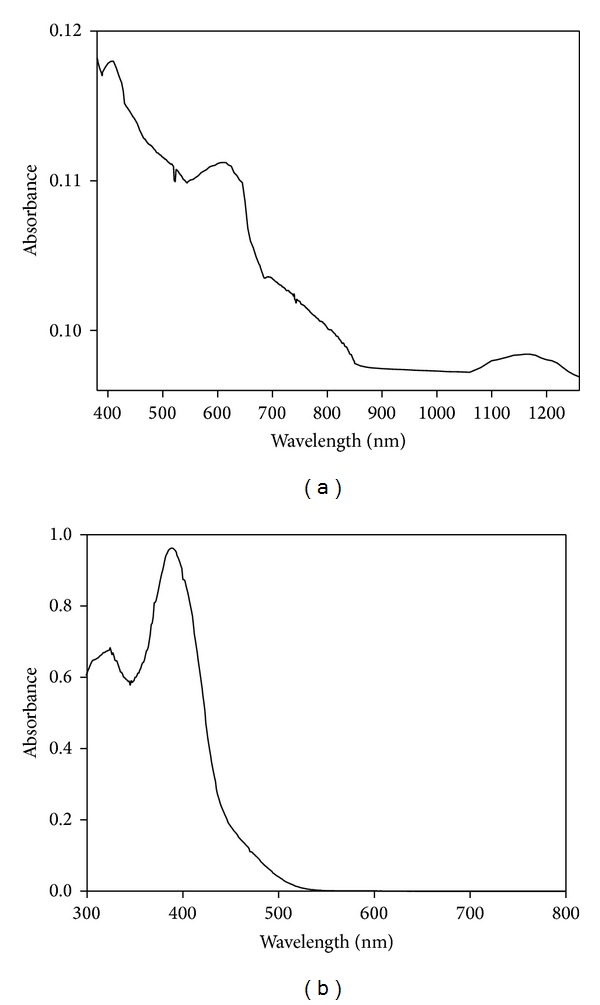
Electronic spectra of (a) [Ni(L4)2] and (b) [Zn(L4)2]·H2O.
3.4. Thermal Analysis
The thermogravimetric analysis (TGA) of the metal complexes was carried out within the temperature range from room temperature to 1000°C. The TG-DTA graphs of [Ni(L4)2] and [Zn(L4)2]·H2O complexes are given in Figure 3. The TGA data and their assignments of all the metal complexes are listed in Table 4. Metal complexes decompose gradually with formation of respective metal oxides. The TG graphs of [Ni(L2)2]·H2O, [Ni(L3)2]·H2O, [Zn(L1)2]·H2O, and [Zn(L4)2]·H2O complexes decompose in two to three steps. The first step corresponds to the loss of lattice water molecule in the temperature range between 30 and 140°C with a weight loss of 2-3%. In the second and third steps, the total loss of ligand molecules was observed in the temperature range between 160 and 850°C leaving behind metal oxide as residue. The thermal decomposition of remaining metal complexes ([Ni(L1)2], [Ni(L4)2] [Zn(L2)2] and [Zn(L3)2]) occurs in two to three steps. Upon starting heating, these metal complexes losses of the organic moieties of Schiff base ligands were observed in two to three successive steps within the temperature range of 150–850°C. In all the complexes the final mass loss is due to the formation of metal oxides as residue.
Figure 3.
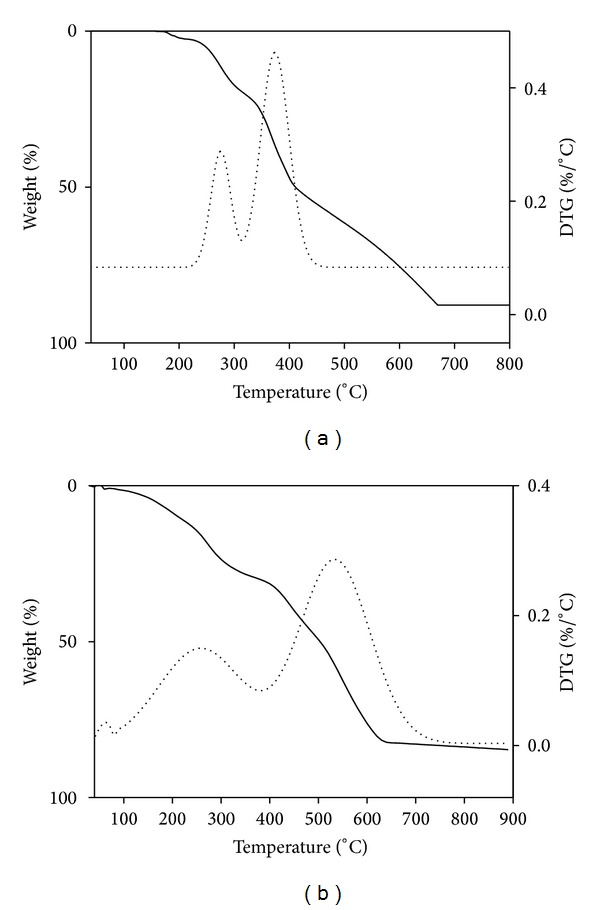
TG-DTG graph of (a) [Ni(L4)2] and (b) [Zn(L4)2]·H2O.
Table 4.
