Abstract
Intestinal epithelial tight junctions (TJs) are a specialized structure that determines the cell polarity and prevents the diffusion of toxins, allergens, and pathogens from the lumen into the tissue. TJs are highly dynamic and its constituent protein complexes undergo continuously remodeling and turnover under tight regulation by numerous extracellular and intracellular factors. RNA-binding proteins (RBPs) and microRNAs (miRNAs) regulate gene expression at the posttranscriptional level and are involved in many aspects of cellular physiology. An increasing body of evidence indicates that RBPs including HuR and CUG-binding protein 1 and miRNAs such as miR-192 modulate the stability and translation of mRNAs encoding TJ proteins and play an important role in the control of intestinal epithelial TJ barrier function. In this mini-review article, we highlight the changes in TJ expression and intestinal epithelial TJ barrier function after activation or inactivation of RBPs and miRNAs and further analyze in some detail the mechanisms through which the stability and translation of TJ mRNAs are regulated by RBPs and miRNAs.
Keywords: permeability, intercellular junctions, intestinal epithelium, mRNA stability and translation, RNA-binding proteins, microRNAs
Introduction
Epithelial cells line the gastrointestinal (GI) mucosa and form an important barrier to a wide array of noxious substances in the lumen. The effectiveness and stability of the GI epithelial barrier depend on specialized structures composing different intercellular junctions including tight junctions (TJs) and adherens junctions (AJs).1,2 In physiological conditions, these intercellular junctional proteins completely surround the subapical region of epithelial cells and maintain the structural integrity and normal function of the gut epithelium. The TJ is the apical-most element of the junctional complex and seals epithelial cells together in a way that prevents even small molecules from leaking between cells. To date, four classes of TJ-transmembrane proteins and > 30 TJ-membrane-associated proteins are identified in mammalian epithelial and endothelial cells.2-4 At the ultrastructural level, the TJ appears as highly organized strands that encircle the apical-lateral boundary. TJs primarily consist of transmembrane proteins such as occludin, tricellulin, and one or more members of the claudin family; these proteins also associate with a cytosolic plaque of proteins that links tightly to the cortical cytoskeleton.
Normal expression of TJ proteins is crucial for maintenance of the gut barrier function, whereas decreased TJ levels or impaired TJ assembly result in the barrier dysfunction and increase GI epithelial paracellular permeability.1-4 In various critical pathological conditions such as trauma, burns, hemorrhage, sepsis, inflammation, and massive surgical operations, disruption of the gut barrier occurs commonly, leading to the translocation of luminal toxic substances and bacteria to the bloodstream. Since the exact mechanism underlying the acute epithelial barrier dysfunction remains obscure, effective therapies to preserve the integrity of the barrier are limited, contributing to death in patients with leaky gut.5,6 Over the past several years, our group and others have demonstrated that posttranscriptional control, particularly altered TJ mRNA turnover and translation by RNA-binding proteins (RBPs) and microRNAs (miRNAs), plays an important role in the regulation of TJ expression in the GI mucosa. In this article, we overview our current understanding regarding the importance of RBPs and miRNAs in posttranscriptional control of TJ expression and epithelial barrier function and also analyze in some detail the mechanisms by which RBPs and miRNAs modulate the stability and translation of TJ mRNAs.
