Abstract
This paper provides an overview of the development and applications of plasmonics-active nanoprobes in biomedical diagnostics. Specific examples of detection techniques using surface-enhanced Raman scattering are presented to illustrate the usefulness and potential of the plasmonics nanoprobes for gene detection and nanobiosensing. The detection of specific target deoxyribonucleic acids sequences using a novel “molecular sentinel” nanoprobe method is presented and discussed in detail.
Index Terms: Gene diagnostics, metallic nanostructures, nanoprobes, near-field detection, plasmonics, Raman, surface-enhanced Raman scattering (SERS)
I. Introduction
For the last few years, there has been an increased interest in using plasmonics-related properties of metallic nanostructures for biosensing. Plasmonics refers to the study of enhanced electromagnetic properties of metallic nanostructures. The term is derived from plasmons, the quanta associated with longitudinal waves propagating in matter through the collective motion of large numbers of electrons. According to classical electromagnetic theory, molecules on or near metal nanostructures experience enhanced fields relative to that of the incident radiation. When a metallic nanostructured surface is irradiated by an incident electromagnetic field (e.g., a laser beam), conduction electrons are displaced into frequency oscillation equal to the incident light. These oscillating electrons, called “surface plasmons,” produce a secondary electric field, which adds to the incident field. When these oscillating electrons become spatially confined, as is the case for isolated metallic nanospheres or otherwise roughened metallic surfaces (nanostructures), there is a characteristic frequency (the plasmon frequency) at which there is a resonant response of the collective oscillations to the incident field. This condition yields intense localized fields that can interact with molecules in contact with or near the metal surface. In an effect analogous to a “lightning rod” effect, secondary fields can become concentrated at high curvature points on the roughened metal surface. Surface plasmons have been associated with important practical applications in surface plasmon resonance (SPR) and surface-enhanced Raman scattering (SERS).
Raman spectroscopy, which involves vibrational transitions that yield very narrow spectral features characteristic of the investigated samples, has long been considered as a powerful tool for the identification of chemical and biological samples as well as the elucidation of molecular structure, surface processes, and interface reactions. Despite these important attributes, Raman applications are often limited in trace detection due to the extremely poor efficiency of the scattering process. However, discoveries in the late 1970s [1], [2] indicated that the Raman scattering efficiency can be enhanced by factors of up to 106 when the sample is adsorbed on or near nanotextured surfaces of special metals such as silver, gold, and transition metals. During a period between the mid 1970s and the early 1980s, this early enthusiasm for the SERS decreased and did not lead to practical applications because the Raman enhancement effect had been observed for only a limited number of molecules. Most studies had involved samples at concentrations between 10−1 and 10−3 M, which are well above the useful concentration ranges for trace analysis, and involved mainly a few highly polarizable small molecules, such as pyridine, benzoic acid, and its derivatives. In 1984, we first reported the general applicability of the SERS as an analytical technique, and the possibility of the SERS measurement for a greater variety of chemicals including homocyclic and heterocyclic polyaromatic compounds [3]. For the following two decades, the development of plasmonics-active SERS substrates and nanoprobes has been an area of extensive fundamental research [4]–[9]. Our laboratory has extensively investigated the SERS technology in the development of plasmonics-active SERS substrates for chemical sensing [10]–[16] and for bioanalysis and biosensing [17]–[27]. These substrates consist of plates, waveguides, or optical fibers having silver-coated dielectric nanoparticles or isolated dielectric nanospheres (30 nm diameter) coated with silver producing half-nanoshells [17], and stochastic nanoposts [10]. The fabrication process involves depositing nanoparticles including polystyrene nanospheres [3], [10]–[14], titanium dioxide [15], alumina [16], and silica [18] on a solid substrate, and then, coating the nanoparticle base with a 50–100 nm layer of silver via vacuum thermal deposition. The nanoparticle-based SERS technology developed in our laboratory has led to a wide variety of analytical applications including sensitive detection of a variety of chemicals of environmental and toxicological significance, including polycyclic aromatic compounds [3], [21], [23], organophosphorus compounds [7], and chlorinated pesticides [9], compounds of biological interest such as deoxyribonucleic acids (DNA) adduct biomakers [5], [15], and diagnostics systems of medical importance [16], [18], [23]–[25]. Certain mechanisms related to the SERS effect, such as the contribution of electromagnetic interactions, have been extensively investigated and are reasonably well understood. Other processes, such as the contribution of chemical effects, are less known and are currently topics of extensive research. The reader is referred to a number of reviews for further details [4]–[9], [28]–[34].
In this paper, an overview of the development and applications of plasmonics-based nanoprobes in biomedical areas is presented. Specific examples of detection techniques and instruments developed in the author’s-laboratory are presented to illustrate the usefulness and potential of the plasmonics nanoprobes for gene diagnostics and nanobiosensing. The detection of specific target DNA sequences using a novel “molecular sentinel” (MS) biosensing method is discussed. Due to the possibility of performing simple homogeneous bioassays, the SERS MSs could provide useful diagnostic probes for multiple biological targets. The potential for combining the spectral selectivity and high sensitivity of the SERS process with inherent molecular specificity of MS nanoprobes to diagnose molecular target sequences is discussed.
