Abstract
The radical S-adenosyl-L-methionine (SAM) superfamily is a widely distributed group of iron-sulfur containing proteins that exploit the reactivity of the high energy intermediate, 5’-deoxyadenosyl radical, which is produced by reductive cleavage of SAM, to carry-out complex radical-mediated transformations. The reactions catalyzed by radical SAM enzymes range from simple group migrations to complex reactions in protein and RNA modification. This review will highlight three radical SAM enzymes that catalyze reactions involving modified guanosines in the biosynthesis pathways of the hypermodified tRNA base wybutosine; secondary metabolites of 7-deazapurine structure, including the hypermodified tRNA base queuosine; and the redox cofactor F420.
1. Radical SAM proteins
The radical S-adenosyl-L-methionine (SAM) superfamily is a growing list of enzymes that utilize radical intermediates formed by reductive cleavage of SAM to abstract H-atoms and initiate chemical transformations that are otherwise unlikely by polar mechanisms. Sofia and co workers defined the radical SAM superfamily on the basis of a conserved CX3CX2C motif that is common to all members of the family [1]. More recent studies have demonstrated that reductive cleavage of SAM, which is a hallmark of the group, also occurs in proteins that do not retain the exact sequence signature [2,3].
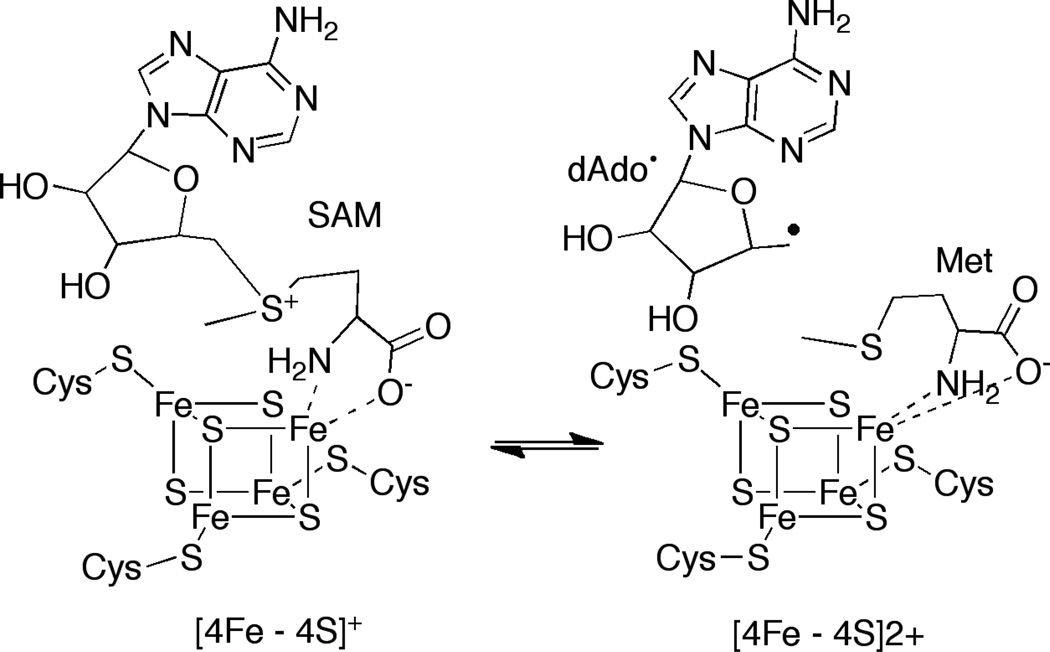
The three conserved Cys residues of the CX3CX2C motif in radical SAM proteins coordinate three of the iron atoms of the cubane [4Fe-4S] cluster. SAM binds the fourth iron through its α-amino and α-carboxylate moieties [4,5] (Fig. 1). The [4Fe-4S] cluster in SAM radical proteins has a +2/+1 redox couple; in its +1 state the cluster reductively cleaves SAM leading to formation of 5’-deoxyadenosyl radical (dAdo•) and methionine. The highly reactive oxidant, dAdo•, abstracts a hydrogen atom to initiate catalysis. While 5’-dAdo• has not been detected directly, its presence has been inferred by monitoring H-atom transfers to/from the 5’-position of the cofactor, or by substitution of SAM with 3’,4’-anhydro-SAM, which leads to allylic stabilization of the unstable 5’-dAdo• [6]. The +1 oxidation state of the cluster is obtained in vitro by reduction with dithionite or with reducing systems such as flavodoxin/flavodoxin reductase.
Figure 1.
Radical SAM proteins catalyze reductive cleavage of SAM to generate dAdo•, which is required for catalysis.
Radical SAM proteins have been divided into three classes on the basis of the fate of dAdo• [7]. Class I proteins catalyze reversible cleavage of SAM and reform the cofactor at the end of each catalytic cycle. The dAdo• produced by Class II enzymes abstract a H-atom from a glycyl residue on a cognate protein; the glycyl radical abstracts a H-atom to initiate catalysis and is reformed at the end of the catalytic cycle. Class III proteins cleave SAM irreversibly, but do not generate a cofactor that can support multiple turnover, as is observed for the Class II enzymes.
This review will focus on three radical SAM proteins that catalyze key steps in the conversion of guanosine-based molecules to the hypermodified RNA bases queuosine or wybutosine, 7-deazapurine containing secondary metabolites such as sangivamycin, and the redox cofactor coenzyme F420. Each of these enzymes retains the Cys cluster, which is conserved in all radical SAM proteins. Although very few mechanistic details are known about them, the presence of the conserved radical SAM cluster suggests catalysis is initiated by H-atom abstraction from their respective substrates. The remainder of this review will discuss the biosynthetic pathways that contain these enzymes and provide plausible mechanistic imperatives for each.
2. 7-Deazapurines
Pyrrolopyrimidine functional groups adorn a large number of compounds from marine or terrestrial sources, or as modified bases in tRNA (see [8] for review). While the biological role of this structurally diverse group of compounds is poorly understood, herbicidal, antibacterial, antifungal, and antineoplastic activities have been demonstrated in vitro, garnering interest in the use of these compounds as therapeutic agents.
