Abstract
We have demonstrated for the first time the feasibility of multiplex detection using the surface-enhanced Raman scattering-based molecular sentinel (MS) technology in a homogeneous solution. Two MS nanoprobes tagged with different Raman labels were used to detect the presence of the erbB-2 and ki-67 breast cancer biomarkers. The multiplexing capability of the MS technique was demonstrated by mixing the two MS nanoprobes and tested in the presence of single or multiple DNA targets.
1. Introduction
The ability to simultaneously detect multiple oligonucleotide sequences is critical for many medical applications such as early diagnosis, high-throughput screening and systems biology research. In particular, the development of practical and sensitive detection techniques with multiplexing capability can lead to improved accuracy for cancer diagnosis, since a variety of molecular alterations in multiple genes are usually involved in tumorigenesis and progression of various cancers. With this important medical application, there has been an increased number of research activities aimed at developing multiplex detection technologies [1–3].
Recent interest in the functionalization of metallic (e.g. silver and gold) nanoparticles with DNA oligonucleotides [4–8] has led to the development of novel analytic tools based on surface plasmon resonance (SPR) [7, 9, 10], metal-quenched [11–13] or metal-enhanced [14] fluorescence, and surface-enhanced Raman scattering (SERS) [15–19] for the detection of nucleic acid targets of interest. These newly developed nanobiotechnologies utilize the unique optical and plasmonics related properties of metal nanoparticles as signal transducers for reporting oligonucleotide hybridization events. In addition, it has been reported that oligonucleotidefunctionalized nanoparticles exhibit remarkably sharp melting profiles when compared to unmodified oligonucleotides [20]. By taking advantage of this unusual hybridization property, several nanoparticle-based DNA detection methods with improved sensitivity have been previously reported [10, 16, 17]. Many techniques require labeling the targets and several washing steps following the reactions, which can complicate detection procedures.
For the past years, our laboratory has devoted extensive research efforts on the development of SERS-based technology for chemical [21–24] and biological sensing [19, 25–31]. By utilizing plasmonics-active nanoparticles, a variety of chemicals and biomolecules have been successfully detected when adsorbed on the surface of nanoparticles [21, 23–25, 27–29, 32]. Moreover, we have also developed a novel ‘molecular sentinel’ (MS) nanoprobes to detect the presence of the HIV gene in a homogeneous assay [19]. Figure 1 schematically illustrates the operating principle of the MS nanoprobe. The MS nanoprobe is composed of a DNA hairpin probe (~30–45 nucleotides (nt) in length) and a metal (e.g., silver) nanoparticle. One end of the hairpin probe is tagged with a SERS-active label as a signal reporter. At the other end, the probe is modified with a thiol group, which is designed to conjugate covalently to the silver nanoparticle via thiol–metal interaction. The sequence within the loop region of the hairpin probe is complementary to a specific target gene sequence of interest. Hairpin DNA probes have been previously used in fluorescence-based molecular beacon systems [33]. In the absence of the target, the Raman label is in close proximity to the metal surface due to the stem-loop configuration (‘closed’ state), and a strong SERS signal is produced upon laser excitation. The metal nanoparticle is used as a signal enhancing platform for the SERS signal associated with the label. The enhancement is due to a nanostructured metal surface scattering process (nano-enhancers) which increases the intrinsically weak normal Raman scattering. The enhancement mechanism for SERS comes from intense localized fields arising from surface plasmon resonance in metallic (e.g. Ag, Au, Cu) nanostructures with sizes in the order of tens of nanometers and from chemical effects at the metal surface [34, 35]. Various models explaining important properties and characteristics of the surface plasmon effect have been reported [36, 37]. Theoretical studies of electromagnetic effects have shown that the SERS enhancement (G) falls off as G = [r/(r + d)]12 for a single analyte molecule located a distance d from the surface of a metal nanoparticle of radius r. The electromagnetic SERS enhancement strongly decreases with increased distance, due to the decay of a dipole over the distance (1/d)3 to the fourth power, thus resulting in a total intensity decay of (1/d)12. Therefore in designing the SERS MS nanoprobe, the hairpin configuration has the Raman label in contact or close proximity (<1 nm) to the nanoparticles thus inducing a strong SERS signal (figure 1, left). However, in the presence of the specific DNA target, hybridization between the target and DNA probe disrupts the stem-loop configuration and spatially separates the Raman label from the metal surface (‘open’ state) as depicted in figure 1 (right). Since the SERS enhancement (G) depends strongly on the distance d between the Raman label and the surface of the metal nanoparticle (G is proportional to d−12), the SERS signal is significantly reduced (quenched) in the open state of the MS nanoprobes.
