Abstract
7-Carboxy-7-deazaguanine (CDG) synthase (QueE) catalyzes the complex heterocyclic radical-mediated conversion of 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) to CDG in the third step of the biosynthetic pathway to all 7-deazapurines. Here we present a detailed characterization of QueE from Bacillus subtilis to delineate the mechanism of conversion of CPH4 to CDG. QueE is a member of the radical S-adenosyl-L-methionine (SAM) superfamily, all of which use a bound [4Fe-4S]+ cluster to catalyze the reductive cleavage of SAM cofactor to generate methionine and a 5′-deoxyadenosyl radical (5′-dAdo•), which initiates enzymatic transformations requiring H-atom abstraction. The UV-visible, EPR, and Mössbauer spectroscopic features of the homodimeric QueE point to the presence of a single [4Fe-4S] cluster per monomer. Steady-state kinetic experiments indicate a Km of 20 ± 7 μM for CPH4 and kcat of 5.4 ± 1.2 min-1 for the overall transformation. The kinetically determined Kapp for SAM is 45 ± 1 μM. QueE is also magnesium-dependent and exhibits a Kapp for the divalent metal ion of 0.21 ± 0.03 mM. The SAM cofactor supports multiple turnovers, indicating that it is regenerated at the end of each catalytic cycle. The mechanism of rearrangement of QueE was probed with CPH4 isotopologs containing deuterium at C-6 or the two prochiral positions at C-7. These studies implicate 5′-dAdo• as initiating the ring contraction reaction catalyzed by QueE by abstraction of the H-atom from C-6 of CPH4.
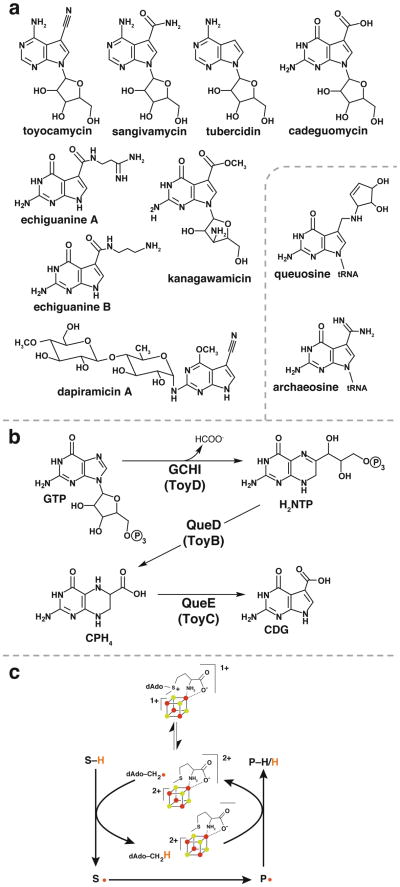
Compounds containing 7-deazapurine moieties are ubiquitous in nature and include structurally diverse nucleoside analogs produced by Streptomyces that have demonstrated antibiotic and antineoplastic activities (see (1, 2) for reviews). The structural diversity in deazapurines arises from substitutions at C-7 and elsewhere in the molecule (see Figure 1a). In addition to secondary metabolites, 7-deazapurines moieties are also found in the hypermodified tRNA nucleosides queuosine (3) and archaeosine (4). Queuosine is located in the wobble position of tRNAs bearing His, Tyr, Asp and Asn with the anticodon 5′-GUN-3′ (5); the hypermodification is almost universally conserved in all kingdoms of life but the precise physiological role of the modification remains to be established (6, 7). Archaeosine is located in the dihydrouridine loop of most archaeal tRNAs and is thought to play a role in stabilizing the tertiary RNA structure (4). Eukaryotes do not have de novo pathways for biosynthesis of queuosine (8) but incorporate queuine (the free base of queuosine) from dietary sources into tRNA (9, 10).
Figure 1.
(a) Examples of 7-deazapurine nucleosides isolated from culture filtrates of soil bacteria are shown. The hypermodified nucleosides queuosine and archaeosine are shown in the inset. (b) The pathway for the biosynthesis of the 7-deazapurine core of pyrrolopyrimidine nucleosides involves 3 steps wherein QueE catalyzes the key ring contraction step leading to the first 7-deazapurine intermediate. (c QueE is a member of the radical SAM superfamily of enzymes (17) and is the subject of studies in the manuscript. (c) General reaction mechanism for radical SAM enzymes that use SAM catalytically.
The 7-deazapurine moiety which is at the core of all pyrrolopyrimidine nucleosides is derived from GTP in three steps (Fig. 1b) (11-14). The first step is the conversion of GTP to 7,8-dihydroneopterintriphosphate (H2NTP), which is catalyzed by GTP cyclohydrolase I (GCH I) (11). Interestingly, this step appears to have been co-opted from primary metabolism as GCH I also catalyzes the first step in the biosynthesis of folic acid (15) and tetrahydrobiopterin (16). The second step is the conversion of H2NTP to 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) catalyzed by CPH4 synthase (QueD or ToyB in Fig. 1b) (13). Next CDG synthase (QueE or ToyC in Fig. 1b) catalyzes a dramatic and unprecedented heterocyclic rearrangement in the third step of the pathway to convert CPH4 to 7-carboxy-7-deazaguanine (CDG) (14). CDG is the first 7-deazapurine in the biosynthetic pathway and likely the precursor to all naturally occurring 7-deazapurine containing molecules.
CDG synthase is a member of the radical S-adenosyl-L-methionine (SAM) superfamily of enzymes (17). Members of the radical SAM superfamily contain conserved cysteine residues that ligate iron present at three vertices of a [4Fe-4S] cluster. While the cysteine residues are typically arranged in the motif CX3CX2C (17), exceptions have been noted (18-20). In Dpn2, for example, the 3 Cys residues that coordinate the Fe/S cluster are distant in primary sequence (20). The fourth iron is coordinated to the carboxylate and amino moieties of SAM (21, 22). In its +1 oxidation state the cluster donates an electron to SAM initiating the reductive cleavage of the C5′-S bond affording methionine and a 5′-deoxyadenosyl radical (5′-dAdo•) (Figure 1c). The 5′-dAdo• abstracts an H-atom from either a site on the substrate molecule or the protein to initiate the catalytic cycle. SAM is reformed at the end of the catalytic cycle in a subset of radical SAM proteins that include lysine-2,3-aminomutase (23) and spore photoproduct lyase (24); however in many others cleavage of SAM is not reversed and 5′-dAdo is a product (see (25-28) for recent reviews).
Here we present spectroscopic and steady-state kinetic characterization of recombinantly expressed CDG synthase from B. subtilis showing that it is indeed a member of the radical SAM superfamily. Furthermore, studies with site-specifically deuterated substrates afford detailed insights into the mechanism of the complex, radical mediated ring contraction catalyzed by the enzyme.
Experimental Procedures
Cloning, expression, purification, and reconstitution of B. subtilis QueE
QueE from B. subtilis was cloned into pET28a for expression of His6 -tagged, recombinant protein, expressed in BL21(DE3) E. coli cells (Novagen), and purified as described previously (14) with the following modifications. The eluent from the affinity chromatography step was loaded onto an Econo-Pac 10DG column (BioRad), which had been pre-equilibrated in 0.05 M PIPES•NaOH (pH 7.4) buffer containing 10 mM DTT. QueE was eluted with 4 mL of the same buffer and an aliquot (1 mL) was frozen at −80 °C for future use as non-reconstituted QueE. The remainder (∼3 mL) was reconstituted with iron and sulfide as follows. To a solution of QueE (∼1 mM), 15 μL of 1 M FeCl3, and 15 μL of 1 M Na2S were added rapidly with mixing following each addition. The reconstitution mixture was incubated at ambient temperature for 6 h and then precipitated material was removed by brief centrifugation (1 min). The supernatant was loaded on an Econo-Pac 10DG column pre-equilibrated in buffer containing 0.05 M PIPES•NaOH (pH 7.4) and 10 mM DTT, and QueE was eluted with 4 mL of the same buffer. QueE reconstituted by this protocol was divided into aliquots, frozen in liquid nitrogen, and stored at −80 °C for future use.
