Abstract
Background
Many disease aetiologies have sex specific effects, which have important implications for disease management. It is now becoming increasingly evident that such effects are the result of the differential expression of autosomal genes rather than sex-specific genes. Such sex-specific variation in the response to Trichuris muris, a murine parasitic nematode infection and model for the human parasitic nematode T. trichiura, has been well documented, however, the underlying genetic causes of these differences have been largely neglected. We used the BXD mouse set of recombinant inbred strains to identify sex-specific loci that contribute to immune phenotypes in T. muris infection.
Results
Response phenotypes to T. muris infection were found to be highly variable between different lines of BXD mice. A significant QTL on chromosome 5 (TM5) associated with IFN-γ production was found in male mice but not in female mice. This QTL was in the same location as a suggestive QTL for TNF-α and IL-6 production in male mice suggesting a common control of these pro-inflammatory cytokines. A second QTL was identified on chromosome 4 (TM4) affecting worm burden in both male and female cohorts. We have identified several genes as potential candidates for modifying responses to T. muris infection.
Conclusions
We have used the largest mammalian genetic model system, the BXD mouse population, to identify candidate genes with sex-specific effects in immune responses to T. muris infection. Some of these genes may be differentially expressed in male and female mice leading to the difference in immune response between the sexes reported in previous studies. Our study further highlights the importance of considering sex as an important factor in investigations of immune response at the genome-wide level, in particular the bias that can be introduced when generalizing results obtained from only one sex or a mixed sex population. Rather, analyses of interaction effects between sex and genotype should be part of future studies.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-193) contains supplementary material, which is available to authorized users.
Keywords: Sex-dependent effect, Parasite, Gut, Trichuris muris, QTL, BXD, Mice
Background
Many human diseases differ in prevalence, course and severity between males and females [1, 2]. This sex bias is most obvious in human autoimmune conditions [3] that predominantly affect women. Additionally, many other diseases such as cardiovascular disease, osteoporosis and Alzheimer’s also have differing effects in males and females [1–3]. Males and females differ genetically by only a few genes located on sex-specific chromosomes, which are unlikely to account for the vast range of phenotypic differences. It has become increasingly evident that these sex-specific phenotypes are due to differentially expressed genes present in both sexes [4–6]. Such genotype by sex interactions have been observed in a number of organisms, from insects [7] to mammals [8]. Further, sexually dimorphic gene expression patterns are often tissue specific suggesting that different regulatory interactions might control gene expression in different tissues. This can involve genes exclusively expressed in one sex or genes expressed predominantly in one sex. The latter are often referred to as male or female biased genes, where male biased genes are generally more functionally diverse than female biased genes [9].
Over three billion people worldwide, mainly in developing countries, are thought to be susceptible to stable transmission of one or more of the four geohelminth species, Ascaris lumbricoides, Trichuris trichiura, Ancylostoma duodenale and Necator americanus[10]. T. trichiura, the human whipworm, is a key parasitic nematode that is known to cause considerable morbidity. This nematode has been the focus of much research (reviewed in [11]) using T. muris, the mouse whipworm, as a well-established model system. The immune response to this infection in mice is very well characterised and there is a distinct polarisation of immune response in resistant and susceptible strains of mouse [12, 13]. Resistant animals produce high levels of interleukin 13 (IL-13) and associated T helper type 2 (Th2) cytokines in response to infection, which is essential for parasite expulsion. In contrast, a susceptible animal produces high amounts of interferon γ (IFN-γ) and associated Th1 cytokines that leads to chronic infection. Importantly, the term resistance in this model is used when expulsion of the worms occurs before they become sexually mature and in both resistant and susceptible animals, T. muris establishes within the intestine. Resistance to T. muris has been demonstrated to have a strong genetic component with both H2 (major histocompatibility complex) linked genes and background genes influencing immunity [14, 15].
A known sex bias exists in immunity to T. muris infection [16, 17] whereby female mice are known to mount a stronger Th2 response to infection and are thus more resistant. This phenotype is more readily seen in genetically deficient animals such as the IL-4 KO mouse, although a weaker effect is still detectable in wild type mice [18]. Such sex-specific immune responses are also known in other models of helminth infection [19, 20]. Thus, while differences between sexes in immunity have been well documented, few studies have attempted to identify the underlying genetic variants that may cause such sex-specific responses. To date, a genome-wide analysis of genetic variants whose effects on immune response differ between the sexes has not been conducted in experimental populations.
In this study, we focus on sex-specific genetic effects on immune response phenotypes to T. muris in a population of BXD recombinant inbred (RI) mice. This reference panel consists of experimentally tractable mouse lines capturing a large amount of naturally occurring genetic variation and is ideally suited to integrate and analyse massive phenotypic data sets [21, 22] thus providing a valuable tool to identify loci that contribute to immune phenotypes in T. muris infection. To determine the heritable differences in immune phenotypes to T. muris, we profiled parasite burden and infection-induced cytokine responses in peripheral blood in 20 BXD lines and both parental strains, C57Bl/6J and DBA/2J. Our analysis concentrated on cytokines that are reliably measurable in serum as a reflection of inflammatory/immune events occurring in the infected animal. We used interval mapping and gene-mining tools in GeneNetwork (GN) to identify novel candidate loci involved in promoting immunity without bias to any particular subset of genes. We mapped a significant QTL to an interval on chromosome 5 (TM5) and an additional suggestive QTL to an interval on chromosome 4 (TM4). TM5 was male-biased suggesting that expression of a gene or multiple genes within this region are differentially expressed in male and female mice. This has important implications not only for highlighting new genes important in immunity to T. muris and by extension, T. trichiura, but also provides a mechanistic basis for the known sex bias in infection.
Results
Response phenotypes to T. murisare highly variable
The four measured immune phenotypes, worm burden, IFN-γ, Tumour Necrosis Factor α (TNF-α) and IL-6 all showed considerable variation across genotypes (Figure 1). Interestingly, the parental measurements did not represent the spread of the offspring results (Figure 1) and although there were no significant differences between DBA/2 and C57BL/6 mice, there were significant differences between the BXD RI lines (Additional file 1: Table S1). We detected a significant line effect on worm burden (ANOVA, F21,333 = 6.681, p < 0.0001), IFN-γ (ANOVA F21,333 = 2.289, p < 0.001), TNF-α (ANOVA, F21,333 = 1.660, p = 0.03) and IL-6 (ANOVA, F21,330 = 5.119, p < 0.0001).
Figure 1.