II. Experimental
A. Preparation of Silver-Coated Nanosphere Substrates
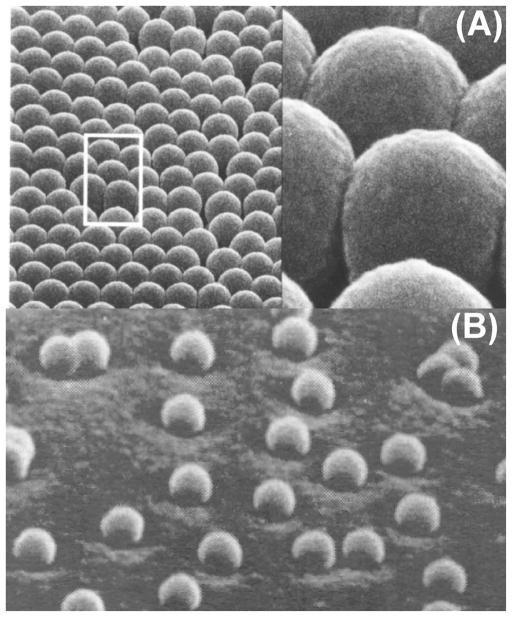
One of the early difficulties in the development of the SERS technique for analytical applications had been the production of surfaces or media that had an easily controlled and reproducible protrusion size (roughness). An approach developed in our laboratory has involved the use of nanospheres spin coated on a solid support in order to produce and control the desired roughness [3], [12]. The nanostructured support is subsequently covered with a layer of silver that provides the conduction electrons required for the surface plasmon mechanisms. Among the techniques based on solid substrates, the methods using simple nanomaterials, such as Teflon or latex nanospheres, appear to be the simplest to prepare. Teflon and latex nanospheres are commercially available in a wide variety of sizes. The shapes of these materials are very regular and their size can be selected for optimal enhancement. These substrates consist of a plate having silver-coated dielectric nanoparticles (see Fig. 1A) or isolated dielectric nanospheres (30 nm diameter) coated with silver producing systems of half-nanoshells, sometimes referred to as crescents (see Fig. 1B) [17]. The effect of the sphere size and metal layer thickness upon the SERS effect can be easily investigated. The results have indicated that, for each sphere size, there is an optimum silver layer thickness for which the maximum SERS signal is observed [12]. The silver coated nanospheres were found to be among the most strongly enhancing substrates investigated with enhancement factors comparable to or greater than those found for electrochemically roughened surfaces.
Fig. 1.
(A) Plasmonics-active substrates based on nanosphere arrays coated with silver. (B) Half-nanoshells consisting of nanospheres coated with silver (bottom).
B. Preparation of Silver Nanoparticles
Silver colloids were prepared according to the standard Lee–Meisel method [35]: 200 mL of 10−3 M AgNO3 aqueous solution was boiled under vigorous stirring, then 5 mL of 35 mM sodium citrate solution were added and the resulting mixture was kept boiling for 1 h. This procedure was reported to yield ~1011 particles/mL of homogenously sized colloidal particles with a diameter of ~35–50 nm and an absorption maximum at 390 nm. The colloidal solutions were stored at 4 °C and protected from room light. Further dilutions of the colloidal solutions were carried out using distilled water.
C. MSs and DNA Targets
The MS probes and target DNA were synthesized by Integrated DNA Technologies (IDT, Coralville, IA) using the phosphoramidite chemistry on a commercial nucleic acid synthesizer (model ABI394) and reverse-phase (RP) high-performance liquid chromatography (HPLC) purified. All the MS probes were modified with 5-carboxytetramethylrhodamine, succin-imidyl ester (TMR) on the 3′-end and a 5′-thiol (-SH) group.
D. SERS MS Nanoprobes
The MS probes were designed and evaluated with the help of the DNA folding software of the Zucker laboratory. We designed a 42-basepair (bp), DNA hairpin incorporating a portion of the Homo sapiens breast cancer 1, early onset (BRCA1) gene (supplementary material). The MS nanoprobe was developed using silver nanoparticles and thiol modified DNA tagged with a Raman label at one end. The nanoparticles were derivatized with DNA probes in glass tubes by incubating 0.5 μM of silver colloids with 5 μM of thiolated MS in phosphate buffer (NaH2 PO4/Na2 HPO4, 10 mM, pH 7.0) overnight. The SERS-MS nanoprobes were purified three times by centrifugation at 8000 r/min for 15 min, resuspended in 10 mM phosphate buffer, pH 7.0, and stored at 4 °C. Hybridization studies were carried out by adding a 5 μL aliquot of 100 μM DNA targets to 40 μL of 0.5 μM, SERS-MS nanoprobe suspension in 10 mM phosphate buffer containing 2 mM MgCl2. The hybridization assay was allowed to react for 2–3 h at room temperature.
III. Applications and Discussion
A. Nanoprobing at Subwavelength Spatial Resolution Using Near-Field SERS
Detecting molecular targets at the nanoscale level is important in many biological sensing applications. Use of conventional optics is severely restricted by the diffraction limit. Near-field scanning optical microscopy (NSOM) is a methodology capable of producing subwavelength image resolution (20–200 nm) with the spectroscopic information afforded by conventional optical spectroscopic techniques [36]. The NSOM combined with a Ra-man microscope is particularly powerful analytical tool. With the use of tapered single-mode fiber probes, near-field Raman spectra have been obtained for dye molecules adsorbed on single SERS-active silver nanoparticles with excitation intensities as low as 10 nW [37]. We have combined NSOM with SERS to obtain spectral, spatial, and chemical information of molecular adsorbates with subwavelength lateral resolution [20], [21]. We showed for the first time the possibility to obtain near-field SERS images with a subwavelength lateral optical resolution of 100 nm, short exposure times, and high SNRs [21]. In these studies, near-field SERS spectra of cresyl fast violet (CFV), brilliant crystal blue (BCB), and rhodamine 6G on silver nanoparticle substrates have been obtained. Spectra from as few as 300 molecules, or less than 10−2 monolayer, adsorbed on about ten silver nano particles have been recorded and the SERS enhancement up to 1013 has been observed [20].