Figure 1.
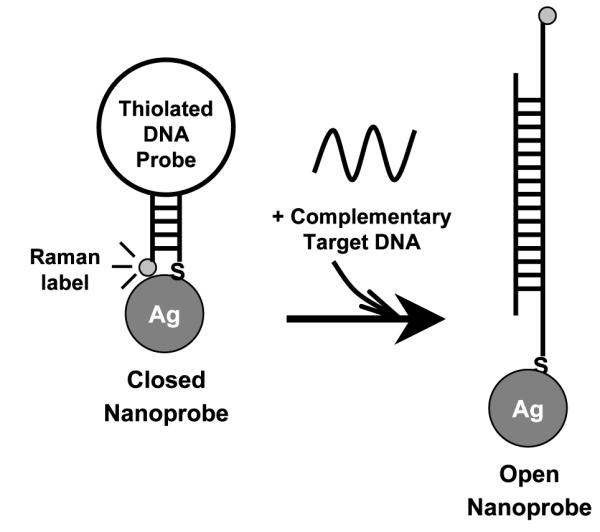
The operating principle of the SERS-based molecular sentinel (MS) nanoprobes. The MS nanoprobe is composed of a Raman-labeled DNA hairpin probe and a silver nanoparticle. In the absence of the complementary target DNA, a strong SERS signal is observed due to the hairpin conformation adopted by the MS nanoprobe (left: closed state). In the presence of the complementary target DNA, the hairpin conformation of the MS nanoprobe is disrupted and the SERS signal is quenched due to the physical separation of the Raman label from the surface of the silver nanoparticle (right: open state).
In this study, we show for the first time the feasibility of multiplex detection using the MS technology in a homogeneous solution without washing or separation steps. We have designed two MS nanoprobes, ERBB2-MS and KI-67-MS, to target the erbB-2 and ki-67 genes, respectively. These two genes, which are well known as critical biomarkers for breast cancer, have been adopted for diagnostic use in clinical practice. The erbB-2 gene (also know as ERBB2 or HER2/neu) encodes a transmembrane glycoprotein, which belongs to the epidermal growth factor receptor (EGFR) family and possesses intrinsic tyrosine kinase activity. Overexpression of ERBB2 is observed in 20%–30% of invasive breast cancers [38, 39]. Ki-67 is known as a cell proliferation marker and can potentially be used as an early predictor for treatment efficacy [40]. The expression of Ki-67 is well regulated throughout the cell cycle. In quiescent cells, Ki-67 expression is low. In proliferating tissues and tumors, Ki-67 is overexpressed [41]. Several studies have shown that the overexpression of ERBB2 is usually associated with the increased expression of Ki-67 in breast cancer [42, 43]. However, the functional relationship between these two genes is not well understood. Simultaneous detection of these two genes can further provide valuable information for insight into the functional relationship between these two genes and may lead to an improved early detection and diagnosis of breast cancer.
2. Materials and methods
Silver nitrate (99.995%) was purchased from Alfa Aesar (Ward Hill, MA). Hydroxylamine hydrochloride, 6-mercapto-1-hexanol and tris-HCl buffer (pH 8.0) were purchased from Sigma-Aldrich. All solution was prepared with deionized water (18 MΩ cm).
The silver nanoparticles used in this study were prepared by reduction of silver nitrate with hydroxylamine hydrochloride at room temperature [44]. Briefly, 10 ml of silver nitrate solution (10−2 M) were added to 90 ml of a hydroxylamine hydrochloride solution (1.67 × 10−3 M) containing 3.3 × 10−3 M NaOH under vigorous stirring for 1 h. The colloidal solutions were then stored at 4 °C and used within a few days. This procedure yields relatively homogeneous distribution of colloidal silver particles with an average diameter of 35 ± 6.3 nm.
