Abstract
Background
Almost 90% of all cases of congenital, non-syndromic, severe to profound inherited deafness display an autosomal recessive mode of transmission (DFNB forms). To date, 47 causal DFNB genes have been identified, but many others remain to be discovered. We report the study of two siblings born to consanguineous Algerian parents and affected by isolated, profound congenital deafness.
Method
Whole-exome sequencing was carried out on these patients after a failure to identify mutations in the DFNB genes frequently involved.
Results
A biallelic nonsense mutation, c.88C > T (p.Gln30*), was identified in EPS8 that encodes epidermal growth factor receptor pathway substrate 8, a 822 amino-acid protein involved in actin dynamics. This mutation predicts a truncated inactive protein or no protein at all. The mutation was also present, in the heterozygous state, in one clinically unaffected sibling and in both unaffected parents, and was absent from the other two unaffected siblings. It was not found in 120 Algerian normal hearing control individuals or in the Exome Variant Server database. EPS8 is an F-actin capping and bundling protein. Mutant mice lacking EPS8 (Eps8−/− mice), which is present in the hair bundle, the sensory antenna of the auditory sensory cells that operate the mechano-electrical transduction, are also profoundly deaf and have abnormally short hair bundle stereocilia.
Conclusion
This new DFNB form is likely to arise from abnormal hair bundles resulting in compromised detection of physiological sound pressures.
Keywords: Epidermal growth factor receptor pathway substrate 8, Congenital deafness, Whole-exome sequencing, Stereocilia bundle, Actin dynamics
Introduction
In developed countries, congenital hearing impairment is thought to have a genetic origin in more than 70% of cases [1]. In about 90% of all cases of non-syndromic (i.e. isolated), severe to profound congenital inherited deafness, the defect is transmitted in an autosomal recessive manner (autosomal recessive non-syndromic deafness or DFNB forms) [2]. Forty-seven causal genes have been identified for DFNB, but many more remain to be identified. Research of genes involved in severe to profound congenital deafness is particularly efficient in geographic regions in which consanguineous marriages are frequent, such as the southern and eastern Mediterranean Basin, including North Africa [3-5]. We thus collected dozens of families affected by severe to profound deafness from the Algerian province of Tiaret, in the Tell Atlas, to the south-west of Algiers (Figure 1A). We used screening tests for mutations or deletions affecting GJB2, which account for 30% to 50% of the congenital profound DFNB cases in countries located around the Mediterranean sea [6], and selected 40 DFNB families without GJB2 mutations identified, for further analysis. We report here on one of these families, in which we identified a new causative deafness gene.
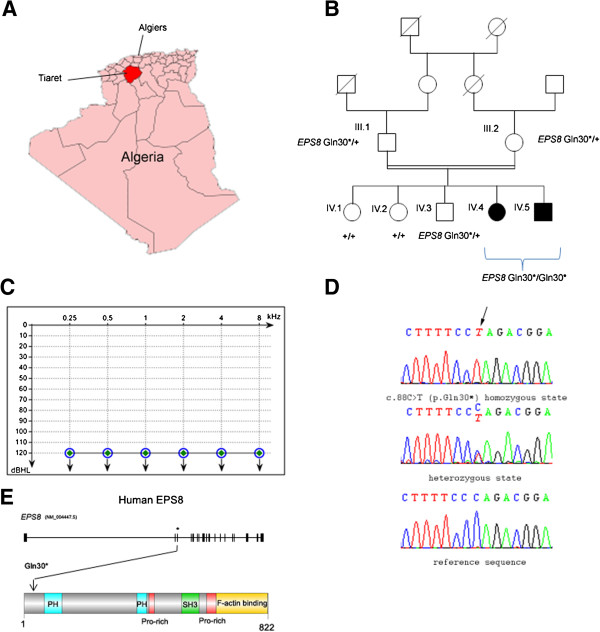
Figure 1.
