Abstract
Background
Glioblastomas with a specific mutation in the isocitrate dehydrogenase 1 (IDH1) gene have a better prognosis than gliomas with wild-type IDH1.
Methods
Here we compare the IDH1 mutational status in 172 contrast-enhancing glioma patients with the invasion profile generated by a patient-specific mathematical model we developed based on MR imaging.
Results
We show that IDH1-mutated contrast-enhancing gliomas were relatively more invasive than wild-type IDH1 for all 172 contrast-enhancing gliomas as well as the subset of 158 histologically confirmed glioblastomas. The appearance of this relatively increased, model-predicted invasive profile appears to be determined more by a lower model-predicted net proliferation rate rather than an increased model-predicted dispersal rate of the glioma cells. Receiver operator curve analysis of the model-predicted MRI-based invasion profile revealed an area under the curve of 0.91, indicative of a predictive relationship. The robustness of this relationship was tested by cross-validation analysis of the invasion profile as a predictive metric for IDH1 status.
Conclusions
The strong correlation between IDH1 mutation status and the MRI-based invasion profile suggests that use of our tumor growth model may lead to noninvasive clinical detection of IDH1 mutation status and thus lead to better treatment planning, particularly prior to surgical resection, for contrast-enhancing gliomas.
Keywords: glioma, growth kinetics, IDH1, mathematical model, patient-specific, proliferation, invasion
See the editorial by Lathia, on pages 763–764.
Since the 2008 discovery of a mutation in the isocitrate dehydrogenase 1 (IDH1) gene in a subset of glioma patients, more than 2 dozen manuscripts have been published about the role of this IDH1 mutation in the natural history of glioma.1–6 This unique mutation in IDH1, which changes arginine at position 132 to histidine, is disproportionately represented in lower-grade gliomas; it is present in >75% of grade II and III gliomas but only ∼10% of grade IV glioblastomas (GBMs).1,7 Furthermore, this mutation in IDH1 is more prevalent in younger patients. Glioma patients with the mutation show significantly longer survival times than those with the wild-type copy of the gene, having a median survival of 3.8 years versus 1.1 years.1,8 Additionally, secondary GBMs are predominantly mutant in IDH1 (83%), while very few primary GBMs (5%) harbor the mutation.7 Since IDH1mut GBMs (and perhaps more contrast-enhancing gliomas) have a significantly better prognosis than IDH1wt GBMs, it is clinically important to have pretreatment (presurgical) predictors of IDH1mut status.
Our laboratory has developed a patient-specific mathematical model of glioma growth that is based on diagnostic and pretreatment MRI scans obtained in the course of normal clinical treatment.9–15 By combining our model formalism with tumor volume measures extracted from these routinely obtained pretreatment MRIs, we are able to estimate patient-specific parameters that quantify the net proliferation rate (ρ) of the glioma cells and their net dispersal or diffusion rate (D). These parameters can be used to characterize the differential role of proliferation versus diffusion in driving the overall tumor growth pattern seen in each patient. The variation in the parameters across patients is consistent with the wide heterogeneity in imaging results and invasive capacity typical of the disease.12
These 2 kinetic parameters can be combined to produce a biological aggressiveness ratio (ρ/D) that quantifies the relative proliferative to invasive nature of each tumor. This measure of biological aggressiveness is predictive of worse prognosis14 and increasing degrees of hypoxia,15 a known feature of tumor aggressiveness. Also, we have previously shown that a high ρ/D (characteristic of more nodular, less diffuse tumors) is more likely to represent a rapidly developing primary GBM that is relatively less invasive, whereas a low ρ/D (characteristic of a more diffuse, less nodular tumor) is associated with a slower developing but more invasive secondary GBM.16 This novel discovery suggests the possibility of predicting primary versus secondary GBM natural histories and, by extension, likely IDH1mutation status based on a single measure quantifiable from routinely obtained MR images alone. Further, a recent game theory-based consideration of the evolutionary role of IDH1 mutation in cellular populations suggested that such a mutation would select for a more invasive overall tumor phenotype.17 Combining these 2 insights suggests the following question: can we predict IDH1 mutation status from image-based analysis of tumor invasion using patient-specific mathematical models of glioma proliferation and invasion kinetics? One of our main hypotheses was that the most diffuse contrast-enhancing gliomas (low ρ/D) would be mutant in IDH1.
To test this hypothesis, we examined the mutational status of IDH1 by mutation-specific immunohistochemistry in a cohort of 172 newly diagnosed, contrast-enhancing glioma patients, 91% of whom were ultimately found to have grade IV tumors. The concept was to distinguish IDH1 mutant tumors (with their associated favorable prognosis) within a cohort of contrast-enhancing gliomas by mapping a favorable molecular feature to patient-specific disease kinetics (ie, net proliferation and invasion rates).
Materials and Methods
Study Inclusion Criteria
A total of 172 patients with suspected contrast-enhancing glioma on pretreatment MRI sequences were selected for this study. Fifty-three of these patients were participants in our ongoing glioma study at the University of Washington (UW) and were either consented prospectively or approved for retrospective study; both procedures were approved by the UW institutional review board. Since contrast-enhancing lesions are more likely to be primary GBM (and thus IDH1wt), we sought to enrich our dataset for contrast-enhancing IDH1muts through a subset of cases included in The Cancer Genome Atlas (TCGA) glioblastoma dataset. Thus, 105 IDH1 wt and 2 IDH1mut GBM patients with appropriate clinical imaging from the public TCGA database (https://tcga-data.nci.nih.gov/tcga/ (last accessed March 3, 2014)) were included in the present study. Additionally, 14 patients from Columbia University were included (2 mutants and 12 wild-types). Of the 53 participants from our UW study included in this report, 5 were noted to have IDH1mut in their UW medical records, and 46 had archived tissue in the UW Department of Neuropathology that required testing for IDH1 mutation status. All 172 participants in this report satisfied the requirement for ρ/D analysis of having pretreatment T1Gd and T2/ fluid-attenuated inversion recovery (FLAIR) MRI scans on the same day. An additional pretreatment MRI study enabling independent estimation of D and ρ was available for only 39 of the 172 study participants.