The MS nanoprobes were prepared as described previously with slight modification [19]. Briefly, silver nanoparticles were incubated with 0.5 μM thiolated DNA hairpin probes containing 0.5 mM MgCl2 and allowed to react for few hours at room temperature. The Ag–DNA conjugates were exposed to 0.1 mM 6-mercapto-1-hexanol (MCH) for 10 min to displace the non-specifically adsorbed DNA and to passivate the silver surface. The MS nanoprobes were then purified three times by centrifugation at 12 000 rpm for 10 min and washed with 20 mM tris-HCl buffer (pH 8.0). The purified nanoprobes were finally resuspended in 20 mM tris-HCl buffer, and stored at 4 °C.
The SERS measurements in this study were performed by using a Renishaw InVia confocal Raman microscope with a 632.8 nm HeNe laser for excitation, and all spectra reported here were smoothed by the Savitzky–Golay filter to remove low-frequency noise while preserving high-frequency Raman spectra. Then the fluorescence background was removed using a numerical algorithm developed in our laboratory, which uses a moving window to locally determine the fluorescence background level.
In this study, the MS hairpins, target oligonucleotides and non-complementary control DNA shown in table 1 were all synthesized by Integrated DNA Technologies (IDT, Coralville, IA). To demonstrate the feasibility of multiplex oligonucleotide detection using the MS technique, the 3′-end of the two DNA hairpins of the ERBB2-MS and KI-67-MS nanoprobes, was modified with two different Raman labels, Cy3 and 5-carboxytetramethylrhodamine (TAMRA), respectively. To effectively separate the Raman labels from the silver nanoparticles upon hybridization to the target DNA, the length of the DNA hairpins was designed to be at least 35 nucleotides (nt), which corresponds to over 10 nm based on the DNA double helix structure. Conjugation of DNA hairpins onto silver nanoparticles was achieved by using an alkyl thiol substituent at the 5′-end.
Table 1.
DNA sequences of the MS hairpins, the corresponding target DNA and non-complementary DNA
| Oligonucleotides | DNA sequencesa |
|---|---|
| ERBB2-MS | 5′-SH-CGCCAT CCACCCCCAAGACCACGACCAGC AGAATATGGCG-Cy3-3′ |
| KI-67-MS | 5′-SH-GCGTAT TCTGCACACCTCTTGACACTCCG ATACGC-TAMRA-3′ |
| ERBB2–Target | 5′-GTTGGCATTCTGCTGGTCGTGGTCTTGGGG GTGGTCTTTG-3′ |
| KI-67–Target | 5′-AGCCAGCCTGCAGCAAGCACTTTGGAGAGCA AATCTGTGCAGAGAGTAACGCGGAGTGTCAAG AGGTGTGCAGAAAATCCAAAGAAGGCTGAGGA CAATG-3′ |
| Non-complementary DNAb | 5′-GCCAGCGTCGAGTTGGTTTGCAGCTCCTGA-3′ |
The underlined sequences indicate the complementary arms of the MS, and the bold sequences represent the target sequences complementary to the loop regions of the MS hairpins.
The non-complementary DNA was used as the negative control DNA for both ERBB2-MS and KI-67-MS nanoprobes.
