Abstract
Neural stem cells contribute to mammalian brain tissue turnover in specific locations throughout life. Differentiation of stem cells is associated with terminal mitosis and cell cycle exit, but it is unclear how the timing and signaling of these are interlinked. Here, we have investigated the cell cycle exit characteristics in comparison with morphological changes during hippocampal stem cell differentiation in an adult mammalian cell line. Our results suggest that the cell-specific gene pathway induction is fast and robust and takes place in one day, whereas the cell cycle exit machinery is slower and takes several days to fully execute. The hippocampal differentiation is associated with epigenetic changes, such as Ezh2 down regulation and histone methylation. A small percentage of stem cells is able to resist differentiation-induced terminal mitosis for weeks in culture, and can be reverted to proliferation by re-adding the mitotic growth factors.
Keywords: Neural stem cell, Differentiation, Cell cycle, Hippocampus
Introduction
Neural stem cells (NSCs) are known to reside in the adult mammalian brain ventricular wall and hippocampus (1, 2). The ventricular wall neurogenic area produces new cells to the rostral migratory stream and olfactory bulb, and the hippocampal NSCs provide differentiated neurons for the dentate gyrus region (3). NSCs are capable of differentiating into all three lineages of neural cells: neurons, astrocytes and oligodendrocytes (2) but the physiological importance of NSCs is unknown. It is hypothesized that at least in specific areas new cells are needed for constant turnover of brain tissue. In addition to the normal brain tissue turnover, NSCs are activated in the case of injury or pathological cell death when new cells are necessary for regeneration. (4, 5). Most NCS studies have focused in the development of functional neurons and neurogenesis (6), but the events of astrocyte differentiation are less well understood.
During embryonic development, the mammalian brain NSCs first generate neurons, after which glial cells are born (7). At the same time, some of the NSCs stay undifferentiated and end up in specific niches, which provide the environment necessary for the quiescent life of a stem cell. The mechanism for staying undifferentiated is not completely understood. Mature neurons and astrocytes are mitotically inactive and do not normally produce new progeny, but cell cycle re-entry of astrocytes can be achieved by affecting the oncogenic or tumor suppressor pathways such as Bmi-1 and Ink4a (8), which are often permanently altered in tumors (9). However, the cell cycle exit or re-entry mechanisms are not well enough understood to allow experimental control of turnover or regeneration.
The mechanisms of terminal differentiation vary from one cell type to another. In many tissues it is accompanied by the increase of Cip/Kip type of cdk inhibitors and epigenetic changes of the chromatin (10, 11), but the role of these factors in the differentiation of NCS to astrocytes and in their cell cycle exit are not fully understood. In this paper we studied the dynamics of differentiation and show that the cell cycle exit of the hippocampal NSCs to astrocytes is slow and gradual, and is accompanied with an increase in p21 cdk inhibitor and changes in histone modifications.
Materials and Methods
Cell lines and culture conditions
Rat adult hippocampal neural stem cells (SCR022 Millipore) were cultured according to the instructions in neural stem cell basal medium (SCM009 Millipore), containing 20 ng/ml FGF2. Cells were cultured on poly-ornithine-laminin coated cell culture plates at 37°C in 5% CO2.
Astrocyte differentiation
To differentiate the stem cells into astrocytes we applied astrocyte differentiation medium (SCM010 Millipore), which contains BMP, LIF and 1% serum, and cultured the cells on poly-ornithine-laminin coated cell culture plates at 37°C in 5% CO2.
Hoechst staining and cell cycle stages
Cells were fixed with 4% PFA for 10min at RT and post-stained with 2 μg/ml Hoechst 33342 (Invitrogen) for 1h at RT. Cell numbers were evaluated based on the number of nuclei by Cellomics high content screening platform (ArrayScan VTI, Thermofisher). Cell cycle bioapplication was applied with the following parameters: fixed threshold 100∼400, background correction 255. In each experiment cells were counted in 49 fields from at least 4 wells per condition. Typically 1,000∼3,000 cells/well were counted thus total number of cells per condition was at least 4,000 cells. For cell cycle curve the nuclear intensity in Ch1 (Hoechst) per cell was taken from the Cellomics and the graph was drawn in Microsoft Excel. Cell cycle curves were derived from the data of 25,000 to 45,000 cells per condition.