Images and Image Analysis
MRI scans were obtained with standard clinical MR pulse sequences for T1- and T2-weighted fast spin-echo images as well as FLAIR images. Slice thickness varied from 1 mm to 6 mm. For contrast-enhanced images, intravenous injection of the contrast agent Prohance (n = 51) preceded T1-weighted imaging. The imaging protocols for the 2 participants included from TCGA were not available. Co-registration of the T1Gd and T2 images for the cell density analysis in Fig. 1 was performed using the Statistical Parametric Mapping Tool in MATLAB.18 The cell density profile was generated by code written in our laboratory.19
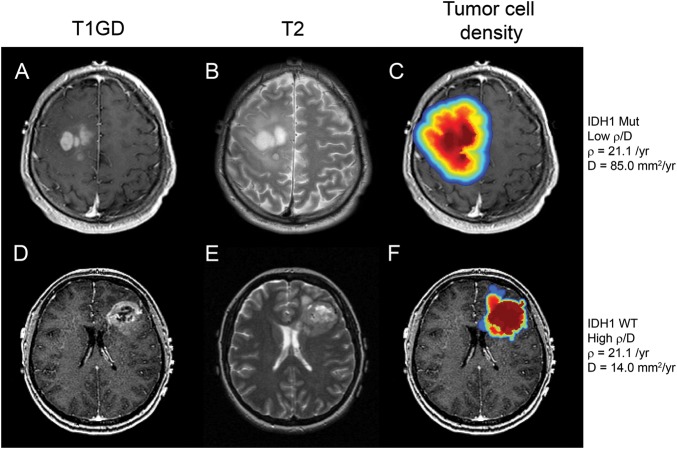
Fig. 1.
MRI scans from 2 patients with similar ρ values, one that is mutant for IDH1 (top row: A,B,C) and one that is IDH1 wild-type (bottom row: D,E,F). All 6 images are pretreatment scans; A and D are T1Gd images, B and E are T2 images, and C and F are false-color images representing tumor cell density overlaid on the T1Gd scans (red, highest cell density; blue, lowest cell density). The growth parameters associated with these GBMs are ρ = 21.1/year, D = 85.0 mm2/year, with ρ/D = 0.25/mm2 for the IDH1mut tumor and ρ = 21.0/year, D = 14.0 mm2/year, ρ/D = 1.5/mm2 for the IDH1wt tumor. The figure illustrates the more diffuse nature of the IDH1 mutant tumor (top row) versus the wild-type (bottom row).
Calculation of Rates of Proliferation (ρ) and Dispersal (D)
Using the mathematical model developed in our laboratory, tumor volumes observed on pretreatment gadolinium-enhanced T1-weighted and T2 MRIs were segmented using MATLAB processing.18,14 The tumor volumes were measured by 2 observers, and each consensus measurement was validated by a third observer. Then,12–16 the mean of each observer's validated volume measurements was used for all analysis. The relationship between T1Gd and T2 volumes and pretreatment radial tumor velocity (change in tumor radius over time) was used to calculate values for D (diffusion, or dispersal, of the tumor outward from the central mass) and ρ (proliferation, the increase in cell density in any given tumor region). These parameters were used to generate patient-specific tumor growth models.
IDH1 Mutation Status
For participants accrued in our UW glioma study, tissue slides were made from archived tumor biopsy samples in paraffin blocks and stained with a primary antibody raised against a synthetic peptide corresponding to IDH1 amino acids 125–137 with histidine at position 132 (DIA-H09, Dianova GmbH). Slides were deparaffinized, and antigen retrieval was in citrate buffer, pH 6.0, with endogenous peroxidase blocked with 3% hydrogen peroxide. Staining with the primary antibody (diluted 1:100) was done for 60 minutes, followed by anti-mouse secondary staining (diluted 1:200) for 40 minutes. Detection was by Vectastain ABC Elite (Vector Laboratories) for 40 minutes. The stained slides were scored by a trained clinical pathologist (D.B.) for the presence or absence of the mutant form of IDH1. For the 5 participants whose IDH1 mutation status was obtained from medical records, similar antibody-based detection was performed clinically at the UW Department of Anatomic Pathology. Scoring was performed by trained clinical pathologists, as noted in the medical reports. IDH1 mutation status for the 2 TCGA patients were retrieved from their publicly available interpreted expression data of mutated IDH1 genes, determined via hybrid-capture performed on an ABI SOLiD platform.
Ki67 Immunohistochemistry
Fixed tissue was available from 58 participants. Sections were cut from archived tumor tissue in formalin-fixed, paraffin-embedded blocks, mounted on glass slides, and processed using automated immunohistochemistry (Bond, Leica) that included deparaffinizing, rehydration, incubation with primary antibody MIB-1 clone (Dako), and application of chromogen. Slides were reviewed on a microscope (BX41, Olympus) with photomicroscopy (DMC76, Leica) of the region with highest labeling density.
Statistics
The Student' t test (2-tailed, unpaired) was performed to determine the statistical significance of any differences in ρ/D, D, ρ, and velocity between the IDH1mut group and the IDHwt group using R version 2.14.2 software (R Foundation for Statistical Computing). Additionally, the crossmatch test was used to examine the difference between 2 multivariate distributions in Figs. 3A and 5. To assess the accuracy of the diagnostic test (IDH1mut = low ρ/D), receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were calculated using MATLAB.18. The log-rank test was used to compare differences in survival rate in the Kaplan-Meier analysis. For all comparisons between IDH1wt and IDH1mut tumor characteristics, we considered a P value ≤.05 to be significant.