Thermal analysis results of metal complexes.
| Compound | Temperature (°C) | Found (cald.) | Assignment |
|---|---|---|---|
| [Ni(L1)2] [Ni(C16H10O3N)2] |
180–420 | 19.38 (18.36) | C6H4ON |
| 421–718 | 68.55 (69.02) | C26H16O5N | |
| >718 | 12.07 (12.62) | NiO | |
|
| |||
| [Ni(L2)2]·H2O [Ni(C17H10O4N)2]·H2O |
30–90 | 2.83 (2.72) | H2O |
| 335–478 | 60.71 (62.05) | C24H14O5N2 | |
| 479–849 | 23.93 (23.61) | C10H6O2 | |
| >849 | 12.53 (11.62) | NiO | |
|
| |||
| [Ni(L3)2]·H2O [Ni(C15H9O3N2)2]·H2O |
30–65 | 3.09 (2.96) | H2O |
| 180–879 | 83.40 (84.73) | C30H18O5N4 | |
| >880 | 13.51 (12.31) | NiO | |
|
| |||
| [Ni(L4)2] [Ni(C16H10O2NS)2] |
171–410 | 45.91 (45.25) | C16H10O2NS |
| 411–670 | 42.14 (42.67) | C16H10ONS | |
| >670 | 11.95 (12.07) | NiO | |
|
| |||
| [Zn(L1)2]·H2O [Zn(C16H10O3N)2]·H2O |
30–135 | 2.45 (2.95) | H2O |
| 194–422 | 24.95 (25.92) | C10H6O2 | |
| 423–790 | 59.48 (58.09) | C22H14O3N2 | |
| >791 | 13.12 (13.04) | ZnO | |
|
| |||
| [Zn(L2)2] [Zn(C17H10O4N)2] |
195–265 | 15.55 (16.01) | C7H4O |
| 266–661 | 69.76 (71.46) | C27H16O4N2 | |
| >662 | 14.69 (12.53) | ZnO | |
|
| |||
| [Zn(L3)2] [Zn(C15H9O3N2)2] |
157–192 | 10.91 (12.93) | C5H3N |
| 193–485 | 28.65 (29.22) | C10H6O2N | |
| 486–835 | 46.03 (44.18) | C15H9O3N2 | |
| >836 | 14.41 (13.67) | ZnO | |
|
| |||
| [Zn(L4)2]·H2O [Zn(C16H10O2NS)2]·H2O |
30–124 | 2.41 (2.79) | H2O |
| 165–348 | 27.41 (26.73) | C10H6O2N | |
| 389–632 | 56.98 (57.84) | C22H14ONS2 | |
| >632 | 13.20 (12.64) | ZnO | |
On the basis of all spectral data (elemental analysis, FTIR, electronic spectra, and thermal analysis), the suggested structures of the complexes are shown in Figure 4.
Figure 4.
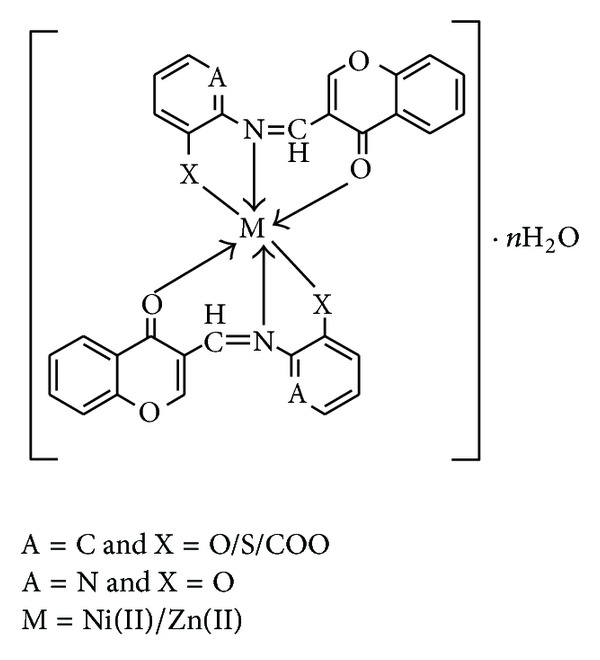
Proposed structure of the metal complexes.
3.5. Powder XRD and SEM Studies
Single crystals of the complexes could not be obtained because of their insolubility in most organic solvents, hence, the powder diffraction data were obtained for structural characterization. The powder XRD patterns of all metal complexes were recorded in the range 2θ = 10–50°. The powder XRD pattern of [Ni(L1)2] and [Zn(L1)2]·H2O are presented in Figure 5. Observed and calculated powder XRD data of [Ni(L1)2] and [Zn(L1)2]·H2O are given in Tables 5(a) and 5(b). Unit cell parameters were found by using trial and error methods. All complexes are triclinic with different unit cell parameters. All metal complexes display sharp crystalline peaks except [Zn(L2)2] and [Zn(L4)2]·H2O; these two complexes do not exhibit well-defined crystalline peaks due to their amorphous nature. The unit cell parameters of metal complexes are as follows: [Ni(L1)2]: a = 8.80 Å, b = 8.52 Å, c = 7.50 Å, α = 99.93°, β = 116.71°, γ = 90.01°, and V = 562 (Å)3; [Ni(L2)2]·H2O: a = 8.24 Å, b = 9.24 Å, c = 7.32 Å, α = 104.12°, β = 124.44°, γ = 98.04°, and V = 557 (Å)3; [Ni(L3)2]·H2O: a = 9.40 Å, b = 9.20 Å, c = 8.46 Å, α = 97.45°, β = 107.38°, γ = 91.45°, and V = 731 (Å)3; [Ni(L4)2]: a = 7.73 Å, b = 8.24 Å, c = 8.49 Å, α = 98.30°, β = 104.76°, γ = 90.55°, and V = 540 (Å)3; [Zn(L1)2]·H2O: a = 9.60 Å, b = 9.94 Å, c = 8.89 Å, α = 102.34°, β = 109.69°, γ = 93.94°, and V = 848 (Å)3; [Zn(L3)2]: a = 8.76 Å, b = 9.04 Å, c = 8.61 Å, α = 105.78°, β = 103.02°, γ = 90.21°, and V = 682 (Å)3. The average crystallite sizes (D) of the metal complexes are calculated from the Scherrer formula [41]:
| (3) |
where λ is the X-ray wavelength, β is the full width at half maximum of prominent intensity peak, and θ is the diffraction angle. The [Ni(L1)2], [Ni(L2)2]·H2O, [Ni(L3)2]·H2O, [Ni(L4)2], [Zn(L1)2]·H2O, and [Zn(L3)2] complexes have an average crystallite size of 70, 17, 16, 27, 42, and 53 nm, respectively, suggesting that the complexes are in nanocrystalline phase.
