Abstract
Colonizing commensal bacteria after birth are required for the proper development of the gastrointestinal tract. It is believed that bacterial colonization pattern in neonatal gut affects gut barrier function and immune system maturation. Studies on the development of faecal microbiota in infants showed that the neonatal gut was first colonized with enterococci followed by other microbiota such as Bifidobacterium. Other studies showed that babies who developed allergy were less often colonized with Enterococcus during the first month of life as compared to healthy infants. Many studies have been conducted to elucidate how bifidobacteria or lactobacilli, some of which are considered probiotic, regulate infant gut immunity. However, fewer studies have been focused on enterococi. In our study, we demonstrate that E. faecalis, isolated from healthy newborns, suppress inflammatory responses activated in vivo and in vitro. We found E. faecalis attenuates proinflammatory cytokine secretions, especially IL-8, through JNK and p38 signaling pathways. This finding shed light on how the first colonizer, E.faecalis, regulates inflammatory responses in the host.
Introduction
Significant controversy exists over the role of Enterococcus, and more specifically E.faecalis, on health. Whereas, in clinical settings with immune-compromised patients, E.faecalis can be considered an opportunistic pathogen [1], it has also been shown to impart beneficial effects to health. A recent in vitro study demonstrated that E. faecalis was inhibitory to C. jejuni MB 4185 infection under simulated broiler caecal condition [2]. An E.faecalis isolated from a healthy adult showed the highest probiotic activity when compared with over 70 other lactic acid bacteria (LAB) isolates, including lactobacilli and bifidobacteria [3]. These contrasting roles suggest an interplay between bacteria and human host that is context-dependent and likely dynamic over time.
Prior to birth, the gut is sterile and bacteria start to colonize after birth. The neonatal gut, with a naïve but competent immune system, represents a valuable context for determining the role of particular bacteria in health. Proper development of the gastrointestinal tract requires timely colonization after birth [4]. Study showed microbiota acquisition in infancy is likely a determinant of early immune programming, subsequent infection, and allergy risk [5]. Among the first wave of microorganisms detected in the stool of infants, enterococci are commonly found on the first day of life [6], [7]. They gradually decrease with concurrent increases in bifidobacteria that appear within 2–3 days in breast fed infants [8]. Babies who developed allergies were less often colonized with Enterococcus during the first month of life as compared to healthy infants [9]. This implies that Enterococcus could have major impact on intestinal immune development in the very early stage of life.
Several factors, such as mode of delivery and gestational age, are known to influence the composition of microbial flora in early stage of life. Preterm infants and infants delivered via cesarean section display a delayed intestinal colonization with smaller species variability and a higher occurrence of potentially pathogenic microorganism [10], [11]. Pre-term births are far more likely to suffer necrotizing enterocolitis (NEC) than are births at term [12]. Interestingly, infants who developed NEC carry less E.faecalis [13]. Previous studies have shown that serum concentrations of IL-8 were elevated in severe cases of NEC from its onset through the first 24 hours [14]. IL-8 is a chemokine that stimulates migration of neutrophils from intravascular to interstitial sites and can directly activate neutrophils and regulate the expression of neutrophil adhesion molecules [15]–[17]. Thus, IL-8 plays an important role in infant infections.
It is also known that cell-wall components from Gram-negative such as lipopolysaccarides as well as host-derived cytokines such as IL-1β and TNF-α, increase IL-8 secretion from IECs through the activation of mitogen activated protein kinase (MAPK) [18], [19]. At least three groups of MAPKs have been identified. These include the extracellular signal-regulated kinases (ERKs), the c-JUN NH2-terminal kinases (JNKs) and p38. P38 was reported to stabilize IL-8 mRNA and its level was increased in the muscularis propria of colonic tissue both in DSS colitis mice and patients with inflammatory bowel disease (IBD) [20]. A p38 inhibitor suppressed inflammation in DSS-induced colitis model by reducing mucosal IL-1β and TNF-α levels [21]. Inhibition of JNK activation correlated with suppression of IL-1β-induced IL-8 secretion in IECs [22]. Study showed activation of the ERK signaling pathway in response to TNF-α in HT-29 cells leads to increased expression of IL-8 [23].