Determination of QueE concentration
Accurate amino acid content, which is required for determining stoichiometry of cofactor, was obtained by amino acid analysis. QueE was prepared for amino acid analysis as follows. QueE (0.1 mL, purified as described above) was passed over an illustra NICK column (GE Healthcare) pre-equilibrated in 10 mM NaOH. QueE was eluted from the column with 0.4 mL of the same, quantified by Bradford assay with BSA as standard, lyophilized, and submitted to the molecular structure Facility at University of California-Davis for amino acid analysis. A correction factor of 0.68 was calculated for the Bradford assay based on the amino acid analysis.
EPR Spectroscopy
EPR samples were prepared by combining QueE with MgSO4, SAM, and sodium dithionite to a final concentration of ∼0.5 mM, 10 mM, 2 mM, and 10 mM, respectively. The mixture was incubated 5 min after the addition of sodium dithionite and frozen in an EPR tube. Sample was stored at −80 °C. Continuous wave (CW) X-band EPR experiments were performed on Bruker ESP-300 spectrometer equipped with a standard rectangular resonator operating in TE102 mode and ESR-900 flow cryostat (Oxford Instruments). Spectra were recorded at 10 K at a microwave frequency of 9.337 GHz. The power and modulation amplitude were 2 mW and 10 Gauss, respectively.
Preparation of 57FeSO4
Metallic 57Fe was obtained from Isoflex USA (San Francisco, CA). The 57Fe powder (0.1 g) was rinsed with 2 mL of CHCl3 and dried under vacuum. The metal was combined with 2.63 mL of 1 M sulfuric acid (to give a ratio of 1.5 moles of acid to 1 mole of iron) and incubated at 60 °C in the anaerobic chamber. The solution was neutralized after 90 min by adding solid sodium bicarbonate to a final concentration of 1 M.
Expression, purification and reconstitution of 57Fe-labeled QueE for Mössbauer spectroscopy
QueE containing 57Fe for Mössbauer spectroscopy was expressed in the presence of the pDB1282 plasmid in E. coli BL21(DE3). The plasmid pDB1282 contains six genes (iscS, iscU, iscA, hscA, hscB, and fdx) from the Azotobacter vinelandii operon, which are important for biogenesis of FeS clusters (29, 30). The plasmid confers ampicillin resistance and the genes are cloned behind an arabinose inducible promoter. Cells containing the QueE expression plasmid were grown minimal in media based on a recipe from Fraenkel and Neinhardt (31) that contained 0.71% Na2HPO4, 1.35% KH2PO4, 0.000147% CaCl2•H2O, 0.0246% MgSO4•7H2O, 0.4% glucose, and 0.06% (NH4)2SO4 along with 34 ⌠g/mL kanamycin and 100 ⌠g/mL ampicillin. When cell density reached OD600 ∼ 0.3, solid arabinose (0.5 g/L) was added to induce transcription of the genes in pDB1282 and 57FeSO4 and cysteine were added to final concentrations 0.05 mM and 0.2 mM, respectively. At OD600 ∼ 0.5 IPTG was added to a final concentration of 0.1 mM to induce expression of QueE. Cells were harvested by centrifugation 5 h after induction with IPTG and frozen in liquid N2. Purification and reconstitution of 57Fe-labeled QueE for Mössbauer spectroscopy was carried out as described above for the unlabeled protein except that 57FeSO4 prepared as described above was included in the incubation mixture. Native or reconstituted 57Fe-QueE was concentrated 2- and 3.5-fold, respectively, in Microcon centrifugal concentrators with YM-10 membranes (Millipore) and frozen in Mössbauer cups with liquid nitrogen.
Mössbauer spectroscopy
Mössbauer spectra were recorded on a spectrometer from WEB research (Edina, MN) operating in the constant acceleration mode in transmission geometry. Spectra were recorded with the temperature maintained at 4.2 K. The sample was kept inside an SVT-400 dewar from Janis (Wilmington, MA), and a magnetic field of 53 mT was applied parallel to the ©-beam. The reported isomer shift is relative to the centroid of the spectrum of a metallic foil of 〈-Fe at room temperature. Data analysis was performed using the program WMOSS from WEB research.
Determination of oligomerization state of QueE
All gel filtration steps were carried out using anaerobically prepared solutions in an anaerobic chamber. QueE (0.1 mL of 0.5 mM protein) was diluted to 0.5 mL with 0.05 M potassium phosphate buffer containing 0.15 M NaCl and loaded onto a HiPrep 16/60 Sephacryl S-200 High Resolution column (GE Healthcare), which was equilibrated in the same buffer. A mixture of molecular weight standards that included ∼20 mg/mL of chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), BSA (67 kDa), and aldolase (158 kDa) in a volume of 0.5 mL was injected separately to calibrate the column. Fractions of the eluent were collected, proteins were detected by SDS-PAGE and elution volume of QueE was compared to those of the standard to obtain molecular weight of the complex.
Preparation and purification of CPH4
CPH4 was prepared enzymatically by incubating GTP in the presence of recombinant GCH I and QueD, which were prepared as described previously (13). The reaction (10 mL) contained 0.02 M PIPES•pH 7.4), 10 mM DTT, 5 mM GTP, 50 μM GCH I and 15 μM QueD. The incubation was carried out in the anaerobic chamber to minimize oxidation of the air-sensitive CPH4 product. After 20 h the mixture was loaded onto a DEAE Sepharose column (2.6 · 12.5 cm) which had been pre-equilibrated in 10 mM ammonium bicarbonate. The column was rinsed with the same buffer and CPH4 was obtained in the flow-through. CPH4 was detected by HPLC analysis (13). Fractions containing CPH4 were pooled, lyophilized, and resuspended in ∼1 mL H2O. The concentration of CPH4 was quantified using the extinction coefficient for tetrahydrobiopterin (32) (Σ297nm = 8.71 · 103 M-1cm-1 at pH 8.0).
Preparation of [6-2H1]-CPH4
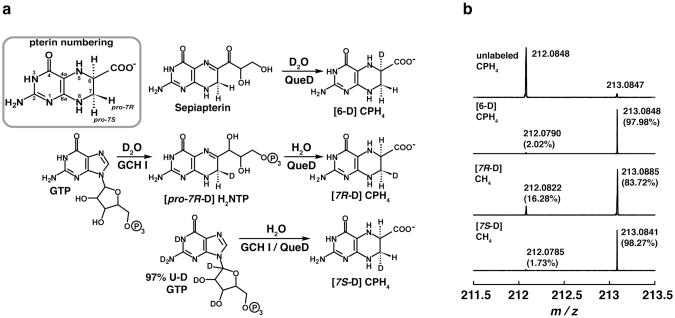
Figure 4 summarizes the strategies that were employed for synthesis of site-specifically deuterated CPH4. Briefly, we have shown previously that E. coli QueD catalyzes the conversion of sepiapterin to CPH4 (13). Therefore, this reaction was used to prepare CPH4 containing deuterium at C-6 by carrying out the reaction in D2O. The reaction (10 mL) contained 5 mM sepiapterin (Sigma), 0.02 M PIPES•NaOH (prepared from a 1 M stock solution in 99.9 % D2O), and 15 μM E. coli QueD that had been lyophilized to remove H2O. The reaction was conducted in 99.9% D2O (Sigma) and allowed to proceed for 20 h. The mixture was lyophilized, dissolved in H2O, and CPH4 was purified over a DEAE Sepharose column as described above for the unlabeled compound.
Figure 4.
Scheme for site-selective deuteration of CPH4 (a). FTICR-MS spectra showing the relative amounts of deuterated vs. non-deuterated CPH4 obtained in the preparation of pure, site specifically deuterated CPH4.
Preparation of [7R-D]-CPH4
CPH4 deuterated at the proR position of C-7 was prepared in two steps by the GCH I-dependent conversion of GTP to H2NTP in D2O, followed by the QueD-dependent conversion of the resulting H2NTP to CPH4 in H2O. The GCH I reaction (8 mL) contained 14.3 mM GTP, and 0.05 M ammonium bicarbonate, and 0.13 mM GCH I that had been buffer exchanged into 99% D2O (Sigma) containing 0.02 M Tris HCl (pH 8.0). After 15 h in the anaerobic chamber GCH I was removed by passing the solution through centrifugal filter devices with a YM-10 membrane filter (Amicon) at 7,000 ·g The flow-through was lyophilized to remove D2O, re-dissolved in H2O and lyophilized a second time. The resulting H2NTP was converted to CPH4 in a reaction mixture (10 mL) that contained 0.02 M PIPES•NaOH (pH 7.4), 10 mM DTT, and 5 μM E. coli QueD. After 18 h the reaction mixture was loaded onto a DEAE Sepharose column and CPH4 was purified as described above.
