Abstract
Background
Tobacco smoking is the main risk factor of chronic obstructive pulmonary disease (COPD) but not all smokers develop the disease. An abnormal pulmonary and systemic inflammatory response to smoking is thought to play a major pathogenic role in COPD, but this has never been tested directly.
Methods
We studied the systemic biomarker and leukocyte transcriptomic response (Affymetrix microarrays) to smoking exposure in 10 smokers with COPD and 10 smokers with normal spirometry. We also studied 10 healthy never smokers (not exposed to smoking) as controls. Because some aspects of COPD may differ in males and females, and the inflammatory response to other stressors (infection) might be different in man and women, we stratified participant recruitment by sex. Differentially expressed genes were validated by q-PCR. Ontology enrichment was evaluated and interaction networks inferred.
Results
Principal component analysis identified sex differences in the leukocyte transcriptomic response to acute smoking. In both genders, we identified genes that were differentially expressed in response to smoking exclusively in COPD patients (COPD related signature) or smokers with normal spirometry (Smoking related signature), their ontologies and interaction networks.
Conclusions
The use of an experimental intervention (smoking exposure) to investigate the transcriptomic response of peripheral leukocytes in COPD is a step beyond the standard case-control transcriptomic profiling carried out so far, and has facilitated the identification of novel COPD and Smoking expression related signatures which differ in males and females.
Introduction
Tobacco smoking is the major risk factor for Chronic Obstructive Pulmonary Disease (COPD) [1]. Yet, only a proportion of smokers, so called “susceptible smokers”, develop the disease [2]. The genetic and epigenetic background of each smoker is likely to regulate the type and intensity of his/her inflammatory response to smoking [1], [3]–[5]. In “susceptible smokers”, this response is thought to be “enhanced”, both in the lungs [6] and in the systemic circulation [7], and is believed to drive disease progression [1], [6]. However, despite the wide acceptance of this notion [1], no previous study has actually studied the “response” to smoking (i.e., the specific inflammatory changes that occur before and after smoking) in susceptible smokers (i.e., patients with COPD) and resistant smokers (i.e., smokers with normal spirometry). Rather, available evidence compares a number of inflammatory markers in these two groups of smokers “after” many years of smoking exposure [6].
To address this gap in knowledge, we compared a number of systemic inflammatory biomarkers and the transcriptome of circulating leukocytes, before and after smoking in susceptible (COPD patients) and resistant smokers. We hypothesized that smoking exposure will induce a different inflammatory signature, at the cellular, protein and/or transcriptome levels, in these two groups of smokers. Importantly, because several previous reports suggest that there may be significant gender differences in the natural history of COPD [8]–[10] and some experimental observations show that the leukocyte transcriptional response to other acute stressors (infection) is different in males and females [11], we recruited participants stratified by sex.
Methods
Data S1 presents an extended explanation of the Methods used.
Design, Participants and Ethics
In this prospective and controlled study, we included 30 volunteers stratified by smoking history, presence of COPD [12] and sex. All COPD patients were clinically stable. The Ethics Committee the Hospital Clinic de Barcelona approved the study protocol and all participants signed their informed consent.
Acute Smoking Exposure Test (ASET)
Current smokers were asked to refrain from smoking for, at least, 8 hours before the ASET. Exhaled carbon monoxide concentration was measured (SC01-STK, CO SmokeCheck Monitor, Care Fusion, San Diego, US) to confirm smoking abstinence. All participants fasted overnight. A catheter was inserted in a peripheral vein and, based on previous studies in healthy subjects [13], [14], smokers were asked to smoke 2 cigarettes (Nobel tar content 9 mg, nicotine 0.7 mg and carbon monoxide 9 mg, Altadis S.A. Spain) in 30 minutes under the direct supervision of an investigator. Venous blood samples were obtained before (T0), 30 (T30) and 180 minutes (T180) after smoking. Plasma and buffy coat were separated immediately by centrifugation at 400 g×6 minutes. Plasma was stored at −70°C until analysis. Erythrocytes were lysed, leukocytes washed, lysed in RLT buffer and stored at −70°C for later RNA extraction. Participants remained at rest for the duration of the test.
Measurements
Forced spirometry
Forced spirometry was measured according to international recommendations [15]. Reference values correspond to a Mediterranean population [16].
Circulating inflammatory markers
Total white blood cells counts were quantified with an ADVIA-2120 system (Siemens, Germany). The plasma concentration of C-Reactive Protein (CRP) was determined by ultra-sensitive quantitative turbidimetric test (Bayer Diagnostics, Germany), and those of interleukin (IL)-6 (IL-6) and IL-8 by high sensitivity ELISA (Invitrogen, USA). Blood carboxyhemoglobin levels were determined with a Rapid Point 500 Blood-Gas-System (Siemens, Germany).
Leukocyte mRNA profile
RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, US) following manufacturer instructions. RNA integrity was assessed with an Agilent 2100 Bionalyzer (Agilent Technologies). All samples had a RIN above 8. The 60 RNA samples were hybridized to Affymetrix HG-U219 array plate, which enables the performance of up to 96 arrays at one time, following Affymetrix's protocols. Microarray data has been deposited in the GEO NCBI database (accession code GSE55962).
Array validation by real time PCR
Selected genes were subjected to real-time quantitative PCR (q-PCR) for independent confirmation of relative expression levels in each group using Taqman assays on demand, actin as a housekeeping gene, and relative quantification was performed with the 2(ΔCP) method [17].
Data analysis
qPCR and systemic biomarker results are summarized as mean ± standard deviation, proportions or range, as appropriate. Clinical variables were compared across groups using unpaired tests (Mann-Whitney, Kruskal-Wallis). Paired tests (Wilcoxon) were used to compare T0 and T180 results within each group.
Differential gene expression was investigated using Rank Prod [18]; probe-sets with FDR (False Discovery Rate) values<0.05 were considered significant. Principal components analysis (PartekH Pro) was used to explore visually global effects on transcription. DAVID (www.david.abcc.ncifcrf.gov) [19], [20] was used to estimate the functional over-representation of the significant genes in the Gene Ontology Biological Process database (www.geneontology.org). Interaction networks were constructed using Ingenuity Pathways Analysis (IPA, www.ingenuity.com) based on the Ingenuity Pathways Knowledge Base (IPKB) [21]. Cytoscape (www.cytoscape.org) was used to analyze and display correlations results graphically [22], [23].
Results
Participant Characteristics
Participants showed the differences in sex, smoking history and lung function expected from the stratified recruitment strategy of the study (Table 1). Of note: (1) age was similar in COPD patients and smokers with normal spirometry; (2) airflow limitation in COPD patients ranged from moderate to severe. Despite that it was similar in males and females with COPD, the latter were older (p = 0.02) and tended to have smoked less (p = 0.07); and (3) cumulative smoking exposure was higher in males with COPD than in male controls (p = 0.01), although this was not observed in females.