Clinical and molecular data in the patients harbouring the nonsense mutation in EPS8. (A) Map of Algeria showing the province of Tiaret (in red). (B) Segregation of the nonsense EPS8 mutation in the family. (C) Air-conduction audiometric curves for patients IV.4 (open blue circles) and IV.5 (closed green diamonds) at the ages of 11 and 7 years, respectively. Identical audiometric curves were obtained for both ears in both patients. (D) DNA sequencing chromatograms showing the mutation (arrow). (E) Schematic representations of the human EPS8 gene and protein. * indicates the position of EPS8 exon 3. The protein (822 amino acids) contains a split pleckstrin homology (PH) domain (in blue), two proline-rich (Pro-rich) domains (in red), an SRC Homology 3 (SH3) domain (in green), and an F-actin-binding domain (in orange).
Materials and methods
Patients
This study was approved by the local Ethical Committees and the Committee for the Protection of Individuals in Biochemical Research as required by French legislation. Written consent for genetic testing was obtained from all family members.
Animals
Mouse cochleas were obtained as previously described [7]. Eyes were collected from adult Macaca fascicularis animals housed at the MIRcen platform (CEA/INSERM, Fontenay-aux-Roses, France). Experiments on animals were performed according to protocols approved by the Animal Use Committees of INSERM, CEA (for Macaca fascicularis), Institut Pasteur, and the ARVO Statement for the Use of Animals in Ophthalmological and Vision Research.
Auditory tests
All family members underwent pure-tone audiometry in a sound-proof room, with recording of air-conduction and bone-conduction thresholds. Air-conduction pure-tone average (ACPTA) threshold in the conversational frequencies (0.5, 1, 2 and 4 kHz) was measured for each ear, and its value for the best ear was used to define the severity of deafness: mild (20 dB < ACPTA ≤ 39 dB), moderate (40 dB < ACPTA ≤ 69 dB), severe (70 dB < ACPTA ≤ 89 dB), or profound (≥90 dB).
Exome sequencing and Sanger sequencing
Whole-exome sequencing and bioinformatic analysis were performed as previously described [7]. Specific oligonucleotides were designed to PCR amplify and sequence the 21 exons of EPS8 by the Sanger technique, using Primer3 (http://http/frodo.wi.mit.edu/primer3/) (Additional file 1: Table S1). EPS8-3 F forward primer: 5′-CGTGGTGATTATAGTGCATTGG-3′ and EPS8-3R reverse primer: 5′-ATCACTGCCTCATTCCAAAC-3′ were used to amplify and sequence EPS8 exon 3.
Immunofluorescence labeling
Dissection and preparation of mouse cochleas were performed as previously described [7]. Eyes obtained from deeply anaesthetized adult macaques perfused with paraformaldehyde, were dissected and processed as previously described [8]. Retinal cryosections were incubated in 10% bovine serum albumin in phosphate buffered saline (PBS) for 1 hour, incubated with the appropriate primary antibody overnight at 4°C, rinsed in PBS, incubated with the appropriate secondary antibody for 1 hour at room temperature, and rinsed again in PBS. The primary antibodies used were anti-EPS8 (BD Transduction Laboratories), anti-synaptophysin (MAB368, Millipore), anti-whirlin (G-8, Santa Cruz Biotechnology, Inc) mouse monoclonal antibodies, and anti-EPS8 (M-238, Santa Cruz Biotechnology, Inc) rabbit polyclonal antibody. Secondary antibody was Alexa Fluor 488 goat anti-rabbit IgG. TRITC-phalloidin (Sigma-Aldrich) and DAPI (1 μg/ml; Sigma-Aldrich) were used to label F-actin and cell nuclei, respectively.