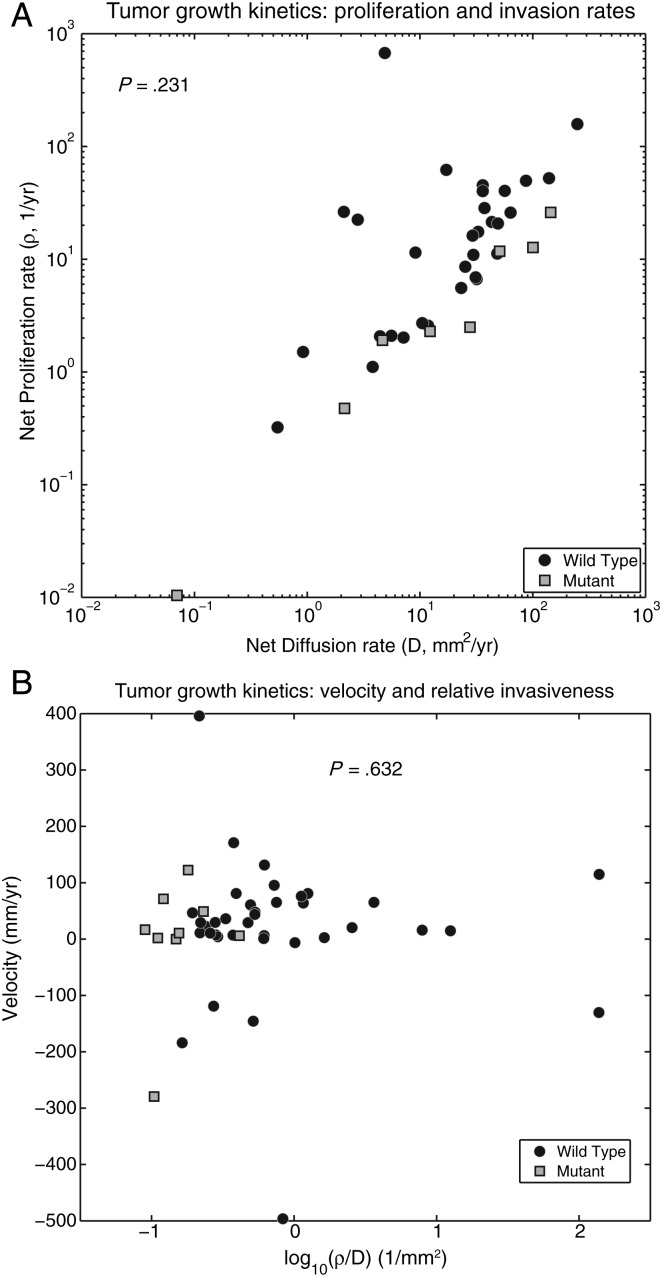
Fig. 3.
(A) A plot of ρ versus D for the 39 patients with contrast-enhancing gliomas, from whom these values were obtainable. There is an apparent segregation of the IDH1 mutant tumors from the IDH1 wild-type tumors with IDH1mut tumors having lower ρ values than IDH1wt tumors. The apparent segregation is not significant; crossmatch analysis of the 2 groupings yielded an approximate P value of .231. (B) Tumor growth velocity plotted against ρ/D for both IDH1 wild-type (n = 32) and IDH1 mutant (n = 7) contrast-enhancing gliomas. The IDH1wt tumors have larger ρ/D values and populate the graph both above and below the value of ρ/D = 1, while the IDH1mut tumors have lower ρ/D values and populate the graph below the value of ρ/D = 1, mirroring the differences in ρ/D values that were seen in Fig. 2. There was no statistical difference in mean velocities between IDH1wt and IDH1mut tumors for any given value of ρ/D (IDH1 wt: mean, 59.83 mm/y; median, 34.43; standard deviation 101.5); (IDH1 mut: mean, 48.91 mm/y; median, 60.66; standard deviation 28.2) (P = .632).
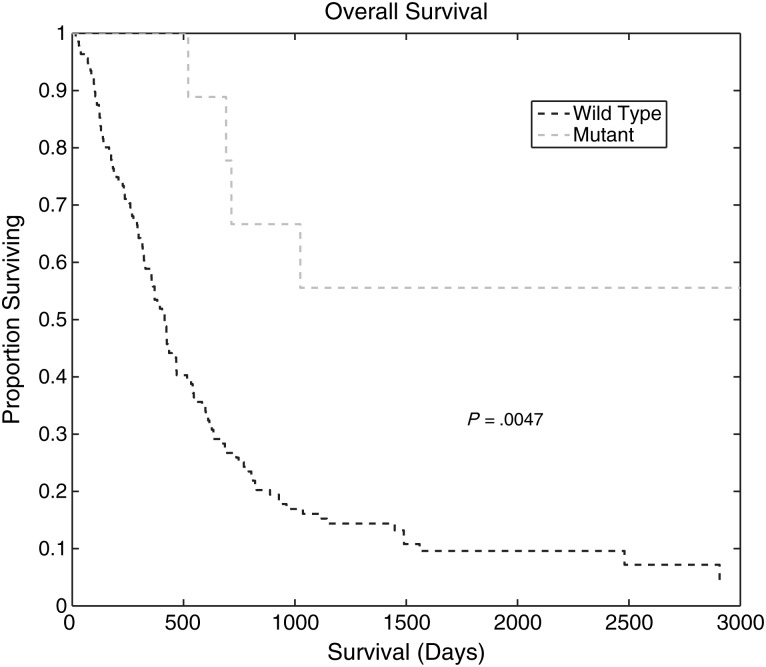
Fig. 5.
Kaplan-Meier survival curves for IDH1wt and IDH1mut glioma patients. The median survival of IDH1wt glioma patients (573 days) was significantly different from that of IDH1mut glioma patients (896 days) (P = .0047).
