Abstract
In this paper, we describe the development and application of a pH-sensitive plasmonics-active fiber-optic nanoprobe suitable for intracellular bioanalysis in single living human cells using surface-enhanced Raman scattering (SERS) detection. The effectiveness and usefulness of SERS-based fiber-optic nanoprobes are illustrated by measurements of intracellular pH in HMEC-15/hTERT immortalized “normal” human mammary epithelial cells and PC-3 human prostate cancer cells. The results indicate that fiber-optic nanoprobe insertion and interrogation provide a sensitive and selective means to monitor cellular microenvironments at the single cell level.
Keywords: Fiber-optic nanoprobe, Intracellular pH measurement
Introduction
Raman spectroscopy is a rapid, non-destructive photon-scattering technique, which provides information regarding vibrational energy levels of analyte molecules and exhibits very narrow spectral features. The resulting molecular specificity and potential for multiplexed detection makes the technique a powerful analytical tool for the identification of chemical and biological species, elucidation of molecular structure, and examination of surface properties. Unfortunately, the broader utility of Raman spectroscopy is limited by the poor efficiency of Raman scattering—even strong scatterers have Raman cross-sections on the order of 10−29 cm2. For comparison, fluorescence cross-sections are often on the order of 10−16 cm2.
One approach, which can greatly improve the efficiency of Raman scattering, is surface-enhanced Raman scattering (SERS), in which analytes are localized near plasmonically active surfaces or substrates [1–8]. When a nanostructured metallic surface is illuminated by light at the proper wavelength, electrons in the conduction band can oscillate at the frequency of the incident light. These electrons, also called “surface plasmons”, generate a secondary electromagnetic field, which can add to the incident field [7, 8]. This resonant response greatly enhances the local electromagnetic field intensity, thereby enhancing the efficiency of Raman scattering (typically 106- to 107-fold, but up to 1015-fold at “hot spots”) for molecules on or near the metal surface [7, 8].
Following early reports of the SERS effect [1–3], our laboratory and others have focused on developing SERS into a powerful analytical tool for analysis of multicomponent samples [9–19]. This capability is one of the technique’s greatest advantages when compared to fluorescence-based analyses.
Biological applications of Raman and SERS
In spite of the low cross-sections exhibited by most molecules, normal Raman spectroscopy has been successfully used for discrimination of cancerous cells from non-cancerous cells [30–32]. Resonance Raman spectroscopy is able to provide subtle detail regarding protein folding, hydration, and ligand binding, especially for heme proteins [33–36], tryptophan [35, 37–39], and tyrosine [34, 35, 37–40]. Although unlabeled silver or gold nanoparticles have limited use in SERS-based biochemical analyses due to their lack of molecular specificity, addition of biochemically responsive labels or ligands to SERS-active silver or gold nanoparticles or nanoshells has demonstrated great potential for intracellular bioanalysis and extracellular labeling [15, 16, 23, 24, 27, 41–51]. SERS-active plasmonic nanostructures have also been used for photothermal treatment of shallow tumors [52–54].
Every analytical technique, however, has its limitations. Normal Raman spectroscopy generally has great difficulty discriminating any specific protein or gene product from another in vivo or in vitro because the underlying building blocks are so similar. The utility of resonance Raman spectroscopy is limited to those cases where an electronic absorption band overlaps a laser line [55, 56]. Unfortunately, such an overlap can result in rapid photodegradation of the analyte [57–60]. Nanoparticle- or nanoshell-based SERS analyses, while theoretically able to provide single-molecule sensitivity, can be limited by the time required for endocytosis or phagocytosis of the SERS-active plasmonic particles, cellular digestion of the biochemically specific labels or ligands anchored to the nanoparticles, and the rate at which the nanoparticles are ejected from the cells. In addition, cellular uptake can shuttle nanoparticles directly into the lysosomal pathway, and intracellular transport can be quite inhomogeneous, thereby limiting the amount and nature of the biochemical information, which can be collected while the SERS-active nanoparticles reside within the cells [44, 61–63]. Plasmonic nanoparticles designed to bind to antigens on the cell membrane can avoid some of these limitations, but such an approach is inherently unable to provide information regarding intracellular processes, which are not reflected by changes in the cell membrane.