Extensive studies have been performed to determine how lactobacilli and bifidobacteria regulate infant gut immunity [24], [25]. However, few studies have focused on E.faecalis, which is the first colonizer in the human GI tract [6], [7]. Here, we demonstrate that E. faecalis, isolated from newborns, can suppress pathogen-mediated inflammatory responses in human IECs as well as DSS-induced inflammation in mice model. E. faecalis attenuates proinflammatory cytokine secretions, especially IL-8, via distinct pathways. These bacteria suppress JNK and p38 as well as disrupt c-JUN-regulated inflammatory responses. These findings shed lights on functions of the first colonizer, E.faecalis, in infant gut protection.
Results
Isolation and identification of bacteria from infants' gut
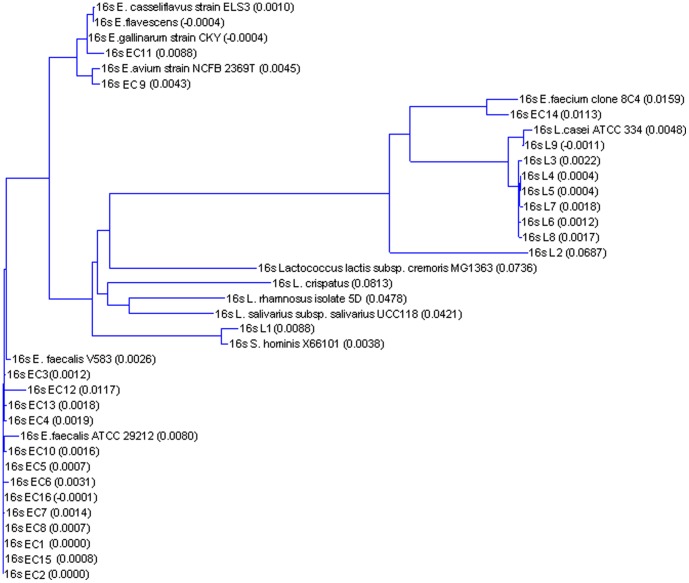
In order to characterize early postnatal gut LABs, we collected feces from 16 healthy infants aged 3 days and 1 month from Indonesia. A total of 25 isolates were expanded and confirmed as LABs based on lactic acid production, a rod or coccal shape, and Gram positivity. Based on their carbohydrate fermentation patterns, nine strains were categorized as Lactobacillus and 16 strains as Enterococcus. 16S rDNA sequence analysis showed that 8/9 of the lactobacilli were Lactobacillus casei and 13/16 of the enterococci were Enterococcus faecalis (Table 1). Thus, we found a restricted diversity of early colonizing species in infants with L.casei and E. faecalis being the prominent early colonizers. A phylogenetic tree, plotted based on a sequence distance method, is provided (Figure 1). From the phylogenetic tree, we could see some enterococcus were closer to lactobacillus than the rest enterococcus.
Table 1. Identification of 25 isolates from infants.
