Abstract
Targeted genome editing using engineered nucleases has rapidly transformed from a niche technology to a mainstream method used by many biological researchers. This widespread adoption has been largely fueled by the emergence of the clustered regularly interspaced short palindromic repeat (CRISPR) technology, an important new platform for generating RNA-guided nucleases (RGNs), such as Cas9, with customizable specificities. RGN-mediated genome editing is facile, rapid and has enabled the efficient modification of endogenous genes in a wide variety of biomedically important cell types and novel organisms that have traditionally been challenging to manipulate genetically. Furthermore, a modified version of the CRISPR-Cas9 system has been developed to recruit heterologous domains that can regulate endogenous gene expression or label specific genomic loci in living cells. Although the genome-wide specificities of CRISPR-Cas9 systems remain to be fully defined, the capabilities of these systems to perform targeted, highly efficient alterations of genome sequence and gene expression will undoubtedly transform biological research and spur the development of novel molecular therapeutics for human disease.
The capability to introduce targeted genomic sequence changes into living cells and organisms provides a powerful tool for biological research as well as a potential avenue for therapy of genetic diseases. Frameshift knockout mutations enable reverse genetics and assignment of function, sequence insertions can be used to fuse genes to epitope tags or other functional domains, such as fluorescent proteins to endogenous gene products, and specific sequence alterations can be used to induce amino acid substitutions for disease modeling, to transfer traits in agricultural crops and livestock, and to correct defective genes for therapeutic applications. For many years, strategies for efficiently inducing precise, targeted genome alterations were limited only to certain organisms (for example, using homologous recombination in yeast or recombineering in mice) and often required drug-selectable markers or left behind ‘scar’ sequences associated with the modification method (for example, residual loxP sites from Cre recombinase-mediated excision). Targeted genome editing using customized nucleases provides a new, general method for inducing targeted deletions, insertions and precise sequence changes in a broad range of organisms and cell types. The high efficiency of genome editing obviates the need for additional sequences, such as drug-resistance marker genes, and therefore the need for additional manipulations to remove them.
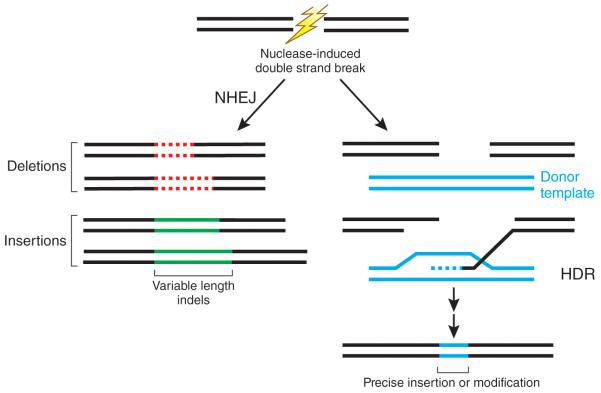
A crucial first step for performing targeted genome editing is the creation of a DNA double-stranded break (DSB) at the genomic locus to be modified1. Nuclease-induced DSBs can be repaired by one of at least two different pathways that are operative in nearly all cell types and organisms: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Fig. 1). NHEJ can lead to the efficient introduction of insertion/deletion mutations (indels) of various lengths, which can disrupt the translational reading frame of a coding sequence or the binding sites of trans-acting factors in promoters or enhancers. HDR-mediated repair can be used to introduce specific point mutations or to insert desired sequences through recombination of the target locus with exogenously supplied DNA ‘donor templates’. When targeted nuclease-induced DSBs are used, the frequencies of these alterations are typically higher than 1% and, in some cases, can be 50% or higher; modifications at these levels enable identification of desired mutations by simple screening, without the need for drug resistance marker selection.
Figure 1. Nuclease-induced genome editing.
Nuclease induced double-strand breaks (DSBs) can be repaired by NHEJ or HDR pathways. Imprecise NHEJ-mediated repair can produce variable-length insertion and deletion mutations at the site of the DSB. HDR-mediated repair can introduce precise point mutations or insertions from a single-stranded or double-stranded DNA donor template.
Early methods for targeting DSB-inducing nucleases to specific genomic sites relied on protein-based systems with customizable DNA-binding specificities such as meganucleases, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). These platforms have enabled important advances, but each has its own set of associated advantages and challenges (Box 1). More recently, a novel platform based on a bacterial CRISPR-Cas system has been developed that is unique and flexible due to its dependence on RNA as the moiety that targets the nuclease to a desired DNA sequence. In contrast to ZFN and TALEN platforms that use protein–DNA interactions for targeting, these RGNs use simple base pairing rules between an engineered RNA and the target DNA site.
Box 1. Meganucleases, ZFNs and TALENs.
Meganucleases, ZFNs, and TALENs have been utilized extensively for genome editing in a variety of different cell types and organisms. Meganucleases are engineered versions of naturally occurring restriction enzymes that typically have extended DNA recognition sequences. Zinc finger nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs) are artificial fusion proteins composed of an engineered DNA binding domain fused to a non-specific nuclease domain from the FokI restriction enzyme. Zinc finger and transcription activator-like effector repeat domains with customized specificities can be joined together into arrays capable of binding to extended DNA sequences.