Our laboratory has recently developed a submicron, pH-sensitive fiber-optic SERS nanoprobe, which is able to circumvent many of the limitations described above. The nanoprobe is functionalized with thiolated ligands or labels, thereby imparting the molecular specificity required for intracellular bioanalysis while incorporating both positive and negative controls in a single experiment. Since the fiber-optic nanoprobe is physically inserted into cells using micromanipulators, concerns regarding rates of nanoparticle uptake and ejection are entirely avoided. The speed of SERS-active nanoprobe insertion, interrogation, and subsequent removal from a cell (often less than 30 s) also decreases the potential for intracellular digestion of the biochemically responsive functionality anchored to the silver island film (AgIF). Fluorescence-based fiber-optic nanoprobes developed for intracellular interrogation possess similar strengths, but lack the multiplexing potential of SERS-based fiber-optic nanoprobes [64–66].
In the current paper, we describe the fabrication and use of a pH-sensitive, SERS-active, submicron-diameter fiberoptic nanoprobe designed for intracellular pH measurement in single living human cells. We have used this nanoprobe in conjunction with a confocal Raman microscope to measure the intracellular pH of tens of PC-3 human prostate cancer cells and HMEC-15/hTERT human mammary epithelial cells, finding that the intracellular pH of both cell lines is ~7.3+0.2/−0.1. Furthermore, the high signal-to-noise ratio of our results and the lack of an aggressive lysosomal response to nanoprobe insertion and interrogation highlight the broader potential of the SERS nanoprobe technique in a systems biology framework as additional biochemically responsive functionality is incorporated onto the nanoprobe for detection of specific gene products, reactive oxygen species, etc.
Experimental
Cell culture
HMEC-15/hTERT cells, provided by Dr. Victoria Seewaldt, were grown in T-75 flasks using MEBM media (Lonza, Charles City, IA, USA). HMEC-15/hTERT cells are, “normal”, non-cancerous human mammary epithelial cell strain immortalized with hTERT and are routinely used as a normal cell control in cancer research studies. PC-3 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in T-75 flasks using F-12/Kaighn’s medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). PC-3 cells are a p53-null human prostate cancer cell line, which is frequently used when investigating anti-cancer drugs, signaling pathways, cell death, etc. Both HMEC-15/hTERT and PC-3 stock cultures were grown to 70% confluence in a 5% CO2 incubator at 37 °C prior to subculturing. HMEC-15/hTERT cells were subcultured at a 1:3 split ratio, and PC-3 cells were subcultured at a 1:6 split ratio.
pH-Sensitive fiber-optic nanoprobe fabrication
The submicron-diameter fiber-optic probes used in this work were fabricated by tapering 400-μm core-diameter optical fibers (CeramOptec Industries, East Longmeadow, MA, USA) using a commercially available pipette puller (P-2000, Sutter Instrument, Novato, CA, USA), producing nanoprobes smaller than 100 nm in diameter. These tapered optical fibers were then coated with a 6-nm mass thickness of 99.99% silver (Kamis, Mahopac Falls, NY, USA) at ~10−7 torr atmospheric pressure using an electron beam evaporator (CVE301EB, Cooke Vacuum Products, South Norwalk, CT, USA), which produces a highly SERS-active AgIF [67–74]. Following AgIF deposition, the nanoprobes were functionalized for 15 s in 10 mM para-mercaptobenzoic acid (pMBA; Sigma, Milwaukee, WI, USA) dissolved in ethanol, which anchored pMBA to the AgIF via a silver-thiol covalent bond. The carboxyl group of pMBA is pH sensitive across the physiologically relevant range [44, 49, 48], thereby rendering the nanoprobe pH sensitive across that range as well.
pH calibration
Standard solutions were prepared at pH values from 6.0 to 7.5 using phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA, USA), nitric acid, and sodium hydroxide. Solution pH was measured using a calibrated digital pH meter with a Ag/AgCl probe (Symphony, VWR, West Chester, PA, USA). Nanoprobes were prepared and functionalized as described above, then immersed in pH-adjusted PBS deposited on a fused silica microscope slide. The confocal microscope (InVia, Renishaw, Gloucestershire, UK) was focused on the nanoprobe tip, and SERS spectra were collected across the range from 1,000 to 1,800 cm−1. Exposure time was 10 s per spectrum, and laser intensity was ~35 mW at 632.8 nm. When corrected for the surface area of the submicron fiber-optic nanoprobe, we estimate that the effective laser power is less than 0.5 mW. The volume examined by the fiber-optic SERS nanoprobe is approximately 0.01 μm3 in the current external-illumination configuration, but can be significantly reduced by through-fiber excitation.
This procedure was repeated ten times for each data point from pH 6.0 to pH 7.5.