Immuncytochemistry
Cells were fixed with 4% PFA for 10min at RT and post-stained with 2 μg/ml Hoechst 33342 (Invitrogen) for 1h at RT. Blocking was done in 5% BSA+0,1 % saponin for 30 min at RT. Ki67 1 : 1000 (ab155580 Abcam), 2.-ry ab alexa-fluor anti-rabbit 488 or 594 (Invitrogen). GFAP 1 : 1000 (ab7260 Abcam), 2.-ry ab alexa-fluor anti-rabbit 488 or 594 (Invitrogen).
For 5-methylCytidine staining cells were fixed with −20°C 100% MeOH for 10 min at RT. Cells were treated with 1N HCl at 37°C for 30 minutes and washed with 0.1M Borate buffer pH 8.5, at RT (Polysciences Europe, 24092-500). Blocking was done in 1% BSA in PBS for 30 minutes at RT. Primary Ab was diluted in blocking buffer and incubated at 4°C o/n. 5-methyl Cytidine: Diagenode MAb-335MEC-100, 1 : 100. Secondary antibody was diluted in blocking buffer and incubated at RT for 1 hour. Alexa anti-mouse 1 : 200. Cells were counterstained in 2 μg/ml Hoechst 33342 (Invitrogen) for 1h at RT. Results were drawn from the Cellomics by Target Activation bio-application. Recorded number: MEAN_TotalIntenCh2.
For methyl-Histone staining cells were fixed with 4% PFA at RT for 15 minutes. Cells were permeabilized with 0.1% TritonX-100 in PBS at RT for 15 minutes. Blocking was done with 1% BSA in PBS for 30 minutes at RT. Primary antibody was diluted in blocking buffer and incubated 4°C o/n. H3K4me3: Diagenode pAb-003-050, 1 : 200. H3K27me3: Diagenode CS-069-050, 1 : 200. Secondary antibody was diluted in blocking buffer at RT for 1 hour. Alexa anti-rabbit 1 : 200. Cells were counterstained in 2 μg/ml Hoechst 33342 (Invitrogen) for 1h at RT. Results were drawn from the Cellomics by Target Activation bioapplication. Recorded number: MEAN_TotalIntenCh2.
BrdU staining
BrdU and staining was performed by BrdU Kit (11296736001 Roche). BrdU pulse was given to the cells for 24h after which the cells were fixed and BrdU Ab staining was performed according to the manufacturer’s instructions.
Quantitative RT-PCR
Hippocampal neural stem cells were cultured in astrocyte differentiation medium. RNA was extracted after 0d, 1d, 2d, 3d, 4d, 7d and 11d of culture (Aurum Total RNA kit 170-6820, Biorad). Total cDNA was transcribed by a reverse-transcriptase (iScript 170-8891, Biorad) and the relative amounts of Gfap and p21 mRNA were measured by Real-Time PCR (IQ supermix 170-8862 and iCycler, Biorad) using TaqMan probes for GFAP, p21 and Gapdh (Mm 01253033_m1, Mm01303209_m1, Mm99999915_g1, Applied Biosystems).
Results and Discussion
Rat adult hippocampal neural stem cell differentiation into the astrocyte lineage initiates rapidly and is accomplished in three days
We wanted to shed light on the events taking place during adult mammalian neural stem cell differentiation. We were specifically interested in the cell cycle exit and the changes in the biology of neural stem cells and investigated the differentiation of hippocampal neural stem cells to astrocytes. We asked how the differentiation is linked to cell cycle exit and what signals are involved. For this, the monolayer cultured rat adult hippocampal neural stem cells were differentiated towards the astrocyte lineage. The cells are able to produce also neurons and oligodendrocytes (12), but we chose this differentiation protocol since it repeatedly gave the most reproducible results and caused rapid changes in cell phenotype and strong expression of astrocyte specific genes.