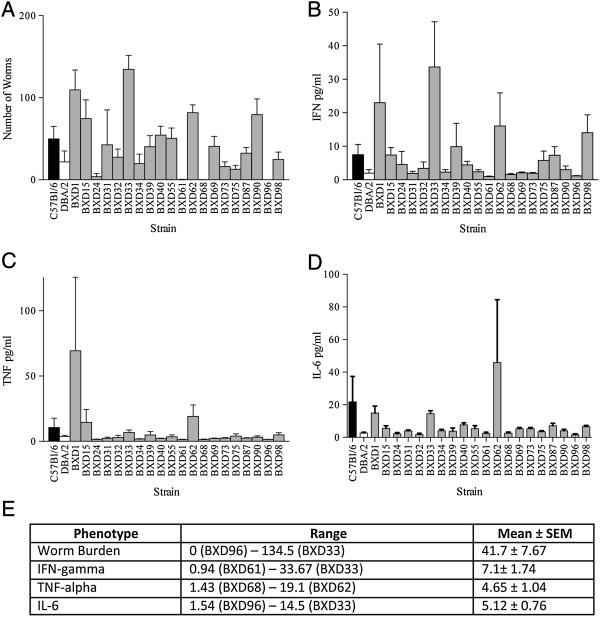
Immune response phenotypes to T. muris . Mean ± SEM of immune response phenotypes to T. muris (A) worm burden, (B) serum IFN-γ, (C) serum TNF-α and (D) serum IL-6 in BXD RI lines (grey bars) and the parental strains C57BL/6 (black bars) and DBA/2 (white bars). (E) Summary of range and mean ± SEM of response phenotypes.
Heritability for each phenotype was calculated by ANOVA with line as the independent variable using batch/age corrected values of our phenotypes. In BXD mice, heritabilities of worm burden by rank (no worms, low numbers, medium numbers and high numbers), IFN-γ, TNF-α and IL-6 were 42%, 22%, 14% and 18%, respectively.
Mapping immune phenotypes
In order to map variation related to worm burden and the levels of cytokines that were produced in response to infection, rather than possible confounding covariates, we used General Linear Models (GLMs) to remove all covariance associated with differences in age, batch of eggs used for infection, and maternal genotype. We found significant effects of age on worm burden (GLM F34,318 = 4.259, p < 0.001), IFN-γ (GLM F34,314 = 2.255, p < 0.001) and TNF-α (GLM F34,314 = 7.532, p < 0.001). Further, sex had a significant effect on worm burden (GLM F1,351 = 3.473, p = 0.063) and IFN-γ (GLM F1,347 = 4.269, p = 0.040) but not TNF-α or IL-6. Maternal genotype had significant effects on worm burden (GLM F1,351 = 23.695, p < 0.001) and IFN-γ (GLM F1,347 = 8.345, p = 0.004) while batch had significant effects on worm burden (GLM F7,345 = 3.379, p = 0.002). Residuals of these models were used to compute adjusted immune phenotypes. First, we mapped adjusted worm burden for the entire population and found a suggestive QTL, TM4, on Ch4 at 120.5-126.5 Mb (LRS = 13.1). An additional suggestive QTL, TM5, was found on Ch5 at 45–55 MB for all adjusted IFN-γ (LRS = 11.0), TNF-α (LRS = 11.8) and IL-6 (LRS = 11.2) (Additional file 2: Figure S1). To investigate sex-specific effects, the data was divided into male and female cohorts and mapped for adjusted worm burden, IFN-γ, TNF-α and IL-6 (Additional file 3: Figure S2 and Additional file 4: Figure S3). As before, there were no significant differences between the parent strains in any phenotype (males or female cohorts) though there were significant differences between lines (Additional file 5: Figure S4 and Additional file 6: Figure S5 and Additional file 7: Table S2). Mapping only male animals significantly improved the strength of linkage to both TM4 (LRS = 18.4) and TM5 (LRS = 22.6) (Figure 2A-D). It was immediately apparent from over-laying the QTL maps from male and female data that worm burden maps similarly for both sexes whereas for IFN-γ, TNF-α and IL-6, male data is most strongly linked to TM5. Additionally, when the QTL maps for IFN-γ, TNF-α and IL-6 were overlaid, the TM5 QTL peaks were all in the same position on Ch5, though it only reached significance with the IFN-γ phenotype (Figure 2E).
Figure 2.
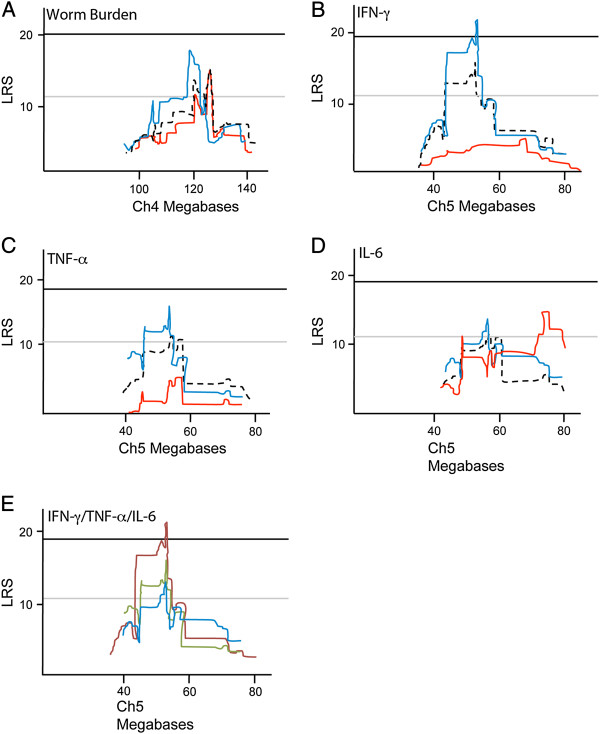
Interval maps for immune response phenotypes in male, female and combined male and female cohorts. QTL for male (blue line) and female (red line) as compared to both sexes together (broken black line) for (A) worm burden (Ch4), (B) IFN-γ, (C) TNF-α, (D) IL-6 (all Ch5) response phenotypes. (E) Overlap of male QTL for INF-γ (red line), TNF-α (green line) and IL-6 (blue line). Upper dark line on maps show significant LRS scores whilst lower lines show suggestive LRS scores.
Both loci were linked to susceptibility. The DBA/2 (D2) allele at TM4 was associated with a 2-fold increase in worm burden when compared with strains having a C57BL/6 (B6) allele (t16 = 2.278, p = 0.04), while a B6 allele at TM5 was associated with a 2-fold increase in IFN-γ production (t19 = 2.421, p = 0.02).
Candidate gene analysis
Candidate genes were selected using the QTLminer module of GN that ranks genes by whether the parent strains have non-synonymous SNPs (nsSNPs) or indels, whether the candidates are expressed in tissues of interest and whether their expression is modulated by cis-eQTL. For each of these four categories a score is assigned to candidates, with a maximum of 4. As GN does not have a gene expression database for the caecum or large intestine, we instead used mRNA expression from naïve T helper cells and leucocytes [23]. However, as worm burden is known to be dependent on gut function [24, 25] we did not reject genes as candidates if they were not expressed in these target cells, provided they had a known biological relevance to the gut. Of the 93 genes within TM4, 16 genes scored 3 or higher out of the possible 4. This was further reduced to ten genes (Table 1) by choosing only candidates located within the QTL for both male and female cohorts (120.5 MB – 126.5 MB), which are therefore likely to be controlled similarly in both males and females.