The engineering of meganucleases has been challenging for most academic researchers because the DNA recognition and cleavage functions of these enzymes are intertwined in a single domain86, 87. By contrast, the DNA binding domains of ZFNs and TALENs are distinct from the FokI cleavage domain88, thereby making it more straightforward to modify the DNA-binding specificities of these nucleases. However, robust construction of engineered zinc finger arrays has also proven to be difficult for many laboratories due to the need to account for context-dependent effects between individual finger domains in an array89. Despite the availability of various publicly available methods designed to simplify the challenge of creating ZFNs90-96, these nucleases have not been engineered by a wide range of laboratories. In contrast to zinc fingers, TALE repeat domains appear to be less affected by context-dependence and can be robustly assembled in a modular fashion to recognize virtually any DNA sequence97 using a simple one-to-one code between individual repeats and the four possible DNA nucleotides98, 99. Although much simpler to design than meganucleases or ZFNs, the assembly of DNAs encoding large numbers of highly conserved TALE repeats can require the use of non-standard molecular biology cloning methods. Many user-friendly methods for making such assemblies have been described in the literature100 but the highly repetitive nature of TALEN-coding sequences also creates barriers to their delivery using certain viral vectors, such as lentiviruses101. Nonetheless, the greater simplicity of TALENs relative to meganucleases and ZFNs has led to their adoption over the past several years by a broader range of scientists. The question of whether to utilize these platforms for a given application must be answered on a case-by-case basis and we refer the reader to recent reviews on these different technologies for additional information86, 88, 100.
In this review, we describe how this RNA-guided system works and how it has been applied to perform genome editing across a wide variety of cell types and whole organisms. We also discuss advantages and limitations of this system, including assessment of off-target effects and recent strategies for improving specificity, and how it can be re-purposed for other applications such as regulation of gene expression and selective labeling of the genome. Finally, we consider the next set of challenges that will need to be addressed for this emerging genome editing platform.
From bacterial CRISPR-Cas immune system to engineered RNA-guided nucleases
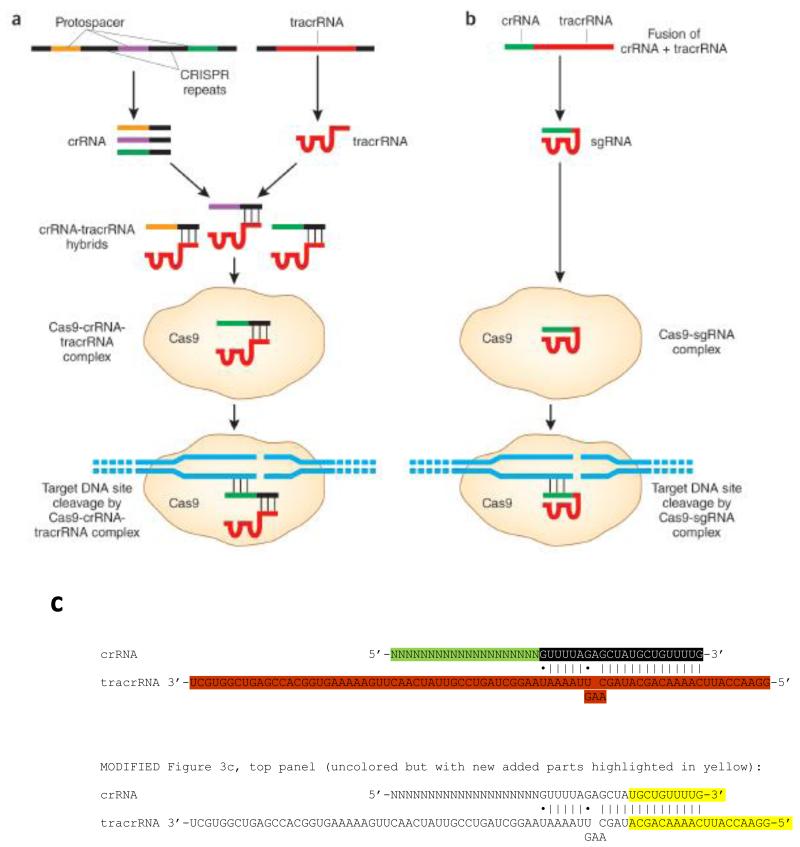
CRISPR systems are adaptable immune mechanisms used by many bacteria to protect themselves from foreign nucleic acids, such as viruses or plasmids2-5. The type II CRISPR system incorporates sequences from invading DNA between CRISPR repeat sequences encoded as arrays within the host genome (Fig. 2a). Transcripts from the CRISPR repeat arrays are processed into CRISPR RNAs (crRNAs) each harboring a variable sequence transcribed from the invading DNA, known as the “protospacer” sequence, as well as part of the CRISPR repeat. Each crRNA hybridizes with a second transactivating CRISPR RNA (tracrRNA) and these two RNAs complex with the Cas9 nuclease6. The protospacer-encoded portion of the crRNA directs Cas9 to cleave complementary target DNA sequences, if they are adjacent to short sequences known as “protospacer adjacent motifs” (PAMs). Protospacer sequences incorporated into the CRISPR locus are not cleaved because they are not present next to a PAM sequence.
Figure 2. Naturally occurring and engineered CRISPR-Cas9 systems.
(a) Naturally occurring CRISPR-Cas systems incorporate foreign DNA sequences into CRISPR arrays, which then produce crRNAs bearing “protospacer” regions that are complementary to the foreign DNA site. crRNAs hybridize to tracrRNAs (also encoded by the CRISPR system) and this pair of RNAs associates with the Cas9 nuclease. crRNA/tracrRNA/Cas9 complexes recognize and cleave foreign DNAs bearing the protospacer sequences. Additional details in the text. (b) and (c) Engineered CRISPR-Cas system consists of a fusion between a crRNA and part of the tracrRNA sequence. This single gRNA then complexes with Cas9 to mediate cleavage of target DNA sites that are complementary to the first (5′) 20 nts of the gRNA and that lie next to a PAM sequence. Additional details in the text.