Table 1. Characterization of participants by sex (mean±SD).
| COPD | Smokers | Non Smokers | |
| N (male/female) | 9 (5/4) | 11 (6/5) | 10 (5/5) |
| Age , years | |||
| Male | 55±5 | 51±2 | 55±6 |
| Female | 68±3* | 58±8 | 58±5 |
| BMI, Kg/m2 | |||
| Male | 26.4±3.4 | 25.0±1.7 | 26.4±1.1 |
| Female | 29.2±1.6 | 24.3±4.2 | 27.5±6.2 |
| Pack/year | |||
| Male | 70±21§ | 24±14 | 0 |
| Female | 40±11 | 34±8 | 0 |
| FEV1/FVC, % | |||
| Male | 61±4.9§ | 77±4 | 81±2.2 |
| Female | 53±4.9§ | 74±2.7 | 78±3.9 |
| FEV1, % reference | |||
| Male | 64±12§ | 93±12 | 96±12 |
| Female | 63±14 | 84±11 | 88±12 |
Asterisk indicate p<0.05 between COPD males and females, § indicates p<0.05 between COPD patients and smoker controls.
Biomarker Response to Smoking
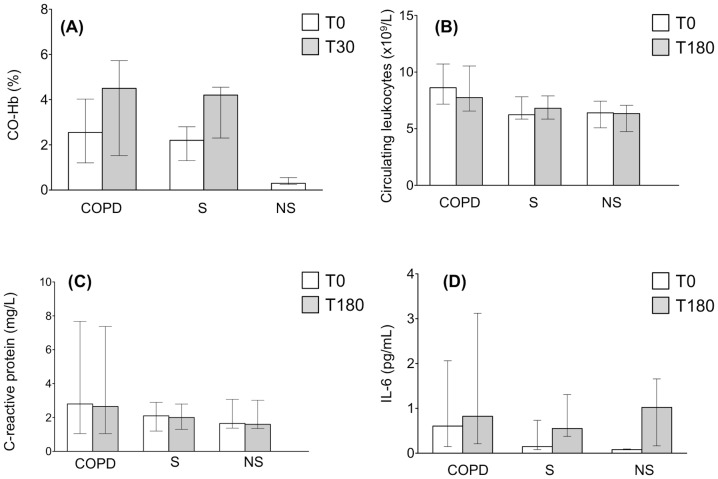
At baseline, carboxyhemoglobin levels were lower (p<0.001) in non-smokers (NS) than in smokers with normal spirometry (S) or with COPD, and they increased further thirty minutes after smoking (Figure 1, panel A). Circulating leukocyte counts at T0 were higher (p = 0.02) in COPD than in controls, and they did not change at T180 in any group (Figure 1, panel B); likewise, we did not observed any significant change after smoking in circulating neutrophils, lymphocytes (CD3+), CD4+ T cells, CD8+ T, CD19+ B cells or NK populations (data not shown). Mean CRP (Figure 2, panel C) and IL-6 levels (Figure 1, panel D) were within the normal range [7] in all groups and time points. IL-8 values were below the lower detection limit in all participants, both before and after smoking (data not shown). Finally, we did not observe significant sex differences in any of these biomarkers, either before or after smoking.
Figure 1. Median (and IQR) values of blood carboxy-hemoglobin (panel A), circulating leukocytes (panel B), C-reactive protein serum levels (panel C) and IL-6 (panel D) in COPD patients, smoker (S) and non-smoker (NS) controls, before (T0) and 30 min.
(T30) or 180 min. (T180) later. For further explanations, see text.
Figure 2.
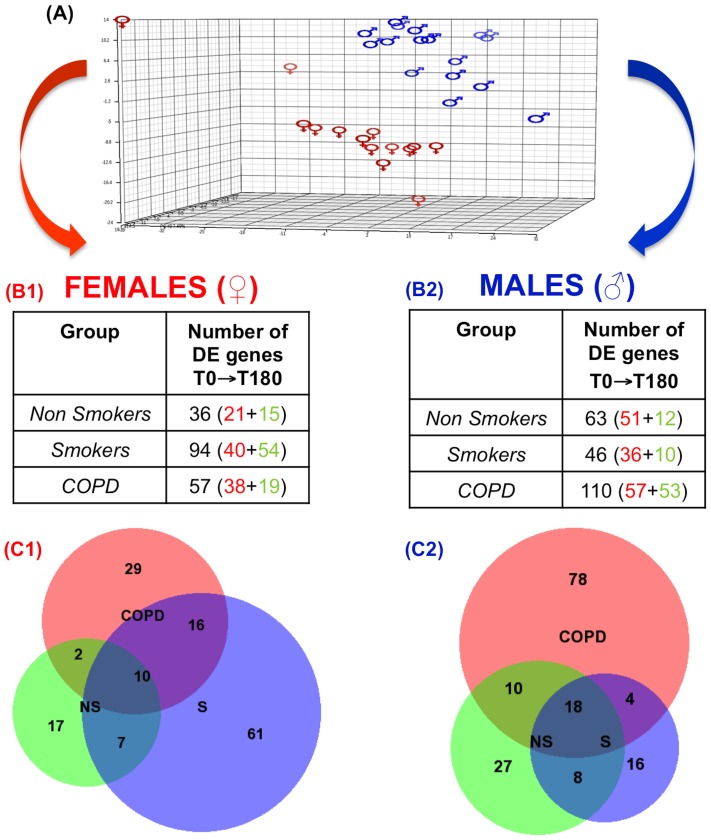
Panel A: Principal component analysis (PCA) of those genes with the largest variability of expression values across the experimental groups (COPD patients, smokers (S) and non-smokers (NS)) at baseline (T0). Because the sex effect observed, further analysis was stratified by sex (right and left columns.) Panels B1 (females) and B2 (males) present the number of differentially expressed (DE) genes from T0 to T180 in the three groups of participants studied. Red and green figures indicate up and down-regulation, respectively. Panels C1 (females) and C2 (males) show a BioVenn diagram (www.cmbi.ru.nl/cdd/biovenn) with the number of DE genes shared between COPD, S and NS controls. For further explanations, see text.
Leukocyte Transcriptomic Response to Smoking
Principal component analysis (PCA) allows the visualization of differences within a multidimensional data set. Here, a PCA of the expression values of those genes that show the largest variability across all experimental groups (COPD, S and NS) at baseline demonstrated a clear segregation of the leukocyte transcriptome profile by sex (Figure 2, Panel A). Hence, further statistical differential analyses were performed separately by gender, as previously described [24]. Figure 2 (Panels B1 and B2) presents the number of differentially expressed (DE) genes between T0 and T180 (red and green figures indicate up or down DE, respectively) in NS, S and COPD stratified by sex (Table S1 presents the complete list of DE genes). Overall, transcriptomic changes were of moderate intensity (log ratio ranged between 0.95 and 0.4). Unsupervised clustering of DE genes in COPD segregated T0 and T180 samples adequately, both in females (n = 57) and males (n = 110) (Figure S1, panels A and B, respectively) indicating that the Rank Prod statistical analysis used here identified clear gene signatures associated with acute smoking exposure.
