Abstract
There is a long history for the bioorganic and biomedical use of N-methyl-pyrrole-derived polyamides (PAs) that are higher homologs of natural products such as distamycin A and netropsin. This work has been pursued by many groups, with the Dervan and Sugiyama groups responsible for many breakthroughs. We have studied PAs since about 1999, partly in industry and partly in academia. Early in this program, we reported methods to control cellular uptake of polyamides in cancer cell lines and other cells likely to have multidrug resistance efflux pumps induced. We went on to discover antiviral polyamides active against HPV31, where SAR showed that a minimum binding size of about 10 bp of DNA was necessary for activity. Subsequently we discovered polyamides active against two additional high-risk HPVs, HPV16 and 18, a subset of which showed broad spectrum activity against HPV16, 18 and 31. Aspects of our results presented here are incompatible with reported DNA recognition rules. For example, molecules with the same cognate DNA recognition properties varied from active to inactive against HPVs. We have since pursued the mechanism of action of antiviral polyamides, and polyamides in general, with collaborators at NanoVir, the University of Missouri-St. Louis, and Georgia State University. We describe dramatic consequences of β-alanine positioning even in relatively small, 8-ring polyamides; these results contrast sharply with prior reports. This paper was originally presented by JKB as a Keynote Lecture in the 2nd International Conference on Medicinal Chemistry and Computer Aided Drug Design Conference in Las Vegas, NV, October 2013.
Introduction
Polyamides (PAs) that recognize and bind the minor groove of DNA have been studied extensively by a number of groups, including those of Dervan [1–3], Sugiyama [4–7], Lee [8–11], Laemmli [12–14], Kodadek [15,16] and others [17–19]. Over the course of that extensive work, a set of binding rules was developed primarily by the Dervan group to allow prediction and control of polyamide-DNA interactions in the minor groove [3,20–34].
We have been engaged in several collaborative N-methylpyrrole/ N-methylimidazole (Py/Im) polyamide projects over the years [35–42]. One project involved design of polyamides to repress cyclooxygenase-2 (COX-2) gene expression by targeting the binding site of ETS (E26 transformation specific) transcription control superfamily member Ets-1 in the COX-2 promoter, followed by study of the detailed thermodynamics of interactions between active PAs and their Ets-1 target [37]. Another project discovered, and is developing, antiviral agents for high-risk, cancer-causing Human Papillomavirus (HPV), and also encompasses understanding the mechanism of action of these antiviral compounds [36,38,41]. During the course of these projects, we have made and studied the chemical, biological and biophysical properties of a number of polyamides of different sizes, from six to twenty-six heterocyclic rings. Especially for larger compounds, many of our results have not followed the reported rules of PA-DNA recognition, so we have pursued the DNA recognition properties of our compounds with chemical, biochemical and biophysical methods [35,37,39,43]. We found excellent antiviral efficacy in human cell and tissue culture with compounds exceeding MW of 3000 [36,38,41], and note that Sugiyama and colleagues have reported no difference in cellular uptake of polyamides from 400–4000 in MW, as long as Im content was kept constant [44].
Testing the effectiveness of polyamides against HPV16, 18 and 31 in cell culture led to some surprising findings, in addition to a number of inventions [41]. In that work, monolayers of human keratinocytes and organotypic raft tissue cultures were used- both support maintenance of high-risk, cancer-causing HPV DNA, a circular, double-stranded molecule of about 8 kb. Levels of viral DNA in monolayer cultures were monitored 48 h after PA treatment using qPCR; viral DNA was measured relative to vehicle-treated controls in dose-dependent experiments with PAs that allowed calculation of IC50 values. The raft cultures were studied for much longer times (up to 19 days after PA treatment in published reports [41], and longer in unpublished studies at NanoVir). Of note, only relatively long polyamides showed antiviral activity, i.e. those compounds expected to bind approximately a full turn or more of DNA. In addition, only a subset of these long polyamides showed activity against HPV. Furthermore, there was a hierarchy of anti-HPV activity: more compounds were active against HPV31 than against HPV16, and more compounds were active against HPV16 than against HPV18. To date, all compounds active against HPV18 were active against both HPV16 and HPV31, and all compounds active against HPV16 were active against HPV31 [41], Small chemical changes to PA structure caused huge differences in activity, even when the nominal DNA recognition sites of the molecules were the same. Therefore, in order to determine the actual binding sites and affinities of the long, antiviral polyamides to wild-type HPV DNA sequences, we employed a variety of methods, including DNase I footprinting [45–48], affinity cleavage (AC) [28,49,50] and capillary electrophoresis (CE) [29,43,45,51–54]. To carry out the footprinting, we studied a number of DNA molecules, about 300–523 base pairs (bp) in length, that are part of the AT-rich, approximately 8 thousand bp double-stranded DNA (dsDNA) genomes of HPV16 and 18 [38,55–62].
Because we observed unexpected effects of β-alanine (β) positioning on antiviral efficacy, and because we had also previously seen unexpected consequences of positional β effects [37,39,41,63], we decided to investigate these effects in more detail with biophysical studies. Solubility limitations prevented some experiments from being carried out on long polyamides, so we worked with smaller polyamides, a series of 6- and 8-ring molecules with their own inherent interest because they are directed to the COX-2 Ets-1 binding site [37,39]. Note that β is reported to be a good substitute for N-methylpyrrole building blocks, one that generally improves or maintains the binding constant between polyamide and DNA [63–70]. Furthermore, incorporation of β or a related “molecular spring” [66] is necessary when working with long polyamides, in order to keep good alignment between hydrogen bonding groups on polyamides with those in the minor groove of DNA. In this area of research, it is therefore common to refer to a polyamide as having eight “rings” even if one or more of those rings has been replaced with β.
Methods
Synthetic chemistry
Synthesis and purification of polyamides was carried out by literature methods [71]. Compounds were characterized by HPLC/ms (ESI+), HRMS, and 500 or 600 MHz 1H NMR, 13C NMR and 2-D NMR techniques [37,39–42].