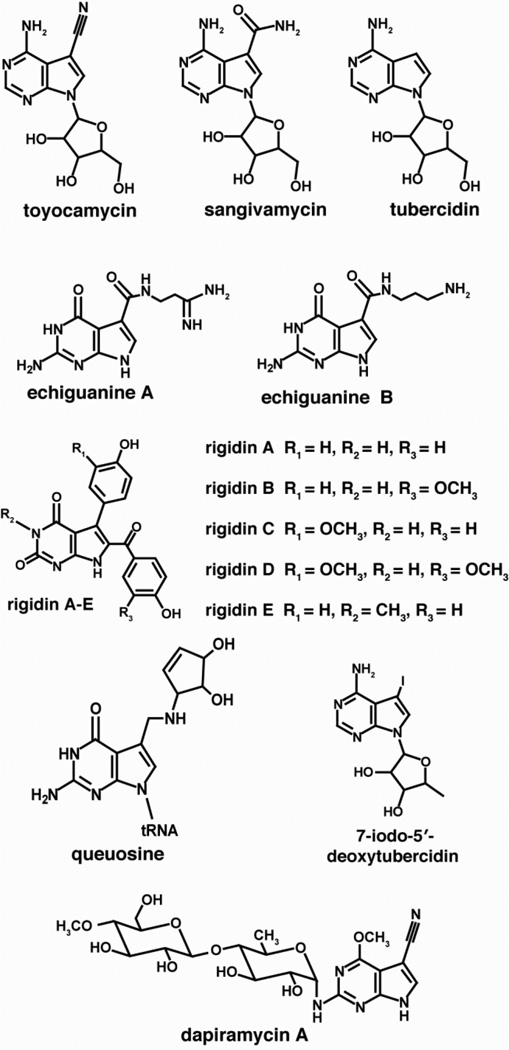
The structural diversity of deazapurine-containing molecules, as illustrated in Fig. 2, results from a large number of substitutions that can occur at the C-7 of the unique pyrrole ring, which is appended to an adenine- or guanine-like six-membered ring. Many of the compounds that have been isolated from marine sources contain modifications of the deazapurine core at positions other than C-7 or on the ribose. Despite the structural differences and diverse biological roles of naturally occurring pyrrolopyrimidines, the biosynthetic pathways that produce all 7-deazapurines are identical up to a central 7-cyano-7-deazaguanine intermediate. The remainder of this review will highlight the biosynthetic pathway for construction of the 7-deaza core structure and discuss CDG synthase, a radical SAM protein that catalyzes the key ring contraction step leading to the unique 7-deazapurine core common to 7-deazapurine containing secondary metabolites and the hypermodified tRNA base queuosine.
Figure 2.
Representative examples of 7-deazapurines showing the structural diversity in pyrrolopyrimidine-based metabolites.
2.1 A “Rosetta stone” for deciphering the biosynthesis of deazapurines
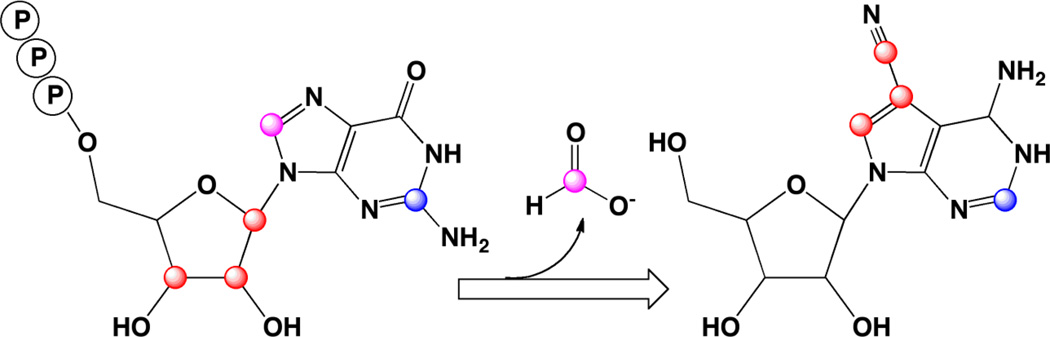
A substantial body of radiotracer feeding experiments had established the source of many of the atoms in 7-deazapurines. The results for toyocamycin are summarized in Fig. 3 (see [8] for an extensive review of the early studies). Parallel studies from two laboratories led to the elucidation of the complete pathway for the biosynthesis of the hypermodified tRNA base, queuosine, and the two structurally related secondary metabolites, sangivamycin/toyocamycin. In all cases that had been examined, the source of the deazapurine moiety was shown to be either adenine or guanine [9–11], whereas the pyrrole ring was derived from ribose (Fig. 3) [10]. Interestingly, GTP cyclohydrolase I (GCH I), which catalyzes the conversion of GTP to 7,8-dihydroneopterin triphosphate in the biosynthesis of folic acid, is induced in S. rimosus simultaneously with the appearance of sangivamycin and toyocamycin in the culture filtrates [12], potentially implicating GCH I or a related homolog in the biosynthesis of deazapurines.
Figure 3.
In the course of biosynthesis from carbon-2 of the starting purine is retained whereas carbon-8 is lost as formate. Carbons 1–3 from ribose of the starting sugar are incorporated into the 5-membered ring and side chain of the 7-deazapurine.
Using a bioinformatics-based approach, deCrecy-Lagard and Iwata-Reuyl searched genome sequences of organisms that produce the deazapurine-containing tRNA base, queuosine [13]. Four genes were identified in this study, which in addition to a GCH I homolog also included a member of the SAM radical superfamily [1]. The GCH I homolog was subsequently shown to catalyze the NADPH-dependent conversion of preQ0 to 7-aminomethyl-7-deazaguanine [14]. The functions of the remaining three orfs were not immediately obvious.
The cluster of genes from S. rimosus involved in the production of sangivamycin and toyoeamyein was identified by taking advantage of co-localization of secondary metabolite biosynthetic genes in Streptomyces [15]. S. rimosus excretes both toyoeamyein and sangivamycin into its culture medium, and Suhadolnik and coworkers had demonstrated the presence of a toyoeamyein nitrile hydratase (TNHase) in this organism [16,17]. TNHase catalyzes the hydration of toyoeamyein to produce sangivamycin. Purification of TNHase followed by sequencing of the N-termini of each of the three subunits of the protein provided sufficient information to identify a gene cluster encoding the biosynthetic pathway to sangivamycin and toyoeamyein. The genes in the cluster, designated toyA-M, serve as a “Rosetta stone” to decipher deazapurine biosynthesis in all bacterial species. The 13 orfs identified in this study included three of the genes that had been identified in the biosynthesis of queuosine [13]. This observation showed, for the first time, that all 7-deazapurines arise through a series of common biosynthetic steps. In addition, the toyoeamyein biosynthesis cluster also contains a functional GTP cyclohydrolase I (GCH I) homolog, suggesting that conversion of GTP to 7,8-dihydroneopterin triphosphate is the first step in the pathway.