Preparation of [7S-D]-CPH4
CPH4 deuterated at the proS position of C-7 was prepared enzymatically by incubating 97% U-D GTP (Cambridge Isotope Laboratories, Inc) in the presence of GCH I and QueD in H2O. The enzymatic production and subsequent purification of the deuterated CPH4 were carried out as described for unlabeled CPH4.
Determination of CPH4 deuteration by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FT-ICR MS)
CPH4 samples were diluted 100-fold in a 1:1 mixture of H2O:acetonitrile containing 0.1% formic acid. FT-ICR MS was carried out in positive ion mode as previously described (14). The level of deuteration was estimated by the ratio of [M+H]+ ion peaks at m/z values of 212.1 and 213.2 for unlabeled and singly deuterated CPH4, respectively.
Preparation of flavodoxin (FldA) and flavodoxin reductase (FPR)
E. coli strains containing the genes for E. coli FldA and FPR that had been cloned into the intein-based expression vector pTYB1 (New England Biolabs, Ipswich, MA), were a generous gift from Prof. Squire Booker (Pennsylvania State University). FldA and FPR were expressed and purified by affinity chromatography to yield native proteins as previously described (33). However, two experiments included in this work were carried out using His6-tagged flavodoxin reductase. The gene encoding E. coli His6-flavodoxin reductase was ordered from Genscript (see Fig. S1 for sequence), excised from the supplied pUC57 plasmid with NdeI and HindIII, and cloned into pET28a to give pET28:FPR. The recombinant protein was expressed in E. coli HMS174 (DE3). E. coli HMS14 (DE3) cells containing pET28:FPR were grown in LB at 37°C with shaking and expression of His6-FPR was induced with the addition of IPTG (0.1 mM) when the cell density had reached OD600nm ∼0.5. At that time solid riboflavin was added to a final concentration of 0.1 mM. Cells were harvested by centrifugation 6 h after induction and frozen in liquid N2. Frozen cells (∼3 g) were resuspended in buffer containing 0.02 M potassium phosphate (pH 7.2), 0.5 M NaCl, 5 mM imidazole, and 1 mM PMSF and sonified at 60% amplitude using a Branson digital sonifier. Lysate was centrifuged for 30 min at 26,500 ·g at 4 °C. The cleared lysate was loaded onto a 1 mL HisTrapHP column (GE Healthcare) that had been pre-equilibrated in buffer containing 0.02 M potassium phosphate (pH 7.2), 0.5 M NaCl, and 5 mM imidazole (buffer A). The column was rinsed with 10 mL of buffer A and the His6-flavodoxin reductase was eluted with a 20 mL linear gradient to 100% buffer B which contained 0.5 M imidazole in buffer A. Fractions were analyzed by SDS-PAGE and those containing the desired protein were combined and dialyzed twice against 4 L of 0.02 M HEPES•NaOH (pH 7.4). Flavodoxin reductase was was quantified by the absorbance of flavin adenine dinucleotide (34) (Σ456nm = 7,100 M-1cm-1).
Enzymatic preparation of SAM
E. coli strain DM22-(pK8), which overexpresses E. coli SAM synthetase, was constructed by Prof. George D. Markham (Fox Chase Cancer Center) (35, 36). E. coli DM22-(pK8) was grown in LB containing 30 μg/mL oxytetracycline at 37 °C with shaking and harvested by centrifugation 12 h after inoculation. The cells (∼12 g) were resuspended in 36 mL of 0.1 M Tris•HCl (pH 8.0) containing 1 mM EDTA and 50 μg/mL lysozyme, and gently stirred at room temperature for 30 min. PMSF was added to a final concentration of 1 mM and cells were sonified at 60% amplitude using a Branson digital sonifier. Cell lysate was centrifuged for 30 min at 26,500 ·g at 4 °C. The cleared lysate was divided into three portions and frozen in liquid nitrogen. SAM was generated enzymatically by incubating an aliquot of the lysate in a buffered solution (0.1 L) containing 0.1 M Tris HCl (pH 8.0), 50 mM KCl, 1 mM EDTA, 20% acetonitrile, 26 mM MgCl2, 13 mM ATP, and 10 mM methionine. After gentle stirring for 5 h at room temperature the reaction was quenched by adjusting the pH to 5.0 with HCl. The mixture was placed on ice for 15 min and centrifuged at 26,500 ·g for 30 minutes at 4 °C to remove precipitated material. The supernatant was diluted to 1 L with 1 mM sodium acetate (pH 5.0) and loaded on a CM-52 cation exchange resin (Whatman) (19.6 cm · 25 cm) that had been charged with 0.2 M sodium acetate (pH 5.0) and equilibrated in 1 mM sodium acetate (pH 5.0). The column was rinsed with 0.7 L of 1 mM sodium acetate and SAM was eluted with a 0.9 L linear gradient from 0 to 0.2 M HCl. The presence and purity of SAM was confirmed by HPLC analysis and fractions containing SAM were combined, lyophilized, resuspended in 4 mL H2O and stored at −80 °C until use.
Preparation of 7-carboxy-7-deazaguanine (CDG)
CDG was prepared by refluxing 50 mg of preQ0 [prepared as described in (37)] in 3.5 mL of 6 M sodium hydroxide for 3 h. The reaction was diluted to 110 mL of 0.35 M ammonium acetate and loaded onto a Q-Sepharose column (1.6 · 20 cm) pre-equilibrated in the same buffer. The column was rinsed with 40 mL of 0.35 M ammonium acetate and CDG was eluted with a gradient to 0.5 M ammonium acetate over 0.4 L. The presence and purity of CDG was confirmed by HPLC analysis and fractions containing CDG were combined, lyophilized and resuspended in 1 mL H2O. The concentration of CDG ultimately obtained was measured by UV-visible absorbance and quantified using the extinction coefficient for the 7-carboxy-7-deazaguanine nucleoside cadeguomycin (Σ298nm = 7,607 M-1cm-1) (38).
Steady-state kinetic analysis of QueE
Steady-state kinetic experiments were conducted to measure the initial rate of QueE activity as a function of [CPH4], [Mg2+], and [SAM]. The reactions contained 0.05 M PIPES•NaOH pH 7.4, 10 mM DTT, 1.2 μM QueE, 24 μM FldA and 2.2 μM FPR. Reactions measuring the dependence of initial activity on [CPH4] and [Mg2+] contained 1.6 mM NADPH but reactions measuring the dependence of initial activity on [SAM] contained 2 mM NADPH. Reactions also contained 1.6 mM SAM (Sigma), 1 mM MgSO4, and 1 mM CPH4 unless the concentration of one of these ingredients was varied. In all cases, reactions (65 μL) were initiated by addition of CPH4 quenched after 2 min by addition of 6.5 μL of 30 % (w/v) TCA. An aliquot (25 μL) was injected directly on an Agilent Zorbax Eclipse C-18 column (4.6 · 250 mm) pre-equilibrated in water. A 30 min gradient from 0 to 30% acetonitrile was used to elute analyte components at a flow rate of 0.75 mL/min. The elution profile was monitored by UV absorbance spectroscopy from 200 to 500 nm using an Agilent 1100 photodiode array detector and the peak area on the UV chromatogram corresponding to CDG was noted.
Dependence of activity on [QueE]
A linear relationship between formation of CDG and concentration of QueE was established by reactions that contained 0.05 M PIPES•NaOH pH 7.4, 10 mM DTT, 1 mM MgSO4, 2 mM SAM (Sigma), 2 mM NADPH, 24 μM FldA, 5 μM FPR, 1 mM CPH4, and 0.7, 1.4, 2.0, or 2.7 μM QueE in a total volume of 0.2 mL. The reactions were initiated by addition of substrate and time points were taken by withdrawing 30 μL at 2.5, 5, and 10 minutes and adding them to 3 μL of 30% (w/v) TCA to quench activity. An aliquot (25 μL) was analyzed by HPLC as described above and the peak area on the UV chromatogram corresponding to CDG was noted.