The type II CRISPR system from Streptococcus pyogenes has been adapted for inducing sequence-specific DSBs and targeted genome editing7. In its simplest and most widely used form, two components must be introduced and/or expressed in cells or an organism to perform genome editing: the Cas9 nuclease; and a ‘guide RNA’ (gRNA), consisting of a fusion of a crRNA and a constant tracrRNA (Fig. 2b). 20 nucleotides at the 5′ end of the gRNA (corresponding to the protospacer sequence of the crRNA; Fig. 2c) direct Cas9 to a specific target DNA site using standard RNA-DNA complementarity base pairing rules. These target sites must lie immediately 5′ of a PAM sequence that matches the canonical form 5′-NGG (although recognition at sites with alternate PAM sequences (e.g. 5′-NAG) has also been reported, albeit at less efficient rates7-9). Thus, with this system, Cas9 nuclease activity can be directed to any DNA sequence of the form N20-NGG simply by altering the first 20 nts of the gRNA to correspond to the target DNA sequence. Type II CRISPR systems from other species of bacteria that recognize alternative PAM sequences and that utilize different crRNA and tracrRNA sequences have also been used to perform targeted genome editing10-12. However, because the most commonly used and extensively characterized system to date is based on the S. pyogenes system, the remainder of this review focuses on this particular platform and its components unless otherwise noted.
Following the initial demonstrations in 2012 that Cas9 could be programmed to cut various DNA sites in vitro7, a flurry of papers published in 2013 showed that this platform also functions efficiently in a variety of cells and organisms. Initial proof-of-principle studies showed that Cas9 could be targeted to endogenous genes in bacteria8, cultured transformed human cancer cell lines and human pluripotent stem cells in culture13-16, as well as in a whole organism, the zebrafish17. Subsequently, Cas9 has been used to alter genes in yeast18, tobacco19, 20, thale cress19, rice21, 22, wheat21, sorghum23, mice24, 25, rats26, rabbits27, frog28, fruit flies29, 30 silkworm31,and roundworms32 (see Table 1 for a list of these published reports).Cas9-induced DSBs have been used to introduce NHEJ-mediated indel mutations as well as to stimulate HDR with both double-stranded plasmid DNA and single-stranded oligonucleotide donor templates. The capability to introduce DSBs at multiple sites in parallel using the Cas9 system is a unique advantage of this platform relative to meganucleases, ZFNs, or TALENs. For example, expression of Cas9 and multiple gRNAs has been used to induce small and large deletions or inversions between the DSBs14,33-35, to simultaneously introduce mutations in three genes in rat cells36, five genes in mouse ES cell clones24 and five genes in the somatic cells of a single zebrafish37.
Table 1. Published examples of cell types and organisms modified by Cas9.
| Cell type or organism | Cas9 form | Cell type | Reference numbers |
|---|---|---|---|
| Human cells | Cas9 nuclease | HEK293FT, HEK293T, HEK293, K562, iPSC, HUES9, HUES1, BJ- RiPS ,HeLa, Jurkat, U2OS |
13-16, 47, 49-51, 54, 58, 61, 72, 83, 102, 103 |
| Cas9 nickase | HEK293FT, HEK293T | 13, 14, 47, 49 | |
| dCas9 (gene regulation) | HEK293FT, HEK293T | 72-74, 76, 83 | |
| dCas9 (imaging) | HEK293T, UMUC3, HeLa |
84 | |
| Mouse or mouse cells | Cas9 nuclease | embryos | 14, 24-26 |
| Cas9 nickase | embryos | 47 | |
| dCas9 (gene regulation) | NIH3T3 | 76 | |
| Rat | Cas9 nuclease | embryos | 26, 36 |
| Rabbit | Cas9 nuclease | embryos | 27 |
| Frog | Cas9 nuclease | embryos | 28 |
| Zebrafish | Cas9 nuclease | embryos | 17, 33, 37, 62, 103 |
| Fruit fly | Cas9 nuclease | embryos | 29, 30, 63 |
| Silkworm | Cas9 nuclease | embryos | 31 |
| Roundworm | Cas9 nuclease | adult gonads | 32, 64-69 |
| Rice | Cas9 nuclease | protoplasts, callus cells |
21, 23 |
| Wheat | Cas9 nuclease | protoplasts | 21 |
| Sorghum | Cas9 nuclease | embryos | 23 |
| Tobacco | Cas9 nuclease | protoplasts, leaf tissue |
19, 20, 23 |
| Thale cress | Cas9 nuclease | protoplasts, seedlings |
19, 23 |
| Yeast | Cas9 nuclease | S. cerevisiae | 18 |
| Bacteria | Cas9 nuclease |
S. pneumonia, E. coli |
8 |
| dCas9 (gene regulation) | E. coli | 71, 72 |
Human Embryonic Kidney (HEK); Induced Pluripotent Stem Cells (iPSCs);
The simplicity of Cas9 targeting has also inspired the generation of large gRNA libraries using array-based oligonucleotide synthesis. These libraries can be engineered to encompass multiple gRNAs for every target gene in a host organism, thereby greatly facilitating forward genetic screens and selections. In contrast to short hairpin RNA libraries, which only mediate gene knockdown, these gRNA libraries have been used with Cas9 nuclease to generate knockout mutations. Libraries comprised of between ~64,000 and ~87,000 distinct gRNAs have been used to demonstrate the capacity for positive and negative forward genetic phenotype screens in both human and mouse cells38-40.