Table 1.
Candidate genes in TM4
| Gene symbol | Description | Mb location | LRS | SNP region |
|---|---|---|---|---|
| Cap1 | CAP, adenylate cyclase-associated protein 1 | 122.53 | 16.81* | 3′ UTR, exon 2, 8 and 10 |
| Mycbp | c-myc binding protein | 123.58 | 16.81* | 3′ UTR |
| Zmpste24 | Zinc metallopeptidase, ste24 homolog | 120.73 | 18.45* | 3′ UTR, exon 1 and 2 |
| Ppt1 | Palmitoyl-protein thioesterase 1 | 122.51 | 16.81* | 3′ UTR and exon 6 |
| Mtf1 | Metal response element binding transcription factor 1 | 124.47 | 16.81* | 3′ UTR, 5′ UTR, exon 11 |
| Macf1 | Microtubule-actin crosslinking factor 1 | 123.02 | 16.81* | Exons 1, 9, 10, 19, 20, 35–40, 43–45, 47–48 |
| Zfp69 | Zinc finger protein 69 | 120.6 | 15.58* | 5′ UTR, exons 3, 4 and 6 |
| Smap2 | Stromal membrane-associated GTPase-activating protein 2 | 120.64 | 15.58* | 5′ UTR |
| Inpp5b | Inositol polyphosphate-5-phosphatase B | 124.41 | 16.81* | Exon 1, 2, 3 and 8 |
| Rspo1 | R-spondin homolog | 124.66 | 9.52 | Intronic only |
*Suggestive LRS = 12.23.
Similarly, QTLminer was used to identify candidate genes in TM5. Again, T helper cells and leucocyte mRNA expression databases were used, as these are the principal cells that would be producing IFN-γ in the serum. Of the 44 genes within this QTL region, only 19 genes had nsSNPs between B6 and D2. Of these 19, seven genes (Table 2) additionally had indels, high expression in target tissues and/or evidence of cis regulation in target tissues.
Table 2.
Candidate genes in TM5
| Gene symbol | Description | Mb location | LRS | SNP region |
|---|---|---|---|---|
| Pi4k2b | Phospohatidylinositol 4-kinase type 2 beta | 53.13 | 18.61* | Exons 1, 2 and 7 |
| Lap3 | Leucine aminopeptidase 2 | 45.88 | 17.45* | Exons 1 and 5 |
| Tapt1 | Transmembrane anterior posterior transformation 1 | 44.56 | 18.45* | 3′ UTR |
| Sepsecs | Sep (O-phosphoserine) tRNA:Sec (selenocysteine tRNA synthase) | 53.03 | 18.61* | 3′ UTR, exon 7 |
| Lcorl | Ligand dependent nuclear receptor corepressor-like | 46.12 | 17.45* | 3′ UTR, exons 1 and 4 |
| Anapc4 | Anaphase promoting complex subunit 4 | 53.22 | 18.61* | Exons 1 and 4 |
| Slit2 | Slit homolog 2 (Drosophila) | 48.37 | 17.45* | 3′ UTR, 5′ UTR, exons 9, 17, 20, 26 and 32 |
| SOD3 | Superoxide dismutase 3 | 52.75 | 18.61* | 3′ UTR |
*Suggestive LRS = 11.89.
Epistasis
We next investigated epistatic interactions (i.e. gene by gene interactions) and found a significant interaction between TM4 and a locus on Ch16, TM16 (Full model LRS = 39.03, p < 0.05) (Figure 3A and C). Those strains with B6 alleles at both QTL have significantly reduced worm burdens (Figure 3B). The genetic correlation between markers at these two loci was not significant (r = 0.051, p = 0.8), thereby excluding the possibility of non-syntenic association [26]. No epistatic interactions with TM5 were found.
Figure 3.
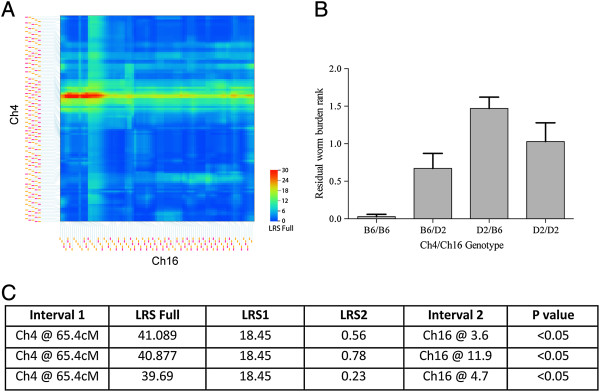
Pair-scan correlation between two chromosomes for worm burden. Pair scan analysis demonstrates significant interactions between TM4 and TM16 for the worm burden phenotype. (A) Enlargement of a significant interaction of TM4 and TM16 from the pair-scan analysis. (B) Histogram illustrating the effect of adjusted worm burden of carrying either the maternal (B6) or paternal (D2) or both alleles at the TM4 and TM16 intervals. (C) LRS scores of TM4 and TM16. *p < 0.05.
To further investigate the interaction effects of TM4 and TM16, we looked for correlations in mRNA expression in T helper cells and leucocytes across BXD lines in genes contained within both intervals using GN databases. TM16 contains 252 genes, of which 58 had nsSNPs between B6 and D2. Again, QTLminer was used to rank candidate genes based on nsSNPs between the parental strains, indels, expression in target tissues or cells and evidence of cis-eQTLs. Of the 252 genes, 16 genes scored highly, of which only seven correlated with expression of genes in TM4. An additional two genes were included (Table 3) that did not score highly but were nonetheless considered to be biologically relevant; Masp1 and Clcn2. Interestingly, Ch4 genes Ppt1 and Inpp5b map as trans-QTL to Ch16 (Figure 4A and B respectively), at the locus that is in epistatic interaction with TM4. To summarize the interactions of genes in TM4 and TM16, we have constructed a model based on the expression and correlation of these genes with the worm burden phenotype (Figure 4C).
Table 3.