Stoichiometry of CDG produced to SAM in the reaction catalyzed by QueE
The reaction mixtures contained 0.05 M PIPES•NaOH (pH 7.4), 10 mM DTT, 1 mM MgSO4, 10 μM SAM (Sigma), 2 mM NADPH, 2.4 μM QueE, 12 μM FldA, 2 μM FPR, and 1 mM CPH4 in a total reaction volume of 0.15 mL. The reaction was initiated by addition of substrate and time points were taken by withdrawing 30 μL at 30, 60, 90 and 120 min and combining with 3 μL of 30% (w/v) TCA to quench the reaction. A quantity of the quenched reactions (25 μL) was analyzed by HPLC as described above and the peak area on the UV chromatogram corresponding to CDG was noted.
Analysis of deuterium content in 5′-dAdo from site-specifically deuterated CPH4
QueE reactions were conducted using unlabeled, C6-, C7-R, or C7-S deuterated CPH4 and analyzed by LC-MS to examine incorporation of substrate deuterium into 5′-dAdo and CDG. The reactions contained 0.05 M PIPES•NaOH (pH 7.4), 10 mM DTT, 2 mM MgSO4, 4 mM SAM (produced enzymatically as described above), 2 mM NADPH, 19 μM QueE, 20 μM FldA, 20 μM His6-FPR, and 2 mM CPH4 (either unlabeled, [6-D], [7R-D], or [7S-D]) in a total reaction volume of 0.1 mL. An additional reaction containing 60% D2O and unlabeled CPH4 was conducted as well to assess the extent to which solvent-derived deuterium can label 5′-dAdo and CDG pools. Reactions were quenched after 20 min with the addition of 10 μL of 30% (w/v) TCA and 0.1 mL was analyzed by LC-MS.
Results and Discussion
Purification, determination of quaternary structure, metal ion content, and spectroscopic characterization
Recombinant QueE from B. subtilis was expressed in E. coli as a His6-fusion protein together with pDB1282, which contains the isc operon encoding genes involved in the biosynthesis of Fe/S clusters in A. vinelandii (29, 30). The pDB1282 construct is commonly included during expression of radical SAM proteins to improve the yield of holoproteins. QueE was purified in an anaerobic chamber by Ni2+-affinity chromatography and emerged from the column in dark brown fractions. The purity was estimated to be at least 95% by SDS-PAGE (Figure S2). Analytical gel filtration reveals a molecular weight between that of ovalbumin (46 kDa) and BSA (67 kDa), consistent with dimerization of the 29 kDa monomers to form a homodimer.
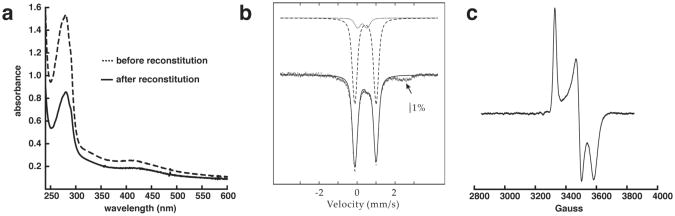
The UV-visible spectrum of the ‘as-isolated’ protein measured in the anaerobic chamber immediately after purification, displays a broad shoulder at ∼410 nm, which is characteristic of proteins containing [4Fe-4S] clusters (see Figure 2a). The three conserved cysteine residues in QueE are in a CX3CX2C motif, which is present in radical SAM proteins (17).
Figure 2.
(a) Representative UV-visible spectra of as isolated (20 μM) and reconstituted (10 μM) QueE. (b) 4.2-K/53-mT Mössbauer spectrum of reconstituted QueE (0.89 mM). The solid line overlaid with the experimental data is a simulation assuming two quadrupole doublets with the following parameters: ™(1) = 0.44 mm/s, ⊗EQ(1) = 1.13 mm/s (80 % of total intensity) and ™(2) = 0.30 mm/s, ⊗EQ(2) = 0.50 mm/s (11 % of total intensity). The individual contributions are shown as dashed and dotted lines, respectively. (c) EPR spectrum of 0.68 mM QueE containing 2 mM SAM, 10 mM sodium dithionite and 10 mM MgSO4. EPR spectra were measured at 10 K with the following settings: microwave frequency 9.337 GHz, microwave power 2 mW, and modulation amplitude 10 Gauss. MgSO4 is not required to observe the EPR signal.
The iron and labile sulfide contents of the protein were determined by ICP-OES and the Beinert method (39), respectively. The “as purified” protein contains 1.5 ± 0.4 equivalents of Fe and 2.4 ± 0.1 equivalents of sulfide. To improve the content of [4Fe-4S] clusters the protein was reconstituted at room temperature in the presence of iron and inorganic sulfide. The A410 / A280 ratio, which reports on Fe-S content, improved from 0.12 to 0.15 upon reconstitution (Figure 2a). Moreover, the reconstitution results in increase in the Fe and sulfide content to 4.2 ± 0.8 and 7.1 ± 1.0, respectively. These observations are consistent with presence of a single 4Fe-4S cluster in QueE.
To determine the types and quantity of Fe/S clusters associated with QueE more rigorously we have used a combination of Mössbauer and EPR spectroscopy on samples enriched with 57Fe. As-isolated, 57Fe-labeled QueE contains 1.4 Fe and 1.9 sulfide per monomer. The combination of EPR and Mössbauer spectroscopies (Figure S3) reveals that as-isolated QueE harbors ∼0.2 equiv [4Fe-4S]2+ clusters and ∼0.2 equiv [2Fe-2S]2+ clusters, in addition to a small amount (less than 0.01 equivalents) of [3Fe-4S]+ clusters. The presence of multiple Fe/S cluster types has been observed previously in other Fe/S-containing enzymes, including radical SAM enzymes, e.g. pyruvate formate lyase activase (24). Reconstitution of as-isolated QueE as described in Materials and Methods leads to a significant uptake of 57Fe and sulfide to 2.8 Fe and 3.9 sulfide per QueE. The 4.2 K/53 mT Mössbauer spectrum of reconstituted QueE is dominated by a quadrupole doublet with an isomer shift of ™ = 0.44 mm/s and quadrupole splitting ⊗EQ= 1.13 mm/s, which are typical of [4Fe-4S]2+ clusters (dashed line in Figure 2b). This quadrupole doublet accounts for ∼80% of the total intensity, which after taking into account the Fe and amino acid analyses, reveals that the 57Fe-reconstituted QueE contains ∼0.6 equivalents of [4Fe-4S]2+ clusters. In addition, the spectrum reveals a distinct shoulder at ∼ +0.6 mm/s. This position is typical of the high-energy line of a quadrupole doublet associated with [2Fe-2S]2+ clusters. This feature can be modeled by a quadrupole doublet (™ = 0.30 mm/s, ⊗EQ= 0.50 mm/s, 11% of total intensity, dotted line in Figure 2b) and suggests the presence of -0.15 [2Fe-2S]2+ clusters per QueE. The added contribution of these two quadrupole doublets is shown as a solid line in Figure 2b. The missing absorption (e.g. the broad features at ∼ -1 mm/s and ∼ 2 - 2.5 mm/s could (at least in part) emanate from Fe/S clusters with S = 1/2 ground state. However, their presence can be confidently ruled out based on the fact that an identical sample is EPR silent (data not shown). Most likely, these features emanate from high-spin Fe(II) complex(es). The broadness of the features suggests the presence of multiple Fe(II) complexes, e.g. Fe(II) coordinated by four thiolate ligands and Fe(II) coordinated by five or six N/O ligands. Although the stoichiometry of [4Fe-4S] clusters (∼0.6 equiv) and total Fe/S clusters (∼0.75 equiv) is less than one in the 57Fe-reconstituted protein, the results suggest that QueE harbors one [4Fe-4S] cluster, which is assigned to the essential radical SAM [4Fe-4S] cluster coordinated by the canonical CX3CX2C motif. The small amount of [2Fe-2S] cluster is presumably due to oxidative degradation.