Biosensor surface plasmon resonance (SPR)
Measurements were performed with Biacore T100 and T200 SPR sensor systems as described [39]. A biotinylated hairpin DNA was attached to sensor chips functionalized with streptavidin. The DNA had the sequence 5'-biotin-CCTTGGCTTCTTTTGAAGCCAAAGG-3', where the bold, underlined region is the dsDNA recognition site for PAs KA1002, KA1007, and KA1055 and the italic T4 region forms the hairpin loop. DNA binding of PAs KA1063 and KA1065, and prior studies of KA1002 and KA1007, were described by a combination of orthogonal methods using several DNA targets; the methods included a novel fluorescence assay [40], SPR as described above and quantitative DNase I footprinting analyzed by CE [51], and these results were reported in concert with other colleagues [37].
Results
One striking result is the relative antiviral activity against HPV16 of four molecules which are isomeric or nearly isomeric. These molecules are NV1020, NV1023, NV1028, and NV1030 as shown below, with their different internal β positions in bold text. The only other difference between these four compounds is at the γ turn. Please note that with over 100 PA compounds tested against HPV16, 18 and 31, we have not seen any advantage or systematic effect of using the chiral γ turn (R)- 2,4-diaminobutyric acid, abbreviated as γ(NH2), in place of γ itself, even though the chiral reagent is reported to impart numerous beneficial properties on shorter PAs (including elimination of the reverse binding mode which we discuss below, and show in Figure 1) [72]. Here, we follow the literature naming conventions for PAs, where residues are referred to by their parent N-methylpyrrole and N-methylimidazole amino acid building blocks. Thus, dIm is the N-terminal des-amino-N-methyl imidazole-2- carboxylic acid, Py is 4-amino-2-carboxy-N-methyl pyrrole, Im is 4-amino-2-carboxy-N-methyl imidazole, γ is gamma-aminobutyric acid, which forms a hairpin turn for polyamides, Dp is dimethylaminopropylamine, NMe2CH2CH2CH2NH2, and Ta is bis(aminopropyl)N-methylamine, NMe(CH2CH2CH2NH2)2:
Figure 1.
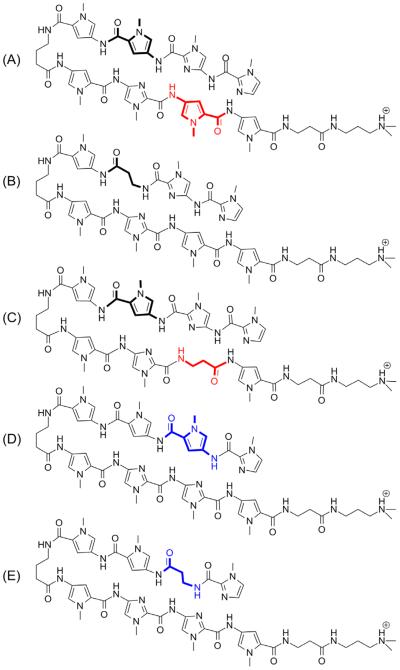
(A) Presumptive NV1028 Polyamide Match Site in the normal, forward binding mode and the corresponding reverse orientation; W = A or T. (B) DNA Sequence and affinity cleavage data (arrows) from a section (7770–7805 bp) of the HPV18 Long Control Region bound to NV1028. The relative heights of the arrows indicate relative affinity cleavage intensities. In the PA diagrams, by convention, Im or dIm is a filled circle, Py is an open circle, β is a diamond, Ta is +), and the γ-turn is a curved line.
In the case of amine tails Ta and. Dp, we have found better anti-HPV activity for Ta analogs much of the time, but there are a number of cases where the Dp analog is more active than its Ta counterpart, so there is no systematic preference for one amino tail over the other. A lack of preference was also found in the antiviral properties of PAs derived from γ and γ(NH2), as will be detailed elsewhere. NV1020, NV1023, NV1028 and NV1030 all recognize and bind identical DNA sites according to literature predictions (with W= A or T, the binding sites are either WGW7 or WWGW7, depending on which binding model one prefers) [24, 27]. However, the antiviral IC50 and IC90 values for these four compounds against HPV16, given in Table 1, fall into two dramatically different groups.
Table 1.
IC50 and IC90 values against HPV16 are shown for nearly isomeric polyamides NV1020, NV1023, NV1028 and NV1030, all with the same putative DNA recognition motif.
| Compound | IC50 HPV16 (μM) | IC90 HPV16 (μM) | n (replicates) |
|---|---|---|---|
| NV1020 | 5 | >>10 | 4 |
| NV1023 | 5 | 10 | 2 |
| NV1028 | 0.10 | 1.3 | 4 |
| NV1030 | 0.13 | 1.3 | 4 |
Interestingly, all compounds in Table 1 are relatively ineffectual against HPV18. For example, NV1028 has anIC50 of 0.7 μM and an IC90> 10 μM against HPV18, while NV1030 has an IC50 of 0.42 μM and an IC90> 10 μM against HPV18. Clearly, we wanted to understand the basis of these differences for such apparently degenerate PAs.
In order to map the position of NV1028 binding on HPV16 and 18, we performed DNase I footprinting following by capillary electrophoresis (CE), and also carried out affinity cleavage/CE [28,49–51] by attaching Fe(II) EDTA to the primary amine of the Ta group using literature methods to give NV1028-Fe (EDTA) [49,68]. A large study on the binding of NV1028 to HPV16 DNA is currently in press [73]. Using affinity cleavage, we identified fourteen binding sites for NV1028 in a section of the HPV18 including part of the LCR, spanning nucleotides 7647 to 157 of the circular, double-stranded DNA viral genome. The complete picture of PA-DNA binding is rather complex for discussion here, so we have provided a subset of data for two overlapping binding sites, a “forward” binding site with one mismatch and a “reverse” binding site with no mismatches (Figure 1A and B). By “forward orientation” we use the standard PA terminology where the N-C direction of the PA hairpin is oriented along the 5'–3' direction of the DNA. For the reverse orientation, the N-C direction of the hairpin is oriented along the 3'–5' direction of the DNA [74].