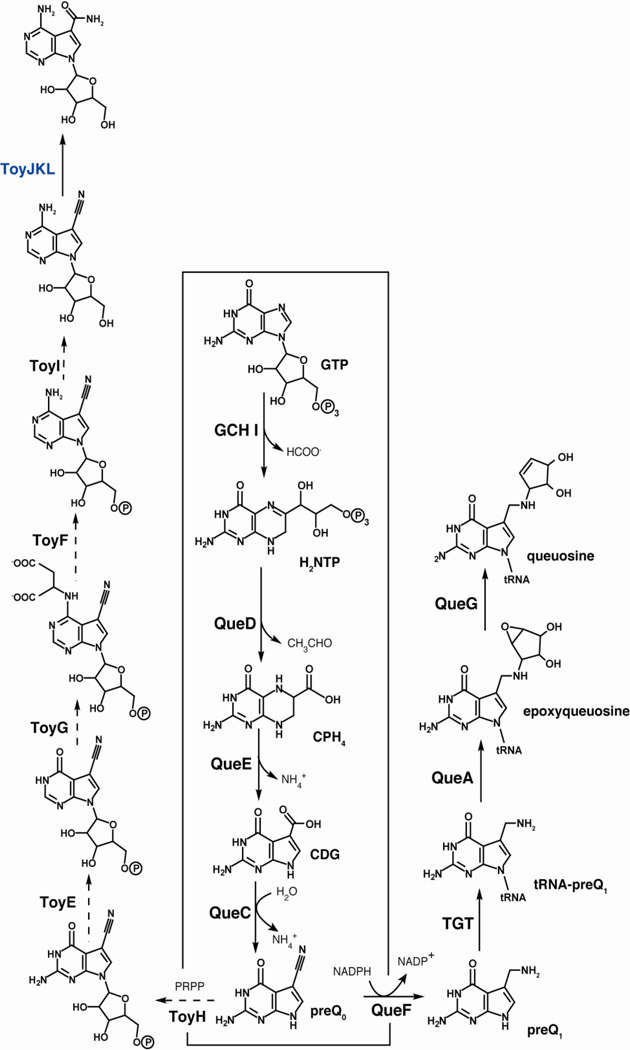
Subsequent functional characterization of the S. rimosus orfs or related homologs from other bacterial species led to the general paradigm for biosynthesis of 7-deazapurines (see Fig. 4). GTP is the starting point and is converted to 7,8-dihydroneopterin triphosphate (H2NTP) by the action of GCH I. A 6-pyruvoyltetrahydropterin synthase homolog (QueD) converts H2NTP to 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) [18]. A member of the radical SAM enzyme superfamily, CDG synthase (QueE), converts CPH4 to 7-carboxydeazaguanine (CDG), which is subsequently converted to 7-cyano-7-deazaguanine (preQ0) [19] by QueC, a previously identified intermediate in the biosynthesis of 7-deazapurines [20]. At this point the pathways toward queuosine, and toyocamycin and sangivamycin diverge. In the case of queuosine, preQ0 undergoes reduction by QueF to preQ1 [14] and is exchanged with a G in the wobble position of tRNA by TGT [20]. The base is further modified by addition of a cyclopentenediol epoxide moiety, which is derived from SAM [21]. The final reaction, conversion of epoxyqueuosine (oQ) to queuosine is catalyzed by oQ reductase (QueG), which requires both Fe-S and B12 [22]. While most of the steps from preQ0 to toyocamycin remain to be confirmed experimentally, TNHase (ToyJKL) has been overexpressed heterologously and activity of the protein has been reconstituted [15].
Figure 4.
Biosynthetic pathways leading to the hypermodified tRNA base queuosine and secondary metabolites sangivamycin/toyocamycin. Steps that have been demonstrated in vitro are denoted by solid arrows.
2.3 Radical-mediated ring contraction catalyzed by CDG synthase
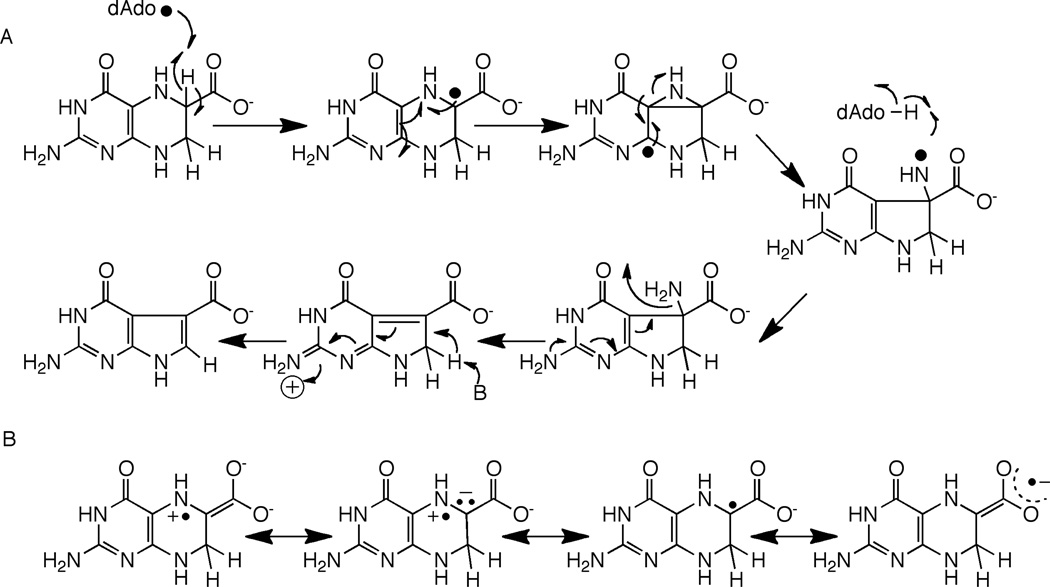
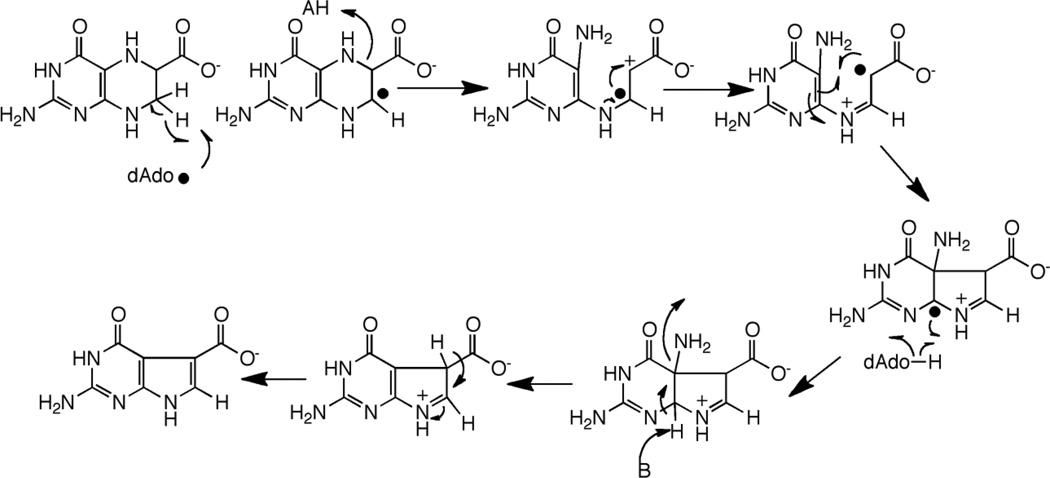
CDG synthase (QueE) catalyzes the conversion of CPH4 to CDG. The activity of CDG synthase was demonstrated with the B. subtilis homolog, and both SAM and dithionite are required for catalytic activity [19]. The ring contraction catalyzed by CDG synthase also requires elimination of the N-7 in the starting material and formation of a new C-C bond. In the context of reactivity of radical SAM proteins, the required transformations can be accomplished by abstraction of a hydrogen atom from C-6 as shown in Fig. 5A. The substrate radical formed from C-H abstraction can rearrange, perhaps through a bridged intermediate, to a nitrogen-centered radical. Transfer of H-atom from 5’-dAdo to the intermediate would lead to formation of the initial product. One can utilize pyrrole ring N-9 to eliminate the amino group and activate the hydrogen at C-7 for general base catalyzed re-aromatization of the system.
Figure 5.
Mechanism of CDG synthase involving H-atom abstraction to generate a substrate based radical (A). In this mechanism it is assumed that the substrate radical abstracts an H-atom from the dAdo to produce the product and that SAM is regenerated. The C-6 radical can be stabilized by capto-dative effects, as illustrated in (B).