Identification of COPD and Smoking related genes
BioVenn diagrams in Figure 2 present the number of DE genes from T0 to T180 that were shared (or not) between groups (NS, S and COPD), both in females (Panel C1) and males (Panel C2). Given that NS did not smoke, genes that were DE from T0 to T180 in this group (36 in females and 63 in males) were consider to represent time-dependent changes, and were therefore excluded from any further analysis investigating the response to smoking in S or COPD. Likewise, we operationally considered that those genes that were DE in response to smoking only in COPD patients represent COPD related genes, whereas those genes DE in response to smoking only in smokers with normal spirometry represent Smoking related genes. Accordingly, we identified 29 COPD related genes in females (78 in males), and 61 Smoking genes in females (16 in males) (Figure 2, Panels C1 and C2, respectively). Table 2 lists the top 10 susceptibility and resistance genes ordered by log ratio after smoking, direction of change and sex.
Table 2. Top 10 COPD a) and Smoking b) related genes ordered by log ratio after smoking, direction of change (up vs. down-regulation) and sex.
| a) | COPD males | COPD females | ||||||
| Up-regulated genes | n# | symbol | Gene Description | Log ratio | n# | symbol | Gene Description | Log ratio |
| 1 | SPAG9 | Sperm associated antigen 9 | 0.69 | 1 | ELMOD2 | ELMO/CED-12 domain containing 2 | 0.71 | |
| 2 | ZNF441 | Zinc finger protein 441 | 0.64 | 2 | ZFAND6 | Zinc finger, AN1-type domain 6 | 0.71 | |
| 3 | BIRC3 | Baculoviral IAP repeat containing 3 | 0.63 | 3 | SNX13 | Sorting nexin 13 | 0.7 | |
| 4 | S1PR1 | Sphingosine-1-phosphate receptor 1 | 0.62 | 4 | PAFAH1B1 | Platelet-activating factor acetylhydrolase 1b | 0.66 | |
| 5 | MGAT4A | Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | 0.61 | 5 | TMEM170B | Transmembrane protein 170B | 0.66 | |
| 6 | CLK1 | CDC-like kinase 1 | 0.6 | 6 | RABEP1 | Rabaptin, RAB GTPase binding effector protein 1 | 0.62 | |
| 7 | KLHL9 | Kelch-like family member 9 | 0.6 | 7 | CCPG1 | Cell cycle progression 1 | 0.61 | |
| 8 | SLFN5 | Schlafen family member 5 | 0.59 | 8 | TMEM41B | Transmembrane protein 41B | 0.6 | |
| 9 | ARRDC3 | Arrestin domain containing 3 | 0.59 | 9 | CLK1 | CDC-like kinase 1 | 0.58 | |
| 10 | SOCS4 | Suppressor of cytokine signaling 4 | 0.59 | 10 | MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 | 0.53 | |
| Down-regulated genes | n# | symbol | Gene Description | Log ratio | n# | symbol | Gene Description | Log ratio |
| 1 | HLX | H2.0-like homeobox | −0.79 | 1 | CXCL1 | Chemokine (C-X-C motif) ligand 1 | −0.94 | |
| 2 | TRIB1 | Tribbles homolog 1 | −0.79 | 2 | FOS | FBJ osteosarcoma oncogene | −0.68 | |
| 3 | CHST11 | Carbohydrate (chondroitin 4) sulfotransferase 11 | −0.72 | 3 | NT5C2 | 5′-nucleotidase, cytosolic II | −0.59 | |
| 4 | FLJ36031 | CCDC71L coiled-coil domain containing 71-like | −0.59 | 4 | BCL2A1 | BCL2-related protein A1 | −0.56 | |
| 5 | STK17B | Serine/threonine kinase 17 b (apoptosis-inducing) | −0.58 | 5 | RBBP6 | Retinoblastoma binding protein 6 | −0.54 | |
| 6 | CD24 | CD24 molecule | −0.58 | 6 | WIPF1 | WAS/WASL interacting protein family, member 1 | −0.53 | |
| 7 | SGK1 | Serum/glucocorticoid regulated kinase 1 | −0.57 | 7 | JUN | Jun proto-oncogene | −0.52 | |
| 8 | IL1R2 | Interleukin 1 receptor, type II | −0.56 | 8 | DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | −0.52 | |
| 9 | BCL2A1 | BCL2-related protein A1 | −0.52 | 9 | RBM6 | RNA binding motif protein 6 | −0.51 | |
| 10 | KYNU | Kynureninase | −0.52 | 10 | TRA2A | Transformer 2 alpha homolog (Drosophila) | −0.51 | |
Additionally, we investigated if baseline differences in gene expression between groups (S vs. COPD) and/or sex (male vs. female) influence the transcriptional response to smoking. Results are explained in detail in the data S1 (and Tables S2 and S3) but, by and large, DE at baseline was only marginally related to the transcriptomic response to smoking.
COPD and Smoking related genes shared by males and females
We identified five COPD related genes shared by males and females: (1) WIPF1 (mean log ratio = −0.479), an important regulator of the immunological synapsis formation and T cell activation[25]; (2) BCL2A1 (mean log ratio = −0.538), that regulates p53tumor suppression induced apoptosis [26]; (3) SGK1 (mean log ratio = −0.512), whose up regulation is involved in hypertension, obesity, diabetes, thrombosis, stroke, fibrosing disease, infertility and tumor growth, and its inhibition mediates skeletal muscle homeostasis and function [27]; (4) ZNF397 (mean log ratio = 0.551), thought to have a role in centromere formation and gene transcription [28]; and, finally, (5) CLK1 (mean log ratio = 0.589), an alternative splicing factor induced by hypoxia [29].
Similarly, we identified two Smoking related genes shared by males and females: POU2AF1 (log ratio = −0.40), a nuclear factor responsible of immunoglobulin transcription regulation [30], and XYLT1 (log ratio = 0.45), the initial enzyme responsible of GAG biosynthesis that has been associated with tissue remodeling [31].
Gene Ontologies Associated with COPD and Smoking related genes
We used DAVID [19], [20] to identify enriched gene ontologies (% FDR<25) associated with the COPD susceptibility and resistance genes identified above. In females, ontologies enriched in COPD related genes (n = 29) included genes that were mostly down regulated (Table 3). Of note, MLL5, JUN and FOS, were involved in most of the ontologies. It is known that FOS and JUN dimerize to form the transcription factor complex AP-1 a key player in the regulation of several biological processes [32]. In keeping with this we observed that enriched ontologies included regulation of transcription control, metabolism of nucleic acids and biosynthetic processes (Table 3) as well as ontologies related with the immune system, such as SMAD signaling and response to extracellular stimulus. On the other hand, ontologies enriched in Smoking related genes in females (n = 61) included both up and down-regulated genes, and were related with the immune system, cell cycle, cellular growth, apoptosis and cell locomotion (Table 4). The former included genes involved in T cell activation (ETS1 and TNFSF14) and antigen presentation control (CYBB and CST7).