Candidate genes within the interactions locus TM16
| Gene | Description | Mb location | SNP region |
|---|---|---|---|
| Abcc1 | ATP-binding cassette, sub-family C (CFTR/MRP), member1 | 14.36 | 3′ UTR and multiple exons (13) |
| Mkl2 | MKL/Myocardin-like 2 | 13.35 | Exons 9, 10, 11, 12, 13 and 15 |
| Gspt1 | G1 to S phase transition 1 | 11.21 | 3′ UTR, exons 1 and 2 |
| Nde1 | Nuclear distribution gene E homolog 1 | 14.16 | Exons 1-4 |
| Dexi | Dexamethasone-induced transcript | 10.53 | 3′ UTR |
| Bcl6 | B-cell leukaemia/lymphoma 6 | 23.96 | Exon 2 |
| Senp2 | SUMO/Sentrin specific peptidase 2 | 22.00 | Exon 1 |
| Myh11 | Myosin, heavy polypeptide 11, smooth muscle | 14.19 | Multiple exons (11) |
| CIIta | Class II transactivator | 10.48 | 3′ UTR, exons 9, 10 and 11 |
| Masp1 | Mannan-binding lectin serine peptidase 1 | 23.45 | Exon 16 |
| Clcn2 | Chloride channel 2 | 20.70 | Exons 1 and 2 |
Figure 4.
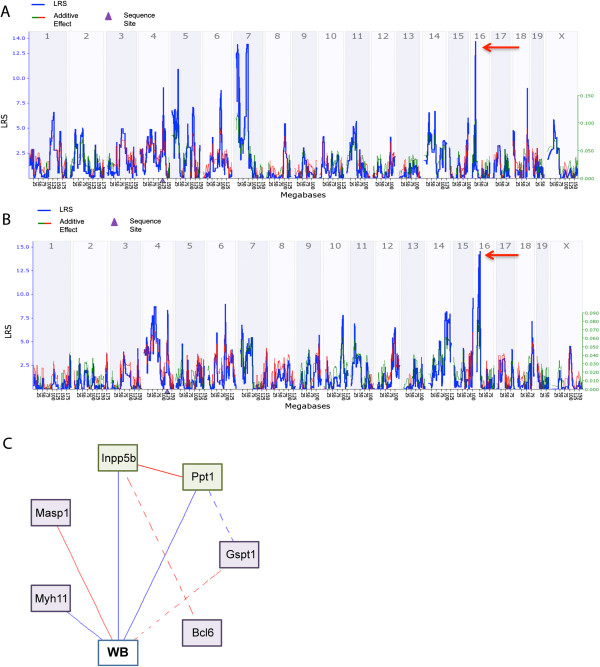
Correlations and networks for genes in TM4 and TM16 . (A) Interval map for Ppt1 gene expression. This TM4 gene modulates gene expression in the TM16 region (red arrow). (B) Interval map for the TM4 gene Innpb5 that also modulates gene expression in the TM16 region (red arrow). (C) Hypothetical gene network modulating worm burden based on gene correlations between TM4 and TM16. Solid blue lines represent correlations between -0.7 and -0.5, broken blue lines between -1.0 and -0.7. Solid and broken red lines represent the equivalent positive correlations. Phenotypes are in white boxes; TM4 genes are in green boxes whilst TM16 genes are in purple boxes. p ≤ 0.01 in all cases.
Interactions between Worm Burden and IFN-γ phenotypes
Our worm burden phenotype and IFN-γ phenotypes positively correlate with each other (Figure 5A) as expected; it is known that high worm burdens at day 35 post infection (p.i.) are associated with an increased production of IFN-γ. Correlations between the expression of genes in TM4 and TM5 in T helper cell and leucocytes were identified on GN and have been summarised in a network graph (Figure 5B).
Figure 5.
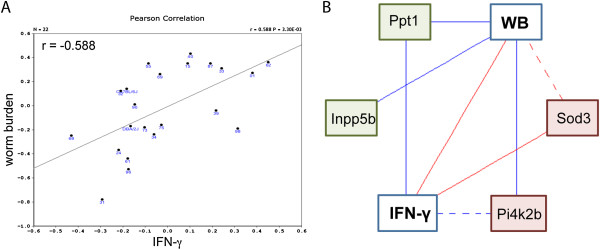
Correlation and networks for worm burden and IFN-γ phenotype genes. (A) Spearman rank correlation (in box) of worm burden with IFN-γ response phenotype. (B) Hypothetical gene network modulating worm burden based on gene correlations between genes in TM4 and TM5. Solid blue lines represent correlations between -0.7 and -0.5, broken blue lines between -1.0 and -0.7. Solid and broken red lines represent the equivalent positive correlations. Phenotypes are in white boxes, TM4 genes are in green boxes and TM5 genes are in red boxes. p ≤ 0.01 in all cases.
Discussion
Sex bias in many diseases has been well documented, particularly with autoimmune diseases, and is attributable in part to the different immune responses that male and females are able to mount. Such sex-dependent effects are potentially caused by differential expression of autosomal genes [8, 9], yet the underlying genetics of this sex-dependence has hitherto been poorly explored. However, if we wish to develop effective interventions we need to understand the genetics of sex-dependent immune responses.
In an effort to address the paucity of data on genetic architecture underlying sex-specific immune response at a genome-wide level we identified loci TM4 and TM5 associated with susceptibility parameters of infection: worm burden and the production of IFN-γ, a Th1 cytokine. Moreover, suggestive QTL can be found in exactly the same position as TM5 when looking at two other susceptibility associated cytokines, IL-6 and TNF-α, suggesting a common control of all these pro-inflammatory cytokines. TM5 was only apparent when analysing male data whereas TM4, associated with worm burden, was apparent in both male and female cohorts. This would suggest that worm burden is controlled similarly in both sexes, whereas the production of the pro-inflammatory cytokines in response to T. muris is differentially controlled in male and female mice, in particular given that this QTL is not located on chromosome X or Y. This phenomenon is also seen in other disease systems, such as coronary heart disease and kidney diseases [3] highlighting the importance of sex on disease outcome. Another recent QTL study utilising the T. muris system [27] has been conducted using an intercross approach between AKR and BALB/c mice with different parameters of infection considered (absolute worm burden and antibody response). Thus, our study here has identified different QTL associated with sex-dependent immune responses, rather than QTL associated with colitis as in the intercross study [27].
Parasite niche in the host
T. muris is a parasite that lives in close association with its host, embedding into the caecal epithelium and therefore causing damage to the integrity of the gut. It is further known that the intestinal epithelium plays a key role in expulsion [24], as resistant animals have a faster rate of epithelial turnover than susceptible animals. Furthermore, increasing the turnover rate in susceptible animals enables expulsion. This process will require a fine balance between apoptosis and proliferation within the gut. Our study has identified several genes from three different chromosomes that play a role in epithelial barrier integrity and host responses to damage.