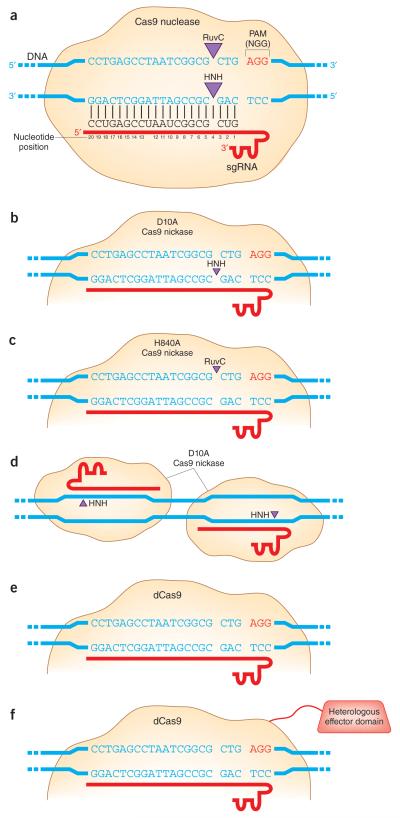
Variant Cas9 nickases that cut one strand, rather than both strands, of the target DNA site have also been shown to be useful for genome editing. Introduction of a D10A or H840A mutation into the RuvC1- or HNH-like nuclease domains present in Cas9 (Fig. 3a)41, 42 results in the generation of nickases that cut either the complementary or non-complementary DNA target strands, respectively, in vitro7, 12, 43 (Fig. 3b and 3c). Consistent with previous studies performed with ZFN-derived nickases44-46, Cas9 nickases can, at some sites, induce HDR with reduced levels of concomitant NHEJ-mediated indels13, 14. However, although at some sites Cas9 nickase can induce HDR with efficiencies similar to what is observed with the Cas9 nuclease from which it is derived13, 14, these rates can also be greatly reduced at other sites47, 48. Importantly, the frequencies of indel mutations introduced by nickases has also been reported to be high at certain sites13, 47-49. Although the precise DNA repair pathways by which these various alterations are induced remain as yet undefined, one potential mechanism that has been postulated is that passage of a replication fork through a nuclease-induced nick site might result in a DNA DSB. Additional studies with Cas9 nickases will be needed to better understand locus-dependent differences in the efficiencies of HDR and indel mutation induction.
Figure 3. Cas9-based systems for altering gene sequence or expression.
(a) Cas9 nuclease creates double strand breaks at DNA target sites with complementarity to the 5′ end of a gRNA. (b) Cas9 nickase created by mutation of the RuvC nuclease domain with a D10A mutation. This nickase cleaves only the DNA strand that is complementary to and recognized by the gRNA. (c) Cas9 nickase created by mutation of the HNH nuclease domain with a H840A mutation. This nickase cleaves only the DNA strand that does not interact with the sRNA. (d) Paired nickase strategy for improving Cas9 specificity. D10A Cas9 nickase directed by a pair of appropriately oriented gRNAs leads to induction of two nicks that, if introduced simultaneously, would be expected to generate a 5′ overhang. (e) Catalytically inactive or ‘dead’ Cas9 (dCas9) that can be recruited by a gRNA without cleaving the target DNA site. (f) Catalytically inactive dCas9 bearing dual D10A/H840A mutations fused to a heterologous effector domain.
Determining the specificities of RNA-guided Cas9 nucleases
Although RGNs generally cleave their intended target sites reliably, an important question is to what extent these nucleases induce off-target cleavage events (and therefore unwanted NHEJ-induced indel mutations). To assess RGN specificity, several groups have created variant gRNAs bearing one to four nucleotide mismatches in the complementarity region and then examined the abilities of these molecules to direct Cas9 nuclease activity in human cells at reporter gene50 or endogenous gene14, 51 target sites. These studies showed that mismatches are generally better tolerated at the 5′ end of the 20 nt targeting region of the gRNA than at the 3′ end; this result is consistent with previous experiments performed in vitro and in bacterial cells, which suggested that the 8 – 12 bps at the 3′ end of the targeting sequence (a.k.a. the seed sequence) are crucial for target recognition7, 8, 14, 52, 53. However, the effects of single and double mismatches are not always predictable based on their location within the gRNA targeting region; some mismatches in the 5′ end can have dramatic effects whereas some in the 3′ end do not significantly affect Cas9 activity50. In addition, not all nucleotide substitutions at a given position necessarily have equivalent effects on activity51.
A reciprocal, and perhaps more relevant, approach for studying specificity is to assess the activities of Cas9 at potential off-target genomic DNA target sites, (i.e. sites that have a few nucleotide differences compared to the intended target). A number of studies have examinedpotential off-target sites that differ at one to six positions from the on-target site in human cells47, 48, 50, 51, 54. Collectively, these reports have found cases of off-target mutations at sites that differ by as many as five positions within the protospacer region50 and/or that have an alternative PAM sequence of the form NAG51. Interestingly, indel mutation frequencies at these off-target sites can be high enough (>2-5%) to detect using the relatively insensitive T7 Endonuclease I (T7E1) mutation mismatch assay and to sometimes be comparable to the on-target site mutation frequency48, 50. In addition, more sensitive deep sequencing assays have been used to identify lower frequency off-target mutations48, 55, 56. It is important to note that all of these directed studies only examined a subset of the much larger number of potential off-target sites in the genome. For example, any given 20 nt protospacer will have typically have hundreds to thousands of potential off-target sites that differ at four or five positions, respectively, in 6 × 109 bps of random DNA. In addition, although it has been suggested that higher GC content at the RNA:DNA hybridization interface might potentially help to stabilize Cas9:gRNA complexes, high rates of mutagenesis have been observed for off-target sites with as little as 30% matched GC-content9, 50.
A somewhat more comprehensive strategy for examining Cas9 specificities is to identify off-target sites from a partially degenerate library of variants that is based on the intended on-target sequence. One recent report identified sites from such libraries based on their abilities to be bound by a catalytically inactive form of Cas9 fused to a transcriptional activation domain (see further discussion below)49. This study found sites that were mismatched by as many as three (and possibly more) positions relative to the on-target site49. These results are similar to those of another study, which used in vitro selection for Cas9 nuclease cleavage activity to identify potential off-target sites from a partially degenerate library of target site variants. Some of the off-target sites identified by these in vitro selections (with up to four mismatches) were also shown to be mutated in human cells9.
A recent study using whole exome sequencing did not find evidence of Cas9-induced off-target mutations in three modified human K562 cell line clones57. Although the authors acknowledge that the high false negative result rate associated with exome sequencing analysis limits interpretation of this data, these results do suggest that with careful target selection it may be possible to isolate Cas9-edited cells with otherwise intact exomes. Additional examples with deeper sequencing coverage and whole genome (rather than whole exome) sequencing will be needed to determine how readily cells that do not have off-target mutations can be isolated. The ability to do so would encourage broader research application of Cas9 technology. However, it is worth noting that deep sequencing the genomes of individual cell clones is neither a sensitive nor an effective approach for defining the full genomewide spectrum of Cas9 off-target sites.