Table 3. Gene ontology enrichment analysis, by sex. Females ontologies/genes associated with COPD.
| Term | Genes | % FDR | p value |
| GO:0009991 response to extracellular stimulus | FOS, MLL5, JUN, KLF4 | 7 | 5.20E-03 |
| GO:0045893 positive regulation of transcription, DNA-dependent | FOS, MLL5, JUN, RBM4, KLF4 | 9 | 6.74E-03 |
| GO:0051254 positive regulation of RNA metabolic process | FOS, MLL5, JUN, RBM4, KLF4 | 9 | 6.94E-03 |
| GO:0060395 SMAD protein signal transduction | FOS, JUN | 12 | 0.01 |
| GO:0045941 positive regulation of transcription | FOS, MLL5, JUN, RBM4, KLF4 | 15 | 0.01 |
| GO:0010628 positive regulation of gene expression | FOS, MLL5, JUN, RBM4, KLF4 | 17 | 0.01 |
| GO:0007611 learning or memory | FOS, JUN, PAFAH1B1 | 18 | 0.01 |
| GO:0045935 positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | FOS, MLL5, JUN, RBM4, KLF4 | 21 | 0.02 |
| GO:0051173 positive regulation of nitrogen compound metabolic process | FOS, MLL5, JUN, RBM4, KLF4 | 23 | 0.02 |
| GO:0010557 positive regulation of macromolecule biosynthetic process | FOS, MLL5, JUN, RBM4, KLF4 | 24 | 0.02 |
Bold italic text indicates up regulated genes and normal text indicates down regulated genes in response to smoking.
Table 4. Gene ontology enrichment analysis, by sex. Females ontologies/genes associated with Smoking.
| Term | Genes | % FDR | p value |
| GO:0006955 immune response | CYBB, POU2AF1, IL18RAP, CST7 , ETS1, BCL2, LILRB4, TGFBR3, TNFSF14, CLEC4D | 0.7 | 4.50E-04 |
| GO:0043065 positive regulation of apoptosis | ADRB2 , ETS1, MMP9 , KLF10, BCL2, TNFSF14, FADD, RRM2B | 0.9 | 5.70E-04 |
| GO:0043068 positive regulation of programed cell death | ADRB2 , ETS1, MMP9 , KLF10, BCL2, TNFSF14, FADD, RRM2B | 0.9 | 6.00E-04 |
| GO:0010942 positive regulation of cell death | ADRB2 , ETS1, MMP9 , KLF10, BCL2, TNFSF14, FADD, RRM2B | 0.9 | 6.10E-04 |
| GO:0051270 regulation of cell motion | ETS1, IL6ST, MMP9 , BCL2, TGFBR3 | 6 | 4.00E-03 |
| GO:0051272 positive regulation of cell motion | ETS1, IL6ST, MMP9 , BCL2 | 7 | 4.00E-03 |
| GO:0042127 regulation of cell proliferation | ADRB2, FOSL2 , ETS1, IL-6ST, KLF10, BCL2, CHST11, TGFBR3, ING1 | 7 | 5.00E-03 |
| GO:0042981 regulation of apoptosis | ADRB2 , ETS1, MMP9 , KLF10, BCL2, CHST11, TNFSF14, FADD, RRM2B | 8 | 5.00E-03 |
| GO:0043067 regulation of programmed cell death | ADRB2 , ETS1, MMP9 , KLF10, BCL2, CHST11, TNFSF14, FADD, RRM2B | 8 | 6.00E-03 |
| GO:0010941 regulation of cell death | ADRB2, ETS1, MMP9 , KLF10, BCL2, CHST11, TNFSF14, FADD, RRM2B | 9 | 6.00E-03 |
| GO:0045926 negative regulation of growth | ADRB2 , BCL2, CDA, ING1 | 9 | 6.00E-03 |
| GO:0005976 polysaccharide metabolic process | XYLT1 , IL6ST, CHST11, MGAM | 10 | 6.00E-03 |
| GO:0048643 positive regulation of skeletal muscle tissue development | ADRB2 , BCL2 | 15 | 0.01 |
| GO:0045844 positive regulation of striated muscle development | ADRB2 , BCL2 | 19 | 0.01 |
| GO:0048636 positive regulation of muscle development | ADRB2 , BCL2 | 19 | 0.01 |
| GO:0051094 positive regulation of developmental process | ADRB2 , ETS1, IL-6ST, KLF10, BCL2 | 21 | 0.01 |
Bold italic text indicates up regulated genes and normal text indicates down regulated genes in response to smoking.
On the other hand, in males COPD related genes (n = 78) were related to immune system, cellular locomotion and cellular activation ontologies (Table 5). Interestingly, some of these ontologies were similar to those identified in females as Smoking related ontologies but with an opposite DE sign (up vs. down-regulation). For instance, as shown in Table 5, genes involved in cell migration were mainly activated in response to smoking in males with COPD but mostly repressed in female S with normal lung function. The ontology enrichment analysis of Smoking related genes in males failed to obtain significant terms because of the small number of genes identified (n = 16) (Figure 2, panel C2).
Table 5. Gene ontology enrichment analysis, by sex. Males ontologies/genes associated with COPD.
| Term | Genes | % FDR | p value |
| GO:0006955 immune response | GPR183 , IL1R2, SH2D1A , KYNU, CLEC4E, THEMIS , SNCA, EOMES, TGFBR3 , CLEC4D, CD24 | 1 | 9.00E-04 |
| GO:0040012 regulation of locomotion | SPAG9, S1PR1 , SNCA, TGFBR3, CBLL1 , TRIB1 | 3 | 1.71E-03 |
| GO:0030334 regulation of cell migration | SPAG9, S1PR1, TGTBR3, CBLL1 , TRIB1 | 10 | 7.06E-03 |
| GO:0051270 regulation of cell motion | SPAG9, S1PR1, TGTBR3, CBLL1 , TRIB1 | 16 | 1.00E-02 |
| GO:0030098 lymphocyte differentiation | GRP183, THEMIS, EOMES , CD24 | 16 | 1.00E-02 |
| GO:0001824 blastocyst development | HOPX, EOMES, TGFBR3 | 23 | 2.00E-02 |
Bold italic text indicates up regulated genes and normal text indicates down regulated genes in response to smoking. Analysis could not be done for ontologies/genes associated with COPD resistance in males due to the small number of DE genes (n = 16) identified in smokers with normal spirometry.