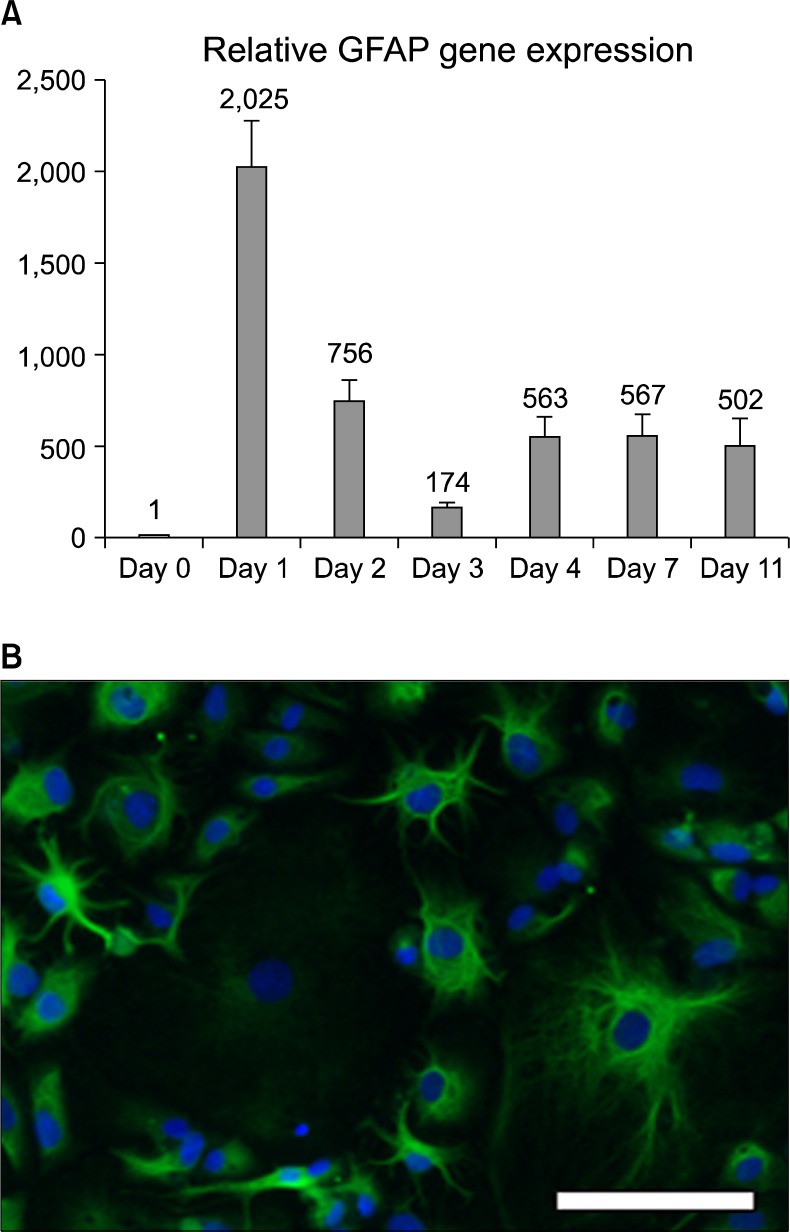
First we measured the intracellular molecular biology landscape of differentiation by quantitative RT-PCR and found rapid changes in transcript levels. After 16h of differentiation in these conditions, the amount of the astrocyte-specific Glial Fibrillary Acidic Protein (GFAP) RNA increased over 2000x, when measured with qPCR and compared to non-differentiated neural stem cells (Fig. 1A, day 1). Thereafter, the GFAP mRNA levels fluctuated and at day 4 of differentiation they stabilized to a level of approximately 500x, representing the GFAP transcription of a differentiated astrocyte. After 3 days of culture in differentiation medium the cells showed astrocyte morphology and strong GFAP immunoreactivity (Fig. 1B).
Fig. 1.
Hippocampal neural stem cell differentiation towards the astrocyte lineage in vitro. A) Analysis of the RNA content of the cells showed that astrocyte specific gene GFAP mRNA amounts already increased one day after the differentiation initiation. After an initial peak and some fluctuation, the mRNA content stabilized to a steady-state level. Error bars represent SEM between triplicates. (B) After differentiation, the hippocampal neural stem cells show astrocyte morphology and are GFAP immunopositive. Scale bar 20 μm.