Results
Image-based Comparison of IDH1 Mutant and IDH1 Wild-type Gliomas
One hundred seventy-two participants with newly diagnosed, contrast-enhancing gliomas were included in our study. Ninety-two percent of these tumors (n = 158) were grade IV, while the remainder (n = 14) were grade II or III. Fig. 1 shows 2 contrast-enhancing tumors, one IDH1mut and the other IDH1wt, with similar net proliferation rates (ρ) but different net dispersal rates (D) and compares the T1Gd and T2 MRI scans as well as model-predicted cell density profiles based on their patient-specific ρ/D. These 2 tumors are generally representative of the average invasion profiles (average ρ/D) for IDH1mut and IDH1wt patients. Contrast-enhancing gliomas that are mutant in IDH1 (IDH1 mut) have lower ρ/D values than contrast-enhancing gliomas that are wild-type IDH1 (IDH1wt). That is, the IDH1mut tumor displays a relatively more diffuse growth pattern (low ρ/D), which is reflected in the relatively slow gradient from the red high-cell density regions to the blue low-cell density regions extending well beyond the edge of the T1Gd abnormality. Thus, the simulated glioma cell density tuned to each patient's ρ/D value clearly reveals the diffuse invasion profile of IDH1mut tumors; most of the IDH1wt tumor has dense cellularity with a steep gradient from high-cell density to low-cell density (high ρ/D), while the IDH1mut tumor has a nearly linear drop-off in cell density out to very low values well beyond the imageable tumor periphery (low ρ/D). A comparison of the T1Gd radii, T2 radii, and age between IDH1 mutants and wild-type participants is displayed in Supplemental Fig. 1.
Comparison of Aggressiveness Index Between IDH1 Mutant and Wild-type Tumors
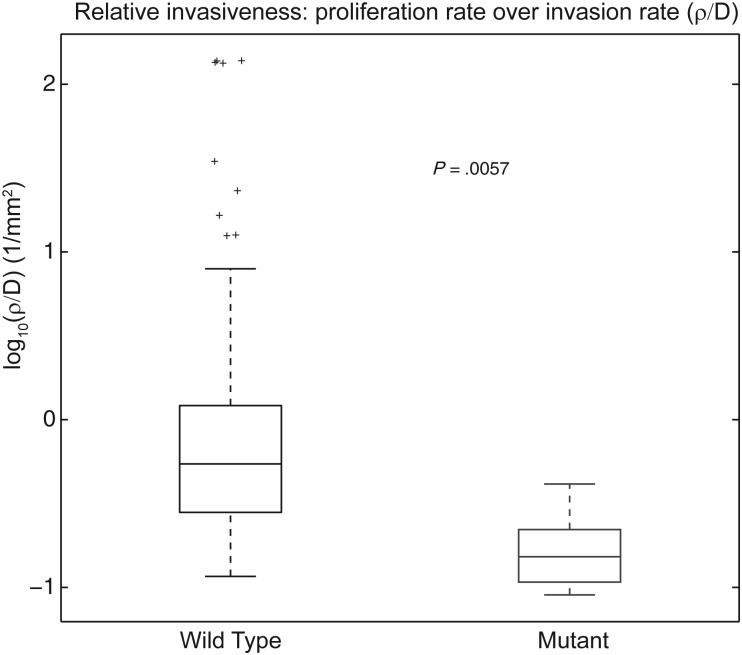
We hypothesized that the more diffuse pattern of growth seen in the IDH1mut tumors would be reflected in low values of the aggressiveness index, defined as ρ/D, the ratio of the predicted patient-specific rate of net proliferation (ρ) to rate of net dispersal (D). We compared the aggressiveness index (ρ/D) between the IDH1wt and IDH1mut contrast-enhancing gliomas in the 53 UW participants, 105 TCGA participants, and 14 Columbia participants with the relevant clinical imaging available. The participants included in our study and the results are shown in Fig 2. IDH1mut gliomas (n = 16) cluster with low ρ/D values, while IDH1wt gliomas (n = 156) have a broad distribution of ρ/D values that are significantly higher than those of the IDH1mut group. IDH1 wild-type tumors have a mean ρ/D value of 5.0409/mm2, while IDH1 mutant tumors have a mean ρ/D value of 0.181/mm2 (P = .0057, t test), suggesting a significant difference in growth patterns between the 2 tumor types. Although this study included primary and secondary enhancing lesions to simulate the perspective of a physician who does not yet know tumor grade, we performed the analysis on the secondary (n = 14) and primary GBM (n = 158) cohorts individually and found the same difference in ρ/D (P = .00036 and P = .018, Supplemental Fig. 2).
Fig. 2.
A plot of ρ/D values for the 42 IDH1 wild-type and 11 IDH1 mutant contrast-enhancing gliomas in this study. The mean values of ρ/D are 5.0409/mm2 for IDH1 wild-type and 0.181/mm2 for IDH1 mutant, which are significantly different (P = .0057).
Measurement of Proliferation, Diffusion, and Tumor Growth Velocity
We were also interested in determining whether the relatively increased diffuse pattern of growth in the IDH1mut participants was the result of a relatively decreased proliferative capacity or an increased invasion rate compared with IDH1wt. Thus, we compared both ρ and D independently with the IDH1 mutational status. Because these measurements of ρ and D require the availability at least 2 sequential pretreatment scans11–15,20,21 separated by at least 5 days, only 39 participants for whom these independent measures were obtained are included in the subsequent analysis.
The ρ and D values for IDH1wt (n = 32) and IDH1mut (n = 7) glioma participants are shown in Fig. 3A. The 2 IDH1 categories appear to segregate within the ρ-D space. IDH1wt tumors are predominantly found in the upper left portion of the graph (high ρ, high D) but are not seen in the lower right portion (low ρ, high D). IDH1mut tumors, on the other hand, are found almost exclusively in the low ρ/high D portion of the graph. Although the relationship between D and ρ was not significantly different between wild types and mutants (P = .231, crossmatch test), when viewed with the ρ/D values for the entire cohort of patients as seen in Fig. 2, the data suggests that IDH1mut tumors characteristically have low ρ and high D values making them relatively diffuse.