Figure 5.
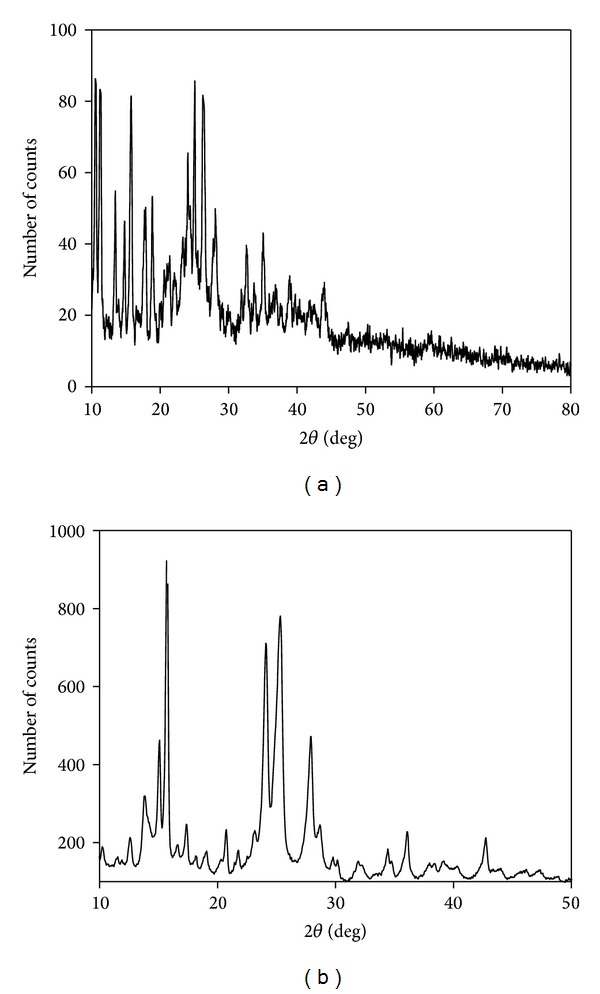
Powder XRD patterns of (a) [Ni(L1)2] (b) [Zn(L1)2]·H2O.
Table 5.
Observed and calculated powder XRD patterns of (a) [Ni(L1)2] (b) [Zn(L1)2]·H2O.
(a)
| S. number | 2θ | Δ2θ | d-spacing | h | k | l | ||
|---|---|---|---|---|---|---|---|---|
| Observed | Calculated | Observed | Calculated | |||||
| 1 | 10.57 | 10.57 | 0 | 8.3627 | 8.3627 | 0 | 1 | 0 |
| 2 | 11.29 | 11.29 | 0 | 7.8314 | 7.8314 | 1 | 0 | 0 |
| 3 | 13.44 | 13.44 | 0 | 6.5805 | 6.5805 | 0 | 0 | 1 |
| 4 | 14.79 | 14.79 | 0 | 5.9847 | 5.9847 | −1 | 1 | 0 |
| 5 | 15.75 | 15.75 | 0 | 5.6205 | 5.6205 | −1 | −1 | 1 |
| 6 | 17.79 | 17.79 | 0 | 4.9805 | 4.9805 | −1 | 1 | 1 |
| 7 | 18.85 | 18.68 | 0.164 | 4.7046 | 4.7455 | 0 | 1 | 1 |
| 8 | 21.16 | 21.22 | −0.06 | 4.1962 | 4.1845 | 1 | 0 | 1 |
| 9 | 22.14 | 22.06 | 0.074 | 4.0121 | 4.0255 | 1 | −1 | 1 |
| 10 | 24.05 | 24.07 | −0.013 | 3.6968 | 3.6948 | −1 | 0 | 2 |
| 11 | 25.04 | 24.99 | 0.054 | 3.5533 | 3.5608 | 1 | 2 | 0 |
| 12 | 26.33 | 26.35 | −0.017 | 3.382 | 3.3799 | −1 | 2 | 1 |
| 13 | 28.1 | 28.04 | 0.061 | 3.1731 | 3.1799 | −1 | 1 | 2 |
| 14 | 32.65 | 32.65 | 0 | 2.7404 | 2.7405 | 2 | 2 | 0 |
| 15 | 35.06 | 35.07 | −0.012 | 2.5575 | 2.5566 | −3 | 1 | 0 |
| 16 | 38.92 | 38.86 | 0.054 | 2.3124 | 2.3155 | −2 | 3 | 1 |
| 17 | 43.89 | 43.95 | −0.058 | 2.0612 | 2.0586 | −1 | 3 | 2 |
| 18 | 45.13 | 45.16 | −0.033 | 2.0076 | 2.0062 | −2 | −3 | 3 |
(b)
| S. number | 2θ values | Δ2θ | d-spacing | h | k | l | ||
|---|---|---|---|---|---|---|---|---|
| Observed | Calculated | Observed | Calculated | |||||
| 1 | 12.58 | 12.58 | 0 | 7.0315 | 7.0315 | 1 | 1 | 0 |
| 2 | 13.81 | 13.81 | 0 | 6.4077 | 6.4077 | −1 | 0 | 1 |
| 3 | 15.73 | 15.73 | 0 | 5.6299 | 5.6299 | 1 | 0 | 1 |
| 4 | 17.35 | 17.35 | 0 | 5.1064 | 5.1064 | −1 | 1 | 1 |
| 5 | 20.75 | 20.77 | −0.017 | 4.2777 | 4.2742 | −1 | 3 | 0 |
| 6 | 24.09 | 24.13 | −0.031 | 3.6907 | 3.6859 | −1 | −3 | 2 |
| 7 | 25.32 | 25.31 | 0.012 | 3.5141 | 3.5158 | 2 | 2 | 0 |
| 8 | 27.93 | 27.97 | −0.038 | 3.1916 | 3.1873 | −1 | −4 | 2 |
| 9 | 30.15 | 30.16 | −0.008 | 2.9620 | 2.9612 | 0 | −3 | 3 |
| 10 | 31.92 | 31.90 | 0.021 | 2.8015 | 2.8033 | 2 | −5 | 2 |
| 11 | 34.43 | 34.45 | −0.017 | 2.6028 | 2.6016 | −1 | 5 | 0 |
| 12 | 36.05 | 36.06 | −0.008 | 2.4892 | 2.4886 | −1 | 0 | 3 |
| 13 | 42.75 | 42.83 | −0.081 | 2.1137 | 2.1099 | 3 | 1 | 2 |
The SEM (Scanning Electron Microscope) is used to evaluate the morphology and particle size of the compounds. The SEM photographs of [Ni(L3)2]·H2O and [Ni(L4)2] are shown in Figure 6. The SEM micrographs show the agglomerate particles of the complexes. In case of [Ni(L3)2]·H2O and [Ni(L4)2] complexes, some agglomerates appear to have tiny needles, while the other agglomerates appear to be of spherical plates like morphologies.