MacF1 is a member of the Wnt signalling pathway [28], a highly conserved signal transduction cascade that is known to play a role in gut epithelium homeostasis [29]. Although Rspo1 does not score highly in the system used to identify candidate genes, it is biologically relevant in the context of T. muris infection as it is involved in the proliferative control of intestinal crypt cells, which will be in close association with the parasite, and enhances Wnt/β-catenin signalling in the intestine [30, 31]. Additionally, QTLminer analysis does not include intronic SNPs that may have regulatory functions, which can be found within the Rspo1 gene. Thus these two genes may be important in T. muris infection by regulating the speed of epithelial turnover in the intestine. Rspo1 has also been shown to ameliorate colitis in mice by improving the mucosal integrity of the gut [32]. Clcn2 is known to be important in regulating intestinal mucosa barrier function [33] and like Rspo1, is important in the recovery of the epithelial barrier in the intestine after damage [34], which will be important in T. muris infection. Not only has mucus been shown to be extremely important in protecting against T. muris infection [35] but also that T. muris worms secrete specific proteins to break down the mucus barrier [36]. As turnover is finely balanced with apoptosis [37] in the gut, and apoptosis has been shown to be important during T. muris infection [25], the candidate gene Ppt1 is of particular interest because it is known to modulate TNF-α induced apoptosis [38]. Accelerated turnover is thought to dislodge T. muris from its niche, pushing it into the lumen of the gut where 'weep and sweep’ mechanisms will then facilitate expulsion. Myh11 is a contractile protein of smooth muscle and so may be important in the expulsion of T. muris worms by peristalsis or by maintaining the integrity of the smooth muscle in the intestine [39].
Damage to the host and repair
Another aspect of T. muris chronic infection is the damage caused by the parasite burrowing through the epithelium. A number of studies on intestinal inflammation have shown a close association of this inflammation with tissue hypoxia. T. muris has been shown to share many phenotypic characteristics of Inflammatory Bowel Diseases (IBD) [40] and as damage is sustained during infection, then it is likely that genes in the hypoxia pathway are likely to be important in protecting the host. SOD3 is an antioxidant enzyme that is known to protect tissues against hypoxic stress [41, 42] and is a primary scavenger of superoxide. It is known that the induction of HIF-1α is regulated closely by oxidant/antioxidant equilibrium involving SOD3[43]. Interestingly, HIF-1α has been shown to be upregulated during chronic T. muris infection (data not shown). Another hypoxia gene highlighted in this study is Mtf1, which induces metallotheionein transactivation [44]. Metallothioneins are a family of small proteins that may have a potential role in IBD [45] and as such may indeed play a role in T. muris infection.
Adaptive immune responses to infection
T. muris infection is controlled largely by the production of Th2 cytokines. Several genes involved in T cell signalling have been identified in this study. MacF1, as well as being important in the homeostasis of the gut epithelium, may also be important as down stream events of MacF1 signalling, such as beta-catenin degradation, are implicated in T cell receptor signalling [46, 47]. Pi4k2b is associated with components of the T cell receptor [48, 49] and so may also play an important role in T. muris infection. CIIta is essential for controlling MHCII expression [50] and so is important in Th2 cell recognition of parasite antigens and subsequent activation. A mutation in this protein is known to upregulate IL-33 dependent differentiation of Th2 cells [51], a process that has been shown to be critical in immunity to T. muris[52]. Senp2 is another member of the Wnt signalling pathway, like MacF1[53]. Additionally, it can conjugate with PPARγ, a nuclear receptor, affecting the transcription of PPARγ response genes [54]. PPARγ is known to have multiple functions in the immune system [55, 56], including protection against IBD.
The loci we identified in this study are susceptibility loci. Susceptible animals mount a strong Th1 response to infection that then facilitates the survival of this parasite. It is not clearly understood why susceptible animals do not mount a Th2 response to infection. Interestingly, two genes identified in this study are known to be negative regulators of immune responses. Slit2 inhibits dendritic cell and neutrophil migration [57–59] whereas Bcl6 is known to negatively regulate Th2 immunity [60, 61]. Thus, it may be that susceptible animals mount strong Th1 responses, at least partly via down-regulation of Th2 immunity through such genes.
Finally, the two response phenotypes, IFN-γ and worm burden, measured in this experiment were also found to positively correlate with each other and further correlations between expression levels in TM4 and TM5 were identified. This has therefore allowed us to construct hypothetical models of interactions between genes on three different chromosomes based on correlation and expression.
Conclusions
Sex-dependence is known to play an important role in the prevalence or severity of diseases and it is becoming increasingly apparent that it is variation within the autosomal genome that cause these wide ranging effects. This study further highlights the importance of sex in parasitic infections and identifies several genes that may be differentially regulated and/or expressed in male and female animals. This not only has implications for research, often dominated by single sex studies, but may also lead to the identification of genes important in T. muris infection that are not considered solely due to a known role in immune responses, a bias that will discount a myriad of genes that may well be critical.
Methods
Mice
C57BL/6JOlaHsd and DBA/2J parent mice were obtained from Harlan and BXD recombinant inbred mice were obtained from the University of Tennessee Health Science Centre, Memphis, TN, USA. All mice were subsequently bred and maintained under specific pathogen-free conditions at the University of Manchester. 353 mice were used in this study (174 males and 179 females) and analysed as groups within lines. Mice in all lines ranged in age from 12–40 weeks and at least 6 males and 6 females from each line were used. All procedures conformed to the requirements of the UK Animals (Scientific Procedures) Act 1986, were subject to local ethical review by the University of Manchester Ethical Review Panel and followed ARRIVE guidelines.
BXD is the largest and best phenotyped genetic model system in mammals, and is derived from two divergent mouse strains (C57Bl/6 J and DBA/2 J, hence BXD), in which different recombination patterns have been inbred (hence recombinant inbred) in over 100 highly diverse lines, each with a fixed recombination pattern of exactly two possible alleles [21, 22]. These strains incorporate 4–5 million segregating single nucleotide polymorphisms, 500,000 insertions and deletions, and 55,000 copy number variants (1 kb to 100 kb). This is sufficient complexity to model the genetics of human populations used in linkage and genome-wide association studies (GWAS).
Parasite maintenance and infection
Stock infections of T. muris were maintained in susceptible mouse strains and adult worms harvested at day 42 p.i. Adult worms were cultured in RPMI 1640 supplemented with 10% FCS, 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (all Invitrogen, UK) and Excretory/Secretory (E/S) antigen and eggs were collected after four hours incubation. T. muris E/S was prepared as follows. The E/S was pelleted to remove eggs, concentrated using a Centriprep YM-10 (Amicon, Gloucester, UK) and then dialysed against PBS. Protein concentration was determined by Nanodrop. Eggs were allowed to embryonate for at least six weeks in dH2O and infectivity established by worm burden in a susceptible mouse strain. Mice were infected with 150–200 embryonated eggs and worm burdens established at day 35 p.i. Seven separate batches of eggs were used for infections, all from the same passage. Parasite specific IgG1 ELISAs as described in [62] were carried out to confirm infection in all animals.