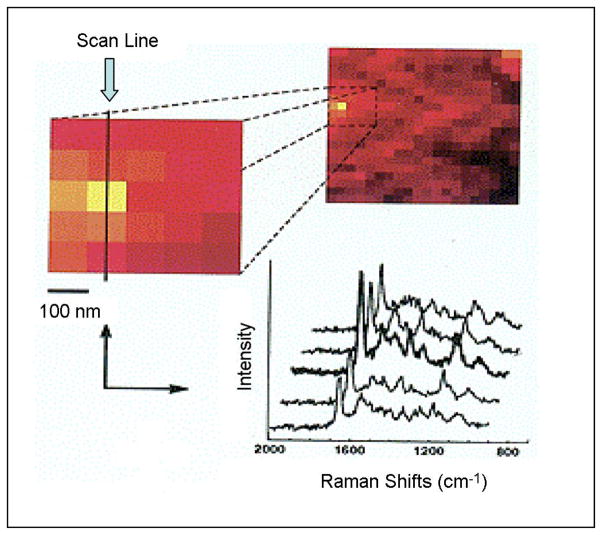
The SERS-NSOM instrumental system consisted of a modified commercial near-field scanning optical microscope (Aurora, Optometric) and a high f-number spectrograph (Hooper f/1.8, Kaiser Optical Systems). Surface-enhanced Raman spectra of DNA labeled with the BCB dye molecules on silver nanoparticle substrates are shown in Fig. 2 [21]. In this figure, the spectra have been obtained at different tip positions on a scan line along an SERS substrate. Spatial variations of near-field optical signals are normally due to strong local topography dependence. If one assumes a uniform distribution of the sample on the surface, variations in SERS intensity can only be attributed to different local enhancements from various locations on the substrate (“hot spots”). Maximum Raman enhancements at hot spots do not necessarily correspond to locations on top of silver nanoparticles but often occur at gaps or “cavities” existing between nanoparticles. Although the existence of hot spots has been discussed previously, no direct experimental observation was reported. In contrast, in this paper, these hot sites are directly visualized due to the recording of sample topography and Raman intensity at each data point [21]. The best proof of changes in peak intensities is illustrated by comparing the whole Raman spectra at distinct lateral positions. This is shown in Fig. 2, where we zoomed in a 500-nm-wide region where we detected the highest intensities and plotted spectra along a scan line of adjacent positions. The glass band at ~1100 cm−1 remains constant for all five positions. On the other hand, the Raman peaks of the BCB show a totally different behavior with intensity changing with locations reflecting the possibility of hot spots. At the first two positions, they show the same intensity, and then, all BCB-DNA bands suddenly increase by 50%. After that, they decrease again to their initial intensity. This change in intensity was the only spectral change we observed. New bands, half-width changes, or band shifts depending on the lateral position could not be observed. The results of this study demonstrated the possibility to probe analyte molecules (SERS-labeled DNA) at subwavelength spatial resolution.
Fig. 2.
Near-field SERS spectra of BCB-labeled DNA molecules on silver island substrates at different tip locations (adapted from [21]).
B. SERS Gene Probes
Over the last few years, there has been a great interest in the development of optical techniques for genomics analysis such as nonradioactive DNA probes for use in biomedical diagnostics, pathogen detection, gene identification, gene mapping, and DNA sequencing. One of the most unambiguous and well-known molecular recognition events is the hybridization of a nucleic acid to its complementary target. We have combined this biorecognition process with the SERS to develop a new generation of DNA-based SERS probes for medical diagnosis. We first reported the development and use of a novel SERS gene probe technology for DNA detection [17]. The feasibility of using surface-enhanced Raman gene probes that exhibit an extremely narrow small spectral bandwidth has been demonstrated [19]. The use of SERS-active labels for primers used in polymerase chain reaction (PCR) amplification of specific target DNA sequences has also been developed in our laboratory. This method has the potential for combining the spectral selectivity and high sensitivity of the SERS technique with the inherent molecular specificity offered by DNA sequence hybridization. Our laboratory has reported the development of SERS-based gene probes and has reported the selective detection of human immunodeficiency virus (HIV) DNA and the breast cancer gene BRCA1 using SERS-active substrates [23], [38]. To demonstrate the SERS gene detection scheme, we used precoated SERS-active solid substrates, on which DNA probes were bound and directly used for hybridization. The effectiveness of this scheme for DNA hybridization and the SERS gene detection involves several considerations. It is important that the unlabeled DNA fragment does not exhibit any significant SERS signal that might interfere with the label signal. The first step involves selection of a label that is SERS-active and compatible with the hybridization platform. An ideal label should exhibit a strong SERS signal when used with the SERS-active substrate of interest. Second, the label should retain its strong SERS signal after being attached to a DNA probe. Finally, one should be able to detect the SERS signal from the labeled probe after hybridization.