Figure 6.

SEM micrograph of (a) [Ni(L3)2]·H2O and (b) [Ni(L4)2].
3.6. Fluorescence Spectra
The fluorescence characteristics of metal complexes were studied at room temperature in solid state. In metal complexes metal to ligand coordination may lead to significant changes of the fluorescence properties of the ligand, including increase or decrease of the intensity, emission wavelength shift, quenching of the fluorescence, or appearance of new emissions [42]. The fluorescence spectra of HL1 ligand and its metal (Ni(II) and Zn(II)) complexes were depicted in Figure 7. The Ni(II) and Zn(II) complexes of HL1 were characterised by emission bands around (522, 569) nm and (458, 644) nm, upon photo excitation at 453 and 357 nm, respectively. The Ni(II) complex of HL2 exhibits emission bands at 449 and 524 nm. Zn(II) complex of HL2 does not show any emission spectra. The Ni(II) and Zn(II) complexes of HL3 were characterised by an emission bands at (504, 570, and 668) and 419 nm, respectively. The Ni(II) and Zn(II) complexes of HL4 exhibit emission bands around 539 and 613 nm, respectively. From the results, red shift and quenching of the metal ions was observed in the case of ligands and its metal complexes.
Figure 7.
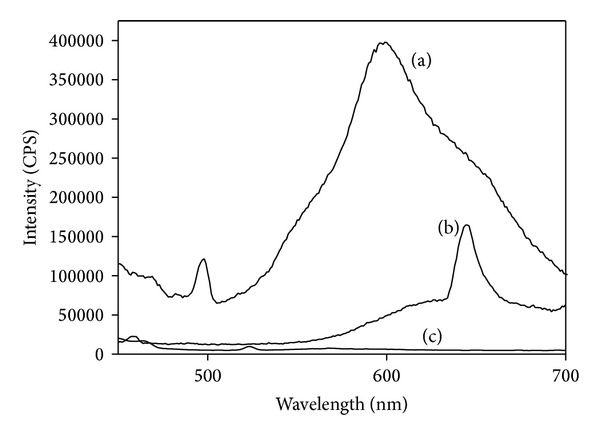
Fluorescence spectra of (a) HL1, (b) [Ni(L1)2], and (c) [Zn(L1)2]·H2O.
3.7. Antimicrobial Activity
The minimum inhibitory concentrations (MIC) of the complexes compared with the ligands and standard drugs are listed in Table 6. The results indicate that the metal complexes displayed more antibacterial activity compared to the parent ligands under similar experimental conditions on same microorganisms except [Ni(L2)2]·H2O and [Zn(L1)2]·H2O complexes. The results indicate that the metal complexes displayed more antibacterial activity compared to the parent ligands under similar experimental conditions on same microorganisms except [Zn(L1)2]·H2O. However, [Ni(L1)2], [Ni(L3)2]·H2O, [Zn(L2)2] and [Zn(L3)2] complexes showed antifungal activity while the remaining complexes did not show any activity. However, [Ni(L1)2], [Ni(L3)2]·H2O, and [Zn(L3)2] complexes showed antifungal activity while the remaining complexes did not show any activity. Among all the metal complexes [Zn(L3)2] complex showed good activity against all bacteria and fungi strains. Increase in the activity of the complexes compared to that of ligands can be explained on the basis of Overtone's concept and Tweedy's chelation theory. The theory states that the polarity of the metal ion is reduced on complexation due to the partial sharing of its positive charge with donor groups. Consequently, the positive charge is delocalized over the whole ring, which causes the improved lipophilicity of the compound through cell membrane of the pathogen [43]. The negative results can be attributed either to the inability of the complexes to diffuse into the bacteria cell membrane and hence they become unable to interfere with its biological activity or they can diffuse and become inactivated by unknown cellular mechanism, that is, bacterial enzymes [44].
Table 6.
MIC values of antimicrobial activity of ligands and their metal complexes (µg/mL).
| Compound | Bacillus subtilis | Staphylococcus aureus | Proteus vulgaris | Klebsiella pneumoniae | Candida albicans |
|---|---|---|---|---|---|
| HL1 | — | — | — | — | — |
| HL2 | 80 | 80 | 80 | 80 | 80 |
| HL3 | — | — | — | — | — |
| HL4 | — | — | — | — | — |
| [Ni(L1)2] | 35 | 40 | 32 | 41 | 75 |
| [Ni(L2)2]·H2O | 70 | 80 | 65 | 85 | — |
| [Ni(L3)2]·H2O | 45 | 45 | 50 | 33 | 33 |
| [Ni(L4)2] | 75 | 80 | — | 78 | — |
| [Zn(L1)2]·H2O | — | — | — | — | — |
| [Zn(L2)2] | 85 | 78 | 80 | 75 | 80 |
| [Zn(L3)2] | 30 | 30 | 33 | 30 | 33 |
| [Zn(L4)2]·H2O | — | 80 | — | 80 | — |
| Kanamycin | 4 | 10 | 8 | 11 | — |
| Clotrimazole | — | — | — | — | 10 |
3.8. Nematicidal Activity
All complexes except [Ni(L3)2]·H2O and [Zn(L3)2] showed very less nematicidal activity against Meloidogyne incognita. However, [Ni(L3)2]·H2O and [Zn(L3)2] complexes exhibited moderate activity indicating 47% and 51% mortality, respectively, after 48 h exposure in 250 μg/mL concentration. However, the activity of the metal complexes depends on concentration and time, that is, activity was higher at high concentrations and increased with time.