Overall, the various studies published to date strongly suggest that off-target sites of RNA-guided Cas9 nucleases can be variable in frequency and challenging to predict. For any given target site, it is not currently possible to predict how many mismatches can be tolerated, nor do we fully understand why some sites are cleaved whereas other are not. We also do not know how genomic and/or epigenomic context might affect the frequency of cleavage. Although some initial evidence suggests that DNA methylation may not inhibit Cas9-based genome editing55, it seems both plausible and likely that chromatin structure could play a role in off-target site accessibility. A more comprehensive understanding of Cas9 off-target effects will have to await the development of unbiased, global measures of Cas9 specificity in cells
Methods for reducing off-target effects of Cas9 nucleases
Even with an incomplete understanding of RNA-guided Cas9 nuclease specificity, researchers have begun to explore various approaches to reduce off-target mutagenic effects. One potential strategy is to test the effects of reducing the concentrations of gRNA and/or Cas9 expressed in human cells. Results with this approach have been mixed with one group observing proportionately larger decreases in rates of off-target mutagenesis relative to on-target mutagenesis for two gRNAs51 and another group observing nearly proportionate decreases for two other gRNAs50. The use of modified gRNA architectures with truncated 3′ ends (within the tracrRNA-derived sequence) or with two extra guanine nucleotides appended to the 5′ end (just prior to the complementarity region) also yielded better on-target to off-target ratios but again with considerably lower absolute efficiencies of on-target genome editing57, 58.
Another proposed approach for improving specificity involves the use of “paired nickases” in which adjacent off-set nicks are generated at the target site using two gRNAs and Cas9 nickase47, 49, 57(Fig. 3d), a strategy analogous to one originally performed with pairs of engineered zinc finger nickases46. Cas9 nickases targeted to sites on opposite DNA strands separated by four to 100 bps can efficiently induce indel mutations or HDR events with a single-stranded DNA oligonucleotide donor template in human and mouse cells47, 49, 57. It has been proposed that the concerted action of paired nickases create a DSB that is then repaired by NHEJ or HDR47, 56. Importantly, paired nickases can reduce Cas9-induced off-target effects of gRNAs in human cells. The addition of a second gRNA and substitution of Cas9 nickase for Cas9 nuclease can lead to lower levels of unwanted mutations at previously known off-target sites of the first original gRNA47. However, an as-yet unanswered question is whether the second gRNA can itself induce its own range of Cas9 nickase-mediated off-target mutations in the genome. Multiple studies have shown that single Cas9 nickases can function on their own to induce indel mutations at certain genomic loci13, 47-49, perhaps because an individual nick might be converted to a DSB when a replication fork passes through the locus59, 60. Thus, one important way to improve the paired nickase system might be to modify it so that the activities of the two gRNAs are strictly co-dependent on each other for genome editing activity—that is, so that each gRNA is only active for genome editing when bound to DNA in close proximity to the other.
Our group has recently shown that off-target effects can be substantially reduced by using gRNAs that have been shortened at the 5′ end of their complementarity regions. These truncated gRNAs (tru-gRNAs) bearing 17 or 18 nucleotides of complementarity generally function as efficiently as full-length gRNAs in directing on-target Cas9 activity but show decreased mutagenic effects at off-target sites and enhanced sensitivity to single or double mismatches at the gRNA/DNA interface48. tru-gRNAs are simple to implement and this strategy avoids the technical challenges associated with expressing multiple gRNAs in a single cell for the paired nickase approach. Importantly, tru-gRNAs might also be used in conjunction with other strategies for improving Cas9 specificity (e.g.—tru-gRNAs have already been shown to improve the specificity of paired nickases48) and to improve the specificities of dCas9 fusion proteins for non-nuclease applications (described below).
Practical considerations for implementing CRISPR-Cas technology
Due to rapid progress in the field, potential users of CRISPR-Cas technology face a variety of choices about how to practice CRISPR-Cas technology. Here we discuss some of the parameters to consider when implementing the methodology.
Choice of Guide RNA Platform
It is important to note that the efficiency of Cas9 activity for any given locus can be influenced by the architecture of guide RNA(s) used. As described above, most recent studies have used a single gRNA that is a fusion of a programmable crRNA and part of the tracrRNA, but earlier studies also used a ‘dual gRNA’ configuration in which the crRNA and tracrRNA are expressed separately. In general, studies using single gRNAs have consistently reported substantially higher editing rates than those using dual gRNAs13, 14, 17, 61. These findings suggest that the single gRNA system may be more active than the double gRNA system, presumably because two components can assemble more efficiently than three components.
In addition, single gRNAs harboring variable lengths of tracrRNA sequence on their 3′ ends have been used by different groups (Supplementary Table 1). Systematic comparisons have generally demonstrated that longer single gRNAs (containing a longer 3′ portion of the tracrRNA sequence) are more active than shorter ones51. The most commonly used single gRNA to date is approximately 100 nts in length (Supplementary Table 1) and has been shown reliably to direct Cas9 to genomic sites in yeast18, tobacco19, 20, thale cress19, rice21, 22, wheat21, sorghum23, mice26, rats26, rabbits27, frog28, fruit flies29, 30, roundworms32, silkworm31 and human somatic and pluripotent stem cells13. The tru-gRNAs described above are shortened versions of this ~100 nt single gRNA.