Abstraction of the hydrogen atom from C-6 of CPH4 to initiate catalysis is attractive from several perspectives. First, a C-6 based radical can be stabilized by delocalization of the unpaired spin density into the carboxylate moiety (Fig 5B). In the analogous product-like radical observed in the steady-state of the reaction catalyzed by lysine 2,3-aminomutase, electron paramagnetic resonance measurements of the magnitudes of the α-H hyperfine coupling constant suggest that as much as 20% of the spin density may be delocalized onto the carboxylate [23,24]. Second, a C-6 based radical intermediate in the CDG synthase reaction can be stabilized captodatively. Captodative stabilization relies on availability of both an electron withdrawing and an electron donating group attached to the carbon bearing the unpaired spin. In the case of the proposed radical, the combined electron withdrawing effect of the carboxylate and the electron donation by the lone pair of the substituted nitrogen adjacent to it may stabilize the intermediate as shown by the resonance forms in Fig. 5B. Captodative effects have been proposed to both stabilize and lower the bond dissociation energies of the glycyl radical in pyruvate formate lyase [25,26].
An alternative mechanism involving abstraction at C-7 can also be proposed [27]. Abstraction of a H-atom from C-7 would permit a reaction cascade that is reminiscent of the group elimination step proposed for some adenosylcobalamin-dependent enzymes, such as ethanolamine ammonia-lyase (see [28] for review). A product-like radical could be quenched by H-atom abstraction from dAdo, as proposed in Fig. 5A for abstraction at C-6.
3. Wybutosine
Wybutosine (yW) and its derivatives are found in position 37 of tRNA that encode Phe in eukaryotes and archea. The modified base was discovered in the course of sequencing tRNAphe by Khorana and coworkers in the Institute for Enzyme Research at the University Wisconsin-Madison [29]. It was one of two unknowns (deemed X and Y) whose presence was noted in the tRNA.
The structure of wybutosine was established by several methods. It was noted early on that mild acid treatment of tRNAphe leads to release of the highly fluorescent base of wybutosine [30]. This treatment provided a convenient method of obtaining sufficient quantities of the base for structural assignment. A variety of NMR, MS, and UV-visible studies yielded the identity of the base [31–33], whose structure was further confirmed by comparison to synthetic standards [34,35]. Examination of archaea and bovine liver led to identification of several additional analogs [36–39].
3.1 Biosynthesis of wybutosine
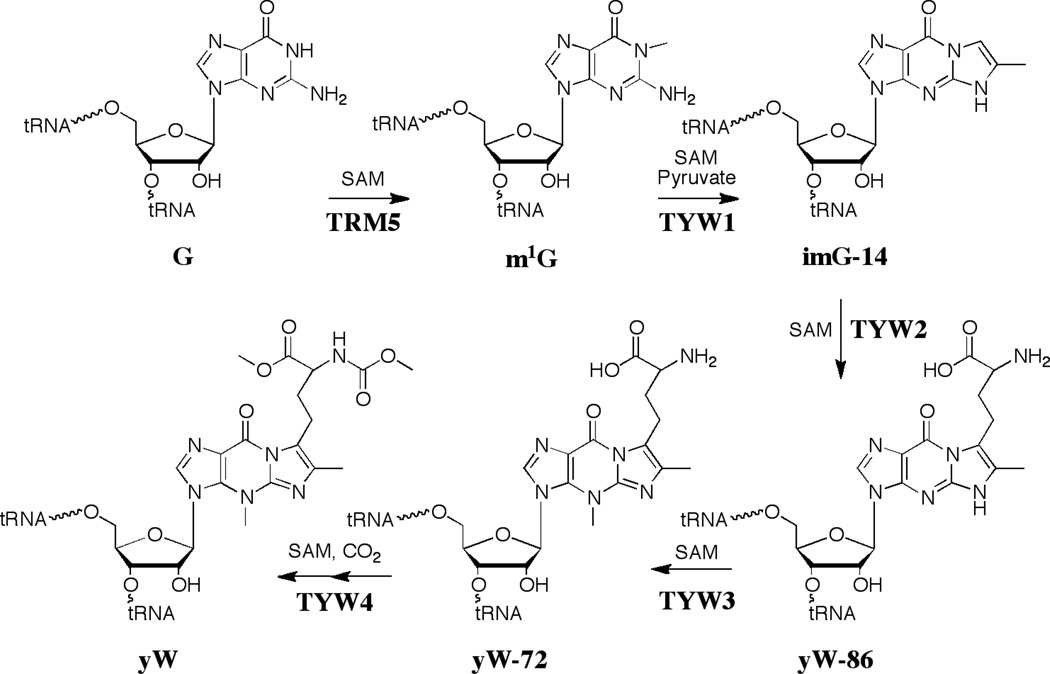
The current paradigm for the biosynthesis of wybutosine is shown in Figure 7. Radiotracer and stable isotope experiments provided significant clues to the biosynthetic pathway. Yeast cells were shown to incorporate radiolabeled guanine into wybutosine, suggesting that guanosine serves as the precursor [40]. The aminocarboxypropyl sidechain was also derived from SAM [41]. NMR studies with 13C labeled tRNAphe led to the insight that the N-methyl groups of the molecule are derived from SAM [42]. The source of the two carbon atoms that are required for the new 5-membered ring was not determined.
Figure 7.
Biosynthetic pathway leading to the hypermodified tRNA base wybutosine (yW). Unlike queuosine, all steps leading to the modified base occur on the tRNA.
The pathway to wybutosine was established by biochemical and bioinformatic methods (see Fig. 7). The first enzyme to be identified in the yeast pathway to Y was YML005w; deletion of the gene led to accumulation of ImG-14. Reconstitution of the pathway from ImG-14 to yW was accomplished as well [43]. An extensive comparative genomic analysis has led to the realization that ImG-14 is the common intermediate in the biosynthesis of wybutosine and all its analogs discovered to date, and that the variation in the form of the modified base that will be found in various organisms can be predicted based on genes downstream of TYW1 [44].
The biosynthetic pathway to wybutosine in yeast has five steps, each of which utilize SAM either as a group donor or for C-H activation. A tour-de-force series of ribonucleome studies by Suzuki and coworkers were crucial to defining the steps [45]. The first step is methylation of the N1 position of guano sine 37 of tRNAphe with SAM catalyzed by Trm5. TYW1 (YPL207w in yeast) was identified by comparative genomic analysis [46] and recent studies from this lab have shown that TYW1 catalyzes the condensation of pyruvate with m1G to generate ImG-14 [47]. The identification of pyruvate as the substrate resolved the long-standing question about the source of the new carbon atoms in wybutsine. TYW2 catalyzes transfer of the aminocarboxypropyl group of SAM to the C-7 position of yW-86. TYW3, which was the first enzyme to be discovered in the pathway, catalyzes the methylation of N-4 of ImG-14 to form yW-72 [48]. yW-72 subsequently undergoes methylation and methoxycarbonylation to form yW. The hydroxywybutosine structural variant of yW found in higher eukaryotes is produced from yW-72 by the 2-oxoglutarate-dependent enzyme TYW5 [49]. The X-ray crystal structures of all but one of the proteins (TYW3) have been solved [50–54].