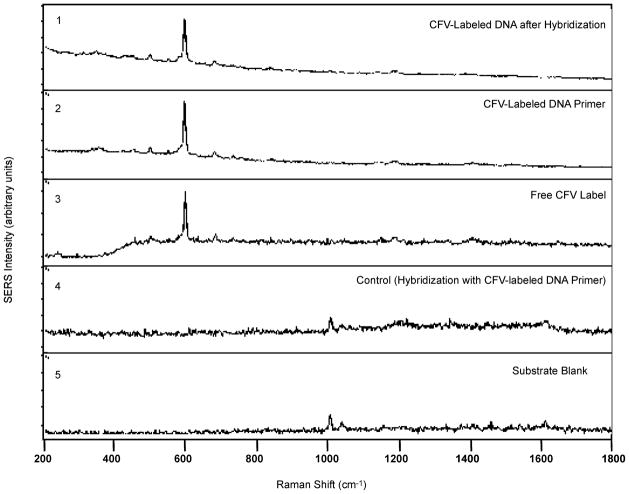
The effectiveness of the SERS detection scheme has been demonstrated using the gag gene sequence of the HIV [19]. Infection with the HIV Type 1 (HIV-1) results in a uniformly fatal disease. Unfortunately, standard HIV serologic tests, including the enzyme-linked immunosorbent assay and the Western blot assay, are not useful in the diagnosis of the HIV infection during early infancy because of the confounding presence of transplacentally derived maternal antibody in the infants’ blood. There is, therefore, a need for a direct nucleic acid based test that detects the presence of HIV viral sequences. This can be achieved by PCR amplification of target DNA sequences. Fig. 3 illustrates the results of the HIV hybridization experiments [19]. Spectrum 1 shows the SERS detection of the CFV-labeled target DNA strand that has been hybridized with the surface-bound capture probe. The signal persists even after rigorous rinsing. For comparison, the detection of a nonhybridized, CFV-labeled single strand of DNA is also shown in spectrum 2. In this case, the DNA strand is the CFV-labeled forward primer sequence, which has no complementarity with the surface-bound capture probe sequence. This sample was simply applied to the polystyrene plate and coated with silver nanoislands; no rinsing step was performed. For standard free CFV comparison, note the strong CFV SERS signal at 590 cm−1 (spectrum 3). In this case, the saturated free CFV solution was simply spotted onto the polystyrene pate and coated with the silver nanoisland layer. There does not appear to be any major alteration in the CFV spectrum as a result of being bound to single- or double-stranded DNA. On the contrary, no CFV signal as observed following the hybridization and rinsing steps when no complementarity existed between the capture probe sequence and the single stranded, CFV-labeled DNA. This is demonstrated by spectrum 4, the hybridization control. In this example, the CFV-labeled DNA was the single-strand forward primer. The labeled primer solution was subjected to the hybridization steps on the capture probe-bound polystyrene plate. Because of the lack of complementarity, the labeled primer was effectively removed from the plate during the ensuing rinsing step. The lack of CFV signal in this example also demonstrates that there is no non-specific binding of the CFV molecule, itself, to the polystyrene plate. The blank illustrated in spectrum 5 demonstrates that the silver-coated polystyrene plate exhibited no major spectral background in the region of interest, even after being treated with the capture probe sequence and the BSA blocking agent. In summary, these results demonstrate the effectiveness of our SERS gene probe technique to biomedical diagnostics.
Fig. 3.
Demonstration of the SERS gene technique for the detection of the HIV-1 gag gene sequnce using hybridization experiments (Adapted from [19]).
C. MS Nanoprobes
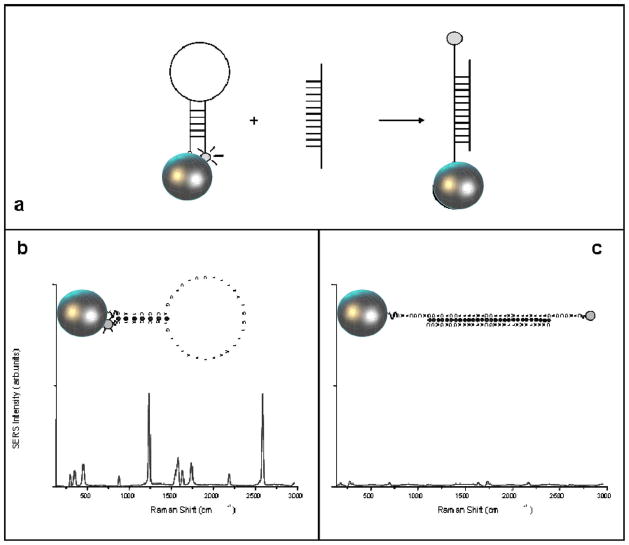
We have developed a novel detection approach that incorporates the “SERS effect modulation” scheme associated with metallic nanoparticles and the DNA hairpin structure [26]. The plasmonics-based MS nanoprobe, refered to as “MS” nanoprobe, comprises of a metal nanoparticle and a stem-loop DNA molecule tagged with a Raman label (Fig. 4). The nanoprobe utilizes the specificity and selectivity of the DNA hairpin probe sequence to detect a specific target DNA sequence of interest. In the normal configuration and in the absence of target DNA, the stem-loop configuration maintains the Raman label in close proximity to the metal nanoparticle, inducing an intense SERS effect that produces a strong Raman signal upon laser excitation. Upon hybridization of a complementary target DNA sequence to the nanoprobe, the stem-loop configuration is disrupted, causing the Raman label to physically separate from the metal nanoparticle, thus quenching the SERS signal.