Targeting range and choice of gRNA target sites
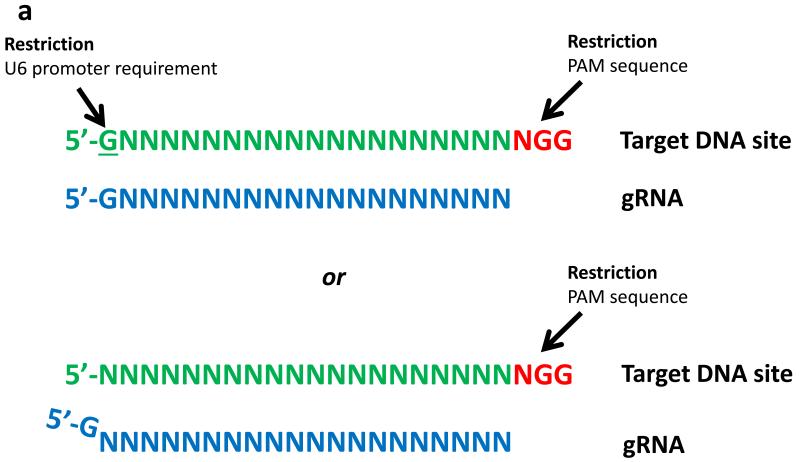
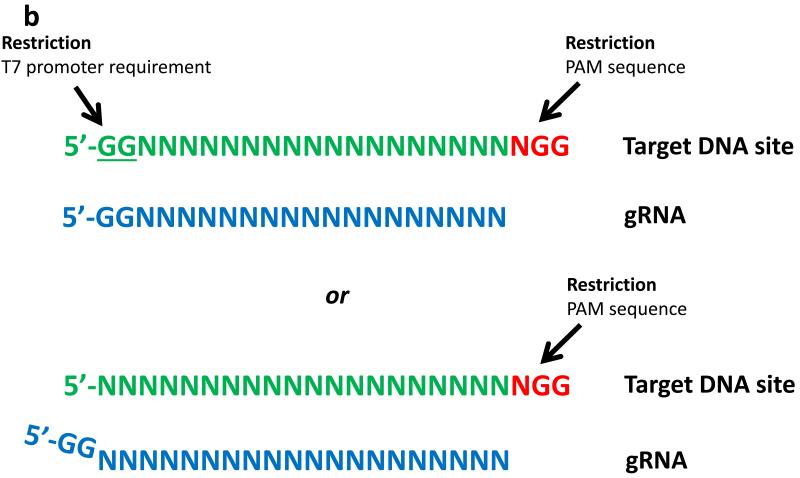
For gRNAs, the choice of promoter used to express these RNAs can limit the options for potential target sites. For example, the RNA polymerase III-dependent U6 promoter or the T7 promoter require a G or GG, respectively, at the 5′ end of the RNA to be transcribed (Fig. 4a); as a result, standard full-length or tru-gRNAs expressed from these promoters are limited to targeting sites that match the forms GN16-19NGG or GGN15-18NGG, sites that each occur every 1 in 32 bps or 1 in 128 bps, respectively, in random DNA. Paired nickase strategies require the identification of two sites on opposite strands of DNA with an appropriate spacing in between (as described above). One strategy to reduce these targeting range restrictions is to choose sites without regard to the identities of the first or first two bases at the 5′ end (i.e..--making gRNAs that are mismatched at these positions). Another potential strategy to bypass these restrictions is to append the required G or GG to the 5′ end of the gRNA, thereby encoding gRNA transcripts that are 1 or 2 bps longer (Fig.4b). Both of these strategies have been used successfully to produce active gRNAs but with variable efficiencies in the genome-editing activities of the Cas9 nucleases47, 54, 57, 62. Larger-scale studies are needed to clarify the effects of using either mismatched or extended gRNAs on the efficiencies and specificities of RGN-mediated cleavage. Several groups have provided web-based software that facilitates the identification of potential CRISPR RGN target sites in user-defined sequences (for example, the ZiFiT Targeter software17, 48 (http://zifit.partners.org) and the CRISPR Design Tool55 (http://crispr.mit.edu/)).
Figure 4. Limitations on targeting range of guide RNAs.
(a) Potential target sites of gRNAs expressed from a U6 promoter. Restrictions are imposed by the requirement for a G at the first 5′ nucleotide of the gRNA and by the need for an NGG adjacent to the target site (top). One strategy to work around the requirement for a 5′ G is to append this nucleotide to the beginning of the gRNA (bottom). (b) Potential target sites of gRNAs expressed from a T7 promoter. Restrictions are imposed by the requirement for a GG at the first two nucleotides of the gRNA and by the need for an NGG adjacent to the target site (top). One strategy to work around the requirement for a 5′ GG dinucleotide is to append GG to the beginning of the gRNA (bottom).
Delivery of CRISPR-Cas Components
RGNs have been delivered to a broad range of cell types and organisms using a variety of delivery methods. In cultured mammalian cells, researchers have used electroporation61, Nucleofection13, 50 and Lipofectamine-mediated transfection13, 14, 50 of non-replicating plasmid DNA to transiently express Cas9 and gRNAs. Lentiviral vectors have also been used to constitutively express Cas9 and/or gRNAs in cultured human 38, 39 and mouse40 cells. In vitro transcribed RNAs and/or plasmid DNA have been injected directly into embryos; for example, in embryos of zebrafish17, fruit flies29, 30, 63, mice24, 26 and rats26. Both plasmid DNA and RNA have also been injected into the gonads of adult roundworms32, 64-68 and in one study purified Cas9 protein complexed with gRNA was injected69. In addition to animal models and cell lines, Cas9 has been used successfully in multiple plant species including wheat, rice, sorghum, tobacco, and thale cress using a range of standard delivery methods including PEG-mediated transformation of protoplasts, Agrobacterium-mediated transfer in embryos and leaf tissue, and/or bombardment of callus cells with plasmid DNA19-21, 23. For most RGN applications, transient expression of gRNAs and Cas9 is typically sufficient to induce efficient genome editing. Although constitutive expression of RGN components might potentially lead to higher on-target editing efficiencies, extended persistence of these components in the cell might also lead to increased frequencies of off-target mutations, a phenomenon that has been previously reported with ZFNs70.