3.2. Radical-mediated ring-formation by TYW1
TYW1 catalyzes condensation of pyruvate with m1G to form imG-14. TYW1 has been classified as a member of the radical SAM super family on the basis of presence of the conserved Cys motif that is characteristic of these proteins. Three additional Cys residues are also conserved. X-Ray crystal structures of TYW1 homologs from Pyrococcus horiskoshii and Methanocaldococcus jannaschii have been solved [50,51]. While neither structure showed occupancy of a putative radical SAM metal center in the active site, the positions of these residues are consistent with a [4Fe-4S] cluster being present in the active protein. Soaking of Fe leads to density consistent with a metal binding site [50]. Interestingly, the structures revealed a second cluster of 3 Cys residues on the opposite side of the active site cleft which may form a second cluster, the identity of which remains to be established. In vivo studies with the variants where the conserved Cys residues that form the radical SAM cluster were mutated show that they are essential for activity [51]. In vitro studies with purified TYW1 have demonstrated that SAM and reductant are required [47].
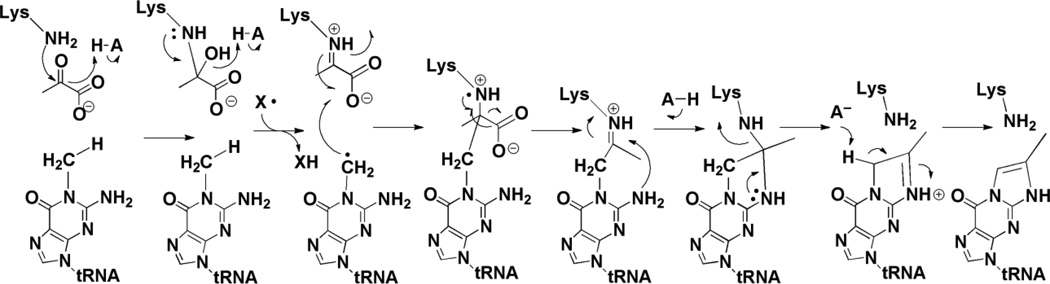
Mass spectrometric analyses of the reaction products obtained with C1, C2 or C3 13C-labeled pyruvate show that C2 and C3 of pyruvate are incorporated into the product whereas C1 is lost. A mechanistic proposal that is consistent with the amino acid conservation and the SAM requirement is shown in Figure 8 (mechanism was adapted from [47]). Pyruvate is proposed to form a Schiff base with the conserved Lys residue (K41 in the M. jannaschii homolog) in the active site of the protein; mutation of the Lys residue to Ala in the yeast homolog leads to accumulation of m1G in vivo [51]. The radical SAM cluster reductively cleaves SAM to generate 5’-dAdo•, which either directly or through an enzyme-based radical, abstracts a H-atom from m1G. The substrate radical adds to C2 of pyruvate, which is followed by homolytic scission of the C1-C2 bond. The resulting intermediate undergoes transimination and deprotonation to form imG-14. The second product, the proposed formyl radical, can either acquire a H-atom or undergo reduction and protonation to produce formate or CO2. Similar decarboxylation reactions have been proposed for pyruvate formate-lyase and coproporphyrinogen synthase [55]. TYW1 is similar to other radical SAM proteins that harbor a second cluster in addition to a radical SAM cluster. In vivo studies have demonstrated that the residues that would be involved in the formation of the second cluster are required for activity, consistent with the intimate role in catalysis proposed here.
Figure 8.
Mechanism of action of TYW1. H-atom abstraction either by dAdo• or another radical generated by it from m1G substrate initiates the radical cascade. A conserved Lys residue is proposed to be involved in formation of Schiff base with pyruvate.
4. Coenzyme F420
Coenzyme F420 (Figure 9) is a low potential redox (−360 mV) cofactor with diverse biological roles ranging from methanogenesis, intermediary metabolism, and DNA photorepair, to production of secondary metabolites. The green fluorescent cofactor, coenzyme F420, so-called for its intense 420 nm absorbance, was first discovered in Mycobacteria [56] and subsequently isolated from Methanobacterium strain M.o.H [57] and structurally characterized [58]. Coenzyme F420 has since been shown to be widely distributed [59].
Figure 9.
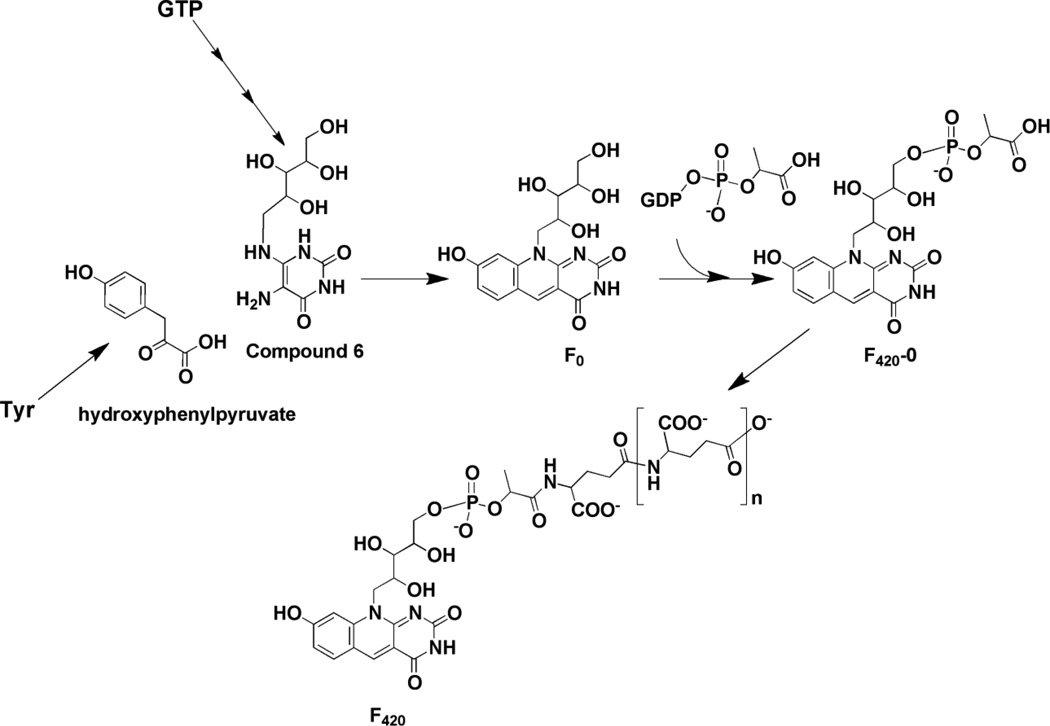
Biosynthesis of coenzyme F420. Compound 6 is an intermediate in the biosynthesis of riboflavin as well. F420 from different organisms can have variable numbers of glutamate residues.
Coenzyme F420 structurally resembles riboflavin but is functionally equivalent to pyridine adenine nucleotide coenzyme [60]. By contrast to riboflavin, which can be a 1- or 2-electron redox cofactor, replacement of N-5 in riboflavin with a methylene renders coenzyme F420 more suited for hydride transfer reactions. A notable exception is its role in photorepair of DNA thymine dimers. Photoactivation of the molecule accesses the excited semiquinone state permitting 1-electron chemistry. This property of coenzyme F420 is exploited in vitro for photoreduction of redox proteins [61].