Fig. 4.
(a) Operating principe of an SERS MS nanoprobe. (b) Strong SERS signal is observed when the MS probe is in the hairpin conformation (closed state). (c) Upon hybridization with the target (open state), the SERS signal is diminished.
The change (or modulation) of the plasmonic effect with the distance between the metallic nanoparticle and the Raman label is the basis in the detection strategy of the MS probes. The plasmonics enhancement is due to electromagnetic effects and chemical effects at the metal surface. Studies of electromagnetic effects have shown that the SERS enhancement, G factor, decreases as G = [r/(r + d)]12 for a single analyte molecule located a distance d from the surface of a metal particle of radius r [39]. Therefore, the electromagnetic SERS enhancement falls off drastically with increasing distance between the label and the metallic nanoparticle, due to the decay of a dipole over the distance (1/d)3 to the fourth power, thus resulting in a total intensity decay of (1/d)12. Because the Raman enhancement field decreases significantly away from the surface, a molecule must be located within a very close range (0–10 nm) of the nanoparticle surface in order to experience the enhanced local field. In the absence of target genes), the stem-loop configuration keeps the Raman label in contact or close proximity (<1 nm) to the nanoparticle, thus inducing in a strong plasmonics enhancement resulting in a strong SERS signal of the label [see Fig. 4(b)]. However, when complementary target DNA was recognized and captured (i.e., hybridized) by the MS nanoprobes, the SERS signal of the labels is drastically decreased, triggering a warning sign of target recognition and capture [Fig. 4(c)]. In other terms, the MS nanoprobes play the role of MSs patrolling the sample solution with their warning light “switched ON” when no significant event occurs. When a target species (e.g., defect genes, specific RNA) is detected, the MSs will lock to them and extinguish their light, thus providing a warning sign.
The SERS MS concept is essentially a label-free technique because the target molecules do not have to be labeled. The technique is also appropriate for detecting the presence of specific DNA sequences in a homogenous solution at room temperature. The SERS MS nanoprobe was used to detect the presence of the HIV gene in a homogeneous assay [26]. The HIV MS nanoprobes that incorporated a partial sequence for a human immunodeficiency, HIV-1 isolate Fbr020 from Thailand reverse transcriptase (pol) gene, were designed with a stem sequence that allowed the formation of stable hairpin structure at room temperature. The HIV-1 MS nanoprobe (5′-HS-(CH2)6)-CCTATCACAACAAAGAGCATACATAGG GATAGG-R6G) consisted of a 42-bp DNA hairpin probe with a seven-bp stem that was modified with rhodamine 6G on the 3′ end and a thiol substituent at the 5′ end that could then be used for covalent coupling to the surface of silver nanoparticles. The underlined sequence represents the complementary arms of the MS. The silver colloidal nanoparticles were prepared according to the citrate reduction method yielding homogenously sized colloids. The GeneAmplimer HIV-1 control reagents and the GeneAmp Gold PCR Reagent Kit were used to amplify a 115-bp sequence in the gag region of the HIV-1 genome. The reagent included both positive and negative HIV-1 DNA control templates. The positive control was a plasmid DNA that contains the entire genome of the HIV isolate, while negative control was a human placenta DNA template that is free from HIV-1 sequence. The forward primers SK 38, (5′-ATAATCCACCTATCCCAGTAGGAGAAAT -3′) and the reverse primer SK 39 (5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′) of the gag region (GeneAmplimer HIV-1, Applied Biosystems) were used. The PCR amplification was carried out in 50 μL of 100 mM Tris–HCl buffer (pH 8.3) containing 50 mM KCl, 2.5 mM MgCl2, 200 μM dNTPs, 10 ng of purified human genomic DNA, 1.25 units of AmpliTag Gold DNA polymerase, and 0.5 μM each of the forward and reverse primers. The amplification was achieved with an initial denaturation step of 95 °C, for 2 min followed by a 30 cycle process that includes a denaturation step for 15 s at 95 °C, an annealing and extension step at 60 °C for 60 s, and a final extension at 72 °C for 2 min. PCR products were stored at −20 °C until required for the hybridization assays. PCR products were analyzed using precast gels stained with ethidium bromide. The hybridization assay using SERS MS nanoprobes was carried out in a 40 μL reaction volume containing 30 μL of 0.5 μM SERS SK 19 MS nanoprobes, and 10 μL of the PCR amplicon. The PCR amplicon and nanoprobe mixture were denatured for 15 s at 95 °C allowed to hybridize at 55 °C for 1 min. SERS measurements were performed using a Renishaw InVia Raman system equipped with a 50 mW, HeNe laser (Coherent, Model 106-1) emitting a 632.8 nm line used as the excitation source. The light from the laser was passed through a line filter, and focused on a sample mounted onto a X–Y–Z translation stage with a 20× microscope objective. The Raman scattered light was collected by the same objective, through a holographic notch filter to block out Rayleigh scattering. An 1800 groove/mm grating was used to provide a spectral resolution of 1 cm−1. Raman scatter was detected by a 1024 × 256 pixel RenCam charge coupled device (CCD) detector. The SERS spectra were acquired with a 10 s integration time and processed with software from Renishaw (WiRE 2.0).