Hippocampal neural stem cell differentiation into astrocytes leads to cell cycle exit of most cells in 4 days
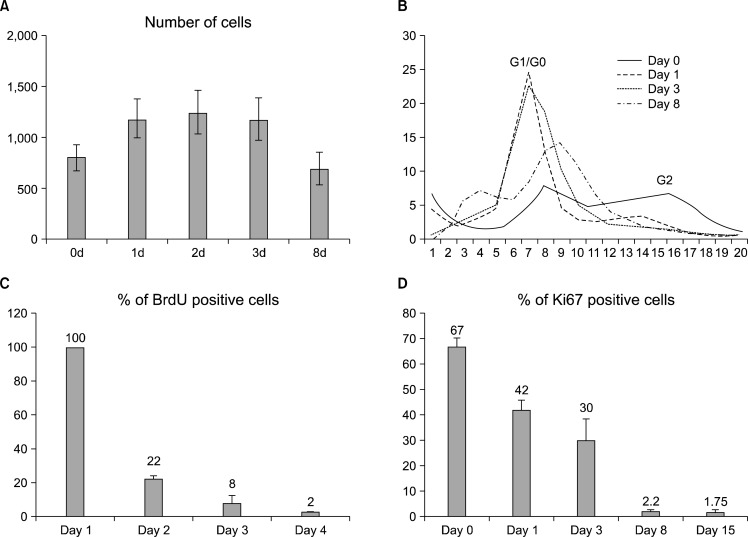
Terminal differentiation of astrocytes is associated with the cell cycle exit of the differentiating cells. We were interested in the timing and mechanisms of this terminal mitosis and followed the cell cycle progression during differentiation. We first analyzed the cell cycle events by counting the number of cells in culture during differentiation. The number increased in the first two time points of 24 h and 48 hours, after which the amount of cells at the population level stayed constant until a drop due to suspected apoptosis around day 8 (Fig. 2A). This indicates that most hippocampal stem cells stop proliferating by 48 h of differentiation.
Fig. 2.
Differentiation-induced changes in proliferation and cell cycle. (A) Cell number increases during the first 24 to 48 h of differentiation and stays constant until a decrease, possibly due to apoptosis. Error bars represent the SEM between triplicates. The development of the cell cycle can be followed in a curve (B), which represents the distribution of cells plotted by the DNA content (Hoechst staining intensity) per cell. The 2N peak representing the cells in the cell cycle phases G1/G0 is initially getting larger, but then decreases at day 8 as the amount of apoptotic nuclei increases. The 4N peak representing cells in G2 phase decreases during the differentiation. By day 8 there are no cells in the 4N peak, indicating that cells have stopped proliferating. 25,000 to 45,000 cells were analyzed in each condition. (C) Analysis of cell proliferation by BrdU and Ki67 show that proliferation decreases during differentiation. After 8 days of differentiation the amount of BrdU+cells is 2%. (D) The amount of Ki67 positive cells decreases indicating the increase of cells that are in G0 cell cycle phase. After 8 and 15 days of differentiation the amount of Ki67+cells is 2% and 1%, respectively. (C&D: more than 10,000 cells/condition were analyzed; error bars represent SEM between Cellomics’ microscope fields).
To confirm this proliferation arrest of the differentiating hippocampal stem cells, we measured the nuclear DNA content of the cell, which doubles during S-phase and thus distinguishes the G0/G1 phases from G2. At day 0 and day1 the cells showed a typical cell cycle profile of proliferating cells with distinct 2N and 4N peaks representing the G1/G0 and G2 cell cycle phases, respectively (Fig. 2B). However, in three days the 4N peak representing the G2 cells had almost vanished and most cells resided in 2N peak representing the cell cycle phases G0/G1 (Fig. 2B) indicating that most cells had gone out of cycle in about three days.
After 8 days in differentiation medium, the number of apoptotic cells was also increased represented by the far left peak showing the cells that contained less DNA than in G1/G0 cells (Fig. 2B). The morphological appearance of the nuclei was disturbed and non-regular (data not shown).
Taken together, the results showed that most differentiating cells responded to the differentiation signals by exiting the cell cycle within 2∼3 days. However, we were interested in the decisions of the individual cells during differentiation and therefore analyzed the cells more closely by monitoring BrdU incorporation in the nucleus, which occurs in the S phase of the cell cycle. As expected, the number of BrdU positive cells decreased during differentiation. According to the results, during the first 24 h of differentiation 100% of the cells were proliferating, but between the first and second days only 22% underwent S-phase, and between the second and third days only 8%. BrdU analysis also showed that after 8d of differentiation 2% of the cells were still able to incorporate BrdU (Fig. 2C). The indirect BrdU result was confirmed by a direct Ki67 antibody staining which indicates the number of cells that are in all other cell cycle phases except G0. The Ki67 analysis (Fig. 2D) supports the BrdU data since the number of Ki67 positive cells systematically decreased during differentiation from 67% in day 0 to 42% and 30% in days 1 and 3, respectively. The Ki67 staining indicated that there were still some (less than 2 %) of cells in cycle after an extended differentiation period of 15 days.