Post-transcriptional Regulation by RBPs and miRNAs on Gut Epithelial Homeostasis
Mammalian intestinal epithelial cells (IECs) elicit rapid changes in gene expression patterns to regulate their survival, adapt to stress, and maintain homeostasis in response to stressful environments. In addition to the stress-modulated gene transcription, changes in posttranscriptional regulation also potently affect the steady-state levels of many transcripts and the levels of the encoded proteins.7-9 It has been well established that posttranscriptional fate of a given mRNA is primarily controlled by the interaction of specific mRNA sequences (cis-elements) with specific trans-acting factors such as RBPs10,11 and miRNAs.12,13 Ribonucleoprotein (RNP) associations regulate the intracellular transport of the mRNA and its association with the translation and decay machineries.7,8 Many labile mRNAs contain relatively long 3′-untranslated regions (UTRs) bearing U- and AU-rich elements (AREs) or GU-rich elements (GREs) that function as determinants of mRNA stability and translation.14-16 Among the RBPs that regulate specific subsets of mRNAs are several RBPs that modulate mRNA turnover (HuR, NF90, AUF1, BRF1, TTP, KSRP) and RBPs that modulate translation (HuR, CUGBP1, TIAR, NF90, TIA-1), collectively known as translation and turnover-regulatory (TTR)-RBPs.17-19 In cells responding to proliferative, immune, and stress-causing stimuli, TTR-RBPs bind to the specific sequences in the 5′- and 3′-UTRs of collections of target mRNAs and govern their turnover and translation rates.18-20
miRNAs are small noncoding RNAs of ~22 nucleotides, which posttranscriptionally repress the expression of target genes and regulate a variety of cellular processes.12,21 To date, 2,042 mature miRNAs are discovered in humans and 1,281 are identified in mice, according to the latest version of the miRNA database (miRBase).22 It has been estimated that > 60% of human genes are the targets of miRNAs.22-24 High-throughput and functional studies show that miRNAs play important roles in many aspects of cellular physiology and development. Generally, miRNAs act by binding to the 3′-UTRs of target mRNAs, destabilizing them and/or inhibiting their translation.12,21 The differential abundance of given miRNAs during disease progression suggests an especially significant role for miRNAs in human pathologies.25-27 Several studies have also revealed that RBPs and miRNAs can function jointly to regulate shared target mRNAs.9,10 For example, HuR recruits let-7/RISC to repress the translation of c-Myc mRNA,28 whereas the RBP Dnd-1 inhibits miRNA access to target mRNA.29
Recently, RBPs and miRNAs have emerged as master regulators of gut epithelial homeostasis by modulating IEC proliferation, apoptosis, migration and cell-to-cell interactions.14-16,25,30,31 RBPs such as HuR, AUF1, and CUGBP1 are highly expressed in IECs and their activities are affected rapidly and dramatically in response to various stressful environments such as inflammation, hypoxia, cancer, and injury.8,9,40 These RBPs regulate expression of several proliferation/apoptosis-associated genes, including p53, nucleophosmin, ATF2, XIAP, JunD, MEK-1, c-Myc, and Stim1 (stromal interaction molecule 1), at the posttranscriptional level, thus playing a critical role in maintaining gut epithelial integrity.14-16,20,32-34 Both miR-29b and miR-222 inhibit intestinal mucosal growth in vitro as well as in vivo by repressing translation of mRNAs encoding CDK2 (cyclin-dependent kinase 2) and CDK4,34,35 whereas miR-503 represses the RBP CUGBP1 translation and modulates IEC apoptosis.36 Moreover, CUGBP1 and miR-222 synergistically repress CDK4 mRNA translation in IECs,34 but miR-195 competes with HuR to modulate Stim1 mRNA stability and regulate IEC migration after wounding.33
Post-transcriptional Regulation of TJs by RBPs
HuR
HuR is one of the best studied TTR-RBPs and has two N-terminal RNA-recognition motifs (RRMs) with high affinity for AREs followed by a nucleocytoplasmic shuttling sequence and a C-terminal RRM that recognizes the poly(A) tail.9,10 Although the exact mechanism underlying target mRNA stabilization and translation by HuR remains largely unknown, this process is intimately associated with the cytoplasmic presence of the HuR RNP. In unstimulated cells, HuR is predominantly located in the nucleus, but it rapidly translocates to the cytoplasm, where it directly interacts with and stabilizes specific mRNAs and/or variably affects their translation in response to various stimuli.17,20,32 Studies from our group revealed that the nucleocytoplasmic shuttling of HuR in IECs is tightly regulated by the levels of cellular polyamines. Decreasing cellular polyamines by inhibiting their biosynthesis elevates cytoplasmic HuR levels, whereas elevating polyamines by ectopic overexpression of ODC gene (encoding key enzyme for polyamine synthesis) decreases cytoplasmic HuR; neither intervention alters whole-cell HuR levels.32,37