The dissociation constants for NV1028 at two HPV18 binding sites (1 and 2) are given in Table 2 and those same two binding sites are illustrated in Figure 1. The Kd values were determined by measuring the protection of the DNA from DNase I digestion as a function of varying NV1028 concentration. To keep an appropriate DNA:PA ratio, the lowest DNA concentration used in the experiment was 200 pM, which is at least five times smaller than the smallest dissociation constant calculated [37,40,48,51,68,75–78].
Table 2.
Dissociation constants from Langmuir and Hill models for NV1028 bound to a section of the HPV18 Long Control Region (LCR).
| Site # | Nucleotide span | Kd (Langmuir) | Kd (Hill) | Hill Coefficient |
|---|---|---|---|---|
| 1 | 7782 – 7791 | 2.2(5) nM | 1.9(4) nM | 1.7 |
| 2 | 7788 – 7797 | 2.4(8) nM | 1.7(5) nM | 2.0 |
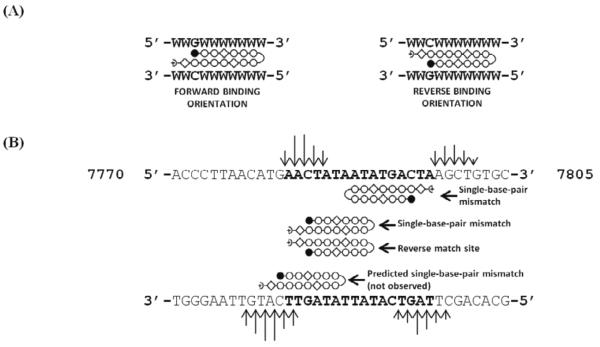
Showing the top strand only, site #1 is 5'-AACTATAATA-3', which is either a perfect match site for NV1028 with reverse binding, or a single mismatch site for NV1028 with forward binding; both possibilities are illustrated in Figure 1B. The perfect match, reverse orientation with the N-terminal Im at the 3' end of the bottom DNA strand and C-terminal Py-γ at the 5' end of the bottom DNA strand is further emphasized by Scheme 1:
Scheme 1.
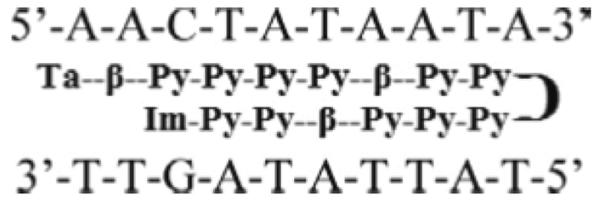
Site #2 is best described as a single mismatch site that is bound by NV1028 in the forward direction (Figure 1B). Thus, for site #2 we have 5'-AATATGACTA-3', where the bold nucleotide indicates the mismatch (in this case, a Py/Py pair is matched with a GC base pair, causing a clash between the minor groove hydrogen bond donor from G and a ring CH, or amide hydrogen bond donor, associated with one of the Py groups). Sites 1 and 2 overlap and cannot likely be occupied by NV1028 simultaneously, but are occupied independently in the ensemble of DNA molecules present under experimental conditions.
Affinity cleavage helps determine orientation of the PA bound to DNA and we used this approach to probe the orientations of the binding sites noted in Table 2 and Figure 1. The predicted forward and reverse PA binding sites for NV1028 are shown in Figure 1A. Affinity cleavage results for NV1028-Fe (EDTA) are shown in Figure 1B, along with PA orientations most consistent with those results. In affinity cleavage, hydroxyl radicals are generated by Fe(EDTA), a Fenton reagent, attached to the C-terminal Ta group of NV1028 through an amide bond. The radicals cleave DNA in a distance-dependent manner, and the different cleavage intensities reflect that distance dependence. The particular helical nature of DNA gives a familiar 3' stagger to the cleavage patterns when cleavage occurs in the minor groove, which is the site of PA binding [79]. These comments help explain the appearance of Figure 1B.
In order to show the raw data used to generate both DNase I and affinity cleavage information by capillary electrophoresis, examples are given in Figure 2A–C.
Figure 2.

Raw capillary electropherograms for DNAse I Footprinting and Affinity Cleavage with NV1028; the target DNA is the HPV18 LCR, nucleotides 7766–7805. (A) Shows the control DNase I fragmentation (0 nM polyamide); (B) illustrates the region protected upon addition of 5 nM NV1028; C shows affinity cleavage where polyamide binds (the black bar above the sequence).
To study the role of internal β residues in DNA recognition and binding thermodynamics, we turned to several series of smaller compounds that are more amenable to a number of thermodynamic techniques than the large, antiviral PAs. Some of these compounds are shown in Figure 3A–E. There are two parent compounds with all internal heterocycles, KA1002 and KA1063. Figure 3A shows KA1002, which has PA sequence dIm-Im-Py-Py-γ-Py-Im-Py-Py-β-Dp, and Figure 3D shows KA1063 with PA sequence dIm-Py-Py-Py-γ-Py-Im-Im-Py-β-Dp. In a series of KA1002 analogs, two compounds were made that replaced one or the other bold Py shown in Figure 3 with β. The first analog shown is KA1007, Figure 3B, where the bold pyrrole in the top strand of hairpin KA1002 was replaced with β, and the second analog is KA1055, shown in Figure 3C, where the bold red Py of KA1002's bottom strand was replaced with β. For additional comparison of the positional effects of β on PA-DNA binding parameters, the bold, blue Py of KA1063 (Figure 3D) was replaced to give KA1065 (Figure 3E).
Figure 3.