Results and discussion
Patients IV.1 and IV.2, who are twelve and eight years old, respectively, were born to first-cousin parents (Figure 1B). Both patients, initially mute, were implanted two years ago. Auditory brainstem responses to click stimuli of various intensities were recorded: no response to these stimuli was detected. More specifically, an elevation of hearing thresholds beyond 120 dB was found for both ears, for pure tones at all frequencies analyzed (250 Hz to 8000 Hz; Figure 1C) indicating profound deafness. Temporal bone CT scan revealed no cochleo-vestibular malformations. The patients did not start walking late (they both started walking at nine months) and did not have any balance problems. Clinical examination failed to detect additional symptoms indicating a syndromic form of deafness, and neither proteinuria nor hematuria was observed. Both parents and the three other siblings did not show any sign of hearing impairment, and audiometric tests carried out on the parents and one of the siblings revealed an absence of hearing threshold elevation for all frequencies tested.
We first sought mutations in the genes most frequently implicated in DFNB: OTOF and MYO15A. No mutations of these genes were found in the two patients. We therefore carried out whole-exome sequencing, with pooled genomic DNA from the two affected children, as previously described [7]. We excluded variants present in the dBSNP132, 1000 genomes, and HapMap databases, with a prevalence greater than 0.01%. As the parents were consanguineous, we hypothesized that the causal mutation would be present in the homozygous state in the patients. We first searched for sequence variants of the coding and non-coding exons and of splice sites in the known DFNB and DFNA (autosomal dominant non-syndromic deafness) genes. No such variants were found (none were detected in the heterozygous state either). We then focused on nonsense, missense, splice site and frame-shifting mutations in all genes, by seeking insertions/deletions and SNPs. By using the same criteria as above, the number of sequence variants found decreased from 6268 to 0 for insertions/deletions (total number of insertion/deletion variants found in this pooled DNA), and from 74007 to 2 for SNPs. One of these biallelic variants was a conservative missense mutation, c.1110A > C (p.E730D), in BNC2 (gene ID: 54796, NM_01763), predicted to be nonpathogenic by PolyPhen-2, SIFT, and Mutation Taster software. The other was a nonsense mutation, c.88C > T (p.Gln30*), in exon 3 of EPS8 (gene ID: 2059, NM_004447.5). An analysis of the segregation of this mutation, based on sequencing of the corresponding DNA fragments in the parents, the two affected children and three unaffected siblings, confirmed the biallelic mutation in the two deaf children and showed that the mutation was present in the heterozygous state in both parents and one sibling, but not in the other two siblings (Figure 1B, D). This mutation was not present in 120 Algerian control individuals or in the Exome Variant Server database. The mutation is expected to result either in an abortive protein truncated at amino acid position 29 or in no protein at all due to nonsense mediated mRNA decay [9]. It is therefore functionally equivalent to the knock-out of the orthologous gene in Eps8−/− mice, which are also profoundly deaf [10,11]. We thus concluded that this mutation is responsible for the profound congenital deafness in the patients. The 39 remaining DFNB families were also investigated for the presence of mutations in EPS8, and we did not find any.
EPS8 is a 822 amino acids protein composed of a pleckstrin homology (PH) domain, a SRC Homology 3 (SH3) domain and a F-actin binding domain (Figure 1E). The C-terminal region has significant sequence homology with the sterile-motif (SAM) domains [12]. Overexpression of EPS8 is associated with some human cancers [13]. The three EPS8-like variants EPS8L1, EPS8L2, and EPS8L3, encoded by different genes, together with EPS8, constitute the EPS8 family of actin regulatory proteins. Of note, mutant mice lacking EPS8L2 are subject to progressive hearing loss [14].
The mechano-electrical transduction of sound stimuli takes place in the two types of auditory sensory cells, the inner and outer hair cells of the cochlea [15,16]. The transduction process involves mechanically gated ion channels located at the tips of specialized, stiff microvilli known as stereocilia. These stereocilia are organized into three rows of increasing height, and together they form the hair bundle projecting from the apical surface of the hair cell. The stereocilia have a cytoskeletal core composed of tightly packed actin filaments. Hair bundle development involves tight control of the differential elongation and thickening of stereocilia [17]. EPS8 is present at the tips of the stereocilia of the inner and outer hair cells (see Figure 2A-C), where it forms a tripartite molecular complex with MYO15A and the PDZ-domain containing protein whirlin [10], both of which are involved in the control of stereocilia length [10]. EPS8, a F-actin capping and also a cross-linking or bundling protein, is involved in F-actin dynamics [18], and knockout mice lacking EPS8 [10,11], like MYO15A- [19] or whirlin [20]-deficient mice, have abnormally short hair cell stereocilia. In addition, inner hair cells of Eps8−/− mice show a defective maturation [11].