4.1 Biosynthesis of Coenzyme F420
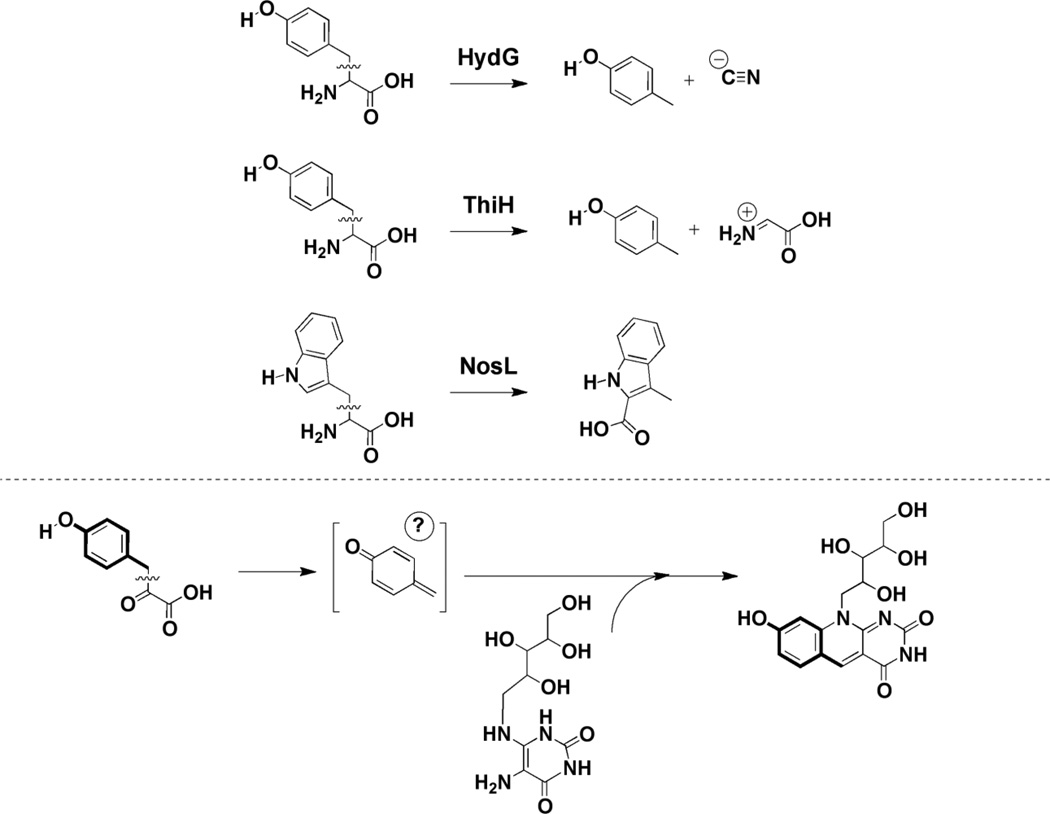
A biosynthetic pathway for F420, which is consistent with all radiotracer experiments and biochemical studies to date, is shown in Fig. 9. F0 has been proposed to form from condensation of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (compound 6) with 4-hydroxyphenylpyruvate [62,63]. Compound 6 is an intermediate in the biosynthetic pathway to riboflavin [64], where it is formed in three steps from GTP; feeding experiments have shown that it is incorporated into deazaflavin without significant dilution of the label [65]. The fluorescent F0 scaffold is elaborated further by addition of phospholactate, which is activated via attachment to GMP by a 2-phospho-L-lactate guanylyltransferase [66]. Variable numbers of glutamyl residues have been noted to cap the lactyl group (see [67] for examples) [58].
4.2 Condensation reaction catalyzed by 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (DS)
The deaza ring structure of coenzyme F420 is formed by a condensation reaction catalyzed by 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (DS). In certain organisms, (such as S. coelicolor or Mycobacterium smegmatis) [62], this protein is encoded by a single gene (sco4429 or fbiC) but in M. jannaschii, two genes (mj0446 and mj1431) encoding the N- and C-terminal halves of the protein are present [63]. Overexpression of either the mycobacterial or methanococcal proteins in E. coli, which do not have a deazaflavin biosynthesis pathway, leads to accumulation of F0 in the growth medium. Production of F0 in cell free extracts of E. coli expressing the M. smegmatis FbiC homolog is stimulated by addition of compound 6 and hydroxyphenylpyruvate (HPP). Early studies could not rule out Tyr as the substrate for the enzyme [65]; however, stimulation of F0 production by HPP is strongly suggestive that hydroxyphenylpyruvate is the substrate [63]. The presence of SAM, dithionite, Fe2+, and inorganic sulfide stimulates production ~ 10-fold, consistent with the radical SAM clusters that are encoded in the sequence.
Protein sequence analysis reveals that DS is a member of the radical SAM superfamily. The sequence contains two radical SAM (CX3CX2C) motifs, one of which is located in the N-terminal half and the other in the C-terminal half of the protein. Both appear to be required for the formation of F0 [63]. The 2-subunit M. jannaschii homolog encodes one such sequence in each subunit, consistent with sequence similarity to the N- and C-terminal halves of FbiC.
The catalytic mechanism of DS remains to be established. It is assumed that one or both of the radical SAM clusters encoded by the protein may be involved in reductive cleavage of SAM to generate 5’-dAdo•, which in turn would activate one of the substrates to facilitate the condensation. A mechanism consistent with this notion has been proposed whereby H-atom abstraction at HPP and Compound 6 are required for initiating catalysis [63]. However, studies of radical SAM proteins involved in the biosynthesis of thiamin (ThiGH) [68], the cyanide ligand of FeFe hydrgenase (HydG) [69], and the 3-methylindole moiety of the antibiotic nosiheptide (NosL) [70] suggest an alternative mechanism. Each of these proteins catalyzes the cleavage of Cα-Cβ bond of Tyr (Fig. 10); in the case of HydG and ThiL, p-cresol has been shown to form [68]. The Cα-Cβ bond of 4-hydroxyphenylpyruvate may be mechanistically similar to the cleavage of the analogous bond in Tyr, and that a reactive p-cresol-like intermediate may be the species involved involved in the condensation reaction. Moreover, by analogy to the mechanistic proposal shown for TYW1, 4-hydroxyphenylpyruvate may form a Schiff base with the protein facilitating the cleavage. Confirmation of these predictions must await in vitro characterization of active DS.
Figure 10.
Similarity of reactions catalyzed by the radical SAM proteins HydG, ThiH, and NosL to that of DS. By analogy to HydG [69] and ThiGH [68], where p-cresol has been shown to be a product, HPP may be activated in a similar manner upon abstraction of a hydrogen atom from the ring hydroxyl and fragmentation.
5. Perspective
The reactions catalyzed by CDG synthase, TYW1, and deazaflavin synthase are only a small fraction of the thousands of transformations attributed to the radical SAM superfamily. As genome sequencing efforts are expanded, this group of enzymes is likely to grow providing fascinating examples of radical-mediated transformations. The challenge in the field is to utilize the growing body of mechanistic, structural, and sequence information to better classify these proteins and predict their function.
Figure 6.
Mechanism for CDG synthase assuming abstraction at C-7. As in the mechanism involving abstraction at C-6 shown in Fig. 5, it is assumed that the substrate radical is quenched by H-atom transfer from dAdo and that SAM is regenerated.