QueE is only active in the presence of SAM and sodium dithionite (14). SAM is required as a source of the 5′-dAdo• radical and sodium dithionite is required to reduce the [4Fe-4S] cluster to the +1 oxidation state (Figure 1c). The EPR spectrum of reconstituted QueE obtained in the presence of sodium dithionite and SAM revealed a paramagnetic species with g-values of 2.003, 1.912, 1.861 (Figure 2c) Interestingly, QueE is only EPR active in the presence of both sodium dithionite and SAM (Figure S4). A similar result was observed with lysine-2,3-aminomutase (40) and it was subsequently shown that the binding of SAM to the [4Fe-4S] cluster in that enzyme increases the reduction potential of the cluster from -479 ± 5 mV to -430 ± 2 mV (41).
Steady-state kinetic analysis of QueE
Steady-state kinetic analysis of QueE was carried out to determine the steady-state kinetic parameters for the optimal concentrations of components to be included in the activity assays. In these experiments, E. coli FldA, FPR, and NADPH were utilized to reductively activate QueE. FldA and FPR are bacterial proteins that contain flavin mononucleotide and flavin adenine dinucleotide, respectively, and catalyze the in vivo transfer of electrons from NADPH to activate a number of enzymes including anaerobic ribonucleotide reductase (42), B12-dependent methionine synthase (34), pyruvate formate lyase activase (43), biotin synthase (44), lipoyl synthase (33), lysine-2,3-aminomutase (45), and AtsB (46). Control experiments showed that dithionite at various concentrations (1-10 mM, data not shown) also activated the protein; however, excess FldA/FPR/NADPH:QueE consistently resulted in higher activities. Therefore, the biological reducing system was used in all subsequent experiments.
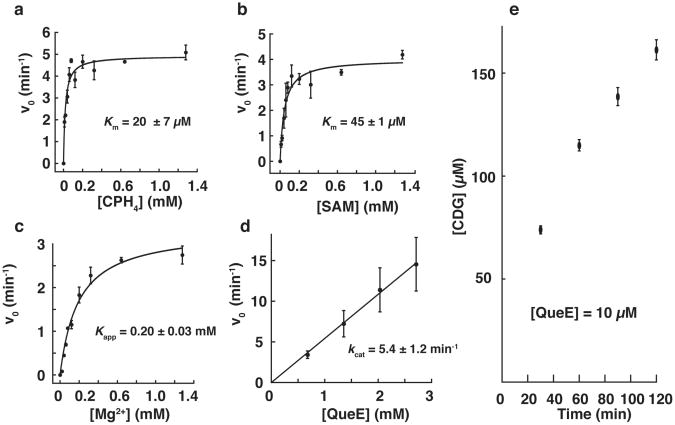
The steady-state kinetic data obtained with QueE showing dependence of activity on either CPH4, SAM, or MgSO4, while the other two components were kept at nearly saturating levels are shown in Figure 3a-c. These experiments reveal Km of 20±7 μM for CPH4 (Figure 3a). A Kapp of 45±1 μM was observed for SAM (Figure 3b). We note that the SAM used in this experiment was commercially obtained and contains a mixture of the biologically active (S,S) along with inactive (R,S) diastereomer. In addition, various additional contaminants, including methylthioadenosine and S-adenosylhomocysteine may be present. Therefore, the actual Kapp for the biologically relevant enantiomer may be even lower.
Figure 3.
Initial rates (v0) of CDG produced by QueE as a function of CPH4 (a), SAM (b), and Mg2+ (c). Data shown in are an average of three experiments and curves were fitted to the average values to obtain Km and Kapp (d) Initial rate (v0) of CDG production as a function of [QueE]; the results are an average of two experiments. (e) [CDG] observed in reactions that contained 10 μM QueE showing SAM is utilized catalytically. Data shown in (e) are an average of three experiments.
QueE requires magnesium metal ion for activity. Control experiments show that ∼11-fold more product is formed when MgCl2 Or MgSO4 (1 mM each) were included in the reaction mixture. Steady-state measurements under saturating concentrations of the substrate and SAM indicate a Kapp∼0.2 mM for magnesium (Fig. 3c). To our knowledge, while electron density consistent with Zn2+ was found in the active site of ThiC (18), a magnesium divalent metal ion requirement has not been demonstrated in other radical SAM proteins.
The turnover number of the enzyme can be estimated under nearly saturating conditions of all assay components. Figure 3d shows rate of formation of CDG as a function of [QueE]. The data shown in the Fig. 3d are representative of the data generally obtained with this enzyme and show a kcat of ∼5.4 ± 1.2 min-1. Turnover numbers for QueE across various preparations generally range from 2.4 to 6.1 min-1. These range of values are in line with turnover numbers observed with other radical SAM enzymes such as BtrN: 2.3 ± 0.22 min-1 (47), lipoate synthase: 0.175 ± 0.10 min-1 (33), pyruvate formate lyase activase: 5 min-1 (48), DesII: 1.0 ± 0.1 min-1 (49), but lower than observed with lysine-2,3-aminomutase (∼27 s-1) (50).
QueE utilizes SAM catalytically
To determine if QueE utilizes SAM in a catalytic capacity or as a reactant, QueE was assayed in a reaction mixture containing a 100-fold excess of CPH4 (1mM) over SAM (10 μM), in the presence of 2.4 μM QueE. As shown in Figure 3e, 150 μM of CDG is produced, which is 15-fold excess over SAM present in the reaction. Therefore, we conclude that SAM is regenerated at the end of each catalytic cycle and that QueE belongs to class I of radical SAM enzymes, which regenerate the cofactor at the end of each catalytic cycle (25). This result, however, does not rule out involvement of an enzyme-based radical that propagates the radical chain from 5′-dAdo• to the substrate and back.
QueE assays with site-specifically deuterated CPH4
H-atom abstraction from CPH4 to initiate the catalytic cycle can occur either at C-6 or C-7, as has been previously proposed (51). To probe the site of H-atom abstraction on the substrate and gain insights into the mechanism of the reaction, isotope transfer experiments were carried out with three monodeuterated CPH4 isotopomers that contain deuterium atoms at C-6 or C-7. The stereospecific deuteration reactions were accomplished enzymatically as described in Materials and Methods and illustrated in Figure 4a. The starting compound for synthesis of deuterated analogs of CPH4 with GCH I and QueD was GTP. Previous studies have shown that the conversion of GTP to H2NTP by GCH I introduces a solvent-derived proton into the pro-7R position (52). The stereoselectivity of the QueD reaction is not known; however, we presumed that it would be similar to its 6-pyruvoyltetrahydropterin synthase (PTPS) which places a solvent derived proton on the si-face of the substrate at C6 (52). QueD is homologous to PTPS and retains the catalytic residues that have been shown to be required for PTPS activity, including the residues that coordinate a catalytic zinc metal ion (53). Moreover, H2NTP is an alternate substrate for QueD, which catalyzes its conversion to CPH4. Additional alternate substrates for QueD include sepiapterin and 6-pyruvoyltetrahydropterin, all which are turned over to CPH4 (13). Therefore, commercially available sepiapterin was used as the starting point for the enzymatic synthesis of [6-D]-CPH4. The three stereoselectively labeled CPH4 isotopologs were prepared by using a combination of deuterated or unlabeled GTP in enzymatic transformations that were carried-out in either H2O or D2O (see Fig. 4a). The deuteration levels of resulting CPH4 were determined by FT-ICR MS (Fig. 4b) to be 97.98% for [6-D], 83.72 % for [7R-D] CPH4, and 98.27 % [7S-D] CPH4.