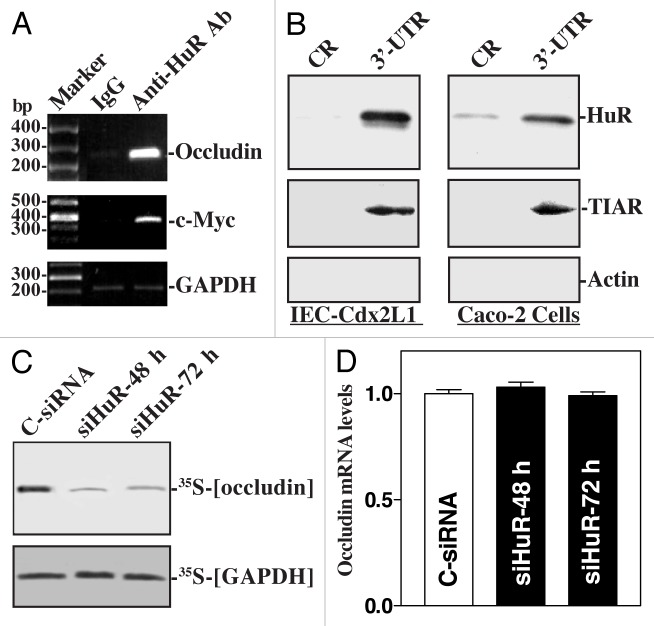
The first evidence showing the role of HuR in the posttranscriptional regulation of TJs in the gut epithelium is obtained from studying occludin expression. Occludin is a transmembrane TJ protein and plays an important role in TJ assembly and function.38 Our previous studies demonstrate that abundance of occludin in the gut epithelium is predominantly regulated at the posttranscriptional level by HuR.39,40 There are several computationally predicted hits of the HuR motif in the occludin 3′-UTR. HuR interacts with the occludin mRNA and enhances its translation (Fig. 1). As shown, the occludin mRNA is a direct target of HuR, since the occludin PCR products were highly enriched in HuR samples compared with control IgG samples. The enrichment of c-Myc PCR product was also examined in this study and served as a positive control, while the amplification of GAPDH PCR products, found in all samples as low-level contaminating housekeeping transcripts (not HuR targets), served to monitor the evenness of sample input. [HuR/occludin mRNA] associations were further tested by using biotinylated transcripts which spanned occludin coding region (CR) and 3′-UTR and show that HuR only associated with the occludin 3′-UTR but did not interact with the occludin CR. Moreover, HuR silencing inhibited occludin protein expression without effect on its mRNA levels. In contrast, ectopic HuR overexpression increases occludin translation.
Figure 1. HuR binds with and represses occludin mRNA translation. (A) Association of endogenous HuR with endogenous occludin mRNA as measured by RNP/immunoprecipitation (IP) assays using either anti-HuR antibody (Ab) or control IgG. (B) HuR and TIAR immunoblots using the pull-down materials by biotinylated transcripts of the occludin coding region (CR) or 3′-UTR. (C) Newly translated occludin protein after HuR by transfecting cells with either siRNA targeting the HuR mRNA coding region (siHuR) or control siRNA (C-siRNA). (D) Levels of occludin mRNA in cells treated as described in panel (C).
HuR association with the occludin mRNA depends on Chk2-dependent HuR phosphorylation.40 Chk2 physically interacts with and modulates HuR function by inducing its phosphorylation.15,40 Reduced HuR phosphorylation by Chk2 silencing or by reduction of Chk2 through polyamine depletion decreases HuR binding to the occludin mRNA and represses occludin translation, whereas Chk2 overexpression enhances [HuR/occludin mRNA] association and stimulates occludin expression. In mice exposed to septic stress induced by cecal ligation and puncture (CLP), Chk2 levels in the intestinal mucosa decreases dramatically, which is associated with a significant decrease in [HuR/occludin mRNA] association, leading to the inhibition of occludin expression and gut barrier dysfunction. In addition, the normal recovery of HuR binding to the occludin mRNA that occurs at 72 and 96 h after CLP is prevented by inhibiting Chk2 through polyamine depletion. Consistently, polyamine depletion also inhibits occludin expression and delays recovery of the gut barrier function after CLP. These results indicate that HuR positively regulates occludin mRNA translation through Chk2-dependent HuR phosphorylation and that this influence is crucial for maintenance of normal gut epithelial barrier function. HuR also stabilizes the claudin-1 mRNA in colon cancer cells,41 thus contributing to its regulatory effect on gut barrier function.
CUGBP1
CUGBP1 (also named CELF1, CUGBP and embryonic lethal abnormal vision-like factor 1) generally interacts with mRNAs through GREs rather than AREs,42 and the CUGBP1 association with mRNAs is shown to enhance mRNA decay and repress the translation of several target transcripts,42,43 although in some instances CUGBP1 also promotes mRNA translation.44 Recently, we reported that in IECs, CUGBP1 represses CDK4 translation in a GRE-dependent fashion and that cellular CUGBP1 abundance is regulated by miRNA-503,34,36 suggesting a regulatory network or “regulon” of CUGBP1 in the intestinal epithelium. Given the multiple posttranscriptional functions of CUGBP1 and the fact that occludin mRNA contains several hits of the CUGBP1 motif, we had investigated if CUGBP1 is implicated in regulating occludin expression and further defined its functional interaction with HuR in this process.