Intracellular pH measurements
SERS-active fiber-optic nanoprobes for intracellular pH measurements were prepared per the fabrication protocol detailed above. Cells were harvested by trypsinization, and trypsinization was halted using 5% MEBM in PBS (for HMEC-15/hTERT cells) or F-12/Kaighn’s medium (for PC-3 cells). The cells were then separated from the trypsinizing solution by centrifugation, resuspended in media, and mixed with growth factor reduced Matrigel™ (BD Biosciences, San Jose, CA, USA) at a 1:1 ratio. Between 150 and 200 μL of cell suspension was deposited on a fused silica microscope slide (Chemglass, Vineland, NJ, USA) and incubated at 37 °C for 90 s to partially gel the suspension. Prior experiments optimizing the incubation time indicated that an incubation time of 60 s was not always sufficient to ensure the degree of gellation necessary to allow intracellular interrogation with the pH-sensitive nanoprobe and that an incubation time of 120 s yielded a gel, which was too thoroughly gelled for efficient interrogation of multiple cells within a single droplet of cell suspension.
Following gellation of the cell suspension on a fused silica microscope slide, the sample was transferred to our confocal Raman microscope for intracellular interrogation. The pH-sensitive nanoprobes were inserted into the cells using micromanipulators, the confocal microscope was focused on the portion of the nanoprobe inserted into the cell, and single 10-s SERS spectra were acquired. The longer acquisition times and spectral smoothing algorithms often required in Raman spectroscopy were not necessary due to the high signal-to-noise ratio associated with the SERS nanoprobes. Our and other groups’ previous work with fiber-optic nanoprobes has indicated that careful insertion, brief interrogation, and removal do not adversely affect cell viability. In addition, decades of work with in vitro fertilization have demonstrated that cell viability can be maintained following careful intracellular insertion of small needles and injection of DNA. Even so, multiple spectra were not acquired due to literature reports of cellular heating and necrosis upon extended excitation of plasmonic nanoparticles in/on cells [52–54].
Results and discussion
pH calibration
Figure 1a shows averaged, normalized SERS spectra from 1,000 to 1,800 cm−1 acquired during calibration of the pH-sensitive nanoprobes from pH 6.0 to pH 7.5. Step size in terms of pH is approximately 0.2 units. The overall consistency of the SERS spectra independent of solution pH illustrates the excellent reproducibility of both the pH-sensitive pMBA layer and the underlying AgIF. The pH-dependent intensity of the bands near 1,400 cm−1 is highlighted in Fig. 1b, with a pH increase from 6.0 to 7.5 substantially increasing the intensity of the band centered near ~1,425 cm−1. An increase in the normalized intensity of this SERS peak with increasing pH is consistent with previous work using pMBA anchored to SERS-active nanoparticles for intracellular pH measurement [44, 46, 48].
Fig. 1.
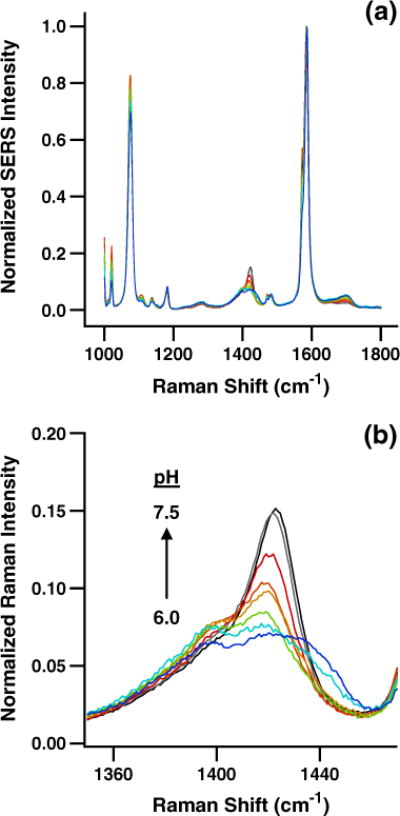
pH calibration data for the pH-sensitive nanoprobe. a SERS spectra of pMBA anchored to the AgIF on the submicron-diameter fiber-optic nanoprobe from pH 6.0 to pH 7.5. b The intensity of the ~1,425 cm−1 carboxylate band for pMBA varies with pH across the physiological range due to its degree of protonation/deprotonation
Figure 2 further illustrates the pH dependence of the SERS intensity of the pMBA carboxylate band at ~1,425 cm−1. At a pH of 6.0, the normalized intensity of the pMBA SERS signal at ~1,425 cm−1 is ~0.07. As the pH of the solution surrounding the SERS-active, pH-sensitive nanoprobe is increased to 7.5, the normalized intensity of the SERS signal at ~1,425 cm−1 increases to ~0.15. Much of the variability present in Fig. 2 is due to nanoprobe vibration and movement during the Raman measurement: The standard deviation of the normalized intensity at ~1,425 cm−1 is typically smaller by a factor of three for pH measurements using pMBA-functionalized AgIFs deposited on glass or quartz slides. We expect that improved vibrational isolation of the confocal microscope, sample stage, etc. would allow the reproducibility of intracellular measurements with fiber-optic SERS nanoprobes to approach that of AgIFs deposited on glass or quartz slides.