3. Results and discussion
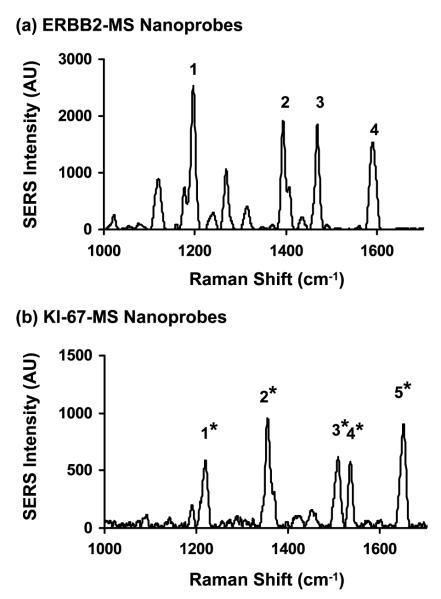
The Raman spectra from the ERBB2-MS and the KI-67-MS nanoprobes are shown in figures 2(a) and (b), respectively. The high intensity of the major SERS bands in each case indicates that with the stem-loop configuration intact, the Raman labels at the 3′-end of the two molecular sentinels (Cy3 and TAMRA) were near the surface of silver nanoparticles (closed state) and the Raman signals were plasmonically enhanced. The major Raman peaks of the two labels are marked in figure 2 and used throughout this paper to illustrate the changes in SERS spectra upon hybridization to specific DNA sequences. It is noteworthy that even though the fluorescence spectra of these two dyes Cy3 and TAMRA strongly overlap, their Raman spectra are clearly distinguishable from one another due to the narrow SERS peaks. This feature underlines the advantage of the SERS-based MS nanoprobes over fluorescence-based assays for multiplex detection.
Figure 2.
SERS spectra of the ERBB2-MS (a) and KI-67-MS (b) nanoprobes in 20 mM tris-HCl buffer (pH 8.0) containing 5 mM MgCl2. The major Raman bands are labeled as follows: ERBB2-MS: (1) 1197 cm−1, (2) 1393 cm−1, (3) 1468 cm−1, (4) 1590 cm−1. KI-67-MS: (1*) 1218 cm−1, (2*) 1354 cm−1, (3*) 1508 cm−1, (4*) 1535 cm−1, (5*) 1650 cm−1.
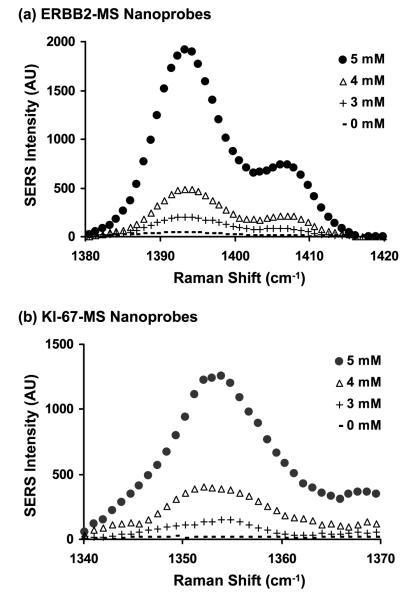
We have investigated the effect of MgCl2 concentration on the stem-loop configuration of the MS nanoprobes. It has been indicated that DNA hairpins can be effectively formed in the presence of MgCl2, since magnesium ions can stabilize the double-stranded stem regions [45, 46]. Figures 3(a) and (b) respectively show that the SERS intensity of the ERBB2-MS nanoprobes at 1393 cm−1 (peak #2) and the KI-67-MS nanoprobes at 1354 cm−1 (peak #2*) increases with increasing MgCl2 concentration up to 5 mM in 20 mM tris-HCl buffer (pH 8.0). When the concentration of MgCl2 was over 5 mM, precipitation of MS nanoprobes was observed, which may be due to the fact that the maximum ionic strength has been reached.
Figure 3.
The effect of MgCl2 concentration on the SERS intensity of one of the major Raman bands (peak #2: 1393 cm−1) from the ERBB2-MS nanoprobes (a) and (peak #2*: 1354 cm−1) from the KI-67-MS nanoprobes (b). (●): 5 mM, (▵): 4 mM, (+): 3 mM, (−): 0 mM.