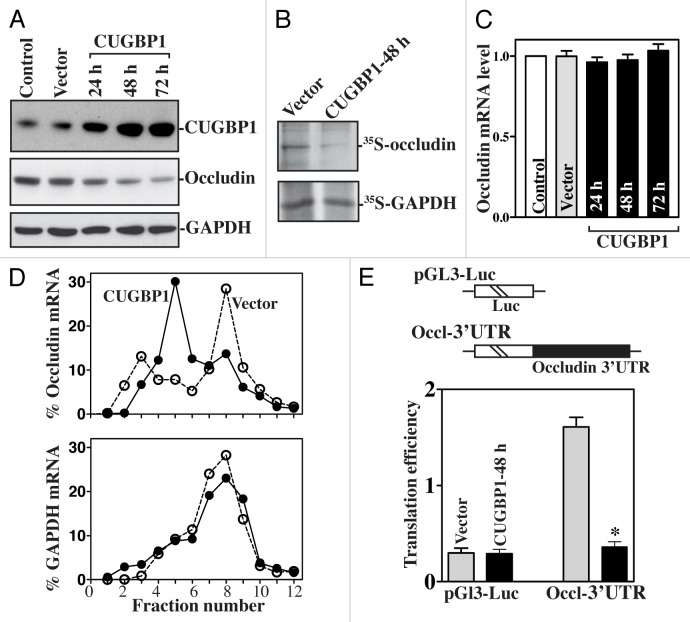
Our studies demonstrate that CUGBP1 interacts with the occludin mRNA via its 3′-UTR and represses occludin translation in human IECs.45 As shown in Figure 2, ectopic CUGBP1 overxpression was associated with a potent inhibition in occludin expression, but not in the levels of ZO-1 and β-catenin. This inhibitory effect of CUGBP1 on occludin expression occurs at the translation level, since ectopic CUGBP1 overexpression did not decrease the levels of total occludin mRNA but it repressed the rate of nascent occludin protein synthesis. To further define the role of CUGBP1 in the regulation of occludin translation, we also examined the relative distribution of occludin mRNA in individual fractions from polyribosome gradients after CUGBP1 overexpression. Although increasing the levels of CUGBP1 did not affect global polysomal profiles, the abundance of occludin mRNA associated with actively translating fractions 7–12 decreased dramatically in CUGBP1-transfected cells with a significant shift of occludin transcripts to low-translating fractions 4–6. In contrast, housekeeping GAPDH mRNA distributed similarly in all two groups. To examine whether the translational effect of CUGBP1 on occludin mRNA was exerted through GREs, we used a firefly luciferase reporter gene construct containing the occludin GRE within the 3′-UTR (Occl-3′UTR) and negative control vector pGL3-Luc. The basal levels of Occl 3′-UTR reporter gene activity were higher than those of control pGL3-Luc, suggesting that the occludin 3′-UTR may have a translation-enhancing element. Inhibition of occludin translation by CUGBP1 is mediated through its 3′-UTR, since ectopic CUGBP1 overexpression decreased the levels of the Luc-Occl 3′-UTR reporter gene activity. On the other hand, CUGBP1 silencing increases occludin expression by enhancing its translation. We also examined changes in the levels of claudin-2, claudin-3 and claudin-5 proteins (three major members of the claudin family in IECs) after decreasing or increasing the levels of CUGBP1 and found that neither CUGBP1 silencing nor ectopic CUGBP1 overexpression altered the levels of these claudin family proteins. Consistent with these findings, the CUGBP1-regulated expression of occludin also modulated the intestinal epithelial barrier function in an in vitro model. Increasing the levels of occludin by silencing CUGBP1 enhanced barrier function, as indicated by an increase in transepithelial electrical resistance (TEER) values and a decrease in the levels of paracellular flux of FITC-dextran. In contrast, lowering occludin levels by CUGBP1 overexpression disrupted barrier function. These results clearly show that CUGBP1 represses occludin translation through a process involving the occludin 3′-UTR, resulting in dysfunction of the epithelial barrier.
Figure 2. CUGBP1 overexpression inhibits occludin mRNA translation via its 3′-UTR. (A) Immunoblots of CUGBP1 and occludin proteins in cells transfected with the vector expressing CUGBP1 or control empty vector for different times. (B) Newly translated occludin protein after transfection with the CUGBP1 expression vector for 48 h. (C) Levels of occludin mRNA in cells treated as described in (A). (D) Distributions of occludin (top) and GAPDH (bottom) mRNAs in each gradient fraction of polysomal profiles after CUGBP1 overexpression. (E) Changes in occludin translation efficiency as measured by occludin 3′-UTR-luciferase reporter assays. *P < 0.05 compared with cells transfected with control vector.