3.9. DPPH Radical Scavenging Activity
The antioxidant activity of the complexes was tested by measuring their ability to donate an electron to the free radical compound DPPH, monitoring changes in absorption at 517 nm using UV-VIS spectrophotometer. IC50 values were calculated and compared with the standard. IC50 values of the metal complexes are tabulated in Table 7. All complexes showed very less activity except [Ni(L1)2], [Ni(L4)2], and [Zn(L4)2]·H2O complexes. Among all the complexes [Zn(L4)2]·H2O (IC50 = 0.69 μg/mL) complex exhibited comparable activity to that of standard drug BHT (butylated hydroxyl toluene) (IC50 = 0.67 μg/mL).
Table 7.
IC50 values (µg/mL) of DPPH radical scavenging activity of ligands and their metal complexes.
| Compound | IC50 (µg/mL) |
|---|---|
| HL3 | 1.27 |
| HL4 | 0.40 |
| [Ni(L1)2] | 1.21 |
| [Ni(L4)2] | 1.09 |
| [Zn(L4)2]·H2O | 0.69 |
| BHT | 0.67 |
3.10. Cytotoxic Activity
The cytotoxicity of Schiff base ligands and their metal complexes were carried out using MTT assay. IC50 is employed to stand for the cytotoxicities of the compounds against the cancer cell lines; the smaller the IC50 value in the same condition is, the higher the cell growth inhibitory potency. DMSO when used as control without test sample, it does not exhibit any cytotoxic activity against all cancer cell lines. Cis-platin is used as positive control and it showed the highest cytotoxicity. The IC50 values were calculated after 48 h of incubation with compounds and are listed in Table 8. As shown in Table 8, all the ligands and their metal complexes showed lower cytotoxicity against selected cancer cell lines compared to the cis-platin. HL3 ligand showed enhanced activity against COLO 205 cell lines compared to all ligands and metal complexes (IC50 = 15.2 μg/mL). The lowest IC50 values (33.1 and 22.6 μg/mL) are observed for Zn(II) complex of HL3 ligand against raw and MCF-7 cell lines among all the ligands and metal complexes indicating its significant potency against two cancer cell lines. The results revealed that pyridine ring may be playing an important role in the cytotoxicity of the compounds.
Table 8.
IC50 values (µg/mL) of cytotoxic activity of ligands and their metal complexes.
| Compound | Raw | MCF-7 | COLO 205 |
|---|---|---|---|
| HL1 | 46.8 | 24.5 | 20.1 |
| HL2 | 56.1 | 34.2 | 39.1 |
| HL3 | 52.8 | 29.2 | 15.2 |
| HL4 | 46.9 | 30.4 | 68.1 |
| [Ni(L1)2] | 224.8 | 67.1 | 54.6 |
| [Ni( L2)2]·H2O | 36.3 | 26.5 | 60.9 |
| [Ni(L3)2]·H2O | 53.0 | 36.9 | 71.4 |
| [Ni(L4)2] | 58.6 | 35.6 | 49.9 |
| [Zn(L1)2]·H2O | 44.0 | 40.1 | 54.7 |
| [Zn(L2)2] | 34.0 | 52.3 | 43.9 |
| [Zn(L3)2] | 33.1 | 22.6 | 42.0 |
| [Zn(L4)2]·H2O | 42.5 | 39.2 | 53.5 |
| Cis-platin | 1.5 | 1.7 | 5.6 |
3.11. DNA Cleavage Activity
pUC19 DNA was chosen for the DNA cleavage activity of the complexes. The studies were done using gel electrophoresis. Its naturally occurring supercoiled form (Form I) when nicked gives rise to an open circular relaxed form (Form II) and upon further cleavage results in the linear form (Form III). When subjected to gel electrophoresis, Form I migrates relatively faster while the nicked form (Form II) migrates slowly and linearized Form III migrates between Form I and Form II [45]. In the present study gel electrophoresis experiments were performed in the presence and absence of an oxidizing agent H2O2 to investigate the mechanism of nucleolytic activity of the complexes. The activity was greater for the complexes in the presence of H2O2. Control (DNA alone) does not show activity (Figure 8(a)). When FeSO4 is used as standard, it shows complete DNA cleavage. In the absence of H2O2 all complexes show partial nucleolytic activity. Probably this may be due to the redox behaviour of the metal ions. These results indicated the important role of the metal ions in cleavage studies. In the presence of H2O2, (Figure 8(b)) absence of marker bands was observed in all the complexes except [Ni(L3)2]·H2O and [Zn(L3)2]; [Zn(L4)2]·H2O complexes indicate the complete DNA cleavage activity. In the case of [Ni(L3)2]·H2O, [Zn(L3)2], and [Zn(L4)2]·H2O complexes a decrease in the intensity of bands was observed compared to the control. This is probably due to the partial cleavage of the DNA. The DNA cleavage activity of the complexes in the presence of H2O2 may be due to the reaction of hydroxy radicals with DNA. The general oxidative mechanisms of the DNA cleavage studies were reported by several research groups [46–48]. Many literature reports infer that the compound was to cleave the DNA; it can be concluded that the compound inhibits the growth of the pathogenic organism by cleaving the genome [49].
Figure 8.