These results are intriguing since they suggest that even if the stem cells mostly respond to the differentiation signal as a population and exit the cell cycle, some of the cells seem able to maintain their capacity to divide and stay in the cycle. Since we made this observation with two different methods, it is unlikely a technical artifact. However, if some cells resist the terminal differentiation signal, the “mixed signals” of simultaneous differentiation and proliferation typically lead to elimination by apoptosis (13). This is actually in line with our results, where the number of cells in cycle and the total number of cells both decline during differentiation.
Cell cycle exit signaling
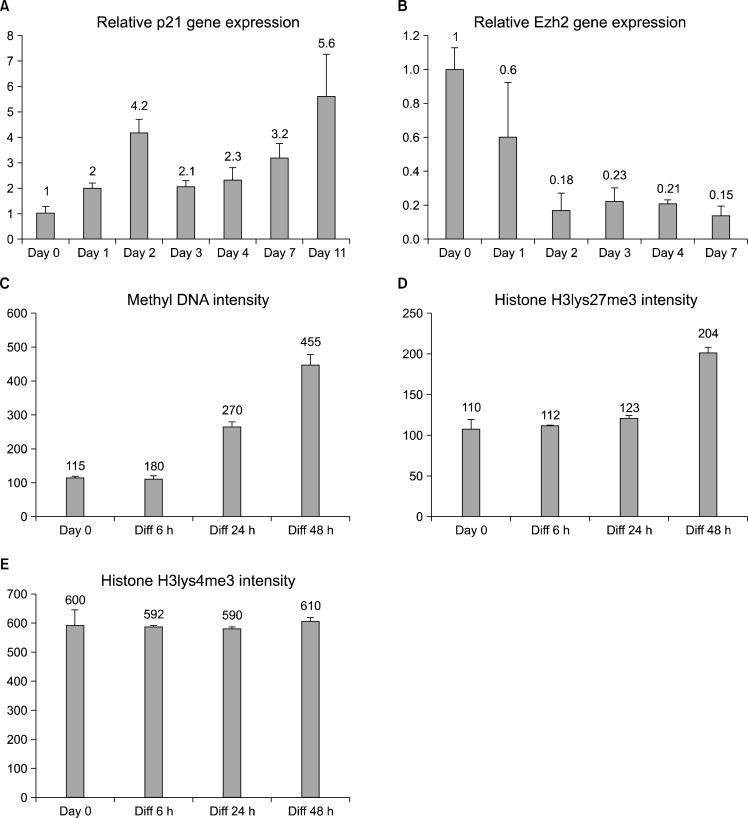
We further studied the timing of the differentiation-induced changes by first analyzing the transcript levels of p21. The cip/kip family of cdk inhibitors is an important cell cycle regulator of terminal differentiation and p21 is a pivotal marker in the cellular proliferation decisions (14), also in NSCs (15). We noticed that p21 increases after the first day of differentiation, but the expression peak was observed at day 2 indicating that the cells are still signaling to execute the cycle exit (Fig. 3A). There were fluctuations also in the levels of p21 RNA induction, but we detected a trend of gradual increase to a 5.5-fold induction by day 11 of differentiation. The qPCR result shows that p21 cell cycle regulator induction is slower and less robust compared to the GFAP levels of the differentiating hippocampal stem cells. This p21 cell cycle regulation timing corresponds with the cell cycle exit results (Fig. 2) and indicates that the hippocampal stem cell to astrocyte differentiation starts with the expression of tissue specific genes, and is followed by the cell cycle exit.
Fig. 3.
Differentiation-induced changes in cell cycle signaling and epigenetics. (A) qRT-PCR analysis using whole RNA extracted from differentiating hippocampal stem cells show that p21 transcript increases during the 11day experiment, indicating an activation of the cell cycle exit machinery in the cells. (B) Inhibitory polycomb group protein Ezh2 expression decreases during differentiation. Error bars represent the SEM between triplicates. (C, D, E) Immunostaining intensities of methylated cytidine bases of DNA as well as lysine27 and lysine4 trimethylation of Histone 3. On average, the signals of 2,500 nuclei were quantified/condition. Error bars represent the SEM between fields.