Previous work by our group,16 using our patient-specific mathematical models of malignant progression, predicted that low-grade gliomas would need to segregate in a certain subset of ρ-D space to progress to secondary GBMs. That is, the model predicted that there is only a subset of net rates of proliferation and invasion that could create the imaging and histological features of secondary GBMs. Interestingly, the IDH1mut in our current study occupy precisely the same region of ρ-D space predicted in our earlier study16 for secondary GBMs (see Fig. 3A). Since IDH1mut tumors overlap significantly with secondary GBMs, this self-consistency between model predictions of the kinetic rates expected of secondary GBMs and those observed in this study for the IDH1mutants provides promising support for using these models to provide further insight into the kinetics of IDH1mut gliomas in future studies.
IDH1mut gliomas continue to cluster separately from IDH1wt tumors on the extreme low (left) end of the aggressiveness (ρ/D) index (x-axis) while ranging widely across the range of tumor growth velocities (y-axis) (Fig. 3B). There was no statistical difference in mean tumor growth velocity on MRI between IDH1wt and IDH1mut tumors. IDH1 wild-type tumors (both grades included) had a mean growth velocity of 59.83 mm/year (median, 34.43 mm/y; standard deviation, 101.5 mm/y), while IDH1 mutant tumors (both grades included) had a mean growth velocity of 48.91 mm/year (median, 60.66 mm/y; standard deviation 28.2 mm/y) (P = .632, t test). Considering only the grade IV (GBM) participants, we again found no difference in velocity between the 2 tumor types. Grade IV IDH1wt tumors had a mean velocity of 51.32 mm/year (median, 34.37 mm/y; standard deviation, 95.0 mm/y), while grade IV IDH1mut tumors had a mean velocity of 71.20 mm/year (median, 69.71 mm/y; standard deviation, 8.6 mm/y) (P = .287). Thus, despite the differences in genetic background between these 2 tumor types, it appears that once an IDH1mut glioma has progressed to grade IV, it grows as rapidly as a primary GBM that presents as grade IV at diagnosis. It remains an unanswered question why the survival time after diagnosis is nearly 3 times longer for patients with IDH1mut tumors. One may suggest that relatively more diffuse tumors may grow to larger imageable tumor sizes before reaching a fatal tumor burden.
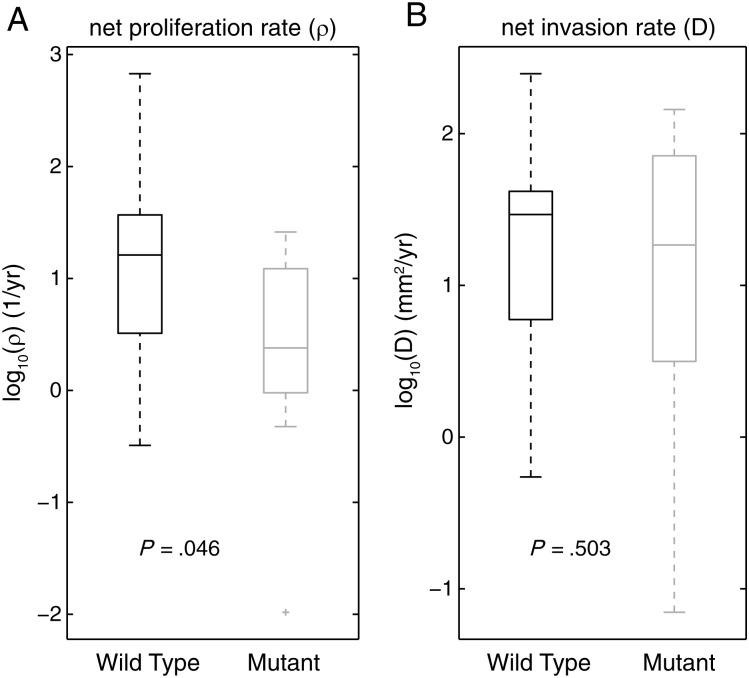
To test whether the growth kinetics of these tumor types were driven more by a cap on proliferation or by increased invasion, we compared the ρ and D values separately between IDH1mut and IDH1wt tumors as shown in Fig. 4. Although there was overlap in the distributions, there was a significant difference between the mean values for proliferation rate (ρ) in the 2 groups (IDH1wt, 29.22/y; IDH1mut, 11.87/y) (P = .046, t test), whereas the difference in the mean values for D (IDH1wt, 41.24 mm2/y; IDH1mut, 53.83 mm2/y) was smaller and not significant (P = .503, t test). This indicates that the driving force for the difference in the aggressiveness index between IDH1wt and IDH1mut tumors may be a relative suppression of the net rate of proliferation in the IDH1mut tumors compared with the net invasion rate. This relative suppression of the net proliferation rate could be explained by either a decreased mitotic rate or an increased cell death rate or both. Although MIB1/Ki67 or similar labeling indices may be used as a proxy for proliferative activity, such histological analysis reflects testing on some small sample of tumor tissue, which is different from the quantifying net proliferation rate for the entire tumor (ρ). Analysis of the MIB1/Ki67 labeling index was quantified for 58 study participants and did not reveal a significant difference between Ki67 in IDH1mut and IDH1wt tumors (Supplement figure 3).
Fig. 4.
Separate plots of ρ values (A) and D values (B) for IDH1 wild-type tumors (n = 32) and IDH1 mutant tumors (n = 7). IDH1wt tumors are shown on the left in both A and B, while IDH1mut tumors are shown on the right in both A and B. In A, the mean proliferation rates, (ρ) are significantly different: IDH1wt (both grades) = 29.22/year, IDH1mut (both grades) = 11.87/year, P = .046. In B, the mean invasion rates (D) are not significantly different: IDH1wt (both grades) = 41.24 mm2/year; IDH1mut (both grades) = 53.83 mm2/year, P = .503.