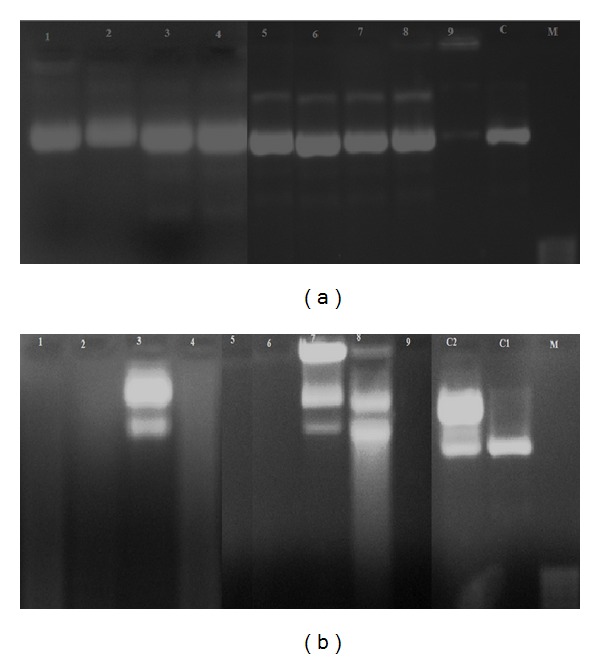
Gel electrophoresis photograph of metal complexes. (a) Gel electrophoresis photograph showing the effects of metal complexes on pUC 19 DNA: lane 1, DNA + [Ni(L1)2]; lane 2, DNA + [Ni(L2)2]·H2O; lane 3, DNA + [Ni(L3)2]·H2O; lane 4, DNA + [Ni(L4)2]; lane 5, DNA + [Zn(L1)2]·H2O; lane 6, DNA + [Zn(L2)2]; lane 7, DNA + [Zn(L3)2]; lane 8, DNA + [Zn(L4)2]·H2O; lane 9, DNA + FeSO4; lane C, DNA alone. (b) Gel electrophoresis photograph showing the effects of metal complexes on pUC 19 DNA in the presence of H2O2: lane 1, DNA + [Ni(L1)2] + H2O2; lane 2, DNA + [Ni(L2)2]·H2O + H2O2; lane 3, DNA + [Ni(L3)2]·H2O + H2O2; lane 4, DNA + [Ni(L4)2] + H2O2; lane 5, DNA + [Zn(L1)2]·H2O + H2O2; lane 6, DNA + [Zn(L2)2] + H2O2; lane 7, DNA + [Zn(L3)2] + H2O2; lane 8, DNA + [Zn(L4)2]·H2O + H2O2; lane 9, DNA + FeSO4 + H2O2; lane C2, DNA + H2O2; lane C1, DNA alone.
4. Conclusions
Ni(II) and Zn(II) complexes have been synthesized using 3-formyl chromone Schiff bases and characterized by various analytical and spectral data. Based on the electronic spectra, magnetic moment, and elemental analysis data, octahedral geometry was proposed for Ni(II) and Zn(II) complexes. The well-defined crystalline and homogeneous nature of the metal complexes is observed from powder XRD and SEM analyses. The antimicrobial activity data has shown that [Zn(L3)2] complex displayed higher activity among all other metal complexes. The nematicidal activity of metal complexes revealed that [Ni(L3)2]·H2O and [Zn(L3)2] complexes showed moderate activity. [Zn(L4)2]·H2O complex exhibited greater antioxidant activity compared to the remaining metal complexes. All metal complexes exhibited considerable cytotoxic activity against Raw, MCF-7, and COLO 205 cell lines. The DNA cleavage studies of metal complexes showed more prominent activity in the presence of H2O2 compared to that in the absence of H2O2.
Acknowledgments
The authors wish to thank Dr. M.R.P. Reddy, CMET (Centre for the Materials for Electronic Technologies), Hyderabad, for providing TG facility. The authors are thankful to the Department of Biochemistry, SV University, Tirupathi, for antioxidant activity study, Vimta Labs Ltd., Hyderabad, for DNA cleavage studies, and PSG college of Pharmacy, Coimbatore, Tamilnadu, for anticancer activity studies. The authors also wish to thank the Ministry of Human Resource Development for granting the research fellowship to Kavitha Palakuri.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Singh NK, Srivastava A, Sodhi A, Ranjan P. In vitro and in vivo antitumour studies of a new thiosemicarbazide derivative and its complexes with 3d-metal ions. Transition Metal Chemistry. 2000;25(2):133–140. [Google Scholar]
- 2.Raman N, Pothiraj K, Baskaran T. DNA interaction, antimicrobial, electrochemical and spectroscopic studies of metal(II) complexes with tridentate heterocyclic Schiff base derived from 2′-methylacetoacetanilide. Journal of Molecular Structure. 2011;1000(1–3):135–144. [Google Scholar]
- 3.Yuan C, Lu L, Wu Y, et al. Synthesis, characterization, and protein tyrosine phosphatases inhibition activities of oxovanadium(IV) complexes with Schiff base and polypyridyl derivatives. Journal of Inorganic Biochemistry. 2010;104(9):978–986. doi: 10.1016/j.jinorgbio.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Yang Z-Y, Wu J-C. Synthesis, crystal structures, biological activities and fluorescence studies of transition metal complexes with 3-carbaldehyde chromone thiosemicarbazone. European Journal of Medicinal Chemistry. 2010;45(12):5692–5701. doi: 10.1016/j.ejmech.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Shim YS, Kim KC, Chi DY, Lee K-H, Cho H. Formylchromone derivatives as a novel class of protein tyrosine phosphatase 1B inhibitors. Bioorganic and Medicinal Chemistry Letters. 2003;13(15):2561–2563. doi: 10.1016/s0960-894x(03)00479-7. [DOI] [PubMed] [Google Scholar]
- 6.Khan KM, Ambreen N, Mughal UR, Jalil S, Perveen S, Choudhary MI. 3-formylchromones: potential antiinflammatory agents. European Journal of Medicinal Chemistry. 2010;45(9):4058–4064. doi: 10.1016/j.ejmech.2010.05.065. [DOI] [PubMed] [Google Scholar]
- 7.Mhaske G, Nilkanth P, Auti A, Davange S, Shelke S. Aqua medicated, microwave assisted, synthesis of Schiff bases and their biological evaluation. International Journal of Innovative Research in Science, Engineering and Technology. 2014;3:8156–8162. [Google Scholar]
- 8.Hussain MT, Rama NH, Khan KM. A novel unusual isocoumarin derivative, 3H-Furo[3, 4-c] isochromene-1, 5-dione. Letters in Organic Chemistry. 2010;7(7):557–560. [Google Scholar]
- 9.Osowole AA. Synthesis, characterization, in-vitro antibacterial and anticancer studies on some metal(II) complexes of (methylsulfanyl)chromenol Schiff base. Elixir Applied Chemistry. 2011;39:4827–4831. [Google Scholar]