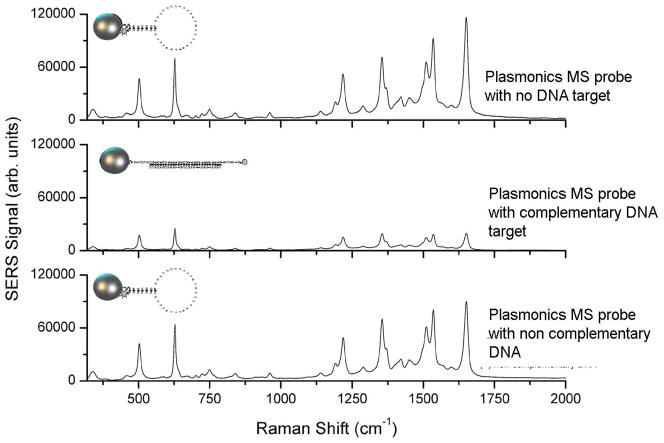
Fig. 5 illustrates the detection of a partial sequence of the HIV-1 gene in a homogenous solution using the MS nanoprobes. In the absence of the target DNA (Fig. 5, top curve), the hairpin conformation of the SERS MS nanoprobes remains stable, maintaining a close proximity of the rhodamine 6G label with the surface of the silver nanoparticles (nanoenhancers). With this stable stem-loop configuration, a strong SERS signal from rhodamine 6G is detected. On the other hand, in the presence of a complementary HIV-1 target sequence (Fig. 5, middle curve), the SERS HIV-1 MS nanoprobes recognize and hybridize to the target DNA, thus opening the stem loop, causing physical separation of the rhodamine 6G label from the surface of the silver nanoparticles. As a result, the SERS signal is greatly diminished. It is noteworthy that the presence of noncomplementary target DNA template (Fig. 5, bottom curve) did not significantly affect the SERS signal indicating that the hairpin configuration of the SERS HIV-1 MS nanoprobes was not disrupted. The results of this study demonstrated that the MS technology can reliably detect specific nucleic acid sequences in a homogenous solution. Using the most intense SERS intensity band at 1521 cm−1 as the marker band for rhodamine 6G, we estimated the SERS quenching efficiency to be ~75% upon hybridization of the SER HIV-1 MS nanoprobe to the HIV-1 DNA target. This result demonstrates the specificity and selectivity of SERS MS nanoprobes and their potential application in selective diagnostics.
Fig. 5.
SERS spectra of HIV-1 plasmonics MS nanoprobe with no target DNA sequence (top curve) and in presence of a noncomplementary DNA target sequence (negative diagnostic: bottom curve) and a complementary HIV-1 DNA target (positive diagnostic: middle curve) (Adapted from [26]).
The MS approach is unique in the context of existing methods that involve hybridization of an oligonucleotide probe to a complementary target sequence using fluorescence detection (e.g., molecular beacon). Molecular beacons exploit the principle of fluorescence resonance energy transfer (FRET) between the fluorescent molecule and the quenching molecule by generating a relatively strong fluorescent signal when complementary target sequences are hybridized, thus separating the quencher and the fluorophore. The fluorescence remains low (quenched) in the absence of a complementary sequence. The SERS-MS technique uses the stem-loop structure similar to that of molecular beacons for DNA recognition. However, the detection scheme is fundamentally different from the molecular beacon detection scheme. In the SERS MS system, the metal nanoparticle is used as a signal-enhancing platform for the SERS signal associated with the label. Detection of probe hybridization to a target sequence by a decrease in SERS signal as the probe’s hairpin structure unfolds would appear initially to be a less sensitive approach than the FRET method with a fluorescent or absorbing dye. However, both detection techniques, i.e., increase in fluorescence signal (FRET) or decrease in SERS signal (MS) should have a similar sensitivity under similar signal levels since they both involve emission signal detection modalities. Emission signal detection approaches are more sensitive than absorption detection techniques because the former ones are not limited by detector noise, which are often the limiting factor of absorption techniques. Another factor limiting sensitivity is the fact that, although the Raman signal is enhanced by several orders of magnitude by surface plasmons of nanoparticles, it is often weaker than photoluminescence for many Raman vibrational bands. However, studies in this and other laboratories have demonstrated that under specific conditions, plasmonics enhancement can increase the Raman signal 1011–1015 fold, leading to single-molecule detection, thus comparable to fluorescence detection [22], [39], [40].