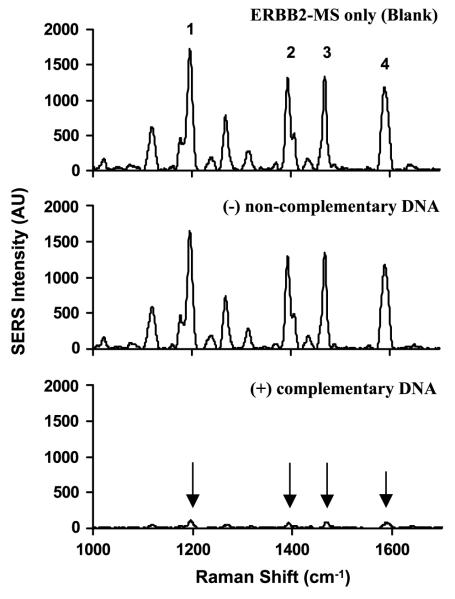
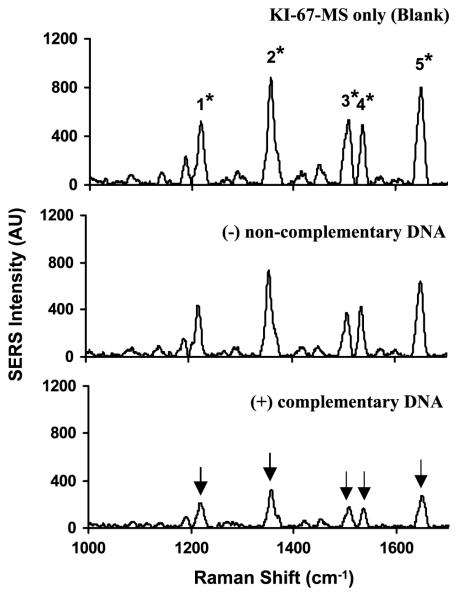
To test the effectiveness of the designed MS nanoprobes, the ERBB2-MS and KI-67-MS nanoprobes were separately incubated with their complementary target DNA in hybridization buffer (20 mM tris-HCl buffer, pH 8.0, 5 mM MgCl2) and allowed to react at 37 °C for 1 h. Figures 4 and 5 respectively show the SERS spectra of the ERBB2-MS and KI-67-MS nanoprobes in the presence or absence of 0.5 μM target DNA. In the case of presence of target DNA (lower spectra), the SERS intensity of the major Raman bands was significantly decreased (marked by arrows), indicating that the Raman labels were physically separated from the silver nanoparticles upon hybridization to their targets when the stem-loop configuration of the hairpin probes was disrupted (open state). On the other hand, in the absence of DNA (blank: upper spectra) or the presence of non-complementary DNA (negative control: middle spectra), the SERS intensity of the major Raman bands remains high, indicating that the MS nanoprobes remain in the stem-loop configuration (closed state).
Figure 4.
SERS spectra of the ERBB2-MS nanoprobes in the presence or absence of complementary DNA targets. Upper spectrum: blank (no target DNA present). Middle spectrum: in the presence of 0.5 μM non-complementary DNA (negative control). Lower spectrum: in the presence of 0.5 μM complementary target DNA (positive diagnostic).
Figure 5.
SERS spectra of the KI-67-MS nanoprobes in the presence or absence of complementary DNA targets. Upper spectrum: blank (no target DNA present). Middle spectrum: in the presence of 0.5 μM non-complementary DNA (negative control). Lower spectrum: in the presence of 0.5 μM complementary target DNA (positive diagnostic).