To confirm that the difference in survival for the IDH1wt and IDH1mut tumor participants in our cohort was comparable to that reported in the literature, we performed a Kaplan-Meier analysis of the 2 participant groups. Figure 5 shows that the survival rates for the 2 groups are significantly different (P = .0047, log-rank test). The median survival for IDH1wt glioma participants was 573 days, while that for IDH1mut glioma participants was 896 days.
ROC Analysis of ρ/D as Predictor of IDH1 Mutation Status
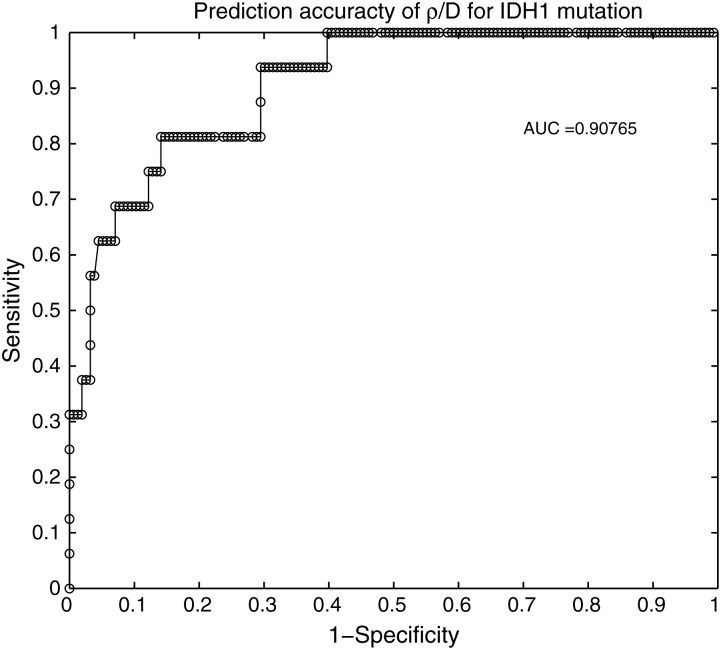
The separation of ρ/D values between IDH1wt and IDH1mut contrast-enhancing gliomas seen in Fig. 2 prompted us to perform a ROC analysis to test the ability of ρ/D value for predicting the mutational status of IDH1 in newly diagnosed contrast-enhancing lesions. Specifically, noting that IDH1mut tumors cluster amongst the more diffuse (small ρ/D) patients, we analyzed the predictive value of ρ/D as a test for IDH1mut status (Fig. 6). The optimal cutoff suggests that we have achieved 81.3% sensitivity and 85.9% specificity in predicting that the lesion is IDH1mut in patients with ρ/D < 0.2381/mm2. The resulting area under the curve (AUC) is 0.9077, indicating a robust ability of ρ/D to discriminate between IDH1 mutants from wild-type tumors (an AUC of 1.0 indicates 100% predictive accuracy). Optimum accuracy achieved is 93.6%. Furthermore, although there was an optimal cutoff differentiating IDH1wt from IDH1mut that balances sensitivity and specificity, there was a clear cutoff of ρ/D above which there were no IDH1 mutants (60.3% sensitivity, 100% specificity, ρ/D>0.414/mm2). Supplemental Fig. 4 displays cross-validation supporting the robustness of ρ/D as a predictive metric for IDH1mut status. Alternatively, we noted that in Fig. 2, high ρ/D tumors were exclusively IDH1wt, which allowed us to test the accuracy of predicting wild-type status using a ρ/D > threshold criteria (Fig. 6). The optimal cutoff suggests that for patients with ρ/D > 0.2313/mm2, we have achieved 85.9% sensitivity and 81.3% specificity in predicting that the lesion is IDH1wt.
Fig. 6.
Receiver operating characteristic (ROC) analysis of ρ/D as a predictor of IDH1 mutation status. True positive rate versus false positive rate for various cutoff values of ρ/D are tested and plotted as sensitivity (y-axis) and an indicator of specificity (x-axis). The resulting area under the curve (AUC) is 0.90765, where an AUC of 1.0 indicates a predictive accuracy of 100%.
Discussion
In this paper, we show that net proliferation and invasion kinetics (ρ, D) derived from our patient-specific mathematical model derived from routine clinical MR imaging can predict IDH1 mutation status in contrast-enhancing gliomas. The ratio of proliferation over invasion (ρ/D) can be viewed as an aggressiveness index or the inverse of the relative invasiveness; we found that IDH1mut gliomas have significantly lower values of ρ/D than IDH1wt gliomas, indicating that IDH1mut tumors are relatively more diffuse and less aggressive than IDH1wt tumors. Further, a ROC analysis demonstrated the predictive power of ρ/D in discriminating mutational status in our cohort of 172 participants.
A recent report, which was similar to what we have concluded, has shown that IDH1 mutation status in GBMs can be predicted from features extracted from MR images.22 Unlike the current study that focuses on all newly diagnosed contrast-enhancing gliomas, the Carrillo et al.22 report was focused exclusively on GBMs and was able to predict the presence of IDH1mut with 94% accuracy (by ROC analysis) using 4 subjective tumor characteristics observable on MR images (size, contrast enhancement, presence or absence of cyst, and presence or absence of satellite lesion). Our ability to predict the presence of IDH1 mutation status with similar accuracy, however, is based on only a single objective parameter obtained from MR images (ρ/D), one that is also less susceptible to interobserver variability. Even when restricting our cohort to only those contrast-enhancing gliomas that were diagnosed as GBM (n = 158), we found 72% accuracy in prediction by ROC analysis (AUC = 0.830).