Strategies to experimentally control for RGN off-target effects
The existence of CRISPR RGN-induced off-target effects and our inability to comprehensively identify all of these alterations on a genome-wide scale requires investigators for now to account for potentially confounding effects of these undesired mutations. Several strategies might be used to rule out off-target mutations as a potential alternative explanation for any phenotypes observed. For example, complementation with re-introduction of a wild-type gene can be used to confirm the effects of knockout mutations. In addition, similar to the strategy of targeting a gene with multiple RNAi hairpins, one could easily create mutations in the same gene using gRNAs targeted to different sites. Presumably, each gRNA will be expected to have a different range of off-target effects and therefore if the phenotype is observed with each of these different gRNAs it would seem unlikely that undesired mutations are the cause. The ease with which multiple gRNAs can be rapidly designed and constructed makes it simple and feasible to implement this latter type of strategy with the Cas9 system. The high efficacy of the Cas9 nuclease for inducing mutations makes it an attractive choice for creating mutant cell lines and whole organisms in spite of the need to account for off-target effects.
Non-nuclease applications of CRISPR-Cas systems
Beyond enabling facile and efficient targeted genome editing, the CRISPR-Cas system has the potential to be used to regulate endogenous gene expression or to label specific chromosomal loci in living cells or organisms. Catalytically inactive or “dead” Cas9 (dCas9), a variant bearing both the D10A and H840A mutations that does not cleave DNA, can be recruited by gRNAs to specific target DNA sites7, 12(Fig. 3e). Targeting of dCas9 to promoters was initially shown to enable repression of gene expression in both Escherichia coli71 and human cells72. Interestingly, dCas9 repressed a bacterial promoter efficiently when recruited with gRNAs that interacted with either strand of sequences located upstream of the promoter; however, when targeting sites downstream of the transcription start point, only gRNAs that interacted with the non-template strand induced dCas9-mediated repression71. dCas9 also provides a general platform for recruitment of heterologous effector domains to specific genomic loci (Fig. 3f). For example, dCas9 fusions to a transcriptional activation domain (VP64 or the p65 subunit of nuclear factor kappa B; NF-κB) or a transcriptional repression domain (the Krüppel associated box (KRAB) domain) have been shown to regulate the expression of endogenous genes in human73-75 and mouse cells76 as well as in bacteria71. Changes in gene expression induced by these dCas9 fusions in human cells thus far appear to be generally lower than those induced by similar TALE-based transcription factors49, 77-80. However, multiplex recruitment of dCas9-based activators using between 2 and 10 sgRNAs targeted to the same promoter can result in substantially higher levels of human gene activation, presumably due to the phenomenon of activator synergy49, 73, 74, 76. This capability of dCas9-based activators to function synergistically is consistent with previous observations for TALE-based activators77, 78 in human cells. It will be interesting to see in future experiments whether dCas9 fusions to histone modifiers and TET proteins can also be used to perform targeted “epigenome editing” including the alteration of specific histone modifications and demethylation of particular cytosine bases in human cells, as has been recently described with TAL effector DNA-binding domains75, 81, 82.
An alternative strategy for tethering heterologous effector domains to DNA-bound dCas9-gRNA complexes is to exploit well-defined RNA–protein interaction pairs. This approach uses two engineered components: a gRNA that has one or two RNA binding sites for the phage MS2 coat protein fused to its 3′ end; and a fusion of MS2 coat protein to an effector domain49. Addition of the MS2 RNA binding sequences to the gRNA does not abolish its ability to target dCas9 to specific DNA sites. Furthermore, co-expression of the MS2 coat protein fusion with the hybrid gRNA and dCas9 has been used to recruit activation domains to a gene promoter in human cells49. Although the activation observed appears to be somewhat less robust than direct fusions to dCas9, this type of configuration might provide additional options and flexibility for multiplex recruitment of effector domains to a specific promoter. For example, one can imagine leveraging multiple gRNAs and MS2 coat protein binding sites on each of gRNA to recruit many copies of different domains to the same promoter.
Thus far, evidence suggests that the effects of the small number of dCas9-activation domain or dCas9-repression domain fusions tested to date can be highly specific in human cells as judged by RNA-seq experiments76, 83; however, this may be because not all binding events lead to changes in gene transcription. It remains to be determined whether the effects of other dCas9 fusions prove to be as specific in their genome-wide effects as these dCas9-based activators and repressors or if, like their nuclease counterparts, they will benefit from enhanced specificity conferred by tru-gRNAs.
In yet another application, it has been demonstrated that an EGFP-dCas9 fusion can be used to visualize DNA loci harboring repetitive sequences such as telomeres with just a single gRNA or non-repetitive loci using 26 to 36 gRNAs tiled across a 5 kb region of DNA84. This imaging strategy provides a powerful tool for studying chromosome dynamics and structure and extends the dCas9 system beyond gene expression-based applications.
Future directions
Progress in the development of Cas9-based technologies over the past eighteen months has been stunning to watch but many interesting questions and applications remain yet to be addressed and explored.
First, methods for expanding the targeting range of RNA-guided Cas9 will be important for inducing precise HDR or NHEJ events as well as for implementing the paired nickase and other multiplex strategies. As noted above, a major source of this targeting range restriction for Cas9, paired Cas9 nickases, and dCas9 fusions is the need for a PAM sequence matching the form NGG. The ability to exploit alternative PAM sequences of the form NAG or NNGG has been noted7, 8, 51 but more experiments are needed to ascertain how robustly these sequences are actually recognized and cleaved. Other gRNA-Cas9 platforms with different PAM sequences isolated from Streptococcus thermophilus, Neisseria meningitidis, and Treponema denticola have also been characterized10, 11, 14 and identification of more of these systems from other species85 could further enhance the targeting range of the platform.