Compounds (A) KA1002; (B) KA1007; (C) KA1055; (D) KA1063; and (E) KA1065.
In contrast to many literature reports [63–70], we found that the replacement of Py by β can have a significantly deleterious effect on PA-DNA binding. Figure 4 contains the relevant SPR data and global kinetic fits of that data. Thus, KA1002 binds DNA with a Kd of 0.3–1.1 nM, depending on conditions (i.e. whether binding isotherms are obtained by fluorescence measurements of dye-labeled DNA in solution, by competition experiments for PA between labeled and unlabeled DNA [37,40] or by SPR at the solid-liquid interface [35,37,39]), but this weakens under those same experimental conditions (SPR, fluorescence or fluorescence competition [35,37,39]) to 35 - 83.4 - 325 nM upon substituting just one Py of the top strand with β to give KA1007 (Figure 3) [37,39]. In contrast, substitution of a Py on the bottom strand with β, giving KA1055, barely alters Kd at all, with a measured value of 0.9 nM [39]. Similarly, the all heterocyclic KA1063 has a Kd of 0.71 nM, but this increases to 70 or 106 nM, depending on measurement conditions, when a single Py is replaced with β to give KA1065 [35,37,39]. The reasons for these strong β effects could not be dissected into thermodynamic components ΔH and TΔS because of solubility limitations and aggregation of the polyamides in isothermal calorimetry experiments, but we hope that new molecular designs will overcome these difficulties. Different aggregation states at different concentrations are likely the cause of the variations in Kd measured by the various methods mentioned here. A slightly smaller, 6-ring PA proved sufficiently soluble to be amenable to complete and fruitful thermodynamic analyses [35,39].
Figure 4.
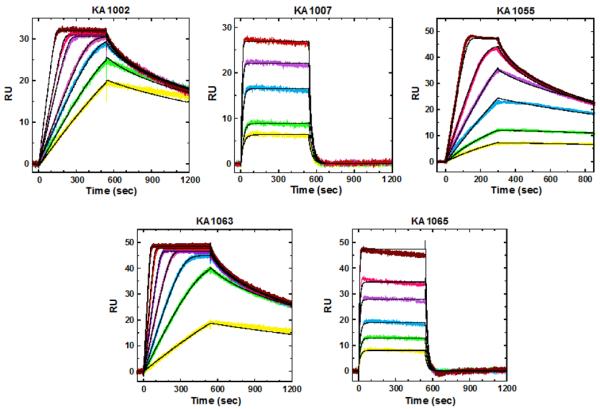
SPR sensorgrams (color) and the global kinetics fits (black overlays) for the interaction of PAs (Figure 3) with the 5'-biotin labeled hairpin DNA sequence, 5'-biotin-CCTTGGCTTCTTTTGAAGCCAAGG-3' [37, 39]. The concentrations from bottom to top are: 1.5, 2, 2.5, 3, 5, 6, 9 nM for KA1002; 5, 10, 20, 40, 60 nM for KA1007; 2, 3, 4, 6, 8, 15 nM for KA1055; 1, 2, 4, 6, 10, 15, 25 and 10, 20, 40, 100, 200, 400 nM for KA1063 and KA1065, respectively.
Discussion
We have found that our long, antiviral polyamides are much more promiscuous at binding DNA than predicted by well-established rules of PA-DNA recognition. This promiscuity is based, we believe, on a large number of favorable enthalpic interactions such that one or more unfavorable interactions can be tolerated with no significant detriment to Kd. We have shown here that single base pair mismatches can still have Kd values in the single digit nM region, similar to perfect match sites. Data on double and triple mismatches have been submitted for publication.
We should point out that the concept of “the binding constant to HPV16 or HPV18” has turned out to have no meaning, because we have discovered a large number of binding sites for various PAs on each HPV genome, essentially all with different binding constants, some with forward orientations, some with reverse orientations, some with orientations yet to be distinguished, and many with one or two mismatches. Furthermore, certain DNA sequences which we expected to bind particular PAs, e.g. NV1028, based on literature rules do not bind NV1028 at all. The details of these complex results will appear shortly for part of the HPV16 genome [73]. However, to date our results have covered about 50% of the HPV16 genome and about 20% of the HPV 18 genome with 1 – 4 polyamides, and the binding sites and affinities in those genomes do not yet show any clear preference or higher affinity for active vs. inactive PAs. Furthermore, antiviral activity does not correlate with the Kd values for PA-DNA binding events that we have characterized. In addition, we found poor correlation for reportedly degenerate aspects of PA sequences, such as β-alanine placement, with anti-HPV activity.
We do not yet have a molecular-level picture of the specific in vivo impact on the negatively supercoiled HPV episome of a large number of bound PA molecules, or how many PA molecules are bound per episome in a given cell. However, we started out to bind very specific viral DNA regions involved in replication and binding viral proteins and ended up with molecules able to bind many more sites on the 8 kb viral genome than expected. It is certainly tempting to invoke this promiscuous binding as a partial explanation of the broad-spectrum anti-HPV activity discovered for a subset of active compounds [41]. We will soon be able to compare the binding patterns for NV1028 on HPV16 and HPV18 DNA over quite a wide range of their genomes, to see if this sheds light on the relatively poor anti-HPV18 activity for this compound and dramatically better anti-HPV18 activity for related compounds.