IV. Conclusion
The SERS spectroscopy is a powerful plasmonics-based technique that has a number of important advantages in biochemical sensing and medical diagnostics. The development of practical and sensitive devices for screening multiple genes related to medical diseases and infectious pathogens is critical for early diagnosis and improved treatments of many illnesses. To achieve the required level of sensitivity and specificity, it is often necessary to use a detection method that is capable of simultaneously identifying and differentiating a large number of biological constituents in complex samples. The multiplex capability, which allows the monitoring of a large number of molecular processes and assays simultaneously, is an important feature in systems biology research, medical diagnostics, and high throughput screening applications. Since Raman spectroscopy is highly compound specific, the potential to spectrally analyze multicomponent samples or to use multiple labels simultaneously is the primary advantage of Raman and SERS over fluorescence-based detection. This is mostly due to the presence of much narrower spectral bands, which allows structural identification and conceivably simultaneous measurement of different analytes in complex samples. Although detection sensitivities achieved by luminescence techniques are excellent, the spectral overlap of the relatively large bandwidth of fluorescence spectra limits the number of labels that can be used simultaneously. Due to the narrow bandwidths of Raman bands, the multiplex capability of Raman and SERS techniques is excellent in comparison to the other spectroscopic alternatives, thus opening new horizons to many applications in medical diagnostics, drug development, and high throughput screening. Potential applications of nanoparticle-based biosensing and diagnostics can involve in vitro and in vivo detection and imaging modalities. The use of nanoparticles for in vitro diagnostics is straight forward since the samples to be analyzed are outside patients and does not involve toxicity issues. However, the use of nanoparticles for in vivo diagnostics or imaging requires that great care be taken to ensure that nanoparticles are safe for use in humans. One approach is to coat the metal nanoparticles with a nontoxic coating (e.g., silica) or biocompatible materials for biomedical applications. Recent research has underlined concerns over the safety of use of nanomaterials and the long-term adverse effect associated with their use in humans. It is, therefore, important to establish the toxicity, safety, and risks involved in the usage of nanoparticles for in vivo applications.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under Grant R01 EB006201.
Biography
Tuan Vo-Dinh received the Ph.D. degree in biophysical chemistry from the Swiss Federal Institute of Technology (ETH) in Zurich, Switzerland, in 1975.
He was the Director of the Center for Advanced Biomedical Photonics, a Group Leader of the Advanced Biomedical Science and Technology Group, and a Corporate Fellow at Oak Ridge National Laboratory (ORNL). In 2006, he joined Duke University, Durham, NC, where he is the Director of the Fitzpatrick Institute for Photonics, R. Eugene and Susie E. Goodson Distinguished Professor of biomedical engineering, and a Professor of chemistry. He is the Editor-in-chief of the NanoBiotechnology, an Associate Editor of the Plasmonics, the Journal of Nanophotonics, and the Ecotoxicology and Environmental Safety, and a Topical Editor of the Polycyclic Aromatic Compounds. His current research interests include the development of advanced technologies for the protection of the environment and the improvement of human health, laser spectroscopy, molecular imaging, medical diagnostics, cancer detection, chemical sensors, biosensors, nanosensors, and biochips.
Dr. Vo-Dinh is the recipient of many awards including seven R&D 100 Awards for most technologically significant advance in research and development for his pioneering research and inventions of innovative chemical and biological sensing technologies.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
References
- 1.Jeanmaire DL, Van Duyne RP. Surface Raman spectroelectro-chemistry.1. Heterocyclic, aromatic, and aliphatic-amines adsorbed on anodized silver electrode. J Electroanal Chem. 1977;84:1–20. [Google Scholar]
- 2.Albrecht MG, Creighton JA. Anomalously intense Raman-spectra of pyridine at a silver. J Amer Chem Soc. 1977;99:5215–5217. [Google Scholar]
- 3.Vo-Dinh T, Hiromoto MYK, Begun GM, Moody RL. Surface-enhanced Raman spectrometry for trace organic-analysis. Anal Chem. 1984;56:1667–1670. [Google Scholar]
- 4.Moskovits M. Surface-enhanced spectroscopy. Rev Mod Phys. 1985;57:783–826. [Google Scholar]
- 5.Wokaun A, Gordon JP, Liao PF. Radiation damping in surface-enhanced Raman-scattering. Phys Rev Lett. 1982;48:957–960. [Google Scholar]
- 6.Schatz GC. Theoretical-studies of surface enhanced Raman-scattering. Acc Chem Res. 1984;17:370–376. [Google Scholar]
- 7.Kerker M. Electromagnetic model for surface-enhanced Raman-scattering (SERS) on metal colloids. Acc Chem Res. 1984;17:271–277. [Google Scholar]
- 8.Chang RK, Furtak TE, editors. Surface-Enhanced Raman Scattering. New York: Plenum; 1982. [Google Scholar]
- 9.Pockrand I. Surface-Enhanced Raman Vibrational Studies at Solid/Gas Interfaces. Berlin, Germany: Springer-Verlag; 1984. [Google Scholar]