Highlights.
Radical SAM proteins catalyze key steps in the biosynthesis of purine-based natural products
Acknowledgements
V.B. is grateful for financial support from NIH (GM72623) and Burroughs Wellcome Fund Career Award in Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sofia H, Chen G, Hetzler B, Reyes-Spindola J, Miller N. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, et al. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamat SS, Williams HJ, Raushel FM. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature. 2011;480:570–573. doi: 10.1038/nature10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An Anchoring Role for FeS Clusters: Chelation of the Amino Acid Moiety of S-Adenosylmethionine to the Unique Iron Site of the [4Fe-4S]. Cluster of Pyruvate Formate-Lyase Activating Enzyme. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 5.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, et al. Spectroscopic Approaches to Elucidating Novel Iron-Sulfur Chemistry in the ”Radical-SAM” Protein Superfamily. Inorg. Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 6.Magnusson O, Reed G, Frey P. Characterization of an allylic analogue of the 5′-deoxyadenosyl radical: an intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry. 2001;40:7773–7782. doi: 10.1021/bi0104569. [DOI] [PubMed] [Google Scholar]
- 7.Frey PA, Booker SJ. Zard SZ, editor. Radical intermediates in the reaction of lysine 2,3-aminomutase. Advances in Free Radical Chemistry, Advances in Free Radical Chemistry. 1999:1–43. [Google Scholar]
- 8.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem. 2012 doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smulson ME, Suhadolnik RJ. The biosynthesis of the 7-deazaadenine ribonucleoside, tubercidin, by Streptomyces tuber cidicus. J Biol Chem. 1967;242:2872–2876. [PubMed] [Google Scholar]
- 10.Suhadolnik RJ, Uematsu T. Biosynthesis of the pyrrolopyrimidine nucleoside antibiotic, toyocamycin. VII. Origin of the pyrrole carbons and the cyano carbon. J Biol Chem. 1970;245:4365–4371. [PubMed] [Google Scholar]
- 11.Uematsu T, Suhadolnik RJ. Nucleoside antibiotics. VI. Biosynthesis of the pyrrolopyrimidine nucleoside antibiotic toyocamycin by Streptomyces rimosus. Biochemistry. 1970;9:1260–1266. doi: 10.1021/bi00807a030. [DOI] [PubMed] [Google Scholar]
- 12.Elstner EF, Suhadolnik RJ. The biosynthesis of the nucleoside antibiotics. IX. Purification and properties of guanosine triphosphate 8-formylhydrolase that catalyzes production of formic acid from the ureido carbon of guanosine triphosphate. J Biol Chem. 1971;246:6973–6981. [PubMed] [Google Scholar]
- 13.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J Biol Chem. 2004;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 14.Van Lanen SG, Reader JS, Swairjo MA, de Crécy-Lagard V, Lee B, Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: discovery of nitrile reductase activity in a common fold. Proc Natl Acad Sci USA. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uematsu H, Suhadolnik R. In vivo and enzymatic conversion of toyocamycin to sangivamycin by Streptomyces rimosus. Arch Biochem Biophys. 1974;162:614–619. doi: 10.1016/0003-9861(74)90223-9. [DOI] [PubMed] [Google Scholar]
- 17.Uematsu T, Suhadolnik R. Toyocamycin nitrile hydrolase. Methods Enzymol. 1975;43:759–762. doi: 10.1016/0076-6879(75)43143-3. [DOI] [PubMed] [Google Scholar]
- 18.McCarty RM, Somogyi A, Bandarian V. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry. 2009;48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty RM, Somogyi A, Lin G, Jacobsen NE, Bandarian V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine 5′-triphosphate in four steps. Biochemistry. 2009;48:3847–3852. doi: 10.1021/bi900400e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada N, Noguchi S, Kasai H, Shindo-Okada N, Ohgi T, Goto T, et al. Novel Mechanism of post-transcriptional modification of tRNA. J Biol Chem. 1979;254:3067–3073. [PubMed] [Google Scholar]
- 21.Slany R, Bösl M, Crain P, Kersten H. A New Function of S-Adenosylmethionine: The Ribosyl Moiety of AdoMet Is the Precursor of the Cyclopentanediol Moiety of the tRNA Wobble Base Queuine. J Biol Chem. 1993;32:7811–7817. doi: 10.1021/bi00081a028. [DOI] [PubMed] [Google Scholar]
- 22.Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci USA. 2011;108:7368–7372. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballinger M, Reed G, Frey P. An organic radical in the lysine 2,3-aminomutase reaction. Biochemistry. 1992;31:949–953. doi: 10.1021/bi00119a001. [DOI] [PubMed] [Google Scholar]
- 24.Ballinger M, Frey P, Reed G. Structure of a substrate radical intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry. 1992;31:10782–10789. doi: 10.1021/bi00159a020. [DOI] [PubMed] [Google Scholar]
- 25.Rauk A, Yu D, Taylor J, Shustov GV, Block DA, Armstrong DA. Effects of structure on alpha C-H bond enthalpies of amino acid residues: relevance to H transfers in enzyme mechanisms and in protein oxidation. Biochemistry. 1999;38:9089–9096. doi: 10.1021/bi990249x. [DOI] [PubMed] [Google Scholar]
- 26.Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Liu W. Complex Biotransformations Catalyzed by Radical S-Adenosylmethionine Enzymes. J Biol Chem. 2011;286:30245–30252. doi: 10.1074/jbc.R111.272690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandarian V, Reed G. Banerjee R. Vitamin B12. New York: John Wiley & Sons, Inc; 1999. Ethanolamine ammonia-lyase; pp. 811–833. [Google Scholar]
- 29.RajBhandary U, Chang SH, Stuart A, Faulkner RD, Hoskinson RM, Khorana HG. Studies on polynucleotides, LXVIII. The primary structure of yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. 1967;57:751–758. doi: 10.1073/pnas.57.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiebe R, Zachau HG. A Specific Modification Next to the Anticodon of Phenylalanine Transfer Ribonucleic Acid. Eur J Biochem. 1968;5:546–555. doi: 10.1111/j.1432-1033.1968.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi K, Furutachi N, Funamizu M, Grunberger D, Weinstein IB. Structure of the fluorescent Y base from yeast phenylalanine transfer ribonucleic acid. J Am Chem Soc. 1970;92:7617–7619. doi: 10.1021/ja00729a035. [DOI] [PubMed] [Google Scholar]
- 32.Thiebe R, Zachau HG, Baczynskyj L, Biemann K, Sonnenbichler J. Study on the properties and structure of the modified base Y+ of yeast tRNA Phe . Biochim Biophys Acta. 1971;240:163–169. doi: 10.1016/0005-2787(71)90653-8. [DOI] [PubMed] [Google Scholar]
- 33.Blobstein SH, Gebert R, Grunberger D, Nakanishi K, Weinstein IB. Structure of the fluorescent nucleoside of yeast phenylalanine transfer ribonucleic acid. Arch Biochem Biophys. 1975;167:668–673. doi: 10.1016/0003-9861(75)90510-x. [DOI] [PubMed] [Google Scholar]
- 34.Funamizu M, Terahara A, Feinberg AM, Nakanishi K. Total synthesis of dl-Y base from yeast phenylalanine transfer ribonucleic acid and determination of its absolute configuration. J Am Chem Soc. 1971;93:6706–6708. doi: 10.1021/ja00753a080. [DOI] [PubMed] [Google Scholar]