Although the Py for β exchange is reported in a great many cases to either improve or leave unchanged the Kd for PA-DNA interactions and to improve properties of intermolecular DNA alkylation efficiency by reactive PAs [63–70], we have found quite a number of cases where swapping β for Py is highly damaging to PA-DNA binding interactions. Most of these studies, whether our groups or by other groups, have been done on relatively similar, 6–8-ring polyamides. This set of seemingly contradictory results is certainly puzzling, and we don't suggest that any prior results from other groups are incorrect. We instead conclude that it may not be possible yet to write completely general rules for PA-DNA recognition when β is involved. Why would this be the case? The β-alanine unit has a very different hydrophobic surface area and set of possible shapes than the PA heterocycles Py and Im, and β may be particularly sensitive to local changes in minor groove width and other DNA structural parameters that are (1) governed by DNA sequence context and (2) altered by complexes between DNA and minor groove-binders [80,81]. In fact, we have already begun to show such sequence context-driven effects to be important for PA-DNA recognition [39]. Therefore, although the small PAs we studied are quite far from our active antiviral compounds in molecular weight, there are definite parallels between 8 ring PAs and 14-ring or larger PAs in the currently unpredictable nature of how Py/β substitutions affect biological and biochemical activities. We hope to expand the understanding of molecular recognition with polyamides so it reaches the levels we have begun to expect from the literature.
Conclusions
Recent work has discovered an entirely novel mechanism of action for polyamides as antiviral agents [36]. That work, largely by CF and TGE, used gene expression arrays, siRNA libraries and pharmacology to show how active antiviral polyamides elicit a DNA Damage Response (DDR) to destroy viral DNA when used to treat HPV-positive, precancerous cells [36]. No such response was triggered by inactive polyamides, nor was the equivalent DDR elicited in HPV-negative cells or in cells having integrated HPV. Evidence was also found of HPV episomal DNA in multiple, unusual super coiled states. These findings lead us to suggest that the interaction of polyamides with viral DNA sequences in the context of a small circular episome is the key to triggering events that ultimately result in viral DNA elimination. An important challenge now is to find the molecular and biophysical links between polyamide-DNA interactions (or other polyamide interactions) and activation of the DDR. We have established that the location of β-alanine building blocks can be much more complex than the literature led us to predict. These complications caused β depending on positioning of this aliphatic building block, and had large consequences for (a) the biophysical parameters of 8-ring polyamides binding to DNA [35,37,39,40] and (b) the ability of nearly identical 14-ring polyamides to show antiviral activity in human cell and tissue culture [36,38,41] against HPV16, 18 and 31.
Acknowledgments
The authors thank NIH NIAID AI083803 to JKB, and AI062182, AI068159, AI062182 and AI068159 to CF for support, AI064200 to WDW, as well as NSF MRI 0959360 for 600 MHz NMR. JKB thanks NanoVir for providing equipment loans for this project. We would like to thank the reviewers for helpful comments.
Footnotes
Publisher's Disclaimer: This article was originally published in a journal by OMICS Publishing Group, and the attached copy is provided by OMICS Publishing Group for the author's benefit and for the benefit of the author's institution, for commercial/research/educational use including without limitation use in instruction at your institution, sending it to specific colleagues that you know, and providing a copy to your institution's administrator.
All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution's website or repository, are requested to cite properly.
Disclosure JKB and CF hold major ownership positions in NanoVir, LLC.
References
- 1.Li BC, Montgomery DC, Puckett JW, Dervan PB. Synthesis of cyclic Py-Im polyamide libraries. J Org Chem. 2013;78:124–133. doi: 10.1021/jo302053v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, et al. Modulation of NF-κB-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci USA. 2012;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho J, Parks ME, Dervan PB. Cyclic polyamides for recognition in the minor groove of DNA. Proc Natl Acad Sci USA. 1995;92:10389–10392. doi: 10.1073/pnas.92.22.10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawamoto Y, Bando T, Kamada F, Li Y, Hashiya K, et al. Development of a new method for synthesis of tandem hairpin pyrrole-imidazole polyamide probes targeting human telomeres. J Am Chem Soc. 2013;135:16468–16477. doi: 10.1021/ja406737n. [DOI] [PubMed] [Google Scholar]
- 5.Zhang HF, Wu YL, Jiang SK, Wang P, Sugiyama H, et al. Recognition by Nonaromatic and Stereochemical Subunit-Containing Polyamides of the Four Watson-Crick Base Pairs in the DNA Minor Groove. Chem Bio Chem. 2012;13:1366–1374. doi: 10.1002/cbic.201200137. [DOI] [PubMed] [Google Scholar]
- 6.Yasuda A, Noguchi K, Minoshima M, Kashiwazaki G, Kanda T, et al. DNA ligand designed to antagonize EBNA1 represses Epstein-Barr virus-induced immortalization. Cancer Sci. 2011;102:2221–2230. doi: 10.1111/j.1349-7006.2011.02098.x. [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara T, Tao ZF, Saito I, Sugiyama H. Synthesis of pyrrole-imidazole-duocarmycin polyamide and its sequence selective DNA alkylation. Nucleic Acids Symp Ser. 1999:249–250. doi: 10.1093/nass/42.1.249. [DOI] [PubMed] [Google Scholar]