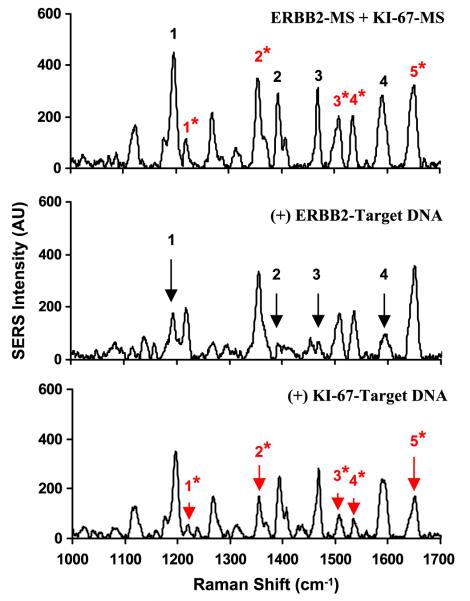
We demonstrated the multiplexing capability of the MS technique by mixing two different and separately prepared ERBB2-MS and KI-67-MS nanoprobes. According to the SERS signal of the individual ERBB2-MS and KI-67-MS nanoprobes shown in figure 2, the SERS intensity from the ERBB2-MS nanoprobes is relatively higher than that from the KI-67-MS nanoprobes. To obtain comparable SERS signals from both MS nanoprobes in the mixture, the ERBB2-MS and KI-67-MS nanoprobes were mixed with volume ratio of ERBB2-MS to KI-67-MS of 1:2 in 20 mM tris-HCl buffer. 5 mM MgCl2 were then added to the mixture sample and allowed to react at room temperature for at least 30 min. The upper spectrum in figure 6 represents the SERS spectrum of the mixture of the two MS nanoprobes. Note that when compared to the SERS spectra of the individual MS nanoprobes (shown in figure 2), all major Raman peaks of the Raman labels used for the ERBB2-MS and KI-67-MS nanoprobes can be easily identified in the new SERS spectrum obtained by mixing the two MS nanoprobes.
Figure 6.
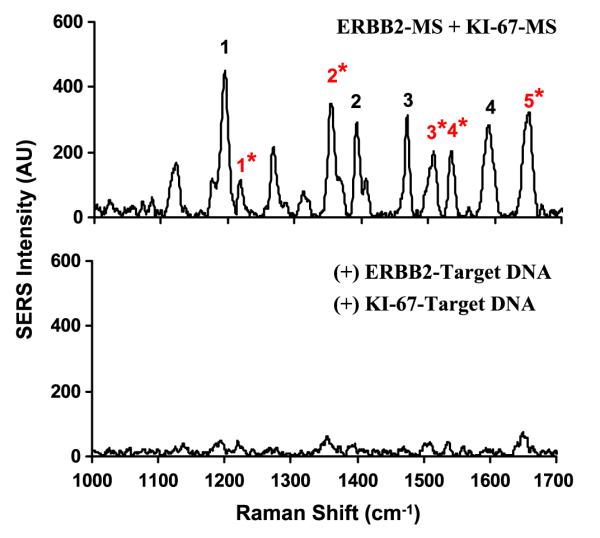
SERS spectra of the composite MS nanoprobes (ERBB2-MS + KI-67-MS) in the presence or absence of target DNA. The major Raman bands from ERBB2-MS are marked with black number, and the major Raman bands from KI-67-MS are marked with red number with (*) sign. Upper spectrum: blank (in the absence of any target DNA). Lower spectrum: in the presence of two target DNA complementary to both MS nanoprobes.
Demonstration of multiplex detection was performed in the presence of both target DNA sequences (0.5 μM for each target) in hybridization buffer, and allowed to react at 37 °C for 1 h. Figure 6 shows that the SERS signal from the composite MS nanoprobes was significantly quenched in the presence of both targets. The SERS intensity of all major Raman peaks (in the lower spectrum) was greatly decreased when both MS nanoprobes hybridized with their complementary DNA targets.
To further demonstrate the specificity and selectivity of the composite MS nanoprobes, the hybridization assays were then performed in the presence of individual complementary DNA target (i.e. only one of the two complementary targets). The middle and lower spectra in figure 7 show the resulted SERS signal from the composite MS nanoprobes targeted to 0.5 μM target DNA complementary to ERBB2-MS and KI-67-MS nanoprobes, respectively. The result indicates that only the SERS peaks associated with the complementary MS nanoprobes was significantly quenched (indicated by arrows) when in the presence of its target DNA. For example, in the middle spectrum only the SERS peaks 1–4, associated with the ERBB2-MS nanoprobes were quenched when the erbB-2 DNA target is present. In this case, only the ERBB2-MS nanoprobes were in an open state. In contrast, the SERS signal of the Raman peaks associated with the second MS nanoprobes (KI-67-MS) remained high, indicating that the second nanoprobes were in a closed state due to the absence of its DNA target. Note that the second MS nanoprobe can be designed to serve as an internal control in multiplex detection due to the specificity and selectivity of the MS technique.
Figure 7.