- 35.Frihart CR, Feinberg AM, Nakanishi K. Synthesis of 4,9-dihydro-4, 6-dimethyl-9-oxo-1H-imidazo [1, 2-a] purine and the “Y” base from Saccharomyces cerevisiae phenylalanine transfer RNA. J. Org. Chem. 1978;43:1644–1649. [Google Scholar]
- 36.Nakanishi K, Blobstein S, Funamizu M, Furutachi N, Van Lear G, Grunberger D, et al. Structure of the “peroxy-Y base” from liver tRNA Phe . Nature New Biol. 1971;234:107–109. doi: 10.1038/newbio234107b0. [DOI] [PubMed] [Google Scholar]
- 37.Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K. Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry. 1973;12:188–193. doi: 10.1021/bi00726a002. [DOI] [PubMed] [Google Scholar]
- 38.Zhou S, Sitaramaiah D, Noon KR, Guymon R, Hashizume T, McCloskey JA. Structures of two new “minimalist” modified nucleosides from archaeal tRNA. Bioorg Chem. 2004;32:82–91. doi: 10.1016/j.bioorg.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Kasai H, Yamaizumi Z, Kuchino Y, Nishimura S. Isolation of hydroxy-Y base from rat liver tRNAPhe. Nuc. Acids Res. 1979;6:993–999. doi: 10.1093/nar/6.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li HJ, Nakanishi K, Grunberger D, Weinstein IB. Biosynthetic studies of the Y base in yeast phenylalanine tRNA. Incorporation of guanine. Biochem Biophys Res Commun. 1973;55:818–823. doi: 10.1016/0006-291x(73)91217-5. [DOI] [PubMed] [Google Scholar]
- 41.Munch HJ, Thiebe R. Biosynthesis of the nucleoside Y in yeast tRNAPhe: incorporation of the 3-amino-3-carboxypropyl-group from methionine. FEBS Lett. 1975;51:257–258. doi: 10.1016/0014-5793(75)80900-8. [DOI] [PubMed] [Google Scholar]
- 42.Smith C, Schmidt PG, Petsch J, Agris PF. Nuclear magnetic resonance signal assignments of purified [13C]methyl-enriched yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1985;24:1434–1440. doi: 10.1021/bi00327a023. [DOI] [PubMed] [Google Scholar]
- 43.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. Embo J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, et al. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 2010;27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noma A, Suzuki T. Ribonucleome analysis identified enzyme genes responsible for wybutosine synthesis. Nucleic Acids Symp Ser (Oxf) 2006:65–66. doi: 10.1093/nass/nrl032. [DOI] [PubMed] [Google Scholar]
- 46.Waas WF, de Crécy-Lagard V, Schimmel P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J Biol Chem. 2005;280:37616–37622. doi: 10.1074/jbc.M506939200. [DOI] [PubMed] [Google Scholar]
- 47.Young AP, Bandarian V. Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry. 2011;50:10573–10575. doi: 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalhor HR, Penjwini M, Clarke S. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Commun. 2005;334:433–440. doi: 10.1016/j.bbrc.2005.06.111. [DOI] [PubMed] [Google Scholar]
- 49.Noma A, Ishitani R, Kato M, Nagao A, Nureki O, Suzuki T. Expanding role of the jumonji C domain as an RNA hydroxylase. J Biol Chem. 2010;285:34503–34507. doi: 10.1074/jbc.M110.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine SI, et al. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr D Biol Crystallogr. 2007;63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, et al. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 52.Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc Natl Acad Sci USA. 2009;106:15616–15621. doi: 10.1073/pnas.0905270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009;37:2910–2925. doi: 10.1093/nar/gkp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato M, Araiso Y, Noma A, Nagao A, Suzuki T, Ishitani R, et al. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 2011;39:1576–1585. doi: 10.1093/nar/gkq919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, et al. Structural and functional comparison of HemN to other radical SAM enzymes. Biol Chem. 2005;386:971–980. doi: 10.1515/BC.2005.113. [DOI] [PubMed] [Google Scholar]
- 56.Cousins FB. The prosthetic group of a chromoprotin from mycobacteria. Biochim Biophys Acta. 1960;40:532–534. doi: 10.1016/0006-3002(60)91396-2. [DOI] [PubMed] [Google Scholar]
- 57.Cheeseman P, Toms-Wood A, Wolfe RS. Isolation and properties of a fluorescent compound, Factor420, from Methanobacterium strain M.o.H. J Bacteriol. 1972;112:527–531. doi: 10.1128/jb.112.1.527-531.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eirich LD, Vogels GD, Wolfe RS. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 59.Isabelle D, Simpson DR, Daniels L. Large-scale production of coenzyme F420-5,6 by using Mycobacterium smegmatis. Appl Environ Microbiol. 2002;68:5750–5755. doi: 10.1128/AEM.68.11.5750-5755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh CT. Naturally occurring 5-deazaflavin coenzymes: Biological redox roles. ACC. Chem. Res. 1986;19:216–221. [Google Scholar]
- 61.Massey V, Hemmerich P. Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavins. Biochemistry. 1978;17:9–16. doi: 10.1021/bi00594a002. [DOI] [PubMed] [Google Scholar]
- 62.Choi K, Kendrick N, Daniels L. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and F0 biosynthesis. J Bacteriol. 2002;184:2420–2428. doi: 10.1128/JB.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graham D, Xu H, White R. Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F(420) biosynthesis. Arch Microbiol. 2003;180:455–464. doi: 10.1007/s00203-003-0614-8. [DOI] [PubMed] [Google Scholar]
- 64.Fischer M, Bacher A. Biosynthesis of flavocoenzymes. Nat Prod Rep. 2005;22:324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 65.Reuke B, Korn S, Eisenreich W, Bacher A. Biosynthetic precursors of deazaflavins. J Bacteriol. 1992;174:4042–4049. doi: 10.1128/jb.174.12.4042-4049.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grochowski LL, Xu H, White RH. Identification and characterization of the 2-phospho-L-lactate guanylyltransferase involved in coenzyme F420 biosynthesis. Biochemistry. 2008;47:3033–3037. doi: 10.1021/bi702475t. [DOI] [PubMed] [Google Scholar]
- 67.Isabelle DW, Daniels L. Structures of coenzyme F420 in Mycobacterium species. Arch Microbiol. 2001;176:37–43. doi: 10.1007/s002030100290. [DOI] [PubMed] [Google Scholar]
- 68.Driesener RC, Challand MR, McGlynn SE, Shepard EM, Boyd ES, Broderick JB, et al. [FeFe]-hydrogenase cyanide ligands derived from S-adenosylmethionine-dependent cleavage of tyrosine. Angewandte Chemie. International Ed. in English. 2010;49:1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 69.Challand MR, Martins FT, Roach PL. Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis inv Escherichia coli. J Biol Chem. 2010;285:5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q, Li Y, Chen D, Yu Y, Duan L, Shen B, et al. Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat Chem Biol. 2011;7:154–160. doi: 10.1038/nchembio.512. [DOI] [PMC free article] [PubMed] [Google Scholar]