- 10.Vo-Dinh T, Meier M, Wokaun A. Surface-enhanced Raman-spectrometry with silver particles on stochastic-post substrates. Anal Chim Acta. 1986;181:139–148. [Google Scholar]
- 11.Vo-Dinh T, Uziel M, Morrison A. Surface-enhanced Raman analysis of benzo[a]pyrene-DNA adducts on silver-coated cellulose substrates. Appl Spectrosc. 1987;41:605–610. [Google Scholar]
- 12.Moody RL, Vo-Dinh T, Fletcher WH. Investigation of experimental parameters for surface-enhanced Raman-scattering (SERS) using silver-coated microsphere substrates. Appl Spectrosc. 1987;41:966–970. [Google Scholar]
- 13.Alak AM, Vo-Dinh T. Surface-enhanced Raman spectrometry of organo phosphorus chemical agents. Anal Chem. 1987;59:2149–2153. doi: 10.1021/ac00144a030. [DOI] [PubMed] [Google Scholar]
- 14.Vo-Dinh T, Alak A, Moody RL. Recent advances in surface-enhanced Raman spectrometry for chemical-analysis. Spectrochim Acta B. 1988;415:605–615. [Google Scholar]
- 15.Bello JM, Stokes DL, Vo-Dinh T. Titanium-dioxide based substrate for optical monitors in surface-enhanced Raman-scattering analysis. Anal Chem. 1989;61:1779–1783. [Google Scholar]
- 16.Bello JM, Stokes DL, Vo-Dinh T. Silver-coated alumina as a new medium for surfaced-enhanced Raman-scattering analysis. Appl Spectrosc. 1989;43:1325–1330. [Google Scholar]
- 17.Vo-Dinh T, Houck K, Stokes DL. Surface-enhanced Raman gene probes. Anal Chem. 1994;66:3379–3383. doi: 10.1021/ac00092a014. [DOI] [PubMed] [Google Scholar]
- 18.Vo-Dinh T. Surface-enhanced Raman spectroscopy using metallic nanostructures. Trends Anal Chem. 1998;17:557–570. [Google Scholar]
- 19.Isola NR, Stokes DL, Vo-Dinh T. Surface enhanced Raman gene probe for HIV detection. Anal Chem. 1998;70:1352–1356. doi: 10.1021/ac970901z. [DOI] [PubMed] [Google Scholar]
- 20.Zeisel D, Deckert V, Zenobi R, Vo-Dinh T. Near-field surface-enhanced Raman spectroscopy of dye molecules adsorbed on silver island films. Chem Phys Lett. 1998;283:381–385. [Google Scholar]
- 21.Deckert V, Zeisel D, Zenobi R, Vo-Dinh T. Near-field surface enhanced Raman imaging of dye-labeled DNA with 100-nm resolution. Anal Chem. 1998;70:2646–2650. doi: 10.1021/ac971304f. [DOI] [PubMed] [Google Scholar]
- 22.Stokes DL, Hueber D, Vo-Dinh T. Towards single-molecule detection with SERS detection using solid substrates. presented at the 1998 Pittsburgh Conf; New Orleans, LA. Mar. 1–5.. [Google Scholar]
- 23.Vo-Dinh T, Allain LR, Stokes DL. Cancer gene detection using surface-enhanced Raman scattering (SERS) J Raman Spectrosc. 2002;33:511–516. [Google Scholar]
- 24.Vo-Dinh T, Yan F, Wabuyele M. Surface-enhanced Raman scattering for medical diagnostics and biological imaging. J Raman Spectrosc. 2005;36:640–647. [Google Scholar]
- 25.Wabuyele M, Yan F, Griffin G, Vo-Dinh T. Hyperspectral surface-enhanced Raman imaging (HSERI) of labeled silver nanoparticles in single cells. Rev Sci Instrum. 2005;76:063710-1–063710-7. [Google Scholar]
- 26.Wabuyele M, Vo-Dinh T. Detection of HIV type 1 DNA sequence using plasmonics nanoprobes. Anal Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 27.Vo-Dinh T, Yan F. Gene detection and multi-spectral imaging using SERS nanprobes and nanostructures. In: Vo-Dinh T, editor. Nanotechnology in Biology and Medicine. New York: Taylor & Francis; 2007. [Google Scholar]
- 28.Etchegoin P, Cohen LF, Hartigan H, Brown RJC, Milton MJT, Gallop JC. Electromagnetic contribution to surface enhanced Raman scattering revisited. J Chem Phys. 2003;119:5281–5289. [Google Scholar]
- 29.Doering WE, Nie SM. Single-molecule and single-nanoparticle SERS: Examining the roles of surface active sites and chemical enhancement. J Phys Chem B. 2002;106:311–317. [Google Scholar]
- 30.Kambhampati P, Child CM, Foster MC, Campion A. On the chemical mechanism of surface enhanced Raman scattering: Experiment and theory. J Chem Phys. 1998;108:5013–5026. [Google Scholar]
- 31.Wokaun A. Surface-enhanced electromagnetic processes. In: Ehrenreich H, Turnbull D, Seitz F, editors. Solid State Physics. Vol. 38. New York: Academic; 1984. p. 223. [Google Scholar]
- 32.Otto A, Mrozek I, Grabhorn H, Akermann W. Surface-enhanced Raman-scattering. J Phys Condens Matter. 1992;4:1143–1212. [Google Scholar]
- 33.Kambhampati P, Child CM, Foster MC, Campion A. On the chemical mechanism of surface enhanced Raman scattering: Experiment and theory. J Chem Phys. 1998;108:5013–5026. [Google Scholar]
- 34.Otto A. What is observed in single molecule SERS, and why? J Raman Spectrosc. 2002;33:593–598. [Google Scholar]
- 35.Lee PC, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86:3391–3395. [Google Scholar]
- 36.Betzig E, Trautman JK, Harris TD, Weiner JS, Kostelak RL. Breaking the diffraction barrier—Optical microscopy on a nanometric scale. Science. 1991;251:1468–1470. doi: 10.1126/science.251.5000.1468. [DOI] [PubMed] [Google Scholar]
- 37.Emory SR, Nie S. Surface-enhanced Raman spectroscopy on single silver nanoparticles. Anal Chem. 1997;69:2631–2635. [Google Scholar]
- 38.Allain LR, Vo-Dinh T. Surface-enhanced Raman scattering detection of the breast cancer susceptibility gene BRCA1 using a silver-coated microarray platform. Anal Chim Acta. 2002;469:149–154. [Google Scholar]
- 39.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS. Single molecule detection using surface-enhanced Raman scattering (SERS) Phys Rev Lett. 1997;78:1667–1670. [Google Scholar]
- 40.Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.