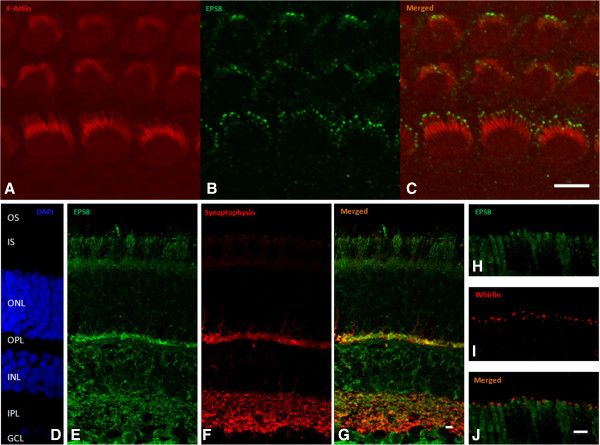
Figure 2.
Immunolocalization of EPS8 in the mouse cochlea and macaque retina. (A-C) Immunolabeling for F-actin (A) and EPS8 (B) in the mouse cochlea. DAPI (D) was used to label cell nuclei. Immunolabeling of EPS8 (E and H), synaptophysin (F) and whirlin (I) in the macaque retina. A polyclonal antibody was used to detect EPS8. (G) Co-immunolabeling of EPS8 and synaptophysin (J) Co-immunolabeling of EPS8 and whirlin. Abbreviations: OS, outer segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GNL, ganglion nuclear layer. Scale bars: 5 μm.
Because mutations of the whirlin gene, WHRN, can cause Usher syndrome of type 2 (USH2D), which associates retinitis pigmentosa to the hearing impairment, we looked for clinical features of retinal defect in the two patients. They did not present defective vision in low-light conditions and ophthalmologic examinations, including fundus autofluorescence, gave normal results (data not shown). As the patients are still young and the abnormal retinal phenotype of USH2D usually appears later in life than in other genetic forms of USH2 [21], we cannot definitively rule out the possibility of a syndromic deafness phenotype. We thus investigated the possible interaction of EPS8 and whirlin in the retina, by assessing the colocalization of these two proteins. Mouse models of Usher syndrome do not faithfully reproduce the visual impairment observed in humans. We therefore assessed colocalization in the adult macaque retina, by immunolabeling experiments based on a technique previously described [8]. Whirlin was detected at the junction between the inner and outer segments of the rod and cone photoreceptors, around the base of the connecting cilium, in the periciliary membrane complex region, as previously reported [8,22], whereas EPS8 was present in small amounts in the inner segment of rod photoreceptors, but was not colocalized with whirlin. By contrast, strong EPS8 immunolabeling was associated with the inner plexiform layer (consisting of the processes of bipolar cells, ganglion cells, and amacrine cells) and the outer plexiform layer of the retina (consisting of the processes of rods and cones, horizontal cells, and bipolar cells), in which it was colocalized with the presynaptic protein synaptophysin. The same EPS8 labeling was observed with two different antibodies (a monoclonal and a polyclonal antibody) directed against the protein (Figure 2 and data not shown). The lack of whirlin and EPS8 colocalization in macaque photoreceptor cells indicates that these two proteins, which interact directly in the hair bundles of cochlear hair cells, are unlikely to interact in the retina. These results are not consistent with mutations in EPS8 causing an Usher-like phenotype in patients, and instead identify this gene as responsible for a new autosomal recessive form of isolated deafness.