- 8.Satam V, Babu B, Chavda S, Savagian M, Sjoholm R, et al. Novel diamino imidazole and pyrrole-containing polyamides: Synthesis and DNA binding studies of mono- and diamino-phenyl-ImPy*Im polyamides designed to target 5'-ACGCGT-3'. Bioorg Med Chem. 2012;20:693–701. doi: 10.1016/j.bmc.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Mackay H, Brown T, Sexton JS, Kotecha M, Nguyen B, et al. Targeting the inverted CCAAT Box-2 of the topoisomerase IIalpha gene: DNA sequence selective recognition by a polyamide-intercalator as a staggered dimer. Bioorg Med Chem. 2008;16:2093–2102. doi: 10.1016/j.bmc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 10.Lacy ER, Le NM, Price CA, Lee M, Wilson WD. Influence of a terminal formamido group on the sequence recognition of DNA by polyamides. J Am Chem Soc. 2002;124:2153–2163. doi: 10.1021/ja016154b. [DOI] [PubMed] [Google Scholar]
- 11.Lacy ER, Cox KK, Wilson WD, Lee M. Recognition of T*G mismatched base pairs in DNA by stacked imidazole-containing polyamides: surface plasmon resonance and circular dichroism studies. Nucleic Acids Res. 2002;30:1834–1841. doi: 10.1093/nar/30.8.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeshima K, Janssen S, Laemmli UK. Specific targeting of insect and vertebrate telomeres with pyrrole and imidazole polyamides. EMBO J. 2001;20:3218–3228. doi: 10.1093/emboj/20.12.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen S, Durussel T, Laemmli UK. Chromatin opening of DNA satellites by targeted sequence-specific drugs. Mol Cell. 2000;6:999–1011. doi: 10.1016/s1097-2765(00)00099-x. [DOI] [PubMed] [Google Scholar]
- 14.Janssen S, Cuvier O, Müller M, Laemmli UK. Specific gain- and loss-of-function phenotypes induced by satellite-specific DNA-binding drugs fed to Drosophila melanogaster. Mol Cell. 2000;6:1013–1024. doi: 10.1016/s1097-2765(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Kodadek T. Investigation of the relative cellular permeability of DNA-binding pyrrole-imidazole polyamides. J Med Chem. 2009;52:4604–4612. doi: 10.1021/jm9002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Yu P, Alluri PG, Kodadek T. Simple reporter gene-based assays for hairpin poly(amide) conjugate permeability and DNA-binding activity in living cells. Mol Bio syst. 2005;1:307–317. doi: 10.1039/b511514k. [DOI] [PubMed] [Google Scholar]
- 17.Correa BJ, Canzio D, Kahane AL, Reddy PM, Bruice TC. DNA sequence recognition by Hoechst 33258 conjugates of hairpin pyrrole/imidazole polyamides. Bioorg Med Chem Lett. 2006;16:3745–3750. doi: 10.1016/j.bmcl.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 18.Blackledge MS, Melander C. Programmable DNA-binding small molecules. Bioorg Med Chem. 2013;21:6101–6114. doi: 10.1016/j.bmc.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao J, Yuan G, Huang W, Chan AS, Lee KL. A convenient method for the synthesis of DNA-recognizing polyamides in solution. J Org Chem. 2000;65:5506–5513. doi: 10.1021/jo000135n. [DOI] [PubMed] [Google Scholar]
- 20.Muzikar KA, Meier JL, Gubler DA, Raskatov JA, Dervan PB. Expanding the repertoire of natural product-inspired ring pairs for molecular recognition of DNA. Org Lett. 2011;13:5612–5615. doi: 10.1021/ol202285y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbach AR, Dervan PB. Toward rules for 1:1 polyamide:DNA recognition. Proc Natl Acad Sci USA. 2001;98:4343–4348. doi: 10.1073/pnas.081070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen DH, Szewczyk JW, Baird EE, Dervan PB. Alternative heterocycles for DNA recognition: an N-methylpyrazole/N-methylpyrrole pair specifies for A.T/T.A base pairs. Bioorg Med Chem. 2001;9:7–17. doi: 10.1016/s0968-0896(00)00219-4. [DOI] [PubMed] [Google Scholar]
- 23.Pilch DS, Poklar N, Baird EE, Dervan PB, Breslauer KJ. The thermodynamics of polyamide-DNA recognition: hairpin polyamide binding in the minor groove of duplex DNA. Biochemistry. 1999;38:2143–2151. doi: 10.1021/bi982628g. [DOI] [PubMed] [Google Scholar]
- 24.Derva PB, Burli RW. Sequence-specific DNA recognition by polyamides. Curr Opin Chem Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 25.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 26.Kielkopf CL, Baird EE, Dervan PB, Rees DC. Structural basis for G.C recognition in the DNA minor groove. Nat Struct Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 27.White S, Baird EE, Dervan PB. On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem Biol. 1997;4:569–578. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- 28.White S, Baird EE, Dervan PB. Effects of the A.T/T.A degeneracy of pyrrole--imidazole polyamide recognition in the minor groove of DNA. Biochemistry. 1996;35:12532–12537. doi: 10.1021/bi960744i. [DOI] [PubMed] [Google Scholar]
- 29.Taylor RD, Asamitsu S, Takenaka T, Yamamoto M, Hashiya K, et al. Sequence-Specific DNA Alkylation Targeting for Kras Codon 13 Mutation by Pyrrole–Imidazole Polyamide seco-CBI Conjugates. Chemistry. 2014;20:1310–1317. doi: 10.1002/chem.201303295. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Minoshima M, Sugiyama H. Base pair recognition of the stereochemically alpha-substituted gamma-turn of pyrrole/imidazole hairpin polyamides. J Am Chem Soc. 2006;128:14905–14912. doi: 10.1021/ja064369l. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Bando T, Sugiyama H. Discrimination of hairpin polyamides with an alpha-substituted-gamma-aminobutyric acid as a 5'-TG-3' reader in DNA minor groove. J Am Chem Soc. 2006;128:8766–8776. doi: 10.1021/ja0580587. [DOI] [PubMed] [Google Scholar]