Immunophenotyping
Adult worm burdens were assessed at day 35 p.i. via longitudinal dissection of caeca and proximal colons. Serum was taken from infected animals at day 35 p.i. Cytokines IFN-γ, TNF-α and IL-6 were measured as they can be detected in sera and provide an indication of the ongoing immune response to T. muris. Commercial Cytometric Bead Array Flex Sets (BD, UK) were used to determine the levels (pg/ml) of IFN-γ, TNF-α and IL-6 in sera according to manufacturer’s instructions. Fluorescence was analysed using a flow cytometer (FACSArray, BD) and cytokine levels were determined using BD CBA software. Linear regression models (GLMs) in SPSS were used to adjust for age, batch and maternal genotype in both worm burden and cytokine levels.
Genetic analysis
QTL analysis was performed using interval mapping [63] as implemented in the WebQTL module of GN [64]. To identify potential candidate genes we used both information from the UCSC genome browser and GO annotation, and QTL miner in GN [65]. Further, we analysed covariation networks and correlated worm burden and IFN-γ phenotypes with BXD genotype databases (leucocyte mRNA and T cell (helper) mRNA) in GN that contain an extensive collection of previously published and unpublished data from BXD [23].
Statistical analysis
We used ANOVA and univariate General Linear Models as implemented in SPSS (v20).
Availability of supporting data
All supporting data available as additional files.
Electronic supplementary material
Additional file 1: Table S1: Table of immune response phenotypes. Multiple comparisons of one-way ANOVA on worm burden, serum IFN-γ, serum TNF-α and serum IL-6 in BXD RI lines and the parental strains C57BL/6 and DBA/2. (CSV 41 KB)
Additional file 2: Figure S1: Interval maps for immune response phenotypes in combined male and female cohorts. QTL for (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey line shows suggestive LRS scores. (TIFF 2 MB)
Additional file 3: Figure S2: Interval maps for immune response phenotypes in male cohorts only. QTL for (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey lines show suggestive LRS scores. (TIFF 2 MB)
Additional file 4: Figure S3: Interval maps for immune response phenotypes in female cohorts only. QTL (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey lines show suggestive LRS scores. (TIFF 2 MB)
Additional file 5: Figure S4: Male immune response phenotypes to T. muris. Mean ± SEM of immune response phenotypes in male mice to T. muris (A) worm burden, (B) serum IFN-γ, (C) serum TNF-α and (D) serum IL-6 in BXD RI lines (grey bars) and the parental strains C57BL/6 (black bars) and DBA/2 (white bars). (E) Summary of range and mean ± SEM of response phenotypes. (TIFF 406 KB)
Additional file 6: Figure S5: Female immune response phenotypes to T. muris. Mean ± SEM of immune response phenotypes in female mice to T. muris (A) worm burden, (B) serum IFN-γ, (C) serum TNF-α and (D) serum IL-6 in BXD RI lines (grey bars) and the parental strains C57BL/6 (black bars) and DBA/2 (white bars). (E) Summary of range and mean ± SEM of response phenotypes. (TIFF 407 KB)
Additional file 7: Table S2: Table of immune response phenotypes in male and female cohorts. Multiple comparisons of one-way ANOVA on worm burden, serum IFN-γ, serum TNF-α and serum IL-6 in BXD RI lines and the parental strains C57BL/6 and DBA/2 in male or female cohorts. (CSV 82 KB)
Acknowledgements
This work was funded by a Wellcome Trust Program Grant (083620Z). The authors would like to thank the staff of the University of Manchester BSF Unit for all technical help. Thanks also go to Dr Joanne Pennock for help with the manuscript.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KH carried out all experimental work and analysis. RH participated in study design and coordination. RG conceived of the study and participated in its design and coordination. All authors read and approved the final manuscript.
Contributor Information
Kelly S Hayes, Email: Kelly.hayes@manchester.ac.uk.
Reinmar Hager, Email: Reinmar.hager@manchester.ac.uk.
Richard K Grencis, Email: Richard.grencis@manchester.ac.uk.
References
- 1.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Regitz-Zagrosek V, Dworatzek E, Kintscher U, Dragun D. Sex and sex hormone-dependent cardiovascular stress responses. Hypertension. 2013;61:270–277. doi: 10.1161/HYPERTENSIONAHA.111.189233. [DOI] [PubMed] [Google Scholar]
- 3.Lockshin MD. Sex differences in autoimmune disease. Lupus. 2006;15:753–756. doi: 10.1177/0961203306069353. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Weiss LA, Abney M, Cook EH, Ober C. Sex-specific genetic architecture of quantitative traits in humans. Nat Genet. 2006;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- 6.Hager R, Cheverud JM, Leamy LJ, Wolf JB. Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol. 2008;8:303–311. doi: 10.1186/1471-2148-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay TFC. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Op Genet Develop. 2004;14:253–257. doi: 10.1016/j.gde.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 10.Pullan RL, Brooker SJ. The global limits and population at risk of soil-transmitted helminth infections in 2010. Parasit Vectors. 2012;5:81–94. doi: 10.1186/1756-3305-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klementowicz JE, Travis MA, Grencis RK. Trichuris muris: a model of gastrointestinal parasite infection. Semin Immunopathol. 2012;34:815–828. doi: 10.1007/s00281-012-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Else KJ, Grencis RK. Cellular immune responses to the murine nematode parasite Trichuris muris. I. Differential cytokine production during acute or chronic infection. Immunology. 1991;72:508–513. [PMC free article] [PubMed] [Google Scholar]
- 13.Else KJ, Hultner L, Grencis RK. Cellular immune responses to the murine nematode parasite Trichuris muris. II. Differential induction of TH-cell subsets in resistant versus susceptible mice. Immunology. 1992;75:232–237. [PMC free article] [PubMed] [Google Scholar]
- 14.Else KJ, Wakelin D. The effects of H-2 and non-H-2 genes on the expulsion of the nematode Trichuris muris from inbred and congenic mice. Parasitology. 1988;96:543–550. doi: 10.1017/S0031182000080173. [DOI] [PubMed] [Google Scholar]
- 15.Else KJ, Wakelin D. Genetic variation in the humoral immune responses of mice to the nematode Trichuris muris. Parasite Immunol. 1989;11:77–90. doi: 10.1111/j.1365-3024.1989.tb00650.x. [DOI] [PubMed] [Google Scholar]