- 10.Khan KM, Ahmad A, Ambreen N, et al. Schiff bases of 3-formylchromones as antibacterial, antifungal, and phytotoxic agents. Letters in Drug Design and Discovery. 2009;6(5):363–373. [Google Scholar]
- 11.Bhatnagar S, Sahi S, Kackar P, et al. Synthesis and docking studies on styryl chromones exhibiting cytotoxicity in human breast cancer cell line. Bioorganic and Medicinal Chemistry Letters. 2010;20(16):4945–4950. doi: 10.1016/j.bmcl.2010.05.108. [DOI] [PubMed] [Google Scholar]
- 12.Gomes A, Neuwirth O, Freitas M, et al. Synthesis and antioxidant properties of new chromone derivatives. Bioorganic & Medicinal Chemistry. 2009;17(20):7218–7226. doi: 10.1016/j.bmc.2009.08.056. [DOI] [PubMed] [Google Scholar]
- 13.Piao LZ, Park HR, Park YK, Lee SK, Park JH, Park MK. Mushroom tyrosinase inhibition activity of some chromones. Chemical and Pharmaceutical Bulletin. 2002;50(3):309–311. doi: 10.1248/cpb.50.309. [DOI] [PubMed] [Google Scholar]
- 14.Patil SA, Unki SN, Kulkarni AD, Naik VH, Badami PS. Synthesis, characterization, in vitro antimicrobial and DNA cleavage studies of Co(II), Ni(II) and Cu(II) complexes with ONOO donor coumarin Schiff bases. Journal of Molecular Structure. 2011;985(2-3):330–338. [Google Scholar]
- 15.Arjmand F, Sayeed F, Muddassir M. Synthesis of new chiral heterocyclic Schiff base modulated Cu(II)/Zn(II) complexes: their comparative binding studies with CT-DNA, mononucleotides and cleavage activity. Journal of Photochemistry and Photobiology B: Biology. 2011;103(2):166–179. doi: 10.1016/j.jphotobiol.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Sudhamani CN, Naik HSB, Naik TRR, Prabhakara MC. Synthesis, DNA binding and cleavage studies of Ni(II) complexes with fused aromatic N-containing ligands. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2009;72(3):643–647. doi: 10.1016/j.saa.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Qin D-D, Yang Z-Y, Wang B-D. Spectra and DNA-binding affinities of copper(II), nickel(II) complexes with a novel glycine Schiff base derived from chromone. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2007;68(3):912–917. doi: 10.1016/j.saa.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Wang B-D, Yang Z-Y, Lü M-H, Hai J, Wang Q, Chen Z-N. Synthesis, characterization, cytotoxic activity and DNA binding Ni(II) complex with the 6-hydroxy chromone-3-carbaldehyde thiosemicarbazone. Journal of Organometallic Chemistry. 2009;694(25):4069–4075. [Google Scholar]
- 19.Li Y, Yang ZY, Liao ZC, Han ZC, Liu ZC. Synthesis, crystal structure, DNA binding properties and antioxidant activities of transition metal complexes with 3-carbaldehyde-chromone semicarbazone. Inorganic Chemistry Communications. 2010;13(10):1213–1216. [Google Scholar]
- 20.Wang B-D, Yang Z-Y, Li T-R. Synthesis, characterization, and DNA-binding properties of the Ln(III) complexes with 6-hydroxy chromone-3-carbaldehyde-(2′-hydroxy) benzoyl hydrazone. Bioorganic and Medicinal Chemistry. 2006;14(17):6012–6021. doi: 10.1016/j.bmc.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Kavitha P, Saritha M, Reddy KL. Synthesis, structural characterization, fluorescence, antimicrobial, antioxidant and DNA cleavage studies of Cu(II) complexes of formyl chromone Schiff bases. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2013;102:159–168. doi: 10.1016/j.saa.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 22.Kavitha P, Chary MR, Singavarapu BVAA, Reddy KL. Synthesis, characterization, biological activity and DNA cleavage studies of tridentate Schiff bases and their Co(II) complexes. Journal of Saudi Chemical Society. 2013 [Google Scholar]
- 23.Kavitha P, Reddy KL. Pd(II) complexes bearing chromone based Schiff bases: synthesis, characterisation and biological activity studies. Arabian Journal of Chemistry. 2012 [Google Scholar]
- 24.Nohara A, Umetani T, Sanno Y. A facile synthesis of chromone-3-carboxaldehyde, chromone-3-carboxylic acid and 3-hydroxymethylchromone. Tetrahedron Letters. 1973;14(22):1995–1998. [Google Scholar]
- 25.Sigg I, Haas G, Winkler T. The reaction of 3-formylchromone with ortho-substituted anilines. Preparation of a Tetraaza [14] annulene. Helevetica Chimica Acta. 1982;65(1):275–279. [Google Scholar]
- 26.Khan KM, Ambreen N, Hussain S, Perveen S, Choudhary MI. Schiff bases of 3-formylchromone as thymidine phosphorylase inhibitors. Bioorganic and Medicinal Chemistry. 2009;17(8):2983–2988. doi: 10.1016/j.bmc.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Stemper JE, Matsen JM. Device for turbidity standardizing of cultures for antibiotic sensitivity testing. Applied Microbiology. 1970;19(6):1015–1016. doi: 10.1128/am.19.6.1015-1016.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanker K, Rohini R, Ravinder V, Reddy PM, Ho Y-P. Ru(II) complexes of N4 and N2O2 macrocyclic Schiff base ligands: their antibacterial and antifungal studies. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2009;73(1):205–211. doi: 10.1016/j.saa.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 29.Trivedi PC. Nematode Diseases in Plants. 1st edition. New Delhi, India: CBS; 1998. [Google Scholar]