We also show that the difference in the biological aggressiveness index (ρ/D) between the 2 tumor types is primarily due to the ability of some IDH1wt contrast-enhancing gliomas to achieve higher rates of proliferation than in IDH1mut tumors (Fig. 4). IDH1wt contrast-enhancing gliomas net proliferation rate (ρ) is 2.46 times that of IDH1mut tumors, but there was no significant difference in diffusion rates (D). This finding is consistent with what is known about primary GBMs, which are predominantly IDH1wt and have high proliferative indices, and is also consistent with a recent report that stable expression of mutant IDH1 in 3 tumor cell lines resulted in decreased proliferation rates compared with wild-type IDH1.23 Since ρ represents the net proliferation rate of the overall tumor mass, it is challenging to find direct histological metrics for comparison. That is, ρ is the net result of birth and death of the tumor cells at a population level that does not have direct histological correlates. Further, the net proliferation rates, that we observed for IDH1mut also had a significant overlap with those observed for IDH1wt tumors, although there was a significant difference in the mean. Thus, the lack of significant difference in our analysis of Ki67 labeling indices may still be consistent with our observations of differences in ρ and may be further explained by the relatively small number of patients for which we had access to tissue.
We were somewhat surprised to find that, although IDH1wt gliomas tend to be more nodular (higher ρ/D) than IDH1mut, the velocity of radial expansion on MRI does not differ from that of IDH1mut gliomas (Fig. 3B). This suggests that IDH1mut tumors do not have an overall difference in imageable growth kinetics in terms of velocity of radial expansion, although they appear more diffuse on MRI than their IDHwt counterparts. However, it turns out that this result was also observed in a recent study of grade II gliomas, which also found no difference in growth velocity between IDH1wt and IDH1mut tumors.24 Although IDH1muts have a similar radial velocity of expansion, they have a much more diffuse pattern of growth. We have previously shown that more diffuse disease was favorable for prognosis.14 Ultimately, these results suggest that brains may accommodate a more diffuse, broadly distributed disease burden than a more nodular (high rho/D) disease. That is, if we compare one IDH1mut and one IDH1wt having a similar velocity of radial expansion but with differing potentials for diffuse invasion, the more invasive IDH1mut tumor would need to have a smaller net proliferation rate (ρ) in order for the velocity of radial expansion to remain the same as the IDH1wt since the velocity is related to D and ρ, 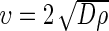 . Thus, although the IDH1mut tumor is more dispersed (larger D), the net rate of doubling the total number of tumor cells (ρ) must be smaller in the mutant tumors, suggesting a longer time period to reach any potentially fatal tumor burden measured in terms of a capacity of tumor cells. These potential explanations also likely involve the age of the patient, as IDH1mut patients tend to be younger. Further research is needed to understand why IDH1mut GBM patients have, on average, a longer survival time as compared with IDH1wt.
. Thus, although the IDH1mut tumor is more dispersed (larger D), the net rate of doubling the total number of tumor cells (ρ) must be smaller in the mutant tumors, suggesting a longer time period to reach any potentially fatal tumor burden measured in terms of a capacity of tumor cells. These potential explanations also likely involve the age of the patient, as IDH1mut patients tend to be younger. Further research is needed to understand why IDH1mut GBM patients have, on average, a longer survival time as compared with IDH1wt.
One potential explanation for our observation of a more diffuse pattern of growth in IDH1mut contrast-enhancing gliomas is that acidification of the external environment leads to tumor growth in a fashion analogous to, but different from, the Warburg effect. This early mutation in IDH1 would constitute an independent mechanism of tumorigenesis, promoting tumor invasion and progression by acidosis, as has been suggested in other cancers as a therapeutic target.25–28 Such a mechanism of tumorigenesis would not preclude the tumor cell from also undergoing a glycolytic shift, but the mutation in IDH1 would precede such an event.29 The acidification hypothesis could be consistent with the slow growth of IDH1mut tumors; they have enough of an advantage to grow and outcompete the neighboring normal cells (and thus develop into a tumor) but not enough to do so as rapidly as an IDH1wt GBM with the different mutation profile that drives its growth.
The acidification theory contributes to the emerging idea of cancer as a metabolic disease, which hypothesizes that mutations in basic metabolic enzymes such as IDH1 often precede, and can indeed be responsible for, the genetic mutations that have been thought to be the primary initiating events in tumorigenesis for several decades. Further, if found to be true, this theory would predict that early IDH1mut gliomas could be ready candidates for metabolically directed therapies, as have been tried with limited success in glioblastoma so far.30,31.
Our previous game-theory model incorporated some of these key elements to predict the evolutionary dynamics of GBM that select for more proliferative phenotypes.17 The results showed that working vascularization plays an important role in the development of GBM, but this does not prevent the emergence of tumor cells with an alternative metabolism. Rapid tumor growth leads to transient hypoxia, forcing selection for cells with either a more diffusive phenotype or a more anaerobic metabolism (such as IDH1mut cells). Our previous model considered a range of parameters to describe the increased fitness resulting from anaerobic metabolism (k) and acidification of the tumor microenvironment (n). The results presented in this paper should make it possible to revisit that model and compare the growth of IDH1wt and IDH1mut tumor cells that have the same ρ/D but different k/n. This would help determine whether a given GBM, which is inherently heterogeneous and thus likely to contain both cell types, is likely to take the route where IDH1mut is the predominant genotype or not.
While the identification of the IDH1 mutation in a subset of gliomas has not yet impacted clinical care, it is a significant prognostic factor and has driven a large amount of insightful basic science into the role of glycolytic metabolism in tumor progression. Recent work presented in abstract form has suggested that those patients with IDH1mut benefit from complete resection of both the enhancing and nonenhancing tumor at time of surgery, while IDH1wt patients derive survival benefit only from resection of the enhancing component of disease.31 Preoperative identification of the minority of patients with enhancing gliomas that are IDH1mut could determine the extent of surgery and result in improved survival. We believe that methods (such as the one presented here) for reliable, noninvasive determination of IDH1 mutation status will become indispensable in the clinic.