- 16.Hepworth MR, Grencis RK. Disruption of Th2 immunity results in a gender-specific expansion of IL-13 producing accessory NK cells during helminth infection. J Immunol. 2009;183:3906–3914. doi: 10.4049/jimmunol.0900577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepworth RM, Hardman MJ, Grencis RK. The role of sex hormones in the development of Th2 immunity in a gender-biased model of Trichuris muris infection. Eur J Immunol. 2010;40:406–416. doi: 10.1002/eji.200939589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bancroft AJ, Artis DA, Donaldson DD, Sypek JP, Grencis RK. Gastrointestinal nematode expulsion in IL-4 knockout mice is IL-13 dependent. Eur J Immunol. 2000;30:2083–2091. doi: 10.1002/1521-4141(200007)30:7<2083::AID-IMMU2083>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Poulin R. Sexual inequalities in helminth infections: a cost of being a male? Am Nat. 1996;147:287–296. doi: 10.1086/285851. [DOI] [Google Scholar]
- 20.Klein SL. Hormonal and immunological mechanisms mediating sex differences in parasite infections. Parasite Immunol. 2004;26:247–264. doi: 10.1111/j.0141-9838.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- 21.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 22.Hager R, Lu L, Rosen GD, Williams RE. Genetic architecture supports mosaic brain evolution and independent brain body size regulation. Nat Commun. 2012;3:1–5. doi: 10.1038/ncomms2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberts R, Chen H, Pommerenke C, Smit AB, Spijker S, Williams RW, Geffers R, Bruder D, Schughart K. Expression QTL mapping in regulatory and helper T cells from the BXD family of strains reveals novel cell-specific genes, gene-gene interactions and candidate genes for auto-immune disease. BMC Genomics. 2011;12:610–623. doi: 10.1186/1471-2164-12-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- 25.Cliffe LJ, Potten CS, Booth CE, Grencis RK. An increase in epithelial cell apoptosis is associated with chronic intestinal nematode infection. Infect Immun. 2007;75:1556–1564. doi: 10.1128/IAI.01375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:1–18. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levison SE, Fisher P, Hankinson J, Zeef L, Eyre S, Ollier WE, McLaughlin JT, Brass A, Grencis RK, Pennock JL. Genetic analysis of the Trichuris muris-induced model of colitis reveals QTL overlap and a novel gene cluster for establishing colonic inflammation. BMC Genomics. 2013;14:127–140. doi: 10.1186/1471-2164-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H-J, Lin C-M, Lin C-S, Perez-Olle R, Leung CL, Liem RKH. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 1933;2006:20. doi: 10.1101/gad.1411206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero JB, Sansom OJ. Wnt signalling and its role in stem cell-driven intestinal regeneration and hyperplasia. Acta Physiol. 2012;204:137–143. doi: 10.1111/j.1748-1716.2011.02288.x. [DOI] [PubMed] [Google Scholar]
- 30.De Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, Van Es JE, Guardavaccaro D, Schasfoort RBM, Mohri Y, Nishimori K, Mohammed S, Heck AJR, Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 31.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, De Vera J, Narushima S, Beck EX, Palencia S, Shinkawa P, Kim K-A, Liu Y, Levy MD, Berg DJ, Abo A, Funk WD. R-spondin1, a novel intestinotrophic mitogen, ameliorates experimental colitis in mice. Gastroenterology. 2007;132:1331–1343. doi: 10.1053/j.gastro.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Nighot PK, Blikslager AT. ClC-2 regulates mucosal barrier function associated with structural changes to the villus and epithelial tight junction. Am J Physiol Gastrointest Liver Physiol. 2010;299:G449–G456. doi: 10.1152/ajpgi.00520.2009. [DOI] [PubMed] [Google Scholar]
- 34.Nighot PK, Moeser AJ, Ryan KA, Ghashghaei T, Blikslager AT. ClC-2 is required for rapid restoration of epithelial tight junctions in ischemic-injured murine jejunum. Exp Cell Res. 2009;315:110–118. doi: 10.1016/j.yexcr.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Hasnain SZ, Wang H, Ghia J-E, Haq N, Deng Y, Velcich A, Grencis RK, Thornton DJ, Khan WI. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. 2010;138:1763–1771. doi: 10.1053/j.gastro.2010.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasnain SZ, McGuckin MA, Grencis RK, Thornton DJ. Serine protease(s) secreted by the nematode Trichuris muris degrade the mucus barrier. PLoS Negl Trop Dis. 2012;6:e1856. doi: 10.1371/journal.pntd.0001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuang S-Q, Kwartler CS, Byanova KL, Pham J, Gong L, Prakash SK, Huang J, Kamm KE, Stull JT, Sweeney HL, Milewicz DM. Rare, nonsynonymous variant in the smooth muscle-specific isoform of myosin heavy chain, MYH11, R247C, alters force generation in the aorta and phenotype of smooth muscle cells. Circ Res. 2012;110:1411–1422. doi: 10.1161/CIRCRESAHA.111.261743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardy C, Sabourdy F, Garcia V, Jalanko A, Therville N, Levade T, Andrieu-Abadie N. Palmitoyl protein thioesterase 1 modulates tumor necrosis factor alpha-induced apoptosis. Biochim Biophys Acta. 2009;1793:1250–1258. doi: 10.1016/j.bbamcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Wallace KN, Dolan AC, Seiler C, Smith EM, Yusuff S, Chaille-Arnold L, Judson B, Sierk R, Yengo C, Sweeney HL, Pack M. Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev Cell. 2005;8:717–726. doi: 10.1016/j.devcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Levison SE, McLaughlin JT, Zeef LAH, Fisher P, Grencis RK, Pennock JL. Colonic transcriptional profiling in resistance and susceptibility to trichuriasis: phenotyping a chronic colitis and lessons for iatrogenic helminthosis. Inflamm Bowel Dis. 2010;16:2065–2079. doi: 10.1002/ibd.21326. [DOI] [PubMed] [Google Scholar]
- 41.Mysore TB, Shinkel TA, Collins J, Salvaris EJ, Fisicaro N, Murray-Segal LJ, Johnson LEA, Lepore DA, Walters SN, Stokes R, Chandra AP, O'Connell PJ, d'Apice AJF, Cowan PJ. Overexpression of glutathione peroxidase with two isoforms of superoxide dismutase protects mouse islets from oxidative injury and improves islet graft function. Diabetes. 2005;54:2109–2116. doi: 10.2337/diabetes.54.7.2109. [DOI] [PubMed] [Google Scholar]