The transcription regulation of differentiation and cell cycle exit of adult hippocampal stem cells has not been thoroughly studied. Some studies on other cell types have addressed the terminal differentiation effects of DNA and histone methylation, factors that influence transcription (16). We investigated the epigenetic status of the adult hippocampal stem cells during astrocyte differentiation by a 7 day transcription analysis of polycomb group protein Ezh2 and short term analyses of methylated DNA and histone methylation antibody staining. These results show that the Ezh2 expression starts to decrease at day 2 of differentiation and stays low thereafter (Fig. 3B), as shown previously in embryonic telencephalic NSCs (11). We also noticed an increase in general DNA methylation and histone3 lysine27 trimethylation, but no changes were detected in the trimethylation of histone3 lysine4 when the intensities of the immunofluorescence stainings were analyzed (average n=2500) (Fig. 3C∼E).
The results show that adult hippocampal stem differentiation cell to astrocytes is accompanied by a general increase in methylation of the DNA and in the inhibitory Histone 3 methylation on lysine 27, but not in the activating Histone 3 methylation on lysine 4. These results suggest that hippocampal differentiation is associated with epigenetic silencing by methylation. However, the expression of the polycomb-type inhibitory gene Ezh2 was shown to decrease during differentiation, indicating that some specific release of transcriptional suppression could also be taking place concurrently.
A small percentage of hippocampal stem cells resists differentiation
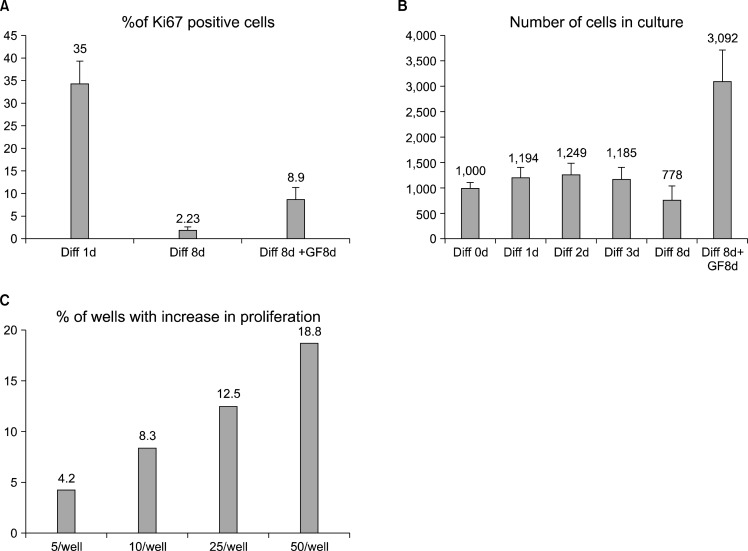
In the cell proliferation analyses (Fig. 2) we identified a small but constant number of BrdU and Ki67 positive cells after 15 days of differentiation. To answer the question whether some cells are still able to proliferate, or whether they are already determined to apoptosis, we differentiated cells for 8 days in differentiation medium and measured the number of cells regularly. After this, the cells were changed back to the FGF2 containing medium, and further cultured for 8 or 15 days. During the last part of the experiment the number of cells and the percentage of Ki67 positive cells increased to 8% (from the lowest detected value of 1∼2%) indicating that some cells had been able to resist the differentiation cues and were able to start to proliferate after 8 days in differentiation conditions (Fig. 4A).
Fig. 4.
A small percentage of hippocampal stem cells delay cell cycle exit. (A) Ki67 staining of cells that were first differentiated for 8 days, before being changed back to FGF2 containing growth factor media, which shows increase of the positive population from 2% to 8%. (B) Counting of the cells during 8d differentiation and subsequent media change shows that the population increases from 600 to 3,000 cells/ well. Error bars represent SEM between triplicate wells. (C) Percentage of the wells, containing cells that were able to resist terminal differentiation. 5, 10, 25 or 50 cells per well were plated in a 48 well plate in order to calculate how many cells are needed to find (at least) one cell that is able to proliferate. In these clonal densities, approximately 0.375 to 0.8% of the initially plated cells were able to return into proliferation, when FGF2 containing growth factor media was reintroduced after an initial 8 days of differentiation.