Fig. 2.
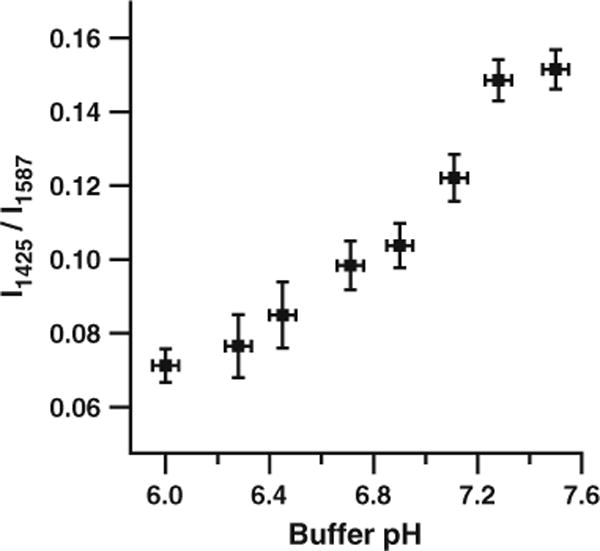
Calibration of the pH-sensitive nanoprobe. A pH calibration curve can be generated using the intensity of the ~1,425 cm−1 pMBA carboxylate band relative to the intensity of the ~1,587 cm−1 combination band. Error bars represent two standard deviations in the y dimension and the resolution of the pH meter in the x dimension
Intracellular pH measurement in single cells
Figure 3 shows images of a SERS-active pH nanoprobe approaching (a) and interrogating (b) a single living cell. Figure 4a shows the averaged, normalized pMBA spectrum acquired from 15 individual HMEC-15/hTERT cells. The characteristic pH-dependent pMBA spectrum of the submicron nanoprobe is quite clear, with a signal-to-noise ratio (SNR) of ~190 for the combination band at ~1,587 cm−1 and a SNR of ~30 for the carboxylate band at ~1,425 cm−1, with a relative standard deviation of approximately 6%. A slight elevation in the baseline is visible due to weak fluorescence from Matrigel™ or from the cells themselves. As noted above, we have chosen to take only one spectrum per cell to limit the potential for cell stress due to photothermal effects.
Fig. 3.
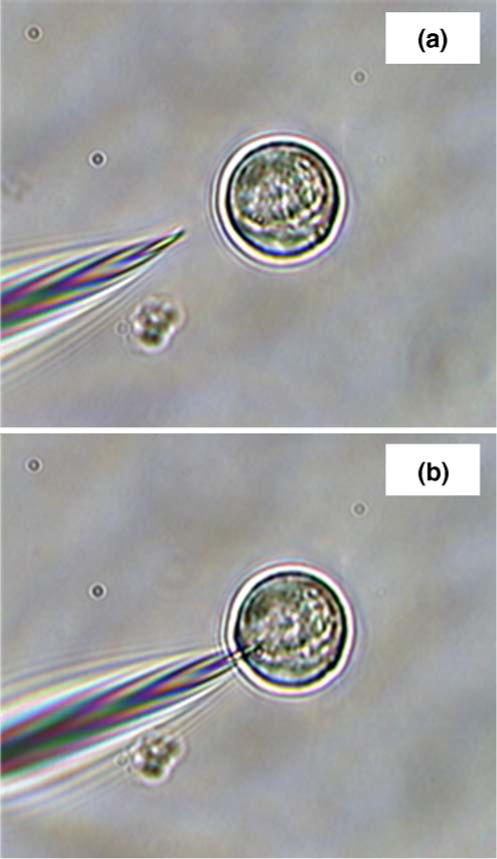
Nanoprobe interrogation of single living cells. SERS-active, pH-sensitive fiber-optic nanoprobes can be quickly and easily inserted into single living cells suspended in a supporting matrix or grown on microscope slides. The approach toward a single cell (a) and insertion for laser interrogation (b) are accomplished through use of micromanipulators
Fig. 4.