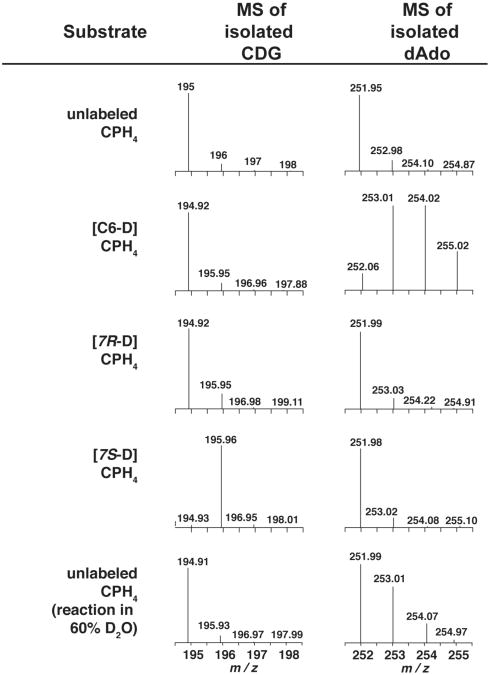
Our hypothesis is that abstraction of a hydrogen atom from the substrate by a 5′-dAdo• initiates the conversion of CPH4 to CDG. Therefore, QueE was incubated with unlabeled and deuterated substrates and the deuterium content of 5′-deoxyadenosine and the extent of deuterium remaining in the product were analyzed. In these experiments, QueE was incubated in the presence of excess unlabeled or deuterated CPH4 (2 mM) for 20 min prior to the reactions being quenched and analyzed by LC-MS for deuterium incorporation into 5′-dAdo and CDG, respectively. While 5′-dAdo is not a product of the QueE reaction, some 5′-dAdo is present during turnover. Occasional abortive cleavage of SAM is another source of 5′-dAdo. Analysis of the extracted ion chromatograms from LC-MS runs show that 5′-dAdo is clearly observable (m/z =252 in Figure S5). Comparison of the mass envelopes for 5′-dAdo when unlabeled or [6-D]-CPH4 are incubated with QueE reveal multiple deuteriums in 5′-dAdo with the latter (see Fig. 5). By contrast, deuterium from the 7S or 7R position of CPH4 is never found in 5′-dAdo (compare MS traces for 5′-dAdo in Fig. 5). Interestingly, however, deuterium is present in CDG (m/z = 195) only when [7S-D]-CPH4 is used as substrate (compare MS traces for CDG in Figure 5). Deuterium from [C6-D]- or [7R-D ]-CPH4 is never retained in the product. The retention of deuterium derived from [7S-D]-CPH4 is consistent with previous radiotracer feeding experiments showing that when [C1-3H]-ribose was present in the growth medium, tritium was retained in toyocamycin (54). In these experiments, the tritiated ribose would have been converted to [C1 ′-3H]-GTP and subsequently to [7S-3H]-CPH4 by the combined actions of GCH I and QueD.
Figure 5.
Mass spectra of CDG (m/z =195.0) and dAdo (m/z =252.1) isolated from turnover of QueE with unlabeled or various site-specifically deuterated CPH4 substrates. An additional experiment was carried out with unlabeled substrate in D2O to determine if reductively cleaved SAM can be quenched by solvent or a solvent exchangeable site (see text).
The MS spectra of 5′-dAdo from the reactions containing [6-D]-CPH4 show clearly that 5′-dAdo is multiply deuterated. Under the conditions employed, 41% of the observed 5′-dAdo was mono-deuterated, 37% was di-deuterated, and 14% was tri-deuterated, while 8% of the 5′-dAdo was not deuterated at all. The presence of multiply deuterated 5′-dAdo is consistent with SAM being used catalytically (Figure 3e). One would imagine that if the initial H-atom abstraction is reversible and/or the product-like radical abstracts an H-atom from the 5′-dAdo, a deuterium isotope effect for transfer of the heavy deuterium isotope would discriminate in favor of protium transfer. At this time it is not possible to confirm a steady-state kinetic isotope effect, as there is no measurable difference between the rates for conversion of saturating levels of unlabeled CPH4 and [6-D]-CPH4 to CDG, suggesting that H-atom abstraction may not be rate-limiting under Vmax conditions (Figure S6).
We were surprised to observe a pool of unlabeled 5′-dAdo in the reactions with the [6D]-CPH4 because based on the MS analysis, unlabeled CPH4 comprises at most 2% of the total substrate pool (Figure 4b). The 8% hydrogen incorporation may reflect in part an isotope effect that favors turnover with unlabeled substrate; in other words, the deuterated substrate is discriminated against. A second possibility, however, is that the protiated pool results from abortive reductive cleavage of the cofactor: 5′-dAdo• formed under some conditions may abstract a hydrogen atom from a solvent exchangeable site. Klinman and coworkers, for example, have observed deuterium incorporation into 5′-dAdo in an abortive SAM-cleavage reaction catalyzed by PqqE, a radical SAM enzyme involved in the biosynthesis of pyrroloquinoline quinone, when the reaction was carried out in D2O (55). To determine the contribution of such abortive cleavage reactions to the unlabeled 5′-dAdo pool an additional QueE reaction was carried out with unlabeled CPH4 in ∼60% D2O (Figure 5). LC-MS analysis revealed that 33% of the 5′-dAdo was mono-deuterated and 58% of the 5′-dAdo remained un-deuterated. We also observed a small quantity of di-deuterated species (∼8%), the source of which is not clear at present, but may be from abortive reactions where there are back H-atom transfers to a solvent exchangeable position. These results, nevertheless, are consistent with the notion that at least some (if not most) of the 8% unlabeled 5′-dAdo that we observe in the presence of [6-D]-CPH4 results from abstraction of an H-atom from a solvent exchangeable pool.
Mechanism of QueE
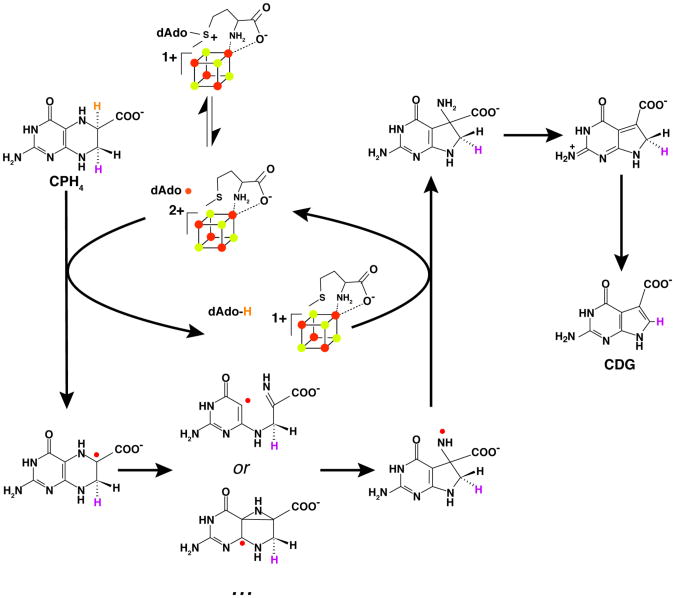
A working model for the reaction catalyzed by QueE which is consistent with all biochemical and spectroscopic data to date is shown in Fig. 6. The deuterium transfer experiments collectively support the model that radical mediated rearrangement of CPH4 to CDG is initiated by direct hydrogen atom abstraction from C-6 of the substrate by 5′-dAdo•, which is formed by reductive cleavage of the SAM cofactor by the +1 oxidation state of the [4Fe-4S] cluster. The proposed C6-centered radical is reminiscent of the α-Lys radical intermediate observed in the reaction catalyzed by LAM (56, 57). The substrate radical in QueE may be stabilized by delocalization of unpaired spin density onto the adjacent carboxylate group; in LAM, simulations of the EPR spectra of the radical indicated that 20% of the spin density is delocalized into the carboxylate moiety (56). The CPH4 radical can be stabilized further by captodative mechanisms. The glycyl radical in pyruvate formate lyase has been proposed to be stabilized by delocalization of the spin density at the α-carbon by combined electron withdrawing and donating effects of the its carbonyl group and the amide nitrogen, respectively (58). The C6-based radical is sandwiched between a carboxylate moiety (electron withdrawing) and a nitrogen atom (electron donating).
Figure 6.
Reaction mechanism proposed for conversion of CPH4 to CDG by QueE. Hydrogen bound to C6 of CPH4 (orange) is abstracted by 5′-dAdo• to initiate the reaction. Homolytic cleavage of the C-N bond of the substrate leads to an imine intermediate, which is proposed to undergo 5-exo- trig ring closure to the new 5-membered ring. Alternatively, the ring rearrangement may proceed via an azacyclopropylcarbinyl intermediate. The proposed nitrogen-centered radical is subsequently quenched by 5′-dAdo to generate 5′-dAdo, which subsequently combines with Met to reform the cofactor. The exocyclic amino group is shown to facilitate the elimination and re-aromatization reactions that are required to form CDG.