SERS spectra of the composite MS nanoprobes (ERBB2-MS + KI-67-MS) in the presence or absence of single target DNA. The major Raman bands from ERBB2-MS are marked with black number, and the major Raman bands from KI-67-MS are marked with red number with (*) sign. Upper spectrum: blank (in the absence of any target DNA). Middle spectrum: in the presence of single target DNA complementary to the ERBB2-MS nanoprobes. Lower spectrum: in the presence of single target DNA complementary to the KI-67-MS nanoprobes. The arrow signs illustrate the decreased SERS intensity of the major Raman bands in the presence of corresponding target DNA.
4. Conclusions
In conclusion, we have demonstrated for the first time the feasibility of using the molecular sentinel technique for qualitative multiplex DNA detection in a homogeneous solution. In this paper, two separately prepared MS nanoprobes were mixed and used to detect specific DNA sequences associated with erbB-2 and ki-67 breast cancer biomarkers present in the sample solution. The results of this study demonstrate the specificity and selectivity of the MS nanoprobes, as well as the ability to use multiple MS nanoprobes for multiplexed DNA detection. Furthermore, the SERS measurements were performed immediately following the hybridization reactions using a homogeneous assay without washing steps, which greatly simplifies the assay procedures. The results of this study demonstrate that the MS nanoprobe technique can provide a useful tool for multiplexed DNA detection in a homogeneous solution for medical diagnostics and high-throughput bioassays.
Acknowledgments
This work was sponsored by the National Institutes of Health (Grant R01 EB006201). The authors acknowledge the assistance of Dr Quan Liu in data processing, including spectral smoothing and background removal.
References
- [1].Dehainault C, Lauge A, Caux-Moncoutier V, Pages-Berhouet S, Doz F, Desjardins L, Couturier J, Gauthier-Villars M, Stoppa-Lyonnet D, Houdayer C. Nucleic Acids Res. 2004;32:e139. doi: 10.1093/nar/gnh137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marras SAE, Kramer FR, Tyagi S. Genet. Anal.: Biomol. Eng. 1999;14:151–6. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- [3].Marras SAE, Tyagi S, Kramer FR. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- [4].Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Nature. 1996;382:609–11. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- [5].Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal. Chem. 2006;78:8313–8. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–9. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- [7].Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J. Am. Chem. Soc. 1998;120:1959–64. [Google Scholar]
- [8].Storhoff JJ, Elghanian R, Mirkin CA, Letsinger RL. Langmuir. 2002;18:6666–70. [Google Scholar]
- [9].Cao YC, Jin R, Thaxton CS, Mirkin CA. Talanta. 2005;67:449–55. doi: 10.1016/j.talanta.2005.06.063. [DOI] [PubMed] [Google Scholar]
- [10].Thompson DG, Enright A, Faulds K, Smith WE, Graham D. Anal. Chem. 2008;80:2805–10. doi: 10.1021/ac702403w. [DOI] [PubMed] [Google Scholar]
- [11].Dubertret B, Calame M, Libchaber AJ. Nat. Biotechnol. 2001;19:365–70. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- [12].Li H, Rothberg LJ. Anal. Chem. 2004;76:5414–7. doi: 10.1021/ac049173n. [DOI] [PubMed] [Google Scholar]
- [13].Ray PC, Fortner A, Griffin J, Kim CK, Singh JP, Yu H. Chem. Phys. Lett. 2005;414:259–64. [Google Scholar]