| Bacterial names | Bacterial source (Infants Age) | Strain ID obtained from 16S rDNA sequencing |
| L1 | 3day | Staphylococcus hominis |
| L2 | 3day | Lactobacillus casei |
| L5 | 3day | Lactobacillus casei |
| L6 | 3day | Lactobacillus casei |
| EC3 | 3day | Enterococcus faecalis |
| EC6 | 3day | Enterococcus faecalis |
| EC9 | 3day | Enterococcus avium |
| EC14 | 3day | Enterococcus faecium |
| EC15 | 3day | Enterococcus faecalis |
| EC16 | 3day | Enterococcus faecalis |
| L3 | 1month | Lactobacillus casei |
| L4 | 1 month | Lactobacillus casei |
| L7 | 1month | Lactobacillus casei |
| L8 | 1month | Lactobacillus casei |
| L9 | 1month | Lactobacillus casei |
| EC1 | 1month | Enterococcus faecalis |
| EC2 | 1month | Enterococcus faecalis |
| EC4 | 1month | Enterococcus faecalis |
| EC5 | 1month | Enterococcus faecalis |
| EC7 | 1month | Enterococcus faecalis |
| EC8 | 1month | Enterococcus faecalis |
| EC10 | 1month | Enterococcus faecalis |
| EC11 | 1 month | Enterococcus gallinarum |
| EC12 | 1 month | Enterococcus faecalis |
| EC13 | 1 month | Enterococcus faecalis |
Figure 1. Phylogeny tree of 25 bacterial strains isolated from new born infants according to their 16sRNA sequences.
Enterococcus faecalis suppress intestinal IL-8 secretion
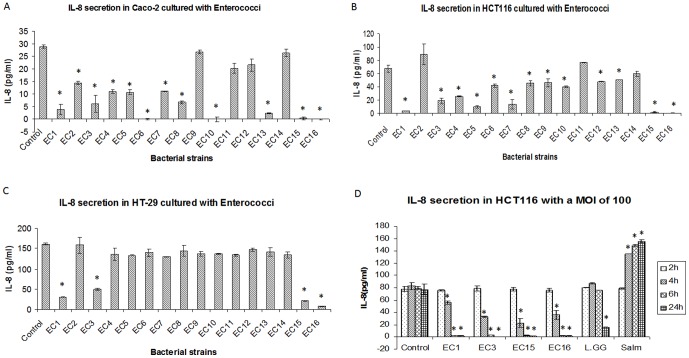
We then examined how these early colonizing bacteria can impact intestinal immunity and epithelial cell signaling. Potential anti-inflammatory effects of infant-isolated enterococci and lactobacilli were investigated by co-culturing them with colorectal cancer-derived IECs (Caco-2, HT29 and HCT116). Supernatants were harvested for IL-8 analysis as a marker for gut inflammation [26]. Although some lactobacilli suppressed IL-8 secretion in Caco-2 cells, none of the Lactobacillus isolates significantly attenuated IL-8 secretion in all the three cell lines (Figure S1A–C). In contrast, the majority of Enterococcus isolates suppressed IL-8 production in Caco-2 and HCT116 cells (Figure 2A, B). Strikingly, four strains, namely EC1, EC3, EC15 and EC16, suppressed IL-8 secretion in all three lines (Figure 2 A–C and the reduction of IL-8 levels was not due to apoptosis induced by E.faecalis (Figure S1D, E). We then tested the kinetics of IL-8 suppression by these four strains in HCT116 cells (Figure 2D). For each of these isolates, the degree of suppression of IL-8 was dependent on bacterial multiplicity of infection (MOI). E. faecalis isolates suppressed IL-8 secretion from 4h at a MOI of 100. However, the same level of suppression was observed much earlier at a MOI of 1000. In contrast, this rapid and robust suppression of IL-8 was not seen with commercial probiotic strains, L. rhamnosus GG (L.gg) (Figure 2D). Salmonella typhimurium (Salm), a known pathogen and stimulator of IL-8 [27], activated IL-8 secretion after 2 h (Figure 2D). These data suggest that early colonizing E.faecalis have potent anti-inflammatory effects as assessed using IL-8 expression as a marker.
Figure 2. IL-8 secretion in IECs.
IL-8 secretions in Caco-2 (A), HCT116 (B) and HT-29 (C) with the treatment of Enterococcus. A total number of 107 cfu/ml bacteria were added into the cells for 6 h. Supernatants were harvested for cytokine assay as described in Materials and Methods. (D) HCT116 cells were co-cultured with E. faecalis (EC1, EC3, EC15, EC16) and L. rhamnosus GG (L.GG) and S. typhimurium (Salm) with a multiplicity of infection (MOI) of 100 for 2 h, 4 h, 6 h and 24 hours. Three independent experiments were compiled to produce the data shown. Data were expressed as mean value ±SD. Student's T-test was used for statistical analysis as described in Materials and Methods. * p<0.05.