Expression Correlation Networks of COPD and Smoking related Genes
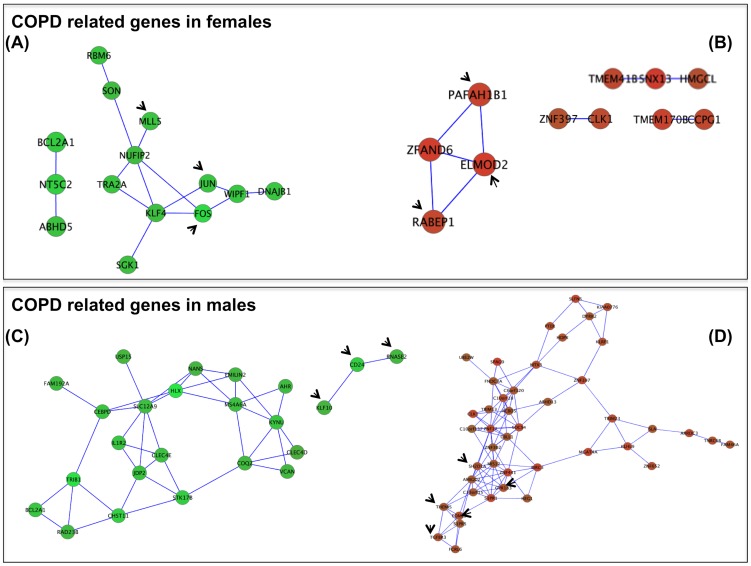
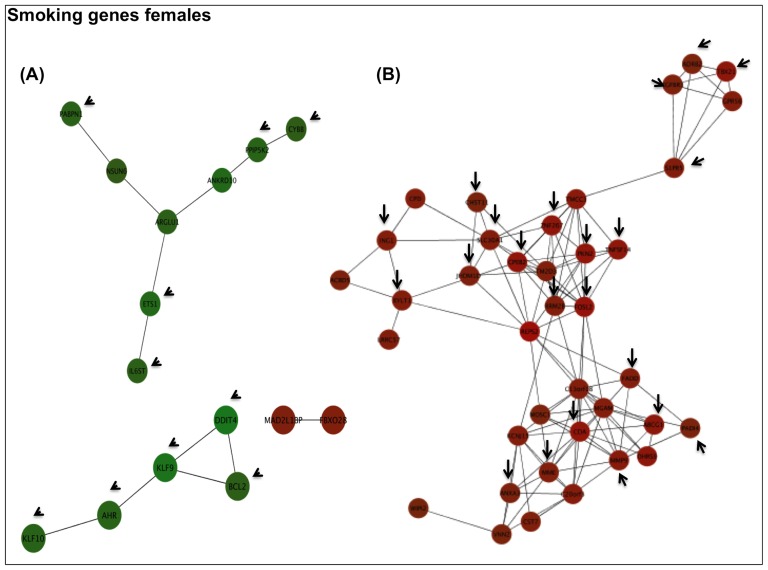
We used Cytoscape (plug-ins Expression Correlation and Bingo) [22] to explore if the expression of the COPD and Smoking related genes identified above were correlated (Pearson r≥0.8) and form networks that have coordinated functionality. We found that, in females, COPD related genes indeed formed several correlation networks of different size (Figure 3, panels A and B). Some of them included genes that form functional cores (arrows) because they belong to the same ontologies. For instance, MLL5, JUN and FOS define a functional core of (repressed) genes (Panel A) that belong to three ontologies: GO60395 (SMAD protein signal transduction, p value = 0.0001), GO6306 (DNA methylation, p value = 0.0003) and GO6305 (DNA alkylation, p value = 0.0003). On the other hand, PAFAH1B1, ELMOD2 and RABEP1 formed a functional core of up-regulated susceptibility genes (panel B) participating in GO16044 (cellular membrane organization, p = 0.001). Overall, these results confirm, expand and support our previous analysis using DAVID. Figure 4 depicts the correlation networks of Smoking related genes in females. Two down-regulated networks were identified containing ontologies related to GO44237 cellular metabolic process (p = 0.015) and GO42221 response to chemical stimulus (p = 0.0005) among others. A large positive correlation network was identified, containing several ontologies as GO7275 multicellular organismal development (p = 0.0039) and GO50794 regulation of cellular process (p = 0.006).
Figure 3. Expression correlation networks (Pearson r≥0.8) of COPD related genes in females (Panels A and B) and males (Panels C and D).
Red and green figures indicate up and down-regulation of expression, respectively. Arrows point genes forming functional clusters identified in the network. For further explanations, see text.
Figure 4. Expression correlation networks (Pearson r≥0.8) of Smoking genes in females.
Arrows indicate genes identified by gene ontology analysis in the network. Red and green indicate up and down-regulation, respectively. For further explanations, see text.
In males, COPD related genes form (Figure 3): (1) a functional core of repressed genes (arrows, Panel C) that belong to “Response to chemical stimulus” (GO42221, p = 0.001); and, (2) a cluster (arrows) of up-regulated genes (Panel D) involved in GO2376 (Immune system process, p = 0.007) and GO30097 (Hemopoiesis, p = 0.0002). The network analysis of Smoking in males was not possible because of the low number of genes identified (n = 16).
Interaction Network Analysis of COPD and Smoking related genes
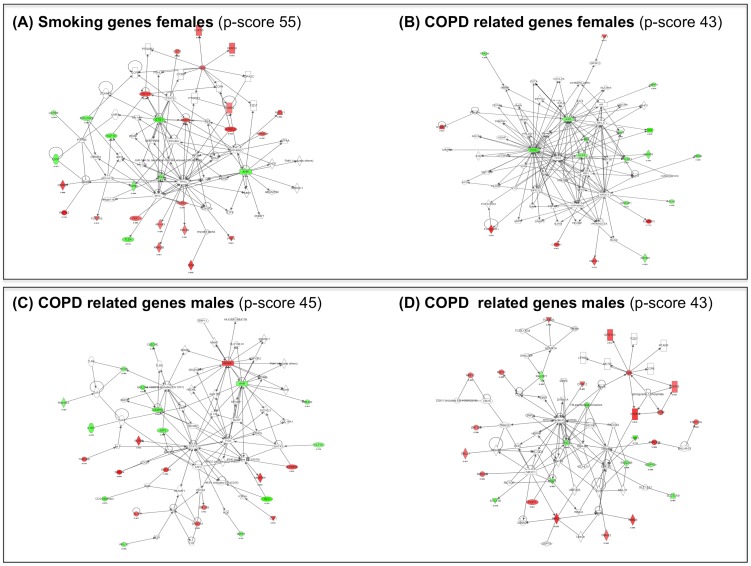
To infer interaction networks of COPD and Smoking related genes we used Ingenuity Pathways Analysis (IPA), a database of known molecule interactions [21]. In females, from the 61 Smoking genes (Figure 2, panel C1) IPA identified one significant network (IPA p-score 55) (Figure 5, panel A) whose main functional terms included organismal injury and abnormalities, cell death and survival and cell morphology. We selected the ETS gene, a central hub in this network, for independent validation by real time PCR (qPCR); the later confirmed the observed microarrays transcriptional change (Figure S2). Likewise, from the 29 COPD related genes identified in females (Figure 2, panel C1), IPA identified a network (p-score 43) enriched in cell death and survival, lipid metabolism and small molecule biochemistry functions (Figure 5, panel B). In this network, we selected FOS and CXCL1 for independent qPCR validation; results (Figure S2) confirmed changes in both CXCL1 (p = 0.01) and FOS (p = 0.07).
Figure 5. Putative interaction networks identified by Ingenuity Pathway Analysis (IPA)[21], and their corresponding scores, in female smoker controls COPD related in females (Panel A), Smoking in females (Panel B) and COPD related in males (Panels C and D).
For further explanations, see text.