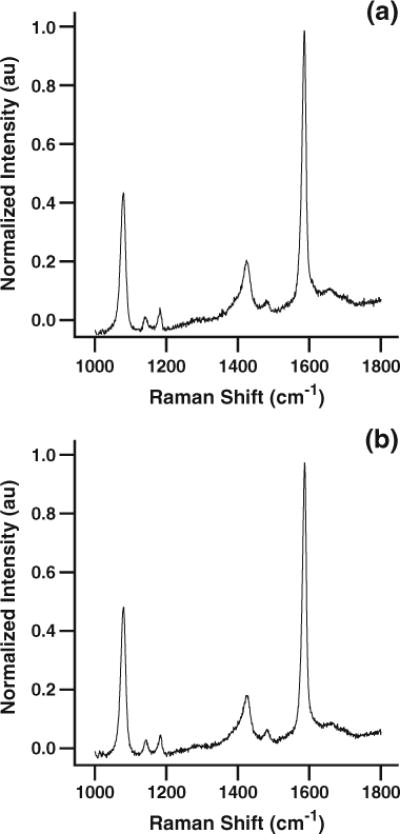
High-SNR SERS spectra acquired by interrogating pH-sensitive nanoprobes in a HMEC-15/hTERT and b PC-3 cells. The intracellular pH of both types of cells is ~7.3, indicating a lack of significant environmental stress and the lack of an aggressive lysosomal response to nanoprobe insertion and interrogation
When corrected for the baseline elevation due to fluorescence by fitting the region between 1,750 and 1,800 wavenumbers with a straight line, extending it across the spectral range and subtracting from the spectrum the normalized intensity of the carboxylate band at ~1,425 cm−1 in Fig. 4a is approximately 0.16±0.01. Comparing this value to the calibration curve in Fig. 2, we conclude that the pH of the immortalized HMEC-15/hTERT cells is 7.3 or higher. This value is consistent with the intracellular pH of cells cultured in pH 7.4 media. Additionally, this high pH indicates that the cells are not under sufficient stress to induce apoptosis, which typically results in intracellular acidification.
Figure 4b shows the averaged, normalized, unsmoothed SERS spectra from nanoprobe measurements in 30 individual PC-3 human prostate cancer cells. The SNR of the SERS peaks at ~1,587 and ~1,425 cm−1 (~175 and ~30, respectively, with relative standard deviations of approximately 6%) again highlights the sensitivity and reproducibility of measurements using the pMBA-functionalized AgIF deposited on the SERS nanoprobe. After correcting for the fluorescence background beneath the SERS spectrum and using the calibration data in Fig. 2, we find that the intracellular pH of the PC-3 cells is 7.3 or higher. As discussed above, this value is consistent with the pH of cells raised in pH 7.4 growth media and is also reasonable for cells which are not currently under significant environmental stress.
The data discussed above illustrate several noteworthy and important results. First, the ease and speed with which the nanoprobe can be manipulated, i.e., inserted into single living cells supported in Matrigel™, demonstrate the suitability of the matrix for live-cell studies. Second, use of the SERS nanoprobe does not produce an apoptotic response in either HMEC-15/hTERT or PC-3 cells. This result, combined with our previous observation of cells undergoing mitosis following interrogation with nanoprobes, indicates that nanoprobe insertion, intracellular bioanalysis, and nanoprobe removal do not significantly affect cell function or result in a self-destructive cellular response. Finally, and perhaps most significantly for development of biochemically functionalized nanoprobes, detection of an intracellular pH near 7.3 indicates the lack of a strong lysosomal response to nanoprobe insertion and interrogation. If a rapid, aggressive lysosomal response were triggered by either insertion or interrogation of the fiber-optic nanoprobe, acidification of the environment immediately around the nanoprobe would be expected as low-pH lysosomes attempted to attack and digest it. This result suggests that the SERS-active nanoprobe provides an effective and practical platform to bypass the limitations of nanoparticle uptake by endocytosis or phagocytosis, as well as the concomitant danger of the biosensing particles being shuttled directly into the lysosomal pathway.
Conclusion
We have used SERS-active fiber-optic nanoprobes to measure the intracellular pH of HMEC-15/hTERT immortalized human mammary epithelial cells and PC-3 human prostate cancer cells. The results indicated the pH value of both cell lines is near that expected for healthy cells in the absence of significant environmental stress. In addition, we have demonstrated that insertion and interrogation of the pH-sensitive, SERS-active fiber-optic nanoprobe induces neither apoptosis nor an aggressive lysosomal response from either of these cell lines. These results demonstrate the robustness of the SERS-active pH nanoprobe for intracellular pH measurements and point toward the potential utility of additional biochemically specific fiber-optic nanoprobes.