Active E. faecalis physiology is not critical for suppression of intestinal inflammation
We then investigated what aspects of E. faecalis may cause reduced IL-8 secretion in IECs. In order to test if intact bacterial physiology was critical for IL-8 suppression, we exposed HCT116 cells to live bacteria or bacteria that had been killed by ultraviolet exposure (UV) or by physical disruption through sonication immediately before use (Figure 3). In each case we found that IL-8 suppression remained intact, indicating that active bacterial physiology was not critical for this activity. We then tested whether physical contact between bacterial membranes and epithelial cells was important. Physically separating live bacteria from the epithelial cells, using a semi-permeable cell culture insert, greatly relieved the IL-8 suppression (Figure 3), indicating that physical contact between the bacteria and epithelial cells was important for this activity. Furthermore, bacteria-conditioned media from bacteria alone or from mammalian cell cocultures resulted in no apparent IL-8 suppression (Figure 3), suggesting that epithelial cells are responding to poorly soluble factor(s), likely present on the exterior cell wall of E. faecalis.
Figure 3. IL-8 secretions in HCT116 cells.
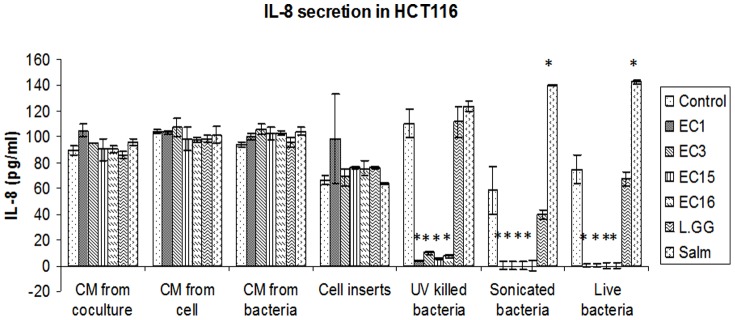
IL-8 production in HCT116 with the treatment of different conditional medium (CM) (from coculture supernatant, from bacteria and from cell supernatant), cell inserts, UV killed and sonicated bacteria. Whole live bacteria were used as control as described in Materials and Methods. Three independent experiments were done and data were expressed as mean value ±SD. Student's T-test was used for statistical analysis as described in Materials and Methods. * p<0.05.
E. faecalis suppresses IL-8 production induced by IL-1β, TNF-α and S. typhimurium
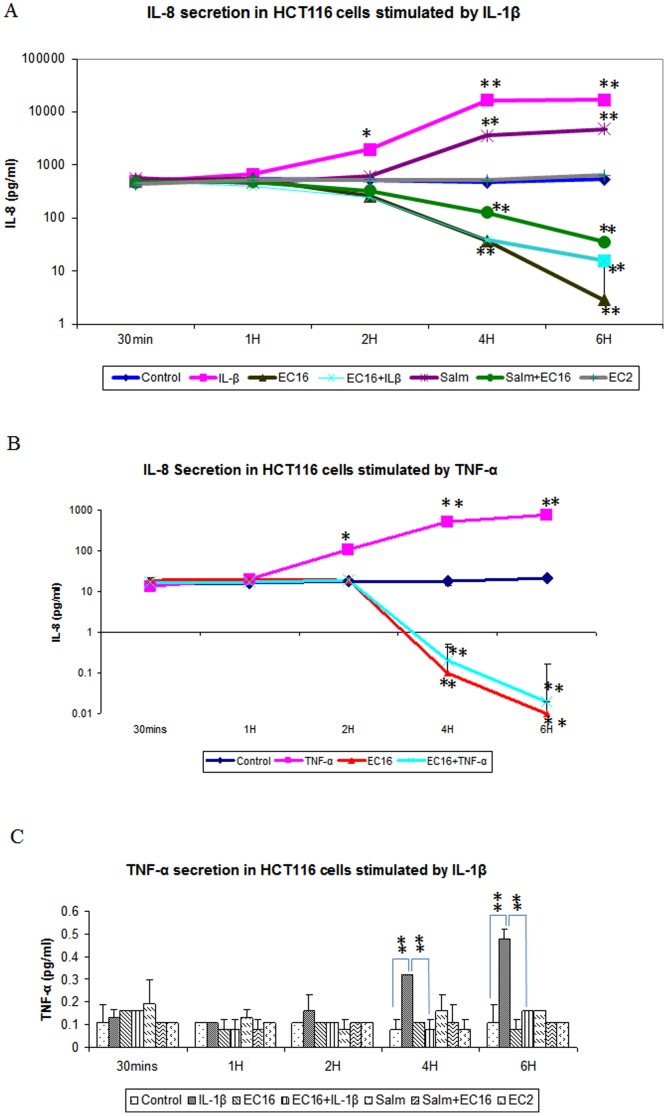