The low number of Smoking genes (n = 16) identified in males did not allow obtaining significant networks in the IPA analysis. By contrast, from the 78 COPD related genes identified in males (Figure 2, panel C2), IPA identified two large networks. The first one (IPA p-score 45) included infectious disease, cellular function and maintenance, hematological system development and function as main functional terms (Figure 5, panel C). The second one (p-score 43) included DNA replication, recombination, cell cycle and cell morphology terms (Figure 5, panel D). From these two networks, we selected HLX, TRIB1 and BIRC3, known to be involved in cell activation, cell migration and apoptosis respectively, for qPCR validation (Figure S2).
Discussion
Despite that an abnormal inflammatory response to smoking is believed to play a major pathogenic role in COPD [1], to our knowledge this is the first study to directly compare the systemic biomarker and transcriptomic leukocyte response to acute smoking exposure in susceptible (i.e. COPD patients) and resistant smokers (i.e., those with normal spirometry). By doing so, our results provide several novel and potentially relevant observations. First, we identified a differential transcriptomic pattern, both at baseline and in response to smoking, in males and females, whose implications go beyond COPD, as discussed below. Second, we identified a number of COPD and Smoking related genes (signatures), both in males and females, and we identified their enriched ontologies and gene interaction networks. Yet, due to the relatively small number of subjects studied, reproducibility in an independent and larger set of COPD patients and smokers is needed.
Previous studies
Several previous studies have investigated the biological response to acute smoking exposure in healthy subjects [13], [14]. In fact, we designed the dose and timing of our ASET (see Methods) mostly based on these previous experiences. Yet, to our knowledge, no previous study has directly contrasted the transcriptomic response of circulating leukocytes to ASET in smokers with or without COPD. Bahr et al [33] compared the basal leukocyte transcriptomic pattern in COPD patients and non-smoker controls, without exposing subjects to smoking or considering potential sex differences. In our study, at baseline, we identified 19 DE genes also identified by Bahr et al [33] (C19orf59, ARHGAP18, C11orf75, CARD16, CLEC4D, FAIM3, FCRL3, GYG1, GZMM, HOPX, MAL, MAPK14, MCTP1, NELL2, RPS23, SAT1, SLC8A1, SNHG8, SPON2). Of note, MLL5, JUN, RABEP1 and ZFAND6 were identified in our study as COPD related genes in females and were also identified by Bhattacharya et al [34] as COPD associated genes in peripheral blood.
Interpretation of findings
Leukocyte transcriptomics changes after acute smoking exposure were relatively low. This might not be surprising if this is framed within the natural history of COPD, which takes decades of smoking exposure to develop. In keeping with this, previous peripheral blood transcriptomic studies in COPD also reported small fold change differences [33], [34].
The fact that our study involved an intervention (ASET) in smokers with and without COPD, that it included never smokers (who served as a control for time-related biological effects), and that we stratified participants by sex prove to be key aspects of the experimental design and of paramount importance to identify different COPD and Smoking related signatures in males and female smokers. In males, the COPD related signature included ontologies related to the immune system (up-regulated) and cellular locomotion (up-regulated), whereas only a few number of genes (which limited ontology enrichment analysis) were associated with Smoking. By contrast, in females the Smoking signature included ontologies that were similar (but of opposite direction) to those identified as COPD related in males, and that their COPD related signature included down-regulation of the MLL5, FOS and JUN genes, all of them involved in the inflammatory response. Collectively, these observations suggest a tighter control of the inflammatory response to smoking in females.
How these sex differences in leukocyte transcriptomics might translate into the clinic is speculative but a number of clinically relevant gender differences have been long recognized in COPD [8]–[10]. For instance, previous studies of our group demonstrated that lung function decline with age was significantly higher in males than in females, both in healthy never smokers as well as in smokers with COPD [35]. A different regulation of the inflammatory response in males and females as that proposed here may well contribute to these observations. In fact, interestingly in the context of this discussion, systemic levels of inflammatory biomarkers appear reduced in females (vs. males) with COPD [36].
The relevance of these observations for other common smoking related diseases, such as cardiovascular disease [37]–[39] or lung cancer [40]–[42], deserves further investigation. In fact, recent publications support similar sex differences in other immune related diseases, such as multiple sclerosis [43], and it is well established that autoimmune diseases are particularly prevalent in females [44]. All in all, therefore, these observations call for a careful exploration and re-analysis of available COPD genetic data by gender.
Finally, we used several network analysis techniques to infer potential interactions among the identified COPD and Smoking related genes and thus to get further insight into the pathobiology of the disease. The expression correlation networks in conjunction with gene ontology identified functional cores of differentially expressed genes. The possible interaction networks between differentially expressed genes identify by IPA (on the basis of already know molecule interactions) showed good IPA p-scores and their main gene hub changes were validated by qPCR. In females, resistance genes networks included genes related to Gpcr signaling (SIPR5, GPR56), inflammation and Th1 responses (ETS-1, MMP9, TBX21), apoptosis (BCL2) and sensing of planar aromatic hydrocarbons that are present in tobacco (AHR), whereas the main hubs in their susceptibly gene networks include the repression of JUN and FOS, both involved also in the regulation of immune responses through the formation of the AP transcription factor. Overall, therefore, these observations suggest again a differential regulation of immune processes in females with or without COPD. On the other hand, in males, we identified two COPD related gene networks that involved, respectively, the response to planar aromatic hydrocarbons (hub gene AHR) and the immune response (hub gene RORA) on the one hand, and Gpcr signaling, ubiquitinization (TRIB1, RAD23B, USP-15) and apoptosis-autophagy (BIRC3, TRIM13, TRIM23) on the other, which were validated by qPCR. Interestingly, other studies have reported alterations in some of these processes in COPD [45]–[47].
Strengths and Limitations
Our study has strengths and limitations. The fact that, despite the widespread assumption that COPD is characterized by an abnormal response to smoking, this is the first study to investigate this possibility specifically at the transcriptional level is a clear strength. Likewise, the use of an active experimental intervention (ASET) is a step beyond the standard transcriptomic case-control studies [48] and has in fact facilitated the identification of novel COPD and Smoking related genes in response to acute tobacco exposure. Finally, the use of network analysis has allowed the description of clear gender differences in the systemic inflammatory response to smoking in COPD patients and Smokers, an observation that may have implications beyond COPD in other smoking-related diseases. Among its limitations, we acknowledge that the transcriptomic changes after ASET were measured only in circulating leukocytes, so it is possible that changes in the pulmonary parenchyma may differ in type and/or intensity; needless to say that ethical and logistic difficulties involved in obtaining lung tissue samples before and after smoking are notorious. Finally, we included in the study 30 volunteers but, when stratified by presence of disease, smoking status and sex, each subgroup includes 5 individuals only. Due to the relatively small number of subjects studied, results need replication in an independent and larger cohort of COPD patients.
Conclusions
This study shows that the transcriptomic response to smoke in circulating leukocytes is different in smokers with and without COPD, and also in males and females.
Supporting Information
Unsupervised clustering of DE genes in COPD segregated T0 and T180 samples adequately, both in females (n = 57) and males (n = 110) (panels A and B, respectively).