Acknowledgments
This research was financially supported by the National Institute of Health (NIH R01 EB006201 and NIH R01 ES014774) and the Department of Justice.
Contributor Information
Jonathan P. Scaffidi, Department of Biomedical Engineering, Duke University, 136 Hudson Hall, P. O. Box 90281, Durham, NC 27708, USA, Fitzpatrick Institute for Photonics, Duke University, 305 Teer Building, P. O. Box 90271, Durham, NC 27708, USA
Molly K. Gregas, Department of Biomedical Engineering, Duke University, 136 Hudson Hall, P. O. Box 90281, Durham, NC 27708, USA, Fitzpatrick Institute for Photonics, Duke University, 305 Teer Building, P. O. Box 90271, Durham, NC 27708, USA
Victoria Seewaldt, Fitzpatrick Institute for Photonics, Duke University, 305 Teer Building, P. O. Box 90271, Durham, NC 27708, USA, Division of Medical Oncology, Duke School of Medicine, DUMC 2628 Room 221A MSRB, Durham, NC 27710, USA.
Tuan Vo-Dinh, Email: tuan.vodinh@duke.edu, Department of Biomedical Engineering, Duke University, 136 Hudson Hall, P. O. Box 90281, Durham, NC 27708, USA, Fitzpatrick Institute for Photonics, Duke University, 305 Teer Building, P. O. Box 90271, Durham, NC 27708, USA, Department of Chemistry, Duke University, 124 Science Drive, P. O. Box 90354, Durham, NC 27708, USA.
References
- 1.Fleischman M, Hendra PJ, McQuillan AJ. Chem Phys Lett. 1974;26:163–166. [Google Scholar]
- 2.Jeanmaire DL, Van Duyne RP. J Electroanal Chem. 1977;84:1–20. [Google Scholar]
- 3.Albrecht MG, Creighton JA. J Am Chem Soc. 1977;99:5215–5217. [Google Scholar]
- 4.Vo-Dinh T, Hiromoto MYK, Begun GM, Moody RL. Anal Chem. 1984;56:1667–1670. [Google Scholar]
- 5.Vo-Dinh T. Surface-enhanced raman spectroscopy. In: Halevi P, editor. Photonic probes of surfaces. Elsevier; New York: 1995. [Google Scholar]
- 6.Vo-Dinh T. Trends in Anal Chem. 1998;17:557–582. [Google Scholar]
- 7.Schatz GC. Accounts Chem Res. 1984;17:370–376. [Google Scholar]
- 8.Kerker M. Accounts Chem Res. 1984;17:271–277. [Google Scholar]
- 9.Campion A, Kambhampati P. Chem Soc Rev. 1998;27:241–250. [Google Scholar]
- 10.Nie S, Emory SR. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 11.Emory SR, Nie S. Anal Chem. 1997;69:2631–2635. [Google Scholar]
- 12.Doering WE, Nie S. Anal Chem. 2003;75:6171–6176. doi: 10.1021/ac034672u. [DOI] [PubMed] [Google Scholar]
- 13.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS. Phys Rev Lett. 1997;78:1667–1670. doi: 10.1103/PhysRevLett.76.2444. [DOI] [PubMed] [Google Scholar]
- 14.Kneipp K, Kneipp H, Deinum G, Itzkan I, Dasari R, Feld MS. Appl Spectrosc. 1998;52:175–178. [Google Scholar]
- 15.Cao YC, Jin R, Mirkin CA. Science. 2002;197:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 16.Cao YC, Jin R, Nam J, Thaxton CS, Mirkin CA. J Am Chem Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 17.Moody RL, Vo-Dinh T, Fletcher WH. Appl Spectrosc. 1987;41:966–970. [Google Scholar]
- 18.Vo-Dinh T, Miller GH, Bello J, Johnson RD, Moody RL, Alak A, Fletcher WH. Talanta. 1989;36:227–234. doi: 10.1016/0039-9140(89)80100-6. [DOI] [PubMed] [Google Scholar]
- 19.Enlow PD, Buncick M, Warmack RJ, Vo-Dinh T. Anal Chem. 1986;58:1119–1123. [Google Scholar]
- 20.Vo-Dinh T, Uziel M, Morrison AL. Appl Spectrosc. 1987;41:605–610. [Google Scholar]