Moreover, CUGBP1 and HuR compete for association with the same occludin 3′-UTR and regulate occludin translation competitively and in opposite directions.40,45 CUGBP1 overexpression decreases HuR binding to occludin mRNA and represses occludin translation, whereas HuR overexpression inhibits CUGBP1 association with occludin mRNA and promotes occludin translation. To study the competitive binding of HuR and CUGBP1 to the occludin mRNA, we further examined the effect of purified GST-HuR or GST-CUGBP1 fusion proteins added to the binding reaction mixture on the occludin 3′-UTR association with HuR and CUGBP1. When increasing concentrations of GST-HuR were added to the binding reaction, the occludin 3′-UTR association with HuR was progressively increased, but its interaction with CUGBP1 was reduced with increasing GST-HuR levels. Neither HuR nor CUGBP1 binding to the occludin 3′-UTR was affected by GST added to the binding reaction. In contrast, increasing the concentrations of GST-CUGBP1 in the binding reaction mixture dose-dependently increased occludin 3′-UTR association with CUGBP1 but decreased the levels of HuR/occludin 3′-UTR complex. These results indicate that HuR and CUGBP1 competitively bind to the occludin 3′-UTR and that CUGBP1 represses occludin mRNA translation by displacing HuR.
TIAR and other RBPs
TIAR (TIA-1 related protein) acts as a translational repressor and plays an important role in the regulation of gene expression, particularly under conditions of cellular stress. TIAR binds to target mRNAs via its RNA recognition motif domains and is implicated in both splicing regulation and translational repression.11 We have reported that the mRNA encoding the membrane-associated TJ protein ZO-1 shows affinity for TIAR, AUF1, and HuR via its 3′-UTR and that increased [TIAR/ZO-1 mRNA] complex represses ZO-1 translation, although the roles of AUF1 and HuR in regulating ZO-1 expression remain to be elucidated.46 Interestingly, the AP-1 transcription factor JunD regulates ZO-1 expression at the levels of transcription and translation; induced JunD inhibits ZO-1 translation by enhancing the interaction of the ZO-1 3′-UTR with TIAR.
Post-transcriptional Regulation of TJs by miRNAs
miRNAs direct mRNA degradation and translational inhibition in IECs and are recently found to play a critical role in the regulation of gut mucosal integrity and epithelial barrier function under pathological conditions.25,30 Active ulcerative colitis in human is associated with the differential expression of various miRNAs including miR-192 that targets the macrophage inflammatory peptide-2α mRNA and modulates gut permeability.25 McKenna et al. quantified the complete miRNA expression profile of the mammalian intestinal mucosa and revealed that miR-192 and let-7 are the most highly expressed in the small and large intestinal mucosa.31 Disruption of miRNA maturation in mice by tissue-specific deletion of Dicer1 gene in IECs decreases goblet cells, increases apoptosis in crypts of both jejunum and colon, and accelerates jejunal cell migration. Interestingly, intestinal epithelial barrier function is also impaired in Dicer1-deficient mice, although the exact changes in TJ expression in this model remain to be investigated.
Treatment with TNF-α causes a rapid increase in expression of miR-122a in the intestinal epithelium in vitro as well as in vivo, which is associated with intestinal TJ barrier dysfunction.30 miR-122a interacts with the occludin mRNA via its 3′-UTR, whereas overexpression of miR-122a in enterocytes enhances occludin mRNA degradation, resulting in increased intestinal TJ permeability. miR-155 binds to the claudin-1 3′-UTR and represses claudin-1 expression post-transcriptionally.47 The claudin-14 mRNA is the target of both miR-9 and miR-374, and these two miRNAs induce claudin-14 mRNA decay and translational repression in a synergistic manner.48 miR-145 represses JAM-1 expression and impairs TJ barrier function.49 In addition, miR-199a-5p regulates urothelial permeability and plays a role in bladder pain syndrome.50
In patients with irritable bowel syndrome (IBS), the enhancement of gut paracellular permeability is also associated with an increase in the levels of several miRNAs, including miR-212 and miR-29b.56,57 These two miRNAs are further shown to regulate intestinal TJ permeability in IBS by repressing expression of zonula occludens 1 (ZO-1) and glutamine synthetase, respectively. Expression of miR-212 in the intestinal mucosa is also increased by excessive ethanol and plays a role in the pathogenesis of gut leakiness in alcoholic liver disease.58 In addition, inflammation in patients with IBS also alters the levels of miR-132, miR-21, miR-24, miR-26b, miR-27a, miR-106a, miR-146a, miR-150, miR-155, miR-181a, miR-326 and miR-221/222 in the gut mucosal tissues,56 but their roles and importance in the regulation of intestinal epithelial TJ permeability remain to be fully elucidated.