To confirm that after 8 days of differentiation some cells were still able to divide and proliferate, we measured the number of the cells in the population during different time points of differentiation and re-addition of the growth factors. The result showed that the cell number tripled in 8 days after the growth factors were added (Fig. 4B). This suggests that hippocampal stem cells do not proliferate in the differentiation conditions, but after being supplied with growth factors, some cells are again able to proliferate.
The measured increase in cell number is in the range of a small percentage of cells starting to double approximately once every day. To analyze the frequency of the proliferation-capable cells more accurately, we plated low numbers of cells sparsely on the 96-well plates. The cells were differentiated for 8 days after which they were cultured in growth factor medium for two weeks. To be able to retrospectively calculate the cells that had not permanently left the cycle, we plated 5, 10, 25, or 50 cells/well, i.e. densities in which the cells are completely separated from each other. After media change, we could see that 4 to 19% of the wells had a growing population of cells (Fig. 4C). This translates to a minimum of 0.375∼0.8% (presuming that only one cell started to proliferate) of the cells which had resisted the terminal cell cycle exit and were able to start proliferation after the switch back to FGF2 containing medium.
These results suggest that not all the cells in the differentiating stem cell population behave similarly. While most cells clearly start to express cell cycle inhibitors like p21 and exit the cell cycle in a few days, some cells retain their proliferation-capable stem cell phenotype. These cells do not proliferate under differentiating conditions, but stay quiescent and still have the ability to enter the proliferative mode when the conditions change become favorable. We hypothesize that if these cells do enter the cell cycle when the conditions favor differentiation, they fail to divide and enter the apoptotic pathway. Unfortunately, at the moment we lack the markers to prospectively identify or characterize the cells that can resist the terminal differentiation. In addition, the pathways controlling this ability are not known at present.
Conclusions
This article describes research findings on the timing of adult mammalian hippocampal stem cell cell cycle exit induced by differentiation. The results indicate that a) the phenotypic differentiation of these cells is fast and starts with a robust induction of differentiation-specific genes, while b) the cell cycle exit is slower and takes place 1∼2 days after initiation of differentiation. The differentiation involves c) changes in the transcriptional level and epigenetic regulation but d) a small percentage of hippocampal cells is able to resist differentiation and terminal cell cycle exit.
Footnotes
Potential Conflict of Interest
The authors have no conflicting financial interest.
References
- 1.Basak O, Taylor V. Stem cells of the adult mammalian brain and their niche. Cell Mol Life Sci. 2009;66:1057–1072. doi: 10.1007/s00018-008-8544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kriegstein A, Alvarez–Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehninger D, Kempermann G. Neurogenesis in the adult hippocampus. Cell Tissue Res. 2008;331:243–250. doi: 10.1007/s00441-007-0478-3. [DOI] [PubMed] [Google Scholar]
- 4.Oya S, Yoshikawa G, Takai K, Tanaka JI, Higashiyama S, Saito N, Kirino T, Kawahara N. Attenuation of Notch signaling promotes the differentiation of neural progenitors into neurons in the hippocampal CA1 region after ischemic injury. Neuroscience. 2009;158:683–692. doi: 10.1016/j.neuroscience.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 5.Carlén M, Meletis K, Göritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabé-Heider F, Yeung MS, Naldini L, Honjo T, Kokaia Z, Shupliakov O, Cassidy RM, Lindvall O, Frisén J. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- 6.Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Moon JH, Yoon BS, Kim B, Park G, Jung HY, Maeng I, Jun EK, Yoo SJ, Kim A, Oh S, Whang KY, Kim H, Kim DW, Kim KD, You S. Induction of neural stem cell-like cells (NSCLCs from mouse astrocytes by Bmi1. Biochem Biophys Res Commun. 2008;27:267–272. doi: 10.1016/j.bbrc.2008.04.068. [DOI] [PubMed] [Google Scholar]
- 9.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 200570:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 10.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retonoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;31:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sher F, Rössler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 12.Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol Cell Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 13.Slack RS, El-Bizri H, Wong J, Belliveau DJ, Miller FD. A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J Cell Biol. 1998;140:1497–1509. doi: 10.1083/jcb.140.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]