Conclusion
To conclude, we report for the first time the identification of one biallelic nonsense mutation in EPS8, in two children affected by profound congenital deafness. This new DFNB form is likely to arise from abnormal hair bundles resulting in compromised detection of physiological sound pressures.
Abbreviations
EPS8: Epidermal growth factor receptor pathway substrate 8; DFNB: Nonsyndromic deafness, autosomal recessive; DFNA: Nonsyndromic deafness, autosomal dominant.
Competing interest
The authors declare that they have no competing interests.
Supplementary Material
Primers for PCR amplification of EPS8 exons
Contributor Information
Asma Behlouli, Email: as_behlouli@yahoo.fr.
Crystel Bonnet, Email: crystel.bonnet@inserm.fr.
Samia Abdi, Email: samiasarabrayan@yahoo.fr.
Aïcha Bouaita, Email: aicha.bouaita@inserm.fr.
Andrea Lelli, Email: andrea.lelli@pasteur.fr.
Jean-Pierre Hardelin, Email: jean-pierre.hardelin@pasteur.fr.
Cataldo Schietroma, Email: cataldo.schietroma@inserm.fr.
Yahia Rous, Email: yahia.rous.2014@gmail.com.
Malek Louha, Email: malek.louha@trs.aphp.fr.
Ahmed Cheknane, Email: ahmed.cheknane.blida@gmail.com.
Hayet Lebdi, Email: hayet.lebdi.1960@gmail.com.
Kamel Boudjelida, Email: boudjelidak@yahoo.fr.
Mohamed Makrelouf, Email: mmakrelouf@gmail.com.
Akila Zenati, Email: akilaze@gmail.com.
Christine Petit, Email: christine.petit@pasteur.fr.
Acknowledgments
We thank the family members for their participation in this study. This work was supported by grants from the LHW-Stiftung, ERC grant “Hair bundle” (ERC-2011-AdG 294570), Foundation BNP Paribas, “Lifesenses” Labex, the Tassili Project and the Algerian government.
References
- van Camp G, Willems PJ, Smith RJ. Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet. 1997;60:758–764. [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13:245. doi: 10.1186/gb-2012-13-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford P, Ayadi H, Blanchard S, Chaib H, le Paslier D, Weissenbach J, Drira M, Petit C. A human gene responsible for neurosensory, non-syndromic recessive deafness is a candidate homologue of the mouse sh-1 gene. Hum Mol Genet. 1994;3:989–993. doi: 10.1093/hmg/3.6.989. [DOI] [PubMed] [Google Scholar]
- Ben Arab S, Hmani M, Denoyelle F, Boulila-Elgaied A, Chardenoux S, Hachicha S, Petit C, Ayadi H. Mutations of GJB2 in three geographic isolates from northern Tunisia: evidence for genetic heterogeneity within isolates. Clin Genet. 2000;57:439–443. doi: 10.1034/j.1399-0004.2000.570607.x. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel I, Mustapha M, Leibovici M, Verpy E, Goodyear R, Liu XZ, Nouaille S, Nance WE, Kanaan M, Avraham KB, Tekaia F, Loiselet J, Lathrop M, Richardson G, Petit C. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci U S A. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abidi O, Boulouiz R, Nahili H, Ridal M, Alami MN, Tlili A, Rouba H, Masmoudi S, Chafik A, Hassar M, Barakat A. GJB2 (connexin 26) gene mutations in Moroccan patients with autosomal recessive non-syndromic hearing loss and carrier frequency of the common GJB2-35delG mutation. Int J Pediatr Otorhinolaryngol. 2007;71:1239–1245. doi: 10.1016/j.ijporl.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Delmaghani S, Aghaie A, Michalski N, Bonnet C, Weil D, Petit C. Defect in the gene encoding the EAR/EPTP domain-containing protein TSPEAR causes DFNB98 profound deafness. Hum Mol Genet. 2012;21:3835–3844. doi: 10.1093/hmg/dds212. [DOI] [PubMed] [Google Scholar]