Mechanisms Underlying RBPs and miRNAs in TJ Expression
The exact mechanisms by which RBPs and miRNAs regulate TJ expression in IECs remain largely unknown. To date, there are no studies available showing the relationship between the modulatory RBPs (such as HuR and CUGBP1) and translation initiation machinery in the control of TJ expression. However, several studies revealed that associations of given TJ mRNA with RBPs or miRNAs alter the mRNA recruitment to processing (P-) bodies.51,52 P-bodies are cytoplasmic RNP foci and contain many proteins and enzymes that are associated with RNA-induced silencing complex (RISC) and mRNA degradation.52 These proteins and enzymes include all four human Ago proteins, GW182 (TNRC6A) together with its two human paralogues (TNRC6B and TNRC6C), two RNA helicases (RCK and MOV10), decapping enzymes (DCP1 and DCP2), mRNA deadenylation factors (such as the CCR4-CAF-1-Not complex), activators of decapping (Dhh1/RCK/p54, Pat1, Scd6/RAP55, Edc3, and the LSm1–7 complex), and exonucleases (such as XRN-1).51,52 The mRNAs within P-bodies are thought to be subject to RNP remodeling to induce their subsequent turnover and to repress translational rates.
CUGBP1 and HuR modulate the occludin mRNA recruitment to P-bodies
To test the possibility that CUGBP1 represses occludin translation by increasing occludin mRNA recruitment to P-bodies, we examined the distribution of CUGBP1 and P-bodies by immunofluorescence and found extensive colocalization of CUGBP1 with the P-body resident proteins Ago2 and RCK in IECs.45 CUGBP1 associates with Ago2, RCK, and hDCP1 (another P-body marker) in an RNA-independent manner and induced CUGBP1 enhances the association of occludin mRNA with P-bodies. Furthermore, CUGBP1 does not repress occludin translation in the absence of RCK or Ago2, since silencing Ago2 or RCK prevented CUGBP1-induced repression of occludin expression. In contrast, increased levels of both CUGBP1 and Ago1 or Ago2 synergistically inhibited occludin expression. In keeping with the suppression of gene expression by Ago proteins, ectopic overexpression of HA-Ago1 or HA-Ago2 alone also modestly represses occludin expression. Interestingly, increasing the levels of HuR alters CUGBP1-induced association of the occludin mRNA with P-bodies, since ectopic HuR overexpression decreased the levels of occludin mRNA associated with Ago2 and RCK and prevented the CUGBP1-induced interaction of occludin mRNA with P-bodies.
By examining the subcytoplasmic localization of occludin mRNA using the reporter construct pMS2-Occludin, we further directly determined if the localization of occludin mRNA in P-bodies plays a role in CUGBP1-induced translational repression of occludin and its regulation by HuR. This construct expresses a chimeric RNA (MS2-Occludin) comprising the occludin 3′-UTR and 24 tandem MS2 RNA hairpins.36,53 Co-transfection of pMS2-occludin together with plasmid pMS2-YFP, which expresses the chimeric fluorescent protein MS2-YFP with a nuclear localization signal, allowed us to track the subcellular localization of the chimeric MS2-occludin RNA by confocal microscopy. The control MS2 RNA is exclusively nuclear in cells due to the presence of the NLS in the chimeric protein MS2-YFP, whereas some MS2-occludin RNA is retained in the cytoplasm, colocalizing with some Ago2 signals in cells transfected with control vector. However, CUGBP1 overexpression increases the co-localization of MS2-occludin RNA and Ago2 signals, suggesting that increasing the levels of CUGBP1 enhances the association of occludin mRNA with P-bodies. Moreover, the colocalization of MS2-occludin RNA and Ago2 signals is lost when cells are co-transfected with the HuR and CUGBP1 expression vectors. Together, these data support a novel regulatory model whereby CUGBP1 represses occludin translation by increasing the recruitment of occludin mRNA to P-bodies, whereas HuR promotes occludin translation by competing for the interaction of CUGBP1 with occludin 3′-UTR, thus preventing the recruitment of occludin mRNA to P-bodies.