- 32.Bando T, Narita A, Iwai A, Kihara K, Sugiyama H. C-H to N substitution dramatically alters the sequence-specific DNA alkylation, cytotoxicity, and expression of human cancer cell lines. J Am Chem Soc. 2004;126:3406–3407. doi: 10.1021/ja0387103. [DOI] [PubMed] [Google Scholar]
- 33.Bando T, Narita A, Saito I, Sugiyama H. Molecular design of apyrroleimidazole hairpin polyamides for effective DNA alkylation. Chemistry. 2002;8:4781–4790. doi: 10.1002/1521-3765(20021018)8:20<4781::AID-CHEM4781>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Iida H, Sugiyama H. Recognition of DNA sequences by pyrrole-imidazole polyamide: Rational drug design in the post-genome era. Yuki Gosei Kagaku Kyokaishi. 2000;58:975–987. [Google Scholar]
- 35.Wang S, Kumar A, Aston K, Nguyen B, Bashkin JK, et al. Wilson, Different thermodynamic signatures for DNA minor groove binding with changes in salt concentration and temperature. Chem. Commun. (Camb) 2013;49:8543–8545. doi: 10.1039/c3cc44569k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards TG, Vidmar TJ, Koeller K, Bashkin JK, Fisher C. DNA damage repair genes controlling human papillomavirus (HPV) episome levels under conditions of stability and extreme instability. PLoS One. 2013;8:e75406. doi: 10.1371/journal.pone.0075406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashkin JK, Aston K, Ramos JP, Koeller KJ, Nanjunda R, et al. Promoter scanning of the human COX-2 gene with 8-ring polyamides: Unexpected weakening of polyamide-DNA binding and selectivity by replacing an internal N-Me-pyrrole with ß-alanine. Biochimie. 2013;95:271–279. doi: 10.1016/j.biochi.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards TG, Helmus MJ, Koeller K, Bashkin JK, Fisher C. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J Virol. 2013;87:3979–3989. doi: 10.1128/JVI.03473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Nanjunda R, Aston K, Bashkin JK, Wilson WD. Correlation of local effects of DNA sequence and position of β-alanine inserts with polyamide-DNA complex binding affinities and kinetics. Biochemistry. 2012;51:9796–9806. doi: 10.1021/bi301327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupureur CM, Bashkin JK, Aston K, Koeller KJ, Gaston KR, et al. Fluorescence assay of polyamide-DNA interactions. Anal Biochem. 2012;423:178–183. doi: 10.1016/j.ab.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards TG, Koeller KJ, Slomczynska U, Fok K, Helmus M, et al. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 2011;91:177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crowley KS, Phillion DP, Woodard SS, Schweitzer BA, Singh M, et al. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg Med Chem Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 43.He G, Koeller KJ, Harris GD, Dupureur CM, Bashkin JK. DNA binding properties of a large antiviral polyamide via capillary electrophoresis. Abstracts of the 243rd American Chemical Society National Meeting; San Diego, CA, USA. 2012. 2012. BIOL-254. [Google Scholar]
- 44.Nishijima S, Shinohara K, Bando T, Minoshima M, Kashiwazaki G, et al. Cell permeability of Py-Im-polyamide-fluorescein conjugates: Influence of molecular size and Py/Im content. Bioorg Med Chem. 2010;18:978–983. doi: 10.1016/j.bmc.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Wilson DO, Johnson P, McCord BR. NonradiochemicalDNase I footprinting by capillary electrophoresis. Electrophoresis. 2001;22:1979–1986. doi: 10.1002/1522-2683(200106)22:10<1979::AID-ELPS1979>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 46.Floreancig PE, Swalley SE, Trauger JW, Dervan PB. Recognition of the Minor Groove of DNA by Hairpin Polyamides Containing a-Substituted-b-Amino Acids. J Am Chem Soc. 2000;122:6342–6350. [Google Scholar]
- 47.Neamati N, Mazumder A, Sunder S, Owen JM, Tandon M, et al. Highly potent synthetic polyamides, bisdistamycins, and lexitropsins as inhibitors of human immunodeficiency virus type 1 integrase. MolPharmacol. 1998;54:280–290. doi: 10.1124/mol.54.2.280. [DOI] [PubMed] [Google Scholar]
- 48.Dabrowiak JC, Goodisman J, Kissinger K. Thermodynamic data from drug-DNA footprinting experiments. Biochemistry. 1990;29:6139–6145. doi: 10.1021/bi00478a005. [DOI] [PubMed] [Google Scholar]
- 49.Farkas ME, Tsai SM, Dervan PB. Alpha-diaminobutyric acid-linked hairpin polyamides. Bioorg Med Chem. 2007;15:6927–6936. doi: 10.1016/j.bmc.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swalley SE, Baird EE, Dervan PB. Discrimination of 5`-GGGG-3`, 5`-GCGC-3`, and 5`-GGCC-3` Sequences in the Minor Groove of DNA by Eight-Ring Hairpin Polyamides. J Am Chem Soc. 1997;119:6953–6961. [Google Scholar]
- 51.He G, Vasilieva E, Bashkin JK, Dupureur CM. Mapping small DNA ligand hydroxyl radical footprinting and affinity cleavage products for capillary electrophoresis. Anal Biochem. 2013;439:99–101. doi: 10.1016/j.ab.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech. 2006;17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 54.Yindeeyoungyeon W, Schell MA. Footprinting with an automated capillary DNA sequencer. Biotechniques. 2000;29:1034–1041. doi: 10.2144/00295st05. [DOI] [PubMed] [Google Scholar]
- 55.Garner-Hamrick PA, Fisher C. HPV episomal copy number closely correlates with cell size in keratinocyte monolayer cultures. Virology. 2002;301:334–341. doi: 10.1006/viro.2002.1558. [DOI] [PubMed] [Google Scholar]
- 56.Stubenrauch F, Laimins LA. Human papillomavirus life cycle: active and latent phases. Semin Cancer Biol. 1999;9:379–386. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- 57.Meyers CM. Artificial system for the production of infectious human papillomavirus. Patent US 5994115 WO 1999004811A1. 1999