- Sahly I, Dufour E, Schietroma C, Michel V, Bahloul A, Perfettini I, Pepermans E, Estivalet A, Carette D, Aghaie A, Ebermann I, Lelli A, Iribarne M, Hardelin JP, Weil D, Sahel JA, El-Amraoui A, Petit C. Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J Cell Biol. 2012;199:381–399. doi: 10.1083/jcb.201202012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Manor U, Disanza A, Grati M, Andrade L, Lin H, di Fiore PP, Scita G, Kachar B. Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol. 2011;21:167–172. doi: 10.1016/j.cub.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampini V, Ruttiger L, Johnson SL, Franz C, Furness DN, Waldhaus J, Xiong H, Hackney CM, Holley MC, Offenhauser N, Di Fiore PP, Knipper M, Masetto S, Marcotti W. Eps8 regulates hair bundle length and functional maturation of mammalian auditory hair cells. PLoS Biol. 2011;9:e1001048. doi: 10.1371/journal.pbio.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, di Fiore PP, Scita G. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6:1180–1188. doi: 10.1038/ncb1199. [DOI] [PubMed] [Google Scholar]
- Maa M-C, Leu T-H. EPS8, an adaptor protein acts as an oncoprotein in human cancer. Carcinogenesis, Dr Kathryn Tonissen (Ed) 2013. ISBN: 978-953-51-0945-7, InTech, DOI: 10.5772/54906. Available from: http://www.intechopen.com/books/carcinogenesis/eps8-an-adaptor-protein-acts-as-an-oncoprotein-in-human-cancer.
- Furness DN, Johnson SL, Manor U, Ruttiger L, Tocchetti A, Offenhauser N, Olt J, Goodyear RJ, Vijayakumar S, Dai Y, Hackney CM, Franz C, Di Fiore PP, Masetto S, Jones SM, Knipper M, Holley MC, Richardson GP, Kachar B, Marcotti W. Progressive hearing loss and gradual deterioration of sensory hair bundles in the ears of mice lacking the actin-binding protein Eps8L2. Proc Natl Acad Sci U S A. 2013;110:13898–13903. doi: 10.1073/pnas.1304644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettiplace R, Hackney CM. The sensory and motor roles of auditory hair cells. Nat Rev Neurosci. 2006;7:19–29. doi: 10.1038/nrn1828. [DOI] [PubMed] [Google Scholar]
- Hudspeth AJ. How the ear’s works work: mechanoelectrical transduction and amplification by hair cells. C R Biol. 2005;328:155–162. doi: 10.1016/j.crvi.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch T, Endlich K, Giese T, Buchler MW, Schmidt J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007;255:205–218. doi: 10.1016/j.canlet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, Rump A, Hardisty RE, Blanchard S, Coimbra RS, Perfettini I, Parkinson N, Mallon AM, Glenister P, Rogers MJ, Paige AJ, Moir L, Clay J, Rosenthal A, Liu XZ, Blanco G, Steel KP, Petit C, Brown SD. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- Audo I, Bujakowska K, Mohand-Said S, Tronche S, Lancelot ME, Antonio A, Germain A, Lonjou C, Carpentier W, Sahel JA, Bhattacharya S, Zeitz C. A novel DFNB31 mutation associated with Usher type 2 syndrome showing variable degrees of auditory loss in a consanguineous Portuguese family. Mol Vis. 2011;17:1598–1606. [PMC free article] [PubMed] [Google Scholar]
- van Wijk E, van der Zwaag B, Peters T, Zimmermann U, te Brinke H, Kersten FF, Marker T, Aller E, Hoefsloot LH, Cremers CW, Cremers FP, Wolfrum U, Knipper M, Roepman R, Kremer H. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet. 2006;15:751–765. doi: 10.1093/hmg/ddi490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for PCR amplification of EPS8 exons