Second, the field urgently needs to develop unbiased strategies to globally assess the off-target effects of Cas9 nucleases or paired nickases in any genome of interest. Such methods will be crucial for evaluating how effectively improvements described to date actually enhance the specificity of the platform. In addition, although tru-gRNAs and paired nickases can reduce off-target effects, it is possible that further improvements will be needed, especially for therapeutic applications. Ideally, these new strategies could be combined with existing approaches, such as the paired nickases and tru-gRNAs. Examples of such improvements might involve using protein engineering to modify Cas9 and/or modifying the nucleotides used by the gRNA to mediate recognition of the target DNA site. Alternatively, the construction of inducible forms of Cas9 and/or gRNAs might provide a means to regulate the active concentration of these reagents in the cell and thereby improve the ratio of on- and off-target effects.
Third, methods for efficient delivery and expression of CRISPR-Cas system components will undoubtedly need to be optimized for each particular cell-type or organism to be modified. For example, some cell types might be refractory to transfection and/or infection by standard viral vectors. A related challenge will be to develop methods that enable tissue-specific, cell-type-specific or developmental stage-specific expression of either the gRNAs and/or the Cas9 nuclease. Strategies that enable efficient multiplex expression of large numbers of gRNAs simultaneously from one vector will also enable this advantage of the technology to be leveraged. Collectively, these advances will be important for research use and therapeutic applications.
Lastly, strategies for shifting the balance away from NHEJ-mediated indel mutations and towards HDR-driven alterations remain a priority for development. Although high rates of HDR can be achieved with the CRISPR RGNs and single-stranded DNA oligonucleotides, competing mutagenic NHEJ also occurs simultaneously. This limitation is particularly problematic when using HDR to induce point mutation changes (as opposed to insertions) in the protospacer part of the target site; these successfully types of altered alleles can still be efficiently re-cut and then mutagenized by NHEJ, thereby reducing the yield of correctly edited sequences. One of the challenges with developing an approach to improve the HDR:NHEJ ratio is that inhibition of NHEJ is likely to be poorly tolerated by most cells, given its central role in normal DNA repair. For therapeutic applications seeking to exploit HDR in particular, reduction or elimination competing NHEJ will be a critically important requirement.
The simplicity, high efficiency and broad applicability of the RNA-guided Cas9 system have positioned this technology to transform biological and biomedical research. The ease with which researchers can now make changes in the sequence or expression of any gene will enable reverse genetics to be performed in virtually any organism or cell type of interest. In addition, the capability to construct large libraries of gRNAs for altering or regulating genes of interest will enable facile comprehensive forward genetic screens. All of these systems can also be multiplexed by expressing multiple gRNAs in a single cell, thereby further extending the complexity of forward and reverse genetic experiments that can be performed. Although the off-target effects of Cas9 remain to be defined on a genome-wide scale, much progress has already been made toward improving specificity and further advances will undoubtedly come rapidly given the intensity of research efforts in this area. All of these recent and future advances in developing and optimizing Cas9-based systems for genome and epigenome editing should propel the technology forward to therapeutic applications, opening the door to treating a wide variety of human diseases.
Supplementary Material
Acknowledgments
J.K.J. is grateful for support from the US National Institutes of Health (NIH) (grants DP1 GM105378 and R01 GM088040), the Defense Advanced Research Projects Agency (grant W911NF-11-2-0056) and The Jim and Ann Orr Massachusetts General Hospital Research Scholar Award. This material is based upon work supported by, or in part by, the U. S. Army Research Laboratory and the U. S. Army Research Office under grant number W911NF-11-2-0056. The authors apologize to colleagues whose studies were not cited due to length and reference constraints.
Footnotes
Competing Interests Statement
J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 3.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 6.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 17.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JF, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 21.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 22.Xie K, Yang Y. RNA-guided Genome Editing in Plants Using A CRISPR-Cas System. Mol Plant. 2013 doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013 doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 27.Yang D, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014 doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama T, et al. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013 doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Z, et al. Highly Efficient Genome Modifications Mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013 doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414–1416. doi: 10.1038/cr.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-Guided Genome Editing for Target Gene Mutations in Wheat. G3 (Bethesda) 2013;3:2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horii T, et al. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ. 2013;1:e230. doi: 10.7717/peerj.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 37.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotech. 2013 doi: 10.1038/nbt.2800. advance online publication. [DOI] [PubMed] [Google Scholar]
- 41.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez CL, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim E, et al. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotech. 2014 doi: 10.1038/nbt.2808. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013 doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013 doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013 doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cortes-Ledesma F, Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Q, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang WY, et al. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo TW, et al. Precise and Heritable Genome Editing in Evolutionarily Diverse Nematodes Using TALENs and CRISPR/Cas9 to Engineer Insertions and Deletions. Genetics. 2013 doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waaijers S, et al. CRISPR/Cas9-Targeted Mutagenesis in Caenorhabditis elegans. Genetics. 2013 doi: 10.1534/genetics.113.156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tzur YB, et al. Heritable Custom Genomic Modifications in Caenorhabditis elegans via a CRISPR-Cas9 System. Genetics. 2013 doi: 10.1534/genetics.113.156075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katic I, Grosshans H. Targeted Heritable Mutation and Gene Conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics. 2013 doi: 10.1534/genetics.113.155754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiu H, Schwartz HT, Antoshechkin I, Sternberg PW. Transgene-Free Genome Editing in Caenorhabditis elegans Using CRISPR-Cas. Genetics. 2013 doi: 10.1534/genetics.113.155879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho SW, Lee J, Carroll D, Kim JS. Heritable Gene Knockout in Caenorhabditis elegans by Direct Injection of Cas9-sgRNA Ribonucleoproteins. Genetics. 2013 doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF. 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012 doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013 doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013 doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013 doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maeder ML, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Pinera P, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 80.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotech. 2013 doi: 10.1038/nbt.2701. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeder ML, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013 doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen B, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silva G, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 89.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 90.Gupta A, et al. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim S, Lee MJ, Kim H, Kang M, Kim JS. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods. 2011;8:7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- 92.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonzalez B, et al. Modular system for the construction of zinc-finger libraries and proteins. Nat Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 96.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 97.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotech. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 99.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 100.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holkers M, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.