- 21.Vo-Dinh T, Alak A, Moody RL. Spectrochim Acta B. 1988;43:605–615. [Google Scholar]
- 22.Vo-Dinh T, Houck TK, Stokes DL. Anal Chem. 1994;66:3379–3383. doi: 10.1021/ac00092a014. [DOI] [PubMed] [Google Scholar]
- 23.Isola N, Stokes DL, Vo-Dinh T. Anal Chem. 1998;70:1352–1356. doi: 10.1021/ac970901z. [DOI] [PubMed] [Google Scholar]
- 24.Wabuyele MB, Vo-Dinh T. Anal Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 25.Chu BCF, Wahl GM, Orgel LE. Nucleic Acids Res. 1983;11:6513–6529. doi: 10.1093/nar/11.18.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckert V, Meisel D, Zenobi R, Vo-Dinh T. Anal Chem. 1998;70:2646–2650. doi: 10.1021/ac971304f. [DOI] [PubMed] [Google Scholar]
- 27.Vo-Dinh T, Allain LR, Stokes DL. J Raman Spectrosc. 2002;33:511–516. [Google Scholar]
- 28.Culha M, Stokes DL, Allain LR, Vo-Dinh T. Anal Chem. 2003;75:6196–6201. doi: 10.1021/ac0346003. [DOI] [PubMed] [Google Scholar]
- 29.Stokes DL, Chi ZH, Vo-Dinh T. Appl Spectrosc. 2004;58:292–298. doi: 10.1366/000370204322886636. [DOI] [PubMed] [Google Scholar]
- 30.Lawson EE, Barry BW, Williams AC, Edwards HGM. J Raman Spectrosc. 1997;28:111–117. [Google Scholar]
- 31.Mahadevan-Jansen A, Richards-Kortum R. Proc IEEE/EMBS. 1997:2722–2728. Oct. 30–Nov. 2. [Google Scholar]
- 32.Kendall C, Stone N, Shepherd N, Geboes K, Warren B, Bennett R, Barr H. J Pathol. 2003;200:602–609. doi: 10.1002/path.1376. [DOI] [PubMed] [Google Scholar]
- 33.Tsubaki M, Srivastava RB, Yu NT. Biochemistry. 1982;21:1132–1140. doi: 10.1021/bi00535a004. [DOI] [PubMed] [Google Scholar]
- 34.Nagai M, Yoneyama Y, Kitagawa T. Biochemistry. 1989;28:2418–2422. doi: 10.1021/bi00432a012. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers KR, Su C, Subramaniam S, Spiro TG. J Am Chem Soc. 1992;114:3697–3709. [Google Scholar]
- 36.Jayaraman V, Rodgers KR, Mukerji I, Spiro TG. Science. 1995;269:1843–1848. doi: 10.1126/science.7569921. [DOI] [PubMed] [Google Scholar]
- 37.Rava RP, Spiro TG. J Am Chem Soc. 1984;106:4062–4064. [Google Scholar]
- 38.Kochendoerfer GG, Kaminaka S, Mathies RA. Biochemistry. 1997;36:13153–13159. doi: 10.1021/bi971541c. [DOI] [PubMed] [Google Scholar]
- 39.Miura T, Takeuchi H, Harada I. J Raman Spectrosc. 2005;20:667–671. [Google Scholar]
- 40.Hildebrandt PG, Copeland RA, Spiro TG, Otlewski J, Laskowski M, Jr, Prendergast FG. Biochemistry. 1988;27:5426–5433. doi: 10.1021/bi00415a007. [DOI] [PubMed] [Google Scholar]
- 41.Michota A, Bukowska J. J Raman Spectrosc. 2003;34:21–25. [Google Scholar]
- 42.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 43.Xu S, Ji X, Xu W, Li X, Wang L, Bai Y, Zhao B, Ozaki Y. Analyst. 2004;129:63–68. doi: 10.1039/b313094k. [DOI] [PubMed] [Google Scholar]
- 44.Talley CE, Jusinski L, Hollars CW, Lane SM, Huser T. Anal Chem. 2004;76:7064–7068. doi: 10.1021/ac049093j. [DOI] [PubMed] [Google Scholar]
- 45.Wabuyele MB, Vo-Dinh T. Anal Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 46.Schwartzberg AM, Oshiro TY, Zhang JZ, Huser T, Talley CE. Anal Chem. 2006;78:4732–4736. doi: 10.1021/ac060220g. [DOI] [PubMed] [Google Scholar]