- 58.Lazarus P, Caruana S. Typing of common human papilloma virus strains by multiplex PCR. Anal Biochem. 1996;243:198–201. doi: 10.1006/abio.1996.0506. [DOI] [PubMed] [Google Scholar]
- 59.Laimins LA. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 60.Coutlee F, Bobo L, Abbass H, Dalabetta G, Hook NE, et al. Detection of HPV-16 in cell lines and cervical lavage specimens by a polymerase chain reaction-enzyme immunoassay assay. J Med Virol. 1992;37:22–29. doi: 10.1002/jmv.1890370105. [DOI] [PubMed] [Google Scholar]
- 61.Park JS, Rader JS, Wu TC, Laimins LA, Currie JL, et al. HPV-16 viral transcripts in vulvar neoplasia: preliminary studies. Gynecol Oncol. 1991;42:250–255. doi: 10.1016/0090-8258(91)90354-8. [DOI] [PubMed] [Google Scholar]
- 62.Bradford CR, Zacks SE, Androphy EJ, Gregoire L, Lancaster WD, et al. Human papillomavirus DNA sequences in cell lines derived from head and neck squamous cell carcinomas. Otolaryngol Head Neck Surg. 1991;104:303–310. doi: 10.1177/019459989110400304. [DOI] [PubMed] [Google Scholar]
- 63.Turner JM, Swalley SE, Baird EE, Dervan PB. Aliphatic/Aromatic Amino Acid Pairings for Polyamide Recognition in the Minor Groove of DNA. J Am Chem Soc. 1998;120:6219–6226. [Google Scholar]
- 64.Baird EE, Dervan PB. Solid Phase Synthesis of Polyamides Containing Imidazole and Pyrrole Amino Acids. J Am Chem Soc. 1996;118:6141–6146. [Google Scholar]
- 65.Herman DM, Baird EE, Dervan PB. Stereochemical Control of the DNA Binding Affinity, Sequence Specificity, and Orientation Preference of Chiral Hairpin Polyamides in the Minor Groove. J Am Chem Soc. 1998;120:1382–1391. [Google Scholar]
- 66.Trauger JW, Baird EE, Dervan PB. Extended hairpin polyamide motif for sequence-specific recognition in the minor groove of DNA. Chem Biol. 1996;3:369–377. doi: 10.1016/s1074-5521(96)90120-9. [DOI] [PubMed] [Google Scholar]
- 67.He G, Vasilieva E, Harris JGD, Koeller KJ, Bashkin JK, et al. Binding Studies of a Large Antiviral Polyamide to a Natural HPV Sequence. Biochimie. 2014 doi: 10.1016/j.biochi.2014.02.011. DOI:10.1016/j.biochi.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rucker VC, Melander C, Dervan PB. Influence of β-alanine on hairpin polyamide orientation in the DNA minor groove. Helvetica Chimica Acta. 2003;86:1839–1851. [Google Scholar]
- 69.Ramos JP, Babu B, Chavda S, Liu Y, Plaunt A, et al. Affinity and kinetic modulation of polyamide-DNA interactions by N-modification of the heterocycles. Biopolymers. 2013;99:497–507. doi: 10.1002/bip.22205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leblanc B, Moss T. DNase I footprinting. Methods Mol Biol. 2009;543:37–47. doi: 10.1007/978-1-60327-015-1_3. [DOI] [PubMed] [Google Scholar]
- 71.Vashisht Gopal YN, Van Dyke MW. Combinatorial determination of sequence specificity for nanomolar DNA-binding hairpin polyamides. Biochemistry. 2003;42:6891–6903. doi: 10.1021/bi027373s. [DOI] [PubMed] [Google Scholar]
- 72.Lee M, Shea RG, Hartley JA, Kissinger K, Pon RT, et al. Molecular recognition between oligopeptides and nucleic acids: sequence-specific binding of the naturally occurring antibiotic (4S)-(+)-anthelvencin A and its (4R)-(−) enantiomer to deoxyribonucleic acids deduced from proton NMR, footprinting, and thermodynamic data. J Am Chem Soc. 1989;111:345–354. [Google Scholar]
- 73.Taylor JS, Schultz PG, Dervan PB. DNA affinity cleaving : Sequence specific cleavage of DNA by Distamycin-EDTA - Fe(II) and EDTA-distamycin Fe(II) Tetrahedron. 1984;40:457–465. [Google Scholar]
- 74.Urbach AR, Love JJ, Ross SA, Dervan PB. Structure of a beta-alanine-linked polyamide bound to a full helical turn of purine tract DNA in the 1:1 motif. J Mol Biol. 2002;320:55–71. doi: 10.1016/S0022-2836(02)00430-8. [DOI] [PubMed] [Google Scholar]
- 75.Dervan PB, Urbach AR. The importance of β-alanine for recognition of the minor groove of DNA. Essays Contemp Chem. 2001:327–339. [Google Scholar]
- 76.Swalley SE, Baird EE, Dervan PB. Effects of γ -Turn and β-Tail Amino Acids on Sequence-Specific Recognition of DNA by Hairpin Polyamides. J Am Chem Soc. 1999;121:1113–1120. [Google Scholar]
- 77.Trauger JW, Baird EE, Mrksich M, Dervan PB. Extension of Sequence-Specific Recognition in the Minor Groove of DNA by Pyrrole-Imidazole Polyamides to 9–13 Base Pairs. J Am Chem Soc. 1996;118:6160–6166. [Google Scholar]
- 78.Han YW, Kashiwazaki G, Morinaga H, Matsumoto T, Hashiya K, et al. Effect of single pyrrole replacement with ß-alanine on DNA binding affinity and sequence specificity of hairpin pyrrole/imidazole polyamides targeting 5'-GCGC-3'. Bioorg Med Chem. 2013;21:5436–5441. doi: 10.1016/j.bmc.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Minoshima M, Bando T, Sasaki S, Fujimoto J, Sugiyama H. Pyrrole-imidazole hairpin polyamides with high affinity at 5'-CGCG-3' DNA sequence; influence of cytosine methylation on binding. Nucleic Acids Res. 2008;36:2889–2894. doi: 10.1093/nar/gkn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tevis DS, Kumar A, Stephens CE, Boykin DW, Wilson WD. Large, sequence-dependent effects on DNA conformation by minor groove binding compounds. Nucleic Acids Res. 2009;37:5550–5558. doi: 10.1093/nar/gkp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hunt RA, Munde M, Kumar A, Ismail MA, Farahat AA, et al. Induced topological changes in DNA complexes: influence of DNA sequences and small molecule structures. Nucleic Acids Res. 2011;39:4265–4274. doi: 10.1093/nar/gkq1362. [DOI] [PMC free article] [PubMed] [Google Scholar]