Several possible mechanisms can be proposed from the CPH4 radical intermediate en route to CDG. One possibility is homolytic cleavage of the C-N bond, which would be followed by ring-opening to form an imine in proximity of the unpaired spin at C4a position of the starting substrate. Such a homolytic cleavage would set up the molecule for a favorable Baldwin 5-exo-trig radical mediated ring closure (59); addition of C-centered radicals to imines is precedented (60). An alternative mechanism involving an azacyclopropyl carbinyl radical can also be proposed; such an intermediate would be strictly analogous to the central radical intermediate in the reaction catalyzed by lysine-2,3-aminomutase (61). In LAM, the intermediate can be stabilized by delocalization of the unpaired spin into the pyridoxal 5′-phosphate cofactor. In QueE, the unpaired spin may be further stabilized by delocalization. In either mechanism, a nitrogen centered radical is formally shown to result. However, it is expected that it will be quickly quenched by H-atom transfer form 5′-dAdo to form 5′-dAdo• and the initial 7-carboxy-7-amino product. The 5′-dAdo• would subsequently recombine with Met to reform SAM and regenerate the +1 oxidation state of the cluster.
The conversion of the 7-carboxy-7-amino intermediate to CDG requires loss of the amino group and aromatization of the 5-membered ring. Loss of an unactivated amino group on the surface would seem difficult. However, we note that QueE activity is magnesium dependent and that the metal ion may have role in catalyzing the elimination. In addition, the loss of ammonia may be facilitated by electron donation by the exocyclic amino group and/or the ring nitrogen atoms. For example, Fig. 6 shows the use of the exocyclic amino group to eliminate ammonia followed by the general base-assisted abstraction of the proR proton from C-7 of the substrate to generate CDG.
The magnesium-dependence observed for the QueE reaction was unexpected and several roles may be envisioned for the divalent cation in the reaction. Mg2+ may coordinate the substrate carboxylate group leading to activation of the C-6 proton by inductive effects akin to the mechanisms proposed for exchange of acetate protons of EDTA in the presence of divalent metal ions (62). Alternatively, as proposed above it can interact with the N-5 of the substrate to facilitate the ring contraction chemistry. At this stage we cannot distinguish between these and other role(s) for the metal ion.
Summary
We have demonstrated that QueE is a member of the radical SAM family and that it catalyzes a complex radical mediated ring-contraction reaction. The isotope transfer experiments implicate direct H-atom abstraction from the substrate by 5′-dAdo• as setting the stage for the complex radical ring contraction chemistry that follows.
Supplementary Material
Acknowledgments
We wish to acknowledge Dr. Arpad Somogyi for the FT-ICR MS measurements.
Funding Statement: Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01 GM72623 to V.B. and NCRR S10 RR23029 to V. H. Wysocki for acquisition of the FT ICR-MS are gratefully acknowledged. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. R.M.M. acknowledges support from Science Foundation Arizona for a Graduate Fellowship and from Biological Chemistry Training grant from NIH (T32 GM008804). In addition, research in the V.B. laboratory is supported (in part) by a Career Award in Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations
- ATP
adenosine 5′-triphosphate
- BSA
bovine serum albumin
- CPH4
6-carboxy-5,6,7,8-tetrahydropterin
- CDG
7-carboxy-7-deazaguanine
- DEAE
diethylaminoethyl
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EPR
electron paramagnetic resonance
- FT-ICR MS
Fourier transform ion cyclotron resonance mass spectrometry
- FldA
flavodoxin
- FPR
ferredoxin
- NADP
oxidoreductase
- GCH I
GTP cyclohydrolase I
- GTP
guanosine-5′-triphosphate
- H2NTP
7,8-dihydroneopterin triphosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HPLC
high-performance liquid chromatography
- ICP-OES
inductively coupled plasma optical emission spectroscopy
- IPTG
isopropyl β-1-thiogalactopyranoside
- LC-MS
liquid chromatography-mass spectrometry
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced form
- PIPES
1,4-piperazinediethanesulfonic acid
- PMSF
phenylmethanesulfonyl fluoride
- PTPS
6-pyruvoyltetrahydropterin synthase
- QueE
CDG synthase
- QueD
CPH4 synthase
- SAM
S-adenosyl-L-methionine (SAM)
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
Supporting Information Available Figures showing sequence of codon optimized FPR (Fig. S1), SDS-PAGE gel of purified QueE (Fig. S2), spectroscopic characterization of 57Fe-labeled QueE (Fig. S3), EPR spectra of QueE obtained in the presence and absence of SAM and dithionite (Fig. S4), extracted ion chromatograms of 5′-dAdo observed in QueE reactions (Fig. S5), and comparison of CDG production in the presence of unlabeled and [6D]-CPH4 (Fig. S6) are in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Suhadolnik RJ. Nucleoside antibiotics. Wiley-Interscience; New York: pp. 298–353. [Google Scholar]
- 2.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasai H, Oashi Z, Harada F, Nishimura S, Oppenheimer N, Crain P, Liehr J, Minden D, McCloskey J. Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid 7-(4,5-cis-dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry. 1975;14:4198–4208. doi: 10.1021/bi00690a008. [DOI] [PubMed] [Google Scholar]
- 4.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro- 4-oxo-7-β-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine) J Biol Chem. 1993;268:10076–10086. [PubMed] [Google Scholar]
- 5.Harada F, Nishimura S. Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry. 1972;11:301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- 6.Kasai H, Nakanishi K, Macfarlane RD, Torgerson DF, Ohashi Z, McCloskey JA, Gross HJ, Nishimura S. The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J Am Chem Soc. 1976;98:5044–5046. doi: 10.1021/ja00432a071. [DOI] [PubMed] [Google Scholar]
- 7.White BN, Tener GM. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su (s) locus. J Mol Biol. 1973;74:635–651. doi: 10.1016/0022-2836(73)90054-5. [DOI] [PubMed] [Google Scholar]
- 8.Farkas WR. Effect of diet on the queuosine family of tRNAs of germ-free mice. J Biol Chem. 1980;255:6832–6835. [PubMed] [Google Scholar]
- 9.Shindo-Okada N, Okada N, Ohgi T, Goto T, Nishimura S. Transfer ribonucleic acid guanine transglycosylase isolated from rat liver. Biochemistry. 1980;19:395–400. doi: 10.1021/bi00543a023. [DOI] [PubMed] [Google Scholar]
- 10.Katze JR, Basile B, McCloskey JA. Queuine, a modified base incorporated posttranscriptionally into eukaryotic transfer RNA: wide distribution in nature. Science. 1982;216:55–56. doi: 10.1126/science.7063869. [DOI] [PubMed] [Google Scholar]
- 11.McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips G, Yacoubi El B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP cyclohydrolase I. J Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty RM, Somogyi A, Bandarian V. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry. 2009;48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty RM, Somogyi A, Lin G, Jacobsen NE, Bandarian V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine 5′-triphosphate in four steps. Biochemistry. 2009;48:3847–3852. doi: 10.1021/bi900400e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burg A, Brown G. The biosynthesis of folic acid. 8 Purification and properties of the enzyme that catalyzes the production of formate from carbon atom 8 of guanosine triphosphate. J Biol Chem. 1968;243:2349–2358. [PubMed] [Google Scholar]
- 16.Buff K, Dairman W. Biosynthesis of biopterin by two clones of mouse neuroblastoma. Mol Pharmacol. 1975;11:87–93. [PubMed] [Google Scholar]