(TIFF)
qPCR validation of array results, RQ = relative quantification of target gene mRNA to Actin mRNA calculated using the comparative CT method. Central hub genes in IPA networks were selected for the validation; a) COPD males (HLX, TRIB1 and BIRC3), b) Healthy smoker females (ETS 1) and c) COPD females (CXCL1 and FOS1).
(TIFF)
Complete list of differentially expressed genes (DE) in COPD patients, smokers (S) and non-smokers (NS) stratified by sex. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Top 10 differentially expressed genes at baseline between COPD patients and Smokers, stratified by sex. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Top 10 differentially expressed genes at baseline due to gender differences in COPD patients and in healthy smokers. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Detailed methods and additional results.
(DOCX)
Acknowledgments
Authors thank all volunteers participating in the study for their willingness to contribute to medical research, Ms. Gemma Sunyer for her excellent technical support during the study, Drs. Sellares, Soler and Ballester, from the Respiratory Department of our hospital, for their help recruiting COPD patients, and the different institutions that financed the study. This work was developed at the Centre de Recerca Biomèdica Cellex, Barcelona, Spain. Affymetrix arrays were performed in the Genomics platform of the IDIBAPS.
Funding Statement
Supported, in part, by Mutua Madrileña PI041966/2012, Fundació Catalana de Pneumologia (Fucap)-Beca Esteve 2011, Instituto de Salud Carlos III-Fondo de Investigaciones Sanitarias (FIS) 10/00523, SEPAR 133/2011. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vestbo J, Hurd SS, Agusti AG, Jones PW, Vogelmeier C, et al. (2013) Global Strategy for the Diagnosis, Management and Prevention of Chronic Obstructive Pulmonary Disease, GOLD Executive Summary. Am J Respir Crit Care Med 187: 347–365 rccm.201204-0596PP [pii]; 10.1164/rccm.201204-0596PP [doi] [DOI] [PubMed] [Google Scholar]
- 2. Fletcher C, Peto R (1977) The natural history of chronic airflow obstruction. Br Med J 1: 1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lomas DA, Silverman EK (2001) The genetics of chronic obstructive pulmonary disease. Respir Res 2: 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silverman EK, Speizer FE, Weiss ST, Chapman HA Jr, Schuette A, et al. (2000) Familial aggregation of severe, early-onset COPD: candidate gene approaches. Chest 117: 273S–274S. [DOI] [PubMed] [Google Scholar]
- 5. DeMeo DL, Carey VJ, Chapman HA, Reilly JJ, Ginns LC, et al. (2004) Familial aggregation of FEF25-75 and FEF25-75/FVC in families with severe, early onset COPD. Thorax 59: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, et al. (2004) The Nature of Small-Airway Obstruction in Chronic Obstructive Pulmonary Disease. N Engl J Med 350: 2645–2653. [DOI] [PubMed] [Google Scholar]
- 7. Agusti A, Edwards LD, Rennard SI, MacNee W, Tal-Singer R, et al. (2012) Persistent Systemic Inflammation is Associated with Poor Clinical Outcomes in COPD: A Novel Phenotype. PLoS ONE 7: e37483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aryal S, Diaz-Guzman E, Mannino DM (2013) COPD and gender differences: an update. Transl Res 162: 208–218 S1931-5244(13)00138-2 [pii]; 10.1016/j.trsl.2013.04.003 [doi] [DOI] [PubMed] [Google Scholar]
- 9. de Torres JP, Casanova C, Hernandez C, Abreu J, guirre-Jaime A, et al. (2005) Gender and COPD in patients attending a pulmonary clinic. Chest 128: 2012–2016. [DOI] [PubMed] [Google Scholar]
- 10.Dransfield MT, Washko GR, Foreman MG, San Jose ER, Reilly J, et al.. (2007) Gender differences in the severity of CT emphysema in COPD. Chest. [DOI] [PubMed]
- 11. McDunn JE, Husain KD, Polpitiya AD, Burykin A, Ruan J, et al. (2008) Plasticity of the systemic inflammatory response to acute infection during critical illness: development of the riboleukogram. PLoS ONE 3: e1564 10.1371/journal.pone.0001564 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, et al. (2007) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 176: 532–555. [DOI] [PubMed] [Google Scholar]
- 13. van der Vaart H, Postma DS, Timens W, Hylkema MN, Willemse BW, et al. (2005) Acute effects of cigarette smoking on inflammation in healthy intermittent smokers. Respir Res 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Vaart H, Postma DS, Timens W, ten Hacken NH (2004) Acute effects of cigarette smoke on inflammation and oxidative stress: a review. Thorax 59: 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. American Thoracic Society Official Statement (1995) Standardization of Spirometry. 1994 Update. Am J Respir Crit Care Med 152: 1107–1136. [DOI] [PubMed] [Google Scholar]
- 16. Roca J, Sanchis J, Agustí-Vidal A, Segarra J, Navajas D, et al. (1986) Spirometric reference values for a mediterranean population. Bull Eur Physiopathol Respir 22: 217–224. [PubMed] [Google Scholar]
- 17. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 10.1006/meth.2001.1262 [doi];S1046-2023(01)91262-9 [pii] [DOI] [PubMed] [Google Scholar]
- 18. Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 10.1016/j.febslet.2004.07.055 [doi];S0014579304009354 [pii] [DOI] [PubMed] [Google Scholar]
- 19. Huang dW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 nprot.2008.211 [pii]; 10.1038/nprot.2008.211 [doi] [DOI] [PubMed] [Google Scholar]
- 20. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- 21. Jimenez-Marin A, Collado-Romero M, Ramirez-Boo M, Arce C, Garrido JJ (2009) Biological pathway analysis by ArrayUnlock and Ingenuity Pathway Analysis. BMC Proc 3 Suppl 4 S6 1753-6561-3-S4-S6 [pii]; 10.1186/1753-6561-3-S4-S6 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smoot M, Ono K, Ideker T, Maere S (2011) PiNGO: a Cytoscape plugin to find candidate genes in biological networks. Bioinformatics 27: 1030–1031 btr045 [pii]; 10.1093/bioinformatics/btr045 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 btq675 [pii]; 10.1093/bioinformatics/btq675 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Menon R, Di DM, Cordiglieri C, Musio S, La ML, et al. (2012) Gender-based blood transcriptomes and interactomes in multiple sclerosis: involvement of SP1 dependent gene transcription. J Autoimmun 38: J144–J155 S0896-8411(11)00117-X [pii]; 10.1016/j.jaut.2011.11.004 [doi] [DOI] [PubMed] [Google Scholar]
- 25.Anton IM, de la Fuente MA, Sims TN, Freeman S, Ramesh N, et al. (2002) WIP deficiency reveals a differential role for WIP and the actin cytoskeleton in T and B cell activation. Immunity 16: : 193–204. S1074761302002686 [pii]. [DOI] [PubMed] [Google Scholar]