Supplementary Material
Funding
James S. McDonnell Foundation, the University of Washington Academic Pathology fund, the National Institutes of Health (U54 CA143970, NS060752, R01 CA16437), the James D. Murray Endowed Chair in the Nancy and Buster Alvord Brain Tumor Center at the University of Washington, the Northwestern Brain Tumor Institute at Northwestern University, and the Zell Scholars Fund and the Wirtz Innovation Fund at Northwestern University.
Supplementary Material
Acknowledgments
We are grateful to Christiane Ulness and Daniel Hamilton of Harborview Medical Center Neuropathology for assistance with tissue acquisition and IDH staining. As always, KRS is eternally grateful to the unwavering support of Dr. E. C. “Buster” Alvord, Jr (1923–2010); may this manuscript continue to honor his memory and foster his scientific legacy.
Conflict of interest statement. None declared.
References
- 1.Parsons DW, Jones S, Zhang XS, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Andronesi OC, Cahill DP. From genomics to the clinic: biological and translational insights of mutant IDH1/2 in glioma. Neurosurg Focus. 2013;34(2):E2. doi: 10.3171/2012.12.FOCUS12355. [DOI] [PubMed] [Google Scholar]
- 3.Balss J, Meyer J, Mueller W, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 5.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-Oncol. 2009;11(4):341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin GL, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloosterhof NK, Bralten LBC, Dubbink HJ, et al. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol. 2011;12(1):83–91. doi: 10.1016/S1470-2045(10)70053-X. [DOI] [PubMed] [Google Scholar]
- 8.Nobusawa S, Watanabe T, Kleihues P, et al. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin Can Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 9.Swanson K. Mathematical Modeling of the Growth and Control of Tumors. Seattle, WA: University of Washington; 1999. [Google Scholar]
- 10.Swanson K, Alvord E. Serial imaging observations and postmortem examination of an untreated glioblastoma: a traveling wave of glioma growth and invasion. Neuro-Oncol. 2002;4(4):340. [Google Scholar]
- 11.Swanson KR, Bridge C, Murray JD, et al. Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J Neurol Sci. 2003;216(1):1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Harpold H, Alvord EJ, Swanson K. The evolution of mathematical modeling of glioma proliferation and invasion. J Neuropathol Exp Neurol. 2007;66(1):1–9. doi: 10.1097/nen.0b013e31802d9000. [DOI] [PubMed] [Google Scholar]
- 13.Rockne R, Rockhill J, Mrugala M, et al. Predicting the efficacy of radiotherapy in individual glioblastoma patients in vivo: a mathematical modeling approach. Phys Med Biol. 2010;55(12):3271–3285. doi: 10.1088/0031-9155/55/12/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CH, Rockhill JK, Mrugala M, et al. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res. 2009;69(23):9133–9140. doi: 10.1158/0008-5472.CAN-08-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szeto MD, Chakraborty G, Hadley J, et al. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 2009;69(10):4502–4509. doi: 10.1158/0008-5472.CAN-08-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson KR, Rockne RC, Claridge J, et al. Quantifying the role of angiogenesis in malignant progression of gliomas: in silico modeling integrates imaging and histology. Cancer Res. 2011;71(24):7366–7375. doi: 10.1158/0008-5472.CAN-11-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basanta D1, Scott JG, Rockne R, et al. The role of IDH1 mutated tumour cells in secondary glioblastomas: an evolutionary game theoretical view. Phys Bio. 2011;8(1):015016. doi: 10.1088/1478-3975/8/1/015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matlab. MA: The MathWorks, Inc; 2011. 7.13.0.564 (R2011b) ed. [Google Scholar]
- 19.Neal ML, Trister AD, Cloke T, et al. Discriminating survival outcomes in patients with glioblastoma using a simulation-based, patient-specific response metric. PLoS One. 2013 doi: 10.1371/journal.pone.0051951. 8(1):e51951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson KR. Quantifying glioma cell growth and invasion in vitro. Math Comput Modelling. 2008;47(5–6):638–648. [Google Scholar]
- 21.Swanson KR, Rostomily RC, Alvord EC. A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: a proof of principle. Br J Cancer. 2008;98(1):113–119. doi: 10.1038/sj.bjc.6604125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrillo JA, Lai A, Nghiemphu PL, et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol. 2012 doi: 10.3174/ajnr.A2950. 33(7):1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bralten LB, Kloosterhof NK, Balvers R, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol. 2011;69(3):455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 24.Gozé C, Bezzina C, Gozé E, et al. 1P19Q loss but not IDH1 mutations influences WHO grade II gliomas spontaneous growth. J Neurooncol. 2012;108(1):69–75. doi: 10.1007/s11060-012-0831-6. [DOI] [PubMed] [Google Scholar]
- 25.Martin NK, Gaffney EA, Gatenby RA, et al. Tumour-stromal interactions in acid-mediated invasion: A mathematical model. J Theor Biol. 2010;267(3):461–470. doi: 10.1016/j.jtbi.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatenby RA, Gawlinski ET, Gmitro AF, et al. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 27.Silva AS, Yunes JA, Gillies RJ, et al. The potential role of systemic buffers in reducing intratumoral extracellular pH and acid-mediated invasion. Cancer Res. 2009;69(6):2677–2684. doi: 10.1158/0008-5472.CAN-08-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smallbone K, Gatenby RA, Maini PK. Mathematical modelling of tumour acidity. J Theor Biol. 2008;255(1):106–112. doi: 10.1016/j.jtbi.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Scott JG, Basanta D, Chinnaiyan P, et al. Production of 2-hydroxyglutarate by isocitrate dehydrogenase 1-mutated gliomas: an evolutionary alternative to the Warburg shift? Neuro Oncol. 2011;13(12):1262–1264. doi: 10.1093/neuonc/nor083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michelakis ED, Sutendra G, Dromparis P, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2012;2(31) doi: 10.1126/scitranslmed.3000677. 31ra34. [DOI] [PubMed] [Google Scholar]
- 31.Beiko J1, Suki D, Hess KR, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. J Clin Oncol. doi: 10.1093/neuonc/not159. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.