- 47.Niidome T, Yamagata M, Okamoto Y, Akiyama Y, Takahashi H, Kawano T, Katayama Y, Niidome Y. J Controlled Release. 2006;114:343–347. doi: 10.1016/j.jconrel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 48.Kneipp J, Kneipp H, Wittig B, Kneipp K. Nano Lett. 2007;7:2819–2823. doi: 10.1021/nl071418z. [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Yu C, Irudayaraj J. Anal Chem. 2007;79:3981–3988. doi: 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Kim S, Choo J, Shin SY, Lee YH, Choi HY, Ha S, Kang K, Oh CH. Anal Chem. 2007;79:916–922. doi: 10.1021/ac061246a. [DOI] [PubMed] [Google Scholar]
- 51.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 52.Hirsch LR, Stafford RJ, Bankson JA, Sershen SR, Rivera B, Price RE, Hazle JD, Halas NJ, West JL. Proc Nat Acad Sci. 2003;100:13549–13554. doi: 10.1073/pnas.2232479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neal DP, Hirsch LR, Halas NJ, Payne JD, West JL. Cancer Lett. 2004;209:171–176. doi: 10.1016/j.canlet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Letfullin RR, Joenathan C, George TF, Zharov VP. Nanomedicine. 2006;1:473–480. doi: 10.2217/17435889.1.4.473. [DOI] [PubMed] [Google Scholar]
- 55.Albrecht AC. J Chem Phys. 1961;34:1476–1484. [Google Scholar]
- 56.Long DA. The raman effect: a unified treatment of the theory of raman scattering by molecules. Wiley; West Sussex: 2001. [Google Scholar]
- 57.Ikeda-Saito M, Argade PV, Rousseau DL. FEBS Lett. 1985;184:52–55. doi: 10.1016/0014-5793(85)80651-7. [DOI] [PubMed] [Google Scholar]
- 58.Wu Q, Balakrishnan G, Pevsner A, Spiro TG. J Phys Chem A. 2003;107:8047–8051. [Google Scholar]
- 59.Barbosa CJ, Vaillancourt FH, Eltis LD, Blades MW, Turner RFB. J Raman Spectrosc. 2002;33:503–510. [Google Scholar]
- 60.Jarvis RM, Goodacre R. FEMS Microbiology Lett. 2004;232:127–132. doi: 10.1016/S0378-1097(04)00040-0. [DOI] [PubMed] [Google Scholar]
- 61.Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 62.Becker C, Hodenius M, Blendinger G, Sechi A, Hieronymus T, Muller-Schulte D, Schmitz-Rode T, Zenke M. J Magn & Magn Mater. 2007;311:234–237. [Google Scholar]
- 63.Shamsaie A, Jonczyk M, Sturgis J, Robinson JP, Irudayaraj J. J Biomed Optics. 2007;12:020502. doi: 10.1117/1.2717549. [DOI] [PubMed] [Google Scholar]
- 64.Tan W, Shi ZY, Smith S, Birnbaum D, Kopelman R. Science. 1992;258:778–781. doi: 10.1126/science.1439785. [DOI] [PubMed] [Google Scholar]
- 65.Vo-Dinh T, Alarie JP, Cullum BM, Griffin GD. Nat Biotechnol. 2000;18:764–767. doi: 10.1038/77337. [DOI] [PubMed] [Google Scholar]
- 66.Lu J, Rosenzweig Z. Fres J Anal Chem. 2000;366:569–575. doi: 10.1007/s002160051552. [DOI] [PubMed] [Google Scholar]
- 67.Schlegel VL, Cotton TM. Anal Chem. 1991;63:241–247. doi: 10.1021/ac00003a010. [DOI] [PubMed] [Google Scholar]
- 68.Van Duyne RP, Hulteen JC, Treichel DA. J Chem Phys. 1993;99:2101–2115. [Google Scholar]
- 69.Roark SE, Rowlen KL. Anal Chem. 1994;66:261–270. [Google Scholar]
- 70.Semin DJ, Rowlen KL. Anal Chem. 1994;66:4324–4331. [Google Scholar]
- 71.Roark SE, Semin DJ, Lo A, Skodje RT, Rowlen KL. Anal Chim Acta. 1995;307:341–353. [Google Scholar]
- 72.Lacy WB, Williams JM, Wenzler LA, Beebe TP, Jr, Harris JM. Anal Chem. 1996;68:1003–1011. [Google Scholar]
- 73.Whitney AV, Van Duyne RP, Casadio F. Proc SPIE. 2005;5993:59930K1–59930K10. [Google Scholar]
- 74.Li X, Xu W, Jia H, Wang X, Zhao B, Li B, Ozaki Y. Thin Solid Films. 2005;474:181–185. [Google Scholar]