- 17.Sofia H, Chen G, Hetzler B, Reyes-Spindola J, Miller N. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol. 2008;4:758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlynn SE, Boyd ES, Shepard EM, Lange RK, Gerlach R, Broderick JB, Peters JW. Identification and characterization of a novel member of the radical AdoMet enzyme superfamily and implications for the biosynthesis of the Hmd hydrogenase active site cofactor. J Bacteriol. 2010;192:595–598. doi: 10.1128/JB.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic Approaches to Elucidating Novel Iron–Sulfur Chemistry in the “Radical-SAM” Protein Superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 22.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An Anchoring Role for FeS Clusters: Chelation of the Amino Acid Moiety of S-Adenosylmethionine to the Unique Iron Site of the [4Fe–4S] Cluster of Pyruvate Formate-Lyase Activating Enzyme. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 23.Baraniak J, Moss ML, Frey PA. Lysine 2,3-aminomutase Support for a mechanism of hydrogen transfer involving S-adenosylmethionine. 1989;264:1357–1360. [PubMed] [Google Scholar]
- 24.Cheek J, Broderick JB. Direct H atom abstraction from spore photoproduct C-6 initiates DNA repair in the reaction catalyzed by spore photoproduct lyase: evidence for a reversibly generated adenosyl radical intermediate. J Am Chem Soc. 2002;124:2860–2861. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 25.Frey PA, Booker SJ. Radical mechanisms of S-adenosylmethionine-dependent enzymes. Adv Protein Chem. 2001;58:1–45. doi: 10.1016/s0065-3233(01)58001-8. [DOI] [PubMed] [Google Scholar]
- 26.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 28.Booker SJ. Anaerobic functionalization of unactivated C-H bonds. Curr Opin Chem Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr Opin Chem Biol. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 30.Frazzon J, Fick JR, Dean DR. Biosynthesis of iron–sulphur clusters is a complex and highly conserved process. Biochem Soc Trans. 2001;30:680–685. doi: 10.1042/bst0300680. [DOI] [PubMed] [Google Scholar]
- 31.Fraenkel DG, Neidhardt FC. Use of chloramphenicol to study control of RNA synthesis in bacteria. Biochim Biophys Acta. 1961;53:96–110. doi: 10.1016/0006-3002(61)90797-1. [DOI] [PubMed] [Google Scholar]
- 32.Pfleiderer W. Chemistry of naturally occurring pterins. In: Blakley RL, Benkovic SJ, editors. Folates and pterins. John Wiley ' Sons, Inc; 1985. pp. 44–114. [Google Scholar]
- 33.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 34.Fujii K, Huennekens FM. Activation of methionine synthetase by a reduced triphosphopyridine nucleotide-dependent flavoprotein system. J Biol Chem. 1974;249:6745–6753. [PubMed] [Google Scholar]
- 35.Takusagawa F, Kamitori S, Misaki S, Markham GD. Crystal structure of S-adenosylmethionine synthetase. J Biol Chem. 1996;271:136–147. [PubMed] [Google Scholar]
- 36.Markham G, DeParasis J, Gatmaitan J. The sequence of metK, the structural gene for S-adenosylmethionine synthetase in Escherichia coli. J Biol Chem. 1984;259:14505–14507. [PubMed] [Google Scholar]
- 37.Quaranta D, McCarty R, Bandarian V, Rensing C. The copper-inducible cin operon encodes an unusual methionine-rich azurin-like protein and a preQ0 reductase in Pseudomonas putida KT2440. J Bacteriol. 2007;189:5361–5371. doi: 10.1128/JB.00377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu R, Okabe T, Namikoshi M, Okuda S, Nishimura T, Tanaka N. Cadeguomycin, A novel nucleoside analog antibiotic. J Antibiot (Tokyo) 1982:279–284. doi: 10.7164/antibiotics.35.279. [DOI] [PubMed] [Google Scholar]
- 39.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 40.Lieder KW, Booker S, Ruzicka FJ, Beinert H, Reed GH, Frey PA. S-Adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance. Biochemistry. 1998;37:2578–2585. doi: 10.1021/bi972417w. [DOI] [PubMed] [Google Scholar]
- 41.Hinckley GT, Frey PA. Cofactor dependence of reduction potentials for [4Fe-4S]2+/1+ in lysine 2,3-aminomutase. Biochemistry. 2006;45:3219–3225. doi: 10.1021/bi0519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianchi V, Eliasson R, Fontecave M, Mulliez E, Hoover DM, Matthews RG, Reichard P. Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem Biophys Res Commun. 1993;197:792–797. doi: 10.1006/bbrc.1993.2548. [DOI] [PubMed] [Google Scholar]
- 43.Blaschkowski HP, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli CoA-acylating pyruvate: flavodoxin and NADPH: flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur J Biochem. 1982;123:563–569. [PubMed] [Google Scholar]
- 44.Birch O, Fuhrmann M, Shaw N. Biotin synthase from Escherichia coli, an investigation of the low molecular weight and protein components required for activity in vitro. J Biol Chem. 1995;270:19158–19165. doi: 10.1074/jbc.270.32.19158. [DOI] [PubMed] [Google Scholar]
- 45.Brazeau BJ, Gort SJ, Jessen HJ, Andrew AJ, Liao HH. Enzymatic activation of lysine 2,3-aminomutase from Porphyromonas gingivalis. Appl Environ Microbiol. 2006;72:6402–6404. doi: 10.1128/AEM.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ. In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry. 2008;47:7523–7538. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoyama K, Numakura M, Kudo F, Ohmori D, Eguchi T. Characterization and Mechanistic Study of a Radical SAM Dehydrogenase in the Biosynthesis of Butirosin. J Am Chem Soc. 2007;129:15147–15155. doi: 10.1021/ja072481t. [DOI] [PubMed] [Google Scholar]
- 48.Külzer R, Pils T, Kappl R, Hüttermann J, Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 49.Szu PH, Ruszczycky MW, Choi SH, Yan F, Liu HW. Characterization and mechanistic studies of DesII: A radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine. J Am Chem Soc. 2009;131:14030–14042. doi: 10.1021/ja903354k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CH, Ballinger MD, Reed GH, Frey PA. Lysine 2,3-aminomutase: rapid mix-freeze-quench electron paramagnetic resonance studies establishing the kinetic competence of a substrate-based radical intermediate. Biochemistry. 1996;35:11081–11084. doi: 10.1021/bi960850k. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q, Liu W. Complex Biotransformations Catalyzed by Radical S-Adenosylmethionine Enzymes. J Biol Chem. 2011;286:30245–30252. doi: 10.1074/jbc.R111.272690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bracher A. Biosynthesis of pteridines NMR studies on the reaction mechanisms of GTP cyclohydrolase I, pyruvoyltetrahydropterin synthase, and sepiapterin reductase. J Biol Chem. 1998;273:28132–28141. doi: 10.1074/jbc.273.43.28132. [DOI] [PubMed] [Google Scholar]
- 53.Burgisser D, Thony B, Redweik U, Hess D, Heizmann C, Huber R, Nar H. 6-Pyruvoyl tetrahydropterin synthase, an enzyme with a novel type of active site involving both zinc binding and an intersubunit catalytic triad motif; site-directed mutagenesis of the proposed active center, characterization of the metal binding site and modelling of substrate binding. J Mol Biol. 1995;253:358–369. doi: 10.1006/jmbi.1995.0558. [DOI] [PubMed] [Google Scholar]
- 54.Suhadolnik RJ, Uematsu T. Biosynthesis of the pyrrolopyrimidine nucleoside antibiotic, toyocamycin. VII. Origin of the pyrrole carbons and the cyano carbon. J Biol Chem. 1970;245:4365–4371. [PubMed] [Google Scholar]
- 55.Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, Britt RD, Klinman JP. Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry. 2009;48:10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballinger M, Frey P, Reed G. Structure of a substrate radical intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry. 1992;31:10782–10789. doi: 10.1021/bi00159a020. [DOI] [PubMed] [Google Scholar]
- 57.Ballinger M, Reed G, Frey P. An organic radical in the lysine 2,3-aminomutase reaction. Biochemistry. 1992;31:949–953. doi: 10.1021/bi00119a001. [DOI] [PubMed] [Google Scholar]
- 58.Rauk A, Yu D, Taylor J, Shustov GV, Block DA, Armstrong DA. Effects of structure on alpha C-H bond enthalpies of amino acid residues: relevance to H transfers in enzyme mechanisms and in protein oxidation. Biochemistry. 1999;38:9089–9096. doi: 10.1021/bi990249x. [DOI] [PubMed] [Google Scholar]
- 59.Baldwin JE. Rules for ring closure. J Chem Soc Chem Commun. 1976:734–736. [Google Scholar]
- 60.Friestad GK. Addition of carbon-centered radicals to imines and related compounds. Tetrahedron. 2001;572:5461–5496. [Google Scholar]
- 61.Frey PA, Reed GH. Pyridoxal-5′-phosphate as the catalyst for radical isomerization in reactions of PLP-dependent aminomutases. Biochim Biophys Acta. 2011;1814:1548–1557. doi: 10.1016/j.bbapap.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 62.Terrill JB, Reilley CN. Base-catalyzed hydrogen-deuterium exchange in bivalent metal-EDTA chelates. Anal Chem. 1966;38:1876–1881. doi: 10.1021/ac50155a050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.