- 26. Vogler M (2012) BCL2A1: the underdog in the BCL2 family. Cell Death Differ 19: 67–74 cdd2011158 [pii]; 10.1038/cdd.2011.158 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lang F, Voelkl J (2013) Therapeutic potential of serum and glucocorticoid inducible kinase inhibition. Expert Opin Investig Drugs 22: 701–714 10.1517/13543784.2013.778971 [doi] [DOI] [PubMed] [Google Scholar]
- 28. Bailey SL, Chang SC, Griffiths B, Graham AN, Saffery R, et al. (2008) ZNF397, a new class of interphase to early prophase-specific, SCAN-zinc-finger, mammalian centromere protein. Chromosoma 117: 367–380 10.1007/s00412-008-0155-7 [doi] [DOI] [PubMed] [Google Scholar]
- 29. Eisenreich A, Zakrzewicz A, Huber K, Thierbach H, Pepke W, et al. (2013) Regulation of pro-angiogenic tissue factor expression in hypoxia-induced human lung cancer cells. Oncol Rep 30: 462–470 10.3892/or.2013.2413 [doi] [DOI] [PubMed] [Google Scholar]
- 30. Luo Y, Roeder RG (1995) Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol 15: 4115–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gotting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K (2000) Molecular cloning and expression of human UDP-d-Xylose:proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-II. J Mol Biol 304: 517–528 10.1006/jmbi.2000.4261 [doi];S0022-2836(00)94261-X [pii] [DOI] [PubMed] [Google Scholar]
- 32. Schonthaler HB, Guinea-Viniegra J, Wagner EF (2011) Targeting inflammation by modulating the Jun/AP-1 pathway. Ann Rheum Dis 70 Suppl 1 i109–i112 70/Suppl_1/i109 [pii]; 10.1136/ard.2010.140533 [doi] [DOI] [PubMed] [Google Scholar]
- 33.Bahr TM, Hughes GJ, Armstrong M, Reisdorph R, Coldren CD, et al.. (2013) Peripheral Blood Mononuclear Cell Gene Expression in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. [DOI] [PMC free article] [PubMed]
- 34. Bhattacharya S, Tyagi S, Srisuma S, DeMeo DL, Shapiro SD, et al. (2011) Peripheral blood gene expression profiles in COPD subjects. J Clin Bioinforma 1: 12 2043-9113-1-12 [pii]; 10.1186/2043-9113-1-12 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohansal R, Martinez-Camblor P, Agusti A, Buist AS, Mannino DM, et al. (2009) The Natural History of Chronic Airflow Obstruction Revisited: An Analysis of the Framingham Offspring Cohort. Am J Respir Crit Care Med 180: 3–10 200901-0047OC [pii]; 10.1164/rccm.200901-0047OC [doi] [DOI] [PubMed] [Google Scholar]
- 36. de Torres JP, Casanova C, Pinto-Plata V, Varo N, Restituto P, et al. (2011) Gender Differences in Plasma Biomarker Levels in a Cohort of COPD Patients: A Pilot Study. PLoS ONE 6: e16021 10.1371/journal.pone.0016021 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roeters van Lennep JE, Westerveld HT, Erkelens DW, van der Wall EE (2002) Risk factors for coronary heart disease: implications of gender. Cardiovasc Res 53: 538–549. [DOI] [PubMed] [Google Scholar]
- 38. Erbel R, Mohlenkamp S, Lehmann N, Schmermund A, Moebus S, et al. (2008) Sex related cardiovascular risk stratification based on quantification of atherosclerosis and inflammation. Atherosclerosis 197: 662–672 S0021-9150(07)00132-3 [pii]; 10.1016/j.atherosclerosis.2007.02.031 [doi] [DOI] [PubMed] [Google Scholar]
- 39. Zhu J, Shearer GM, Norman JE, Pinto LA, Marincola FM, et al. (2000) Host response to cytomegalovirus infection as a determinant of susceptibility to coronary artery disease: sex-based differences in inflammation and type of immune response. Circulation 102: 2491–2496. [DOI] [PubMed] [Google Scholar]
- 40. Gawron A, Hou L, Ning H, Berry JD, Lloyd-Jones DM (2012) Lifetime risk for cancer death by sex and smoking status: the lifetime risk pooling project. Cancer Causes Control 23: 1729–1737 10.1007/s10552-012-9959-0 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryu JS, Jeon SH, Kim JS, Lee JH, Kim SH, et al. (2011) Gender differences in susceptibility to smoking among patients with lung cancer. Korean J Intern Med 26: 427–431 10.3904/kjim.2011.26.4.427 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park SK, Cho LY, Yang JJ, Park B, Chang SH, et al. (2010) Lung cancer risk and cigarette smoking, lung tuberculosis according to histologic type and gender in a population based case-control study. Lung Cancer 68: 20–26 S0169-5002(09)00318-3 [pii]; 10.1016/j.lungcan.2009.05.017 [doi] [DOI] [PubMed] [Google Scholar]
- 43. Menon R, Di DM, Cordiglieri C, Musio S, La ML, et al. (2012) Gender-based blood transcriptomes and interactomes in multiple sclerosis: involvement of SP1 dependent gene transcription. J Autoimmun 38: J144–J155 S0896-8411(11)00117-X [pii]; 10.1016/j.jaut.2011.11.004 [doi] [DOI] [PubMed] [Google Scholar]
- 44. Pennell LM, Galligan CL, Fish EN (2012) Sex affects immunity. J Autoimmun 38: J282–J291 S0896-8411(11)00126-0 [pii]; 10.1016/j.jaut.2011.11.013 [doi] [DOI] [PubMed] [Google Scholar]
- 45. Zeng H, Kong X, Peng H, Chen Y, Cai S, et al. (2012) Apoptosis and Bcl-2 family proteins, taken to chronic obstructive pulmonary disease. Eur Rev Med Pharmacol Sci 16: 711–727. [PubMed] [Google Scholar]
- 46. Ryter SW, Lee SJ, Choi AM (2010) Autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. Expert Rev Respir Med 4: 573–584 10.1586/ers.10.61 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barrero CA, Perez-Leal O, Aksoy M, Moncada C, Ji R, et al. (2013) Histone 3.3 participates in a self-sustaining cascade of apoptosis that contributes to the progression of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 188: 673–683 10.1164/rccm.201302-0342OC [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faner R, Tal-Singer R, Riley JH, Celli B, Vestbo J+, et al.. (2013) Lessons from ECLIPSE: a review of COPD biomarkers. Thorax. In press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unsupervised clustering of DE genes in COPD segregated T0 and T180 samples adequately, both in females (n = 57) and males (n = 110) (panels A and B, respectively).
(TIFF)
qPCR validation of array results, RQ = relative quantification of target gene mRNA to Actin mRNA calculated using the comparative CT method. Central hub genes in IPA networks were selected for the validation; a) COPD males (HLX, TRIB1 and BIRC3), b) Healthy smoker females (ETS 1) and c) COPD females (CXCL1 and FOS1).
(TIFF)
Complete list of differentially expressed genes (DE) in COPD patients, smokers (S) and non-smokers (NS) stratified by sex. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Top 10 differentially expressed genes at baseline between COPD patients and Smokers, stratified by sex. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Top 10 differentially expressed genes at baseline due to gender differences in COPD patients and in healthy smokers. Gene ID, affymetrix probe ID, log ratio and FDR.
(DOCX)
Detailed methods and additional results.
(DOCX)