- 30.Cayrol J, Djian C, Pijarowski L. Study of the nematicidal properties of the culture filtrate of the nematophagous fungus Paecilomyces lilacinus . Revue de Nematologie. 1989;12:331–336. [Google Scholar]
- 31.Braca A, de Tommasi N, di Bari L, Pizza C, Politi M, Morelli I. Antioxidant principles from Bauhinia tarapotensis . Journal of Natural Products. 2001;64(7):892–895. doi: 10.1021/np0100845. [DOI] [PubMed] [Google Scholar]
- 32.Sathiyaraj S, Butcher RJ, Jayabalakrishnan Ch. Synthesis, characterization, DNA interaction and in vitro cytotoxicity activities of ruthenium(II) Schiff base complexes. Journal of Molecular Structure. 2012;1030:95–103. doi: 10.1016/j.saa.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 33.Geary WJ. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coordination Chemistry Reviews. 1971;7(1):81–122. [Google Scholar]
- 34.Vogel AI. A Text Book of Quantitative Inorganic Analysis. 3rd edition. Longman; 1961. [Google Scholar]
- 35.El-Gammal OA, El-Reash GA, Ahmed SF. Structural, spectral, thermal and biological studies on 2-oxo-N′-((4-oxo-4H-chromen-3-yl)methylene)-2-(phenylamino)acetohydrazide (H2L) and its metal complexes. Journal of Molecular Structure. 2012;1007:1–10. [Google Scholar]
- 36.Anitha C, Sheela CD, Tharmaraj P, Raja SJ. Synthesis and characterization of VO(II), Co(II), Ni(II), Cu(II) and Zn(II) complexes of chromone based azo-linked Schiff base ligand. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2012;98:35–42. doi: 10.1016/j.saa.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 37.Padmaja M, Pragathi J, Kumari CG. Synthesis, spectral characterization, molecular modeling and biological activity of first row transition metal complexes with Schiff base ligand derived from chromone-3-carbaldehyde and o-amino benzoic acid. Journal of Chemical and Pharmaceutical Research. 2011;3(4):602–613. [Google Scholar]
- 38.Kannamba B, Reddy KL, AppaRao BV. Removal of Cu(II) from aqueous solutions using chemically modified chitosan. Journal of Hazardous Materials. 2010;175(1–3):939–948. doi: 10.1016/j.jhazmat.2009.10.098. [DOI] [PubMed] [Google Scholar]
- 39.Chandra S, Gupta LK. Spectroscopic and biological studies on newly synthesized nickel(II) complexes of semicarbazones and thiosemicarbazones. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2005;62(4-5):1089–1094. doi: 10.1016/j.saa.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Singh PK, Kumar DN. Spectral studies on cobalt(II), nickel(II) and copper(II) complexes of naphthaldehyde substituted aroylhydrazones. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2006;64(4):853–858. doi: 10.1016/j.saa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Warren BE. X-Ray Diffraction. 2nd edition. New York, NY, USA: Dover; 1990. [Google Scholar]
- 42.Huang X, Xia Y, Zhang H, et al. Synthesis, crystal structure, and fluorescence studies of (1-naphthyl)(pyridyl)urea metal complexes. Inorganic Chemistry Communications. 2008;11(4):450–453. [Google Scholar]
- 43.Sharma AK, Chandra S. Complexation of nitrogen and sulphur donor Schiff’s base ligand to Cr(III) and Ni(II) metal ions: synthesis, spectroscopic and antipathogenic studies. Spectrochimica Acta A: Molecular and Biomolecular Spectroscopy. 2011;78(1):337–342. doi: 10.1016/j.saa.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 44.Yousef TA, Abu El-Reash GM, El-Gammal OA, Bedier RA. Co(II), Cu(II), Cd(II), Fe(III) and U(VI) complexes containing a NSNO donor ligand: synthesis, characterization, optical band gap, in vitro antimicrobial and DNA cleavage studies. Journal of Molecular Structure. 2012;1029:149–160. [Google Scholar]
- 45.Seng H-L, Ong H-KA, Rahman RNZRA, et al. Factors affecting nucleolytic efficiency of some ternary metal complexes with DNA binding and recognition domains. Crystal and molecular structure of Zn(phen)(edda) Journal of Inorganic Biochemistry. 2008;102(11):1997–2011. doi: 10.1016/j.jinorgbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 46.Bharti SK, Patel SK, Nath G, Tilak R, Singh SK. Synthesis, characterization, DNA cleavage and in vitro antimicrobial activities of copper(II) complexes of Schiff bases containing a 2,4-disubstituted thiazole. Transition Metal Chemistry. 2010;35(8):917–925. [Google Scholar]
- 47.Sigman DS, Mazumder A, Perrin DM. Chemical nucleases. Chemical Reviews. 1993;93(6):2295–2316. [Google Scholar]
- 48.Kobayashi T, Kunita M, Nishino S, et al. Release of free nucleobases from oligomers by copper(II)-peroxide adduct. Polyhedron. 2000;19(26-27):2639–2648. [Google Scholar]
- 49.Yousef TA, Abu El-Reash GM, El-Gammal OA, Bedier RA. Synthesis, characterization, optical band gap, in vitro antimicrobial activity and DNA cleavage studies of some metal complexes of pyridyl thiosemicarbazone. Journal of Molecular Structure. 2013;1035:307–317. [Google Scholar]