- 42.Nozik-Grayck E, Suliman HB, Majka S, Albietz J, Van Rheen Z, Roush K, Stenmark KR. Lung EC-SOD overexpression attenuates hypoxic induction of Egr-1 and chronic hypoxic pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2008;295:L422–L430. doi: 10.1152/ajplung.90293.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannan KB, Colorado I, Reino D, Palange D, Lu Q, Qin X, Abungu B, Watkins A, Caputo FJ, Xu D-Z, Semenza GL, Deitch EA, Feinman R. Hypoxia-inducible factor plays a gut-injurious role in intestinal ischemia reperfusion injury. Am J Physiol Gastrointest Liver Physiol. 2011;300:G853–G861. doi: 10.1152/ajpgi.00459.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dube A, Harrisson J-F, Saint-Gelais G, Seguin C. Hypoxia acts through multiple signaling pathways to induce metallothionein transactivation by the metal-responsive transcription factor-1 (MTF-1) Biochem Cell Biol. 2011;89:562–577. doi: 10.1139/o11-063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waeytens A, De Vos M, Laukens D. Evidence for a potential role of metallothioneins in inflammatory bowel diseases. Mediators Inflamm. 2009;2009:729172. doi: 10.1155/2009/729172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 2010;8:e1000296. doi: 10.1371/journal.pbio.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lovatt M, Bijlmakers M-J. Stabilisation of beta-catenin downstream of T cell receptor signalling. PLoS One. 2010;5:e12794. doi: 10.1371/journal.pone.0012794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandis AZ, Srivastava R, Sinha RK, Subrahmanyam G. A type II phosphatidylinositol 4-kinase associates with T cell receptor zeta chain in Con A stimulated splenic lymphocytes through tyrosyl phosphorylation-dependent mechanisms. Mol Immunol. 2005;42:561–568. doi: 10.1016/j.molimm.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava R, Sinha RK, Subrahmanyam G. Type II phosphatidylinositol 4-kinase beta associates with TCR-CD3 zeta chain in Jurkat cells. Mol Immunol. 2006;43:454–463. doi: 10.1016/j.molimm.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Chang CH, Guerder S, Hong SC, Van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/S1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 51.Yang Q, Monticelli LA, Saenz SA, Chi AW-S, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, Huang J, Haczku A, Pear WS, Artis D, Bhandoola A. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humphreys NE, Xu D, Hepworth MR, Liew FY, Grencis RK. IL-33, a potent inducer of adaptive immunity to intestinal nematodes. J Immunol. 2008;180:2443–2449. doi: 10.4049/jimmunol.180.4.2443. [DOI] [PubMed] [Google Scholar]
- 53.Shitashige M, Satow R, Honda K, Ono M, Hirohashi S, Yamada T. Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology. 1961;2008:134. doi: 10.1053/j.gastro.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Chung SS, Ahn BY, Kim M, Kho JH, Jung HS, Park KS. SUMO modification selectively regulates transcriptional activity of peroxisome-proliferator-activated receptor gamma in C2C12 myotubes. Biochem J. 2011;433:155–161. doi: 10.1042/BJ20100749. [DOI] [PubMed] [Google Scholar]
- 55.Straus DS, Glass CK. Anti-inflammatory actions of PPAR ligands: new insights on cellular and molecular mechanisms. Trends Immunol. 2007;28:551–558. doi: 10.1016/j.it.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Szeles L, Torocsik D, Nagy L. PPARgamma in immunity and inflammation: cell types and diseases. Biochim Biophys Acta. 2007;1771:1014–1030. doi: 10.1016/j.bbalip.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Tole S, Mukovozov IM, Huang Y-W, Magalhaes MAO, Yan M, Crow MR, Liu GY, Sun CX, Durocher Y, Glogauer M, Robinson LA. The axonal repellent, Slit2, inhibits directional migration of circulating neutrophils. J Leukocyte Biol. 2009;86:1403–1415. doi: 10.1189/jlb.0609391. [DOI] [PubMed] [Google Scholar]
- 58.Guan H, Zu G, Xie Y, Tang H, Johnson M, Xu X, Kevil C, Xiong W-C, Elmets C, Rao Y, Wu JY, Xu H. Neuronal repellent Slit2 inhibits dendritic cell migration and the development of immune responses. J Immunol. 2003;171:6519–6526. doi: 10.4049/jimmunol.171.12.6519. [DOI] [PubMed] [Google Scholar]
- 59.Prasad A, Kuzontkoski PM, Shrivastava A, Zhu W, Li DY, Groopman JE. Slit2N/Robo1 inhibit HIV-gp120-induced migration and podosome formation in immature dendritic cells by sequestering LSP1 and WASp. PLoS One. 2012;7:e48854. doi: 10.1371/journal.pone.0048854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, De Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 61.Dent AL, Doherty TM, Paul WE, Sher A, Staudt LM. BCL-6-deficient mice reveal an IL-4-independent, STAT6-dependent pathway that controls susceptibility to infection by Leishmania major. J Immunol. 1999;163:2098–2103. [PubMed] [Google Scholar]
- 62.Else KJ, Entwistle GM, Grencis RK. Correlations between worm burden and markers for Th1 and Th2 cell subset induction in an inbred strain of mouse infected with Trichuris muris. Parasite Immunol. 1993;15:595–600. doi: 10.1111/pim.1993.15.10.595. [DOI] [PubMed] [Google Scholar]
- 63.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.GeneNetwork, University of Tennessee; http://www.genenetwork.org
- 65.Alberts R, Schugart K. QTLminer: identifying genes regulating quantitative traits. BMC Bioinformatics. 2010;11:516–518. doi: 10.1186/1471-2105-11-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Table of immune response phenotypes. Multiple comparisons of one-way ANOVA on worm burden, serum IFN-γ, serum TNF-α and serum IL-6 in BXD RI lines and the parental strains C57BL/6 and DBA/2. (CSV 41 KB)
Additional file 2: Figure S1: Interval maps for immune response phenotypes in combined male and female cohorts. QTL for (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey line shows suggestive LRS scores. (TIFF 2 MB)
Additional file 3: Figure S2: Interval maps for immune response phenotypes in male cohorts only. QTL for (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey lines show suggestive LRS scores. (TIFF 2 MB)
Additional file 4: Figure S3: Interval maps for immune response phenotypes in female cohorts only. QTL (A) worm burden (TM4), (B) serum IFN-γ, (C) serum TNF-α, (D) serum IL-6 (all TM5) response phenotypes. Upper red line on maps show significant LRS scores whilst lower grey lines show suggestive LRS scores. (TIFF 2 MB)
Additional file 5: Figure S4: Male immune response phenotypes to T. muris. Mean ± SEM of immune response phenotypes in male mice to T. muris (A) worm burden, (B) serum IFN-γ, (C) serum TNF-α and (D) serum IL-6 in BXD RI lines (grey bars) and the parental strains C57BL/6 (black bars) and DBA/2 (white bars). (E) Summary of range and mean ± SEM of response phenotypes. (TIFF 406 KB)
Additional file 6: Figure S5: Female immune response phenotypes to T. muris. Mean ± SEM of immune response phenotypes in female mice to T. muris (A) worm burden, (B) serum IFN-γ, (C) serum TNF-α and (D) serum IL-6 in BXD RI lines (grey bars) and the parental strains C57BL/6 (black bars) and DBA/2 (white bars). (E) Summary of range and mean ± SEM of response phenotypes. (TIFF 407 KB)
Additional file 7: Table S2: Table of immune response phenotypes in male and female cohorts. Multiple comparisons of one-way ANOVA on worm burden, serum IFN-γ, serum TNF-α and serum IL-6 in BXD RI lines and the parental strains C57BL/6 and DBA/2 in male or female cohorts. (CSV 82 KB)