Interaction of HuR with miR-195 regulates mRNA translocation to P-Bodies
We have recently found that HuR competes with miR-195 to modulate stim1 mRNA stability by altering stim1 mRNA translocation to P-bodies.33 Although product of the stim1 mRNA is not TJ protein, it alters intestinal epithelial integrity and TJ permeability through regulation of Ca2+ influx.54,55 miR-195 and HuR compete for association with the stim1 3′-UTR and regulate stim1 mRNA decay in opposite directions. Interaction of miR-195 with the stim1 3′-UTR destabilizes stim1 mRNA, whereas the stability of stim1 mRNA increases with HuR association. Ectopic miR-195 overexpression enhances stim1 mRNA association with Ago-containing complexes and increases the colocalization of tagged stim1 RNA with P-bodies, whereas this translocation of stim1 mRNA to P-bodies is abolished by HuR overexpression. These findings provide additional evidence that control of the stability and translation of mRNAs by RBPs and miRNAs is mediated through alterations in target mRNA recruitment to P-bodies.
Conclusions and Future Study
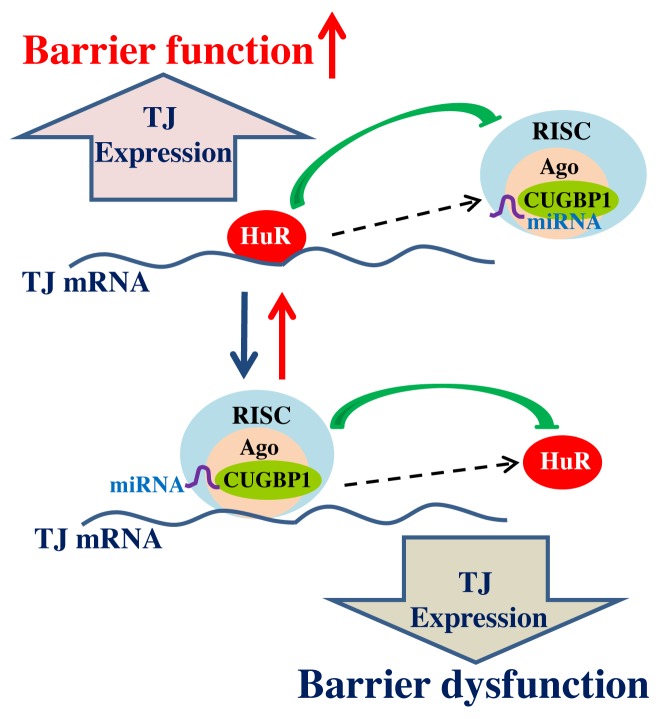
The experimental data summarized in this article provide novel evidence indicating the importance of RBPs and miRNAs in posttranscriptional regulation of TJs, especially in TJ mRNA stability and translation. Based on our studies33,40,45 and others,52,53 we propose a model delineating the posttranscriptional control of TJ gene expression and subsequent epithelial TJ barrier function by RBPs and miRNAs (Fig. 3). In this model, TJ expression is controlled by two factors competing for influence on TJ mRNA stability and translation: the positive factor HuR and negative regulators such as CUGBP1, TIAR, and miRNAs. HuR stabilizes and/or promotes TJ mRNA translation, thus upregulating TJ expression and enhancing the barrier function. In contrast, CUGBP1, TIAR, and miRNAs (including miR-192, miR-122a, miR-155, miR-9, and miR-374) destabilize and/or inhibit TJ mRNA translation, leading to the barrier dysfunction. Importantly, altering the interaction of HuR with the TJ mRNA affects the levels of target mRNA associated with CUGBP1 or miRNA. Available findings further suggest that CUGBP1 and miRNAs destabilize and/or repress TJ mRNA translation by increasing TJ mRNA recruitment to P-bodies, whereas HuR stabilizes and/or promotes TJ mRNA translation by blocking target mRNA translocation to P-bodies via the displacement of CUGBP1 and miRNAs.
Figure 3. Schematic diagram depicting posttranscriptional regulation of TJ gene expression by RBPs and miRNAs. HuR and CUGBP1/miRNAs interact with the TJ mRNA and modulate the production of TJ protein expression. HuR binding may trigger changes in RNA structure that conceal the site of interaction with miRNA/CUGBP1-RISC, thus causing an increase in TJ expression and enhancing the barrier function. Conversely, binding of miRNA/CUGBP1-RISC may trigger a conformational change that hides the site of HuR binding to the TJ mRNA, in turn lowering expression of TJs and leading to barrier dysfunction.