We next tested the ability of E. faecalis to regulate IL-8 production induced by IL-1β (2 ng/ml), TNF-α (200 ng/ml) and S. typhimurium (107 CFU/ml) in Caco-2 and HCT116 cells. We found that E. faecalis EC16 suppressed IL-8 production induced by IL-1β, TNF-α and S. typhimurium after 1 h of incubation. The suppression of IL-8 production inside the cells was observed at 30 mins of treatment (Figure S2A,B). This phenomenon was observed in both HCT116 (Figure 4A,B) and Caco-2 (Figure S2C) cells. Interestingly, E. faecalis EC2 could not suppress IL-8 production either inside or outside of IECs, suggesting distinctive ability of these four E. faecalis strains (EC1, EC3, EC15, EC16) in regulating IL-8 production.
Figure 4. Cytokine secretions in HCT116.
IL-8 (A) and TNF-α (C) production in HCT116 with the treatment of 2 ng/ml of IL-1β and 107 cfu/ml of S. typhimurium and E. faecalis (EC16 and EC2) as described in Materials and Methods. (B) IL-8 production in HCT116 with the treatment of TNF-α and E. faecalis EC16 as described in Materials and Methods. Data was expressed as mean value ±SD. Student's T-test was used for statistical analysis as described in Materials and Methods. *p<0.05, **P<0.01.
E. faecalis suppresses TNF-α expression induced by IL-1β
In addition to IL-8, we tested potential TNF-α regulation by E. faecalis in HCT116 and Caco-2 cells. TNF-α is an important regulator of epithelial inflammation. TNF-α levels are elevated in both human inflammatory bowel diseases and animal models of intestinal inflammation [28]–[30]. As expected, we found that TNF-α secretion from HCT116 (Figure 4C) and Caco-2 (Figure S2D) cells were activated by IL-1β. This increase secretion, however, was attenuated by E. faecalis EC16. Interestingly, ICAM1, IL-2, IL-5, IL-17 and INF-γ induction by IL-1β and S. typhimurium was suppressed by EC16 in Caco-2 cells (Figure S2D).