- [14].Zhang J, Malicka J, Gryczynski I, Lakowicz JR. Anal. Biochem. 2004;330:81–6. doi: 10.1016/j.ab.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536–40. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- [16].Faulds K, Smith WE, Graham D. Anal. Chem. 2004;76:412–7. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- [17].Jin R, Cao YC, Thaxton CS, Mirkin CA. Small. 2006;2:375–80. doi: 10.1002/smll.200500322. [DOI] [PubMed] [Google Scholar]
- [18].McCabe AF, Eliasson C, Prasath RA, Hernandez-Santana A, Stevenson L, Apple I, Cormack PAG, Graham D, Smith WE, Corish P. Faraday Discuss. 2006;132:303–8. doi: 10.1039/b506942d. [DOI] [PubMed] [Google Scholar]
- [19].Wabuyele MB, Vo-Dinh T. Anal. Chem. 2005;77:7810–5. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- [20].Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. J. Am. Chem. Soc. 2003;125:1643–54. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- [21].Alak AM, Vo-Dinh T. Anal. Chem. 1987;59:2149–53. doi: 10.1021/ac00144a030. [DOI] [PubMed] [Google Scholar]
- [22].Bello J, Stokes D, Vo-Dinh T. Anal. Chem. 1989;61:1779–83. [Google Scholar]
- [23].Enlow PD, Vo-Dinh T. Anal. Chem. 1986;58:1119–23. [Google Scholar]
- [24].Vo-Dinh T, Uziel M, Morrison AL. Appl. Spectrosc. 1987;41:605–10. [Google Scholar]
- [25].Allain LR, Vo-Dinh T. Anal. Chim. Acta. 2002;469:149–54. [Google Scholar]
- [26].Deckert V, Zeisel D, Zenobi R, Vo-Dinh T. Anal. Chem. 1998;70:2646–50. doi: 10.1021/ac971304f. [DOI] [PubMed] [Google Scholar]
- [27].Isola NR, Stokes DL, Vo-Dinh T. Anal. Chem. 1998;70:1352–6. doi: 10.1021/ac970901z. [DOI] [PubMed] [Google Scholar]
- [28].Vo-Dinh T, Allain LR, Stokes DL. J. Raman Spectrosc. 2002;33:511–6. [Google Scholar]
- [29].Vo-Dinh T, Houck K, Stokes DL. Anal. Chem. 1994;66:3379–83. doi: 10.1021/ac00092a014. [DOI] [PubMed] [Google Scholar]
- [30].Wabuyele MB, Yan F, Griffin GD, Vo-Dinh T. Rev. Sci. Instrum. 2005;76:063710. [Google Scholar]
- [31].Wang H-N, Yan F, Zhang Y, Vo-Dinh T. Proc. SPIE. 2008;6869:686903. [Google Scholar]
- [32].Vo-Dinh T, Yan F, Wabuyele MB. J. Raman Spectrosc. 2005;36:640–7. [Google Scholar]
- [33].Tan W, Wang K, Drake TJ. Curr. Opin. Chem. Biol. 2004;8:547–53. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- [34].Campion A, Kambhampati P. Chem. Soc. Rev. 1998;27:241–50. [Google Scholar]
- [35].Wolkow RA, Moskovits M. J. Chem. Phys. 1987;87:5858–69. [Google Scholar]
- [36].Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Phys. Rev. Lett. 1997;78:1667–70. [Google Scholar]
- [37].Nie S, Emory SR. Science. 1997;275:1102–6. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- [38].Menard S, Tagliabue E, Campiglio M, Pupa SM. J. Cell. Physiol. 2000;182:150–62. doi: 10.1002/(SICI)1097-4652(200002)182:2<150::AID-JCP3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- [39].Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- [40].Urruticoechea A, Smith IE, Dowsett M. J. Clin. Oncol. 2005;23:7212–20. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- [41].van Dierendonck JH. Cancer Res. 1989;49:2999–3006. [PubMed] [Google Scholar]
- [42].Müunzel P, Marx D, Köchel H, Schauer A, Bock KW. J. Cancer Res. Clin. Oncol. 1991;117:603–7. doi: 10.1007/BF01613296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pavelic ZP. Cancer Res. 1992;52:2597–602. [PubMed] [Google Scholar]
- [44].Leopold N, Lendl B. J. Phys. Chem. B. 2003;107:5723–7. [Google Scholar]
- [45].Culha M, Stokes DL, Griffin GD, Vo-Dinh T. Biosen. Bioelectron. 2004;19:1007–12. doi: 10.1016/j.bios.2003.09.006. [DOI] [PubMed] [Google Scholar]
- [46].Wang H, Li J, Liu H, Liu Q, Mei Q, Wang Y, Zhu J, He N, Lu Z. Nucleic Acids Res. 2002;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]