However, there are still many critical issues that remain to be addressed regarding this model. For example, studies to define the molecular processes responsible for the control of RBP activation and miRNA biogenesis in response to stressful environments and how altered RBPs and miRNAs interact with and modulate expression of other TJ proteins remain to be fully elucidated. Studies using intestinal epithelial tissue-specific RBP and miRNA knockout or transgenic mouse models will provide a better understanding of physiological functions of RBPs/miRNAs in the posttranscriptional regulation of TJ expression and epithelial barrier function. Moreover, experiments investigating the roles and mechanisms of RBPs/miRNAs in the pathogenesis of leaky gut in patients with inflammatory diseases and/or critical surgical conditions are still limited and badly needed for future clinical application.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Merit Review Grants from the Department of Veterans Affairs (to JYW and JNR) and by NIH Grants DK-57819, DK-61972, and DK-68491 (to JYW). J-Y. Wang is a Senior Research Career Scientist, Medical Research Service, US Department of Veterans Affairs.
Footnotes
Previously published online: www.landesbioscience.com/journals/tissuebarriers/article/28320
References
- 1.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 2.Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu Rev Pathol. 2010;5:119–44. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- 3.Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009;124:3–20, quiz 21-2. doi: 10.1016/j.jaci.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris L, Tonutti L, Vannini C, Bazzoni G. Structural organization of the tight junctions. Biochim Biophys Acta. 2008;1778:646–59. doi: 10.1016/j.bbamem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Anders HJ, Andersen K, Stecher B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013;83:1010–6. doi: 10.1038/ki.2012.440. [DOI] [PubMed] [Google Scholar]
- 6.Fukui H. How leaky gut and endotoxemia induce bacterial infection in cirrhosis and gastrointestinal hemorrhage? J Gastroenterol Hepatol. 2011;26:423–5. doi: 10.1111/j.1440-1746.2011.06668.x. [DOI] [PubMed] [Google Scholar]
- 7.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–86. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 9.Gorospe M, Tominaga K, Wu X, Fähling M, Ivan M. Post-Transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front Mol Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372–9. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HS, Headey SJ, Yoga YM, Scanlon MJ, Gorospe M, Wilce MC, Wilce JA. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013;10:579–89. doi: 10.4161/rna.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–37. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–98. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–94. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell. 2007;18:4579–90. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J. 2010;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–35, e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 26.Di Leva G, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–86. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–33. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–64, e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–32. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–19. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, Wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–69. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24:3038–46. doi: 10.1091/mbc.E13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23:151–62. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1. Biochem J. 2008;409:389–98. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–88. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo X, Rao JN, Liu L, Zou T, Keledjian KM, Boneva D, Marasa BS, Wang JY. Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1159–69. doi: 10.1152/ajpgi.00407.2004. [DOI] [PubMed] [Google Scholar]
- 40.Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39:8472–87. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, Bhat AA, Krishnan M, Singh AB, Dhawan P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis. 2013;34:2610–21. doi: 10.1093/carcin/bgt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–7. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–63. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–17. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19:3701–12. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375–80. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 48.Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J. Claudin-14 regulates renal Ca⁺⁺ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J. 2012;31:1999–2012. doi: 10.1038/emboj.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adammek M, Greve B, Kässens N, Schneider C, Brüggemann K, Schüring AN, Starzinski-Powitz A, Kiesel L, Götte M. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertil Steril. 2013;99:1346–55, e5. doi: 10.1016/j.fertnstert.2012.11.055. [DOI] [PubMed] [Google Scholar]
- 50.Monastyrskaya K, Sánchez-Freire V, Hashemi Gheinani A, Klumpp DJ, Babiychuk EB, Draeger A, Burkhard FC. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am J Pathol. 2013;182:431–48. doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–51. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 53.Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17:732–9. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao JN, Rathor N, Zhuang R, Zou T, Liu L, Xiao L, Turner DJ, Wang JY. Polyamines regulate intestinal epithelial restitution through TRPC1-mediated Ca²+ signaling by differentially modulating STIM1 and STIM2. Am J Physiol Cell Physiol. 2012;303:C308–17. doi: 10.1152/ajpcell.00120.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao JN, Rathor N, Zou T, Liu L, Xiao L, Yu TX, Cui YH, Wang JY. STIM1 translocation to the plasma membrane enhances intestinal epithelial restitution by inducing TRPC1-mediated Ca2+ signaling after wounding. Am J Physiol Cell Physiol. 2010;299:C579–88. doi: 10.1152/ajpcell.00066.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vicario M, Martínez C, Santos J. Role of microRNA in IBS with increased gut permeability. Gut. 2010;59:710–2. doi: 10.1136/gut.2009.203695. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775–84. doi: 10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–64. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]