E. faecalis regulates multiple immune-signaling pathways
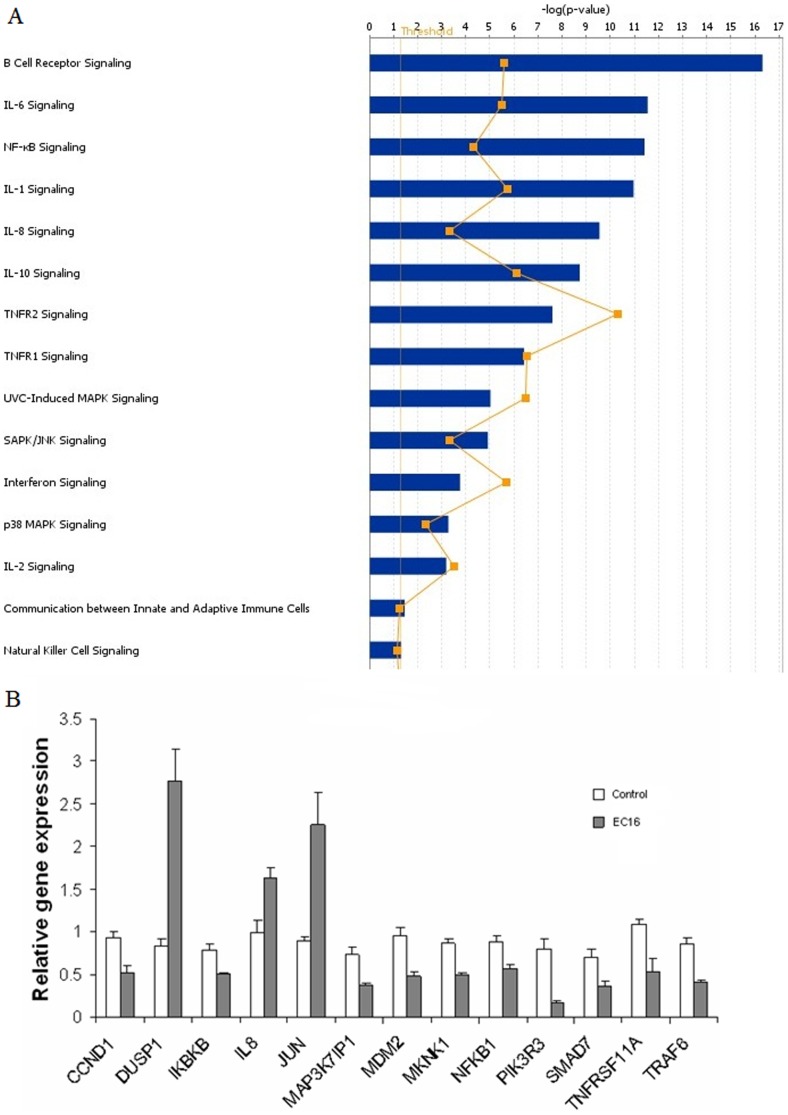
Our finding that E.faecalis could suppress IL-8 expression led us to investigate if these bacteria could regulate other immune-signaling pathways. We tested immune-gene expression by the four strains of E. faecalis (EC1, EC3, EC15, EC16) using cDNA microarray analysis. Potential expression changes for 406 immune-signaling genes were assayed in Caco-2 cells after 6 hour coculture with E. faecalis isolates. Acquired data from array membranes were initially scanned (Figure S3A) and volcano plots were obtained to identify statistically significant gene expression changes (Figure S3B). A partial list of the genes that demonstrate statistically significant expression changes (>1.5 fold) by cDNA microarray analysis is provided in Table 2. From the data of microarray, we found several signaling pathways that may be involved in the anti-inflammatory effects of the four E. faecalis strains. Using Ingenuity Pathway Analysis, we mainly identified cytokine signaling (IL-1, IL-2, IL-6, IL-8 and IL-10 signaling), SAPK/JNK signaling, P38 MAPK signaling and NF-κB signaling are responsible for the responses (Figure 5A). Together these data suggest that E. faecalis may alter multiple immunomodulatory pathways simultaneously.
Table 2. Significantly regulated gene (P<0.05) in Caco-2 cells treated with E. faecalis for 6h measured by cDNA microarray.
| Functional category | Gene population |
| Response protein | GBP1,DUSP1; SLC2A4, NPPB, |
| Receptors | EGFR;IL8RA, TNFRSF11A, P2RX7 |
| Transcription factors | MLLT7, EGR1, ETS1;MAPKAPK2, MAPKAPK3, MAX, MEF2B, MKNK1, SP1 |
| AKT& PI3K family | PIK3R3, TCL1A, TCL1B |
| MAKP family | MAPK13;MAPK7, MAP4K1 |
| Adaptor | TNFAIP3, MDM2; |
| Nuclear factors | NCOA1, SPI1 |
| Signal transduction kinases | IKBKB, MAP3K7IP1, PRKCA |
| Rel/NF-kb | NFKB1, NFKBIL2, NFKB2 |
| Others | SMAD4, SMAD1, BCL2; NFATC2, NCK2, PPP3CB, PPP3CC, CTLA4, SMAD6. |
Note: Bold italic labeled genes are those upregulated. The rest are downregulated genes.
Figure 5. IPA and Realtime PCR analysis.
Ingenuity pathway analysis (A) and Real time PCR (B) on E.faecalis treated HCT116 cells. All pathways and genes showed in the figure were significantly (p<0.05) regulated in HCT116 cells with the treatment of E. faecalis EC16 for 6 h at a MOI of 100. Experiments were done on 3 biological replicates.
From the microarray analysis, we selected 46 genes for further study using TLDA. The gene ID and Taqman probes are listed in Table S1. Each of the four E. faecalis isolates demonstrated a similar pattern on the immune-gene regulation in Caco-2 and HCT116 cells. Data for gene-regulation by isolate EC16 is provided as a representative example (Figure 5B). To our surprise, we found that IL-8 mRNA was unregulated. We also found that DUSP1, a reported MAPK phosphatase that can attenuate MAPK signaling, was strongly upregulated. Other genes involved in MAPK signaling were downregulated in Caco-2 cells, namely MAP3K7IP1, a positive regulator of the MAP kinase cascade [31]; MKNK1, a target of ERK and activator of CREB-mediated proliferation and differentiation [32] and MAPKAPK2, a target of p38 MAP kinase involved in many cellular processes including inflammatory responses (Figure S4A). Other MAPK family members like MAPK7 in Caco-2 cells, MAPK9 in HCT116 cells were also suppressed by E. faecalis (Figure S4B,C). From the above data, we hypothesize that early-colonizing E. faecalis might influence the inflammatory responses in the host by regulating MAPK signaling pathway. Furthermore, consistent with observations from microarray, TLDA results also showed that NF-κB1 and IKBKB were suppressed by E. faecalis in both Caco-2 and HCT116 cells (Figure S4D,E) suggesting that E. faecalis suppressed NF-κB1 signaling pathway at transcriptional level.
E. faecalis suppresses activation of P38, P-JNK and C-JUN
Because factors regulating MAPK signaling were altered upon E.faecalis exposure, we hypothesized that these bacteria may attenuate IL-8 production, at least in part through inhibiting MAPK pathways. To test this, we examine MAPK expression and phosphorylation by immunoblotting. Whereas p-JNK was strongly and transiently activated by IL-1β and S. typhimurium in HCT116 cells within half hour of treatment, E. faecalis suppressed this activation (Figure 6A). Consistent with this finding, cJUN which is downstream of JNK, was activated by IL-1β and S. typhimurium at one hour while suppressed by E. faecalis EC16 (Figure 6B). Similarly, the presence of phosphorylated p38 was increased by incubation with either IL-1β or S. typhimurium at one hour, which was abrogated by coculture with E. faecalis EC16 (Figure 6C). Therefore, E. faecalis can inhibit JNK and p38 signaling as one potential means of suppressing IL-8 production. In contrast, ERK phosphorylation did not change upon the treatment of IL-1β, S. typhimurium and E. faecalis (Figure S5), suggesting that ERK does not impact IL-8 production in these cells. These data suggest that E.faecalis may suppress inflammatory signaling in IECs through reducing the activation of JNK and p38.
Figure 6. Protein expression in HCT116JNK expression (A) at 30 mins, c-JUN (B) and P38 (C) expression at 1 h in HCT116 with the treatment of 2 ng/ml of IL-1β and 107 cfu/ml of S. typhimurium, E. faecalis EC16 as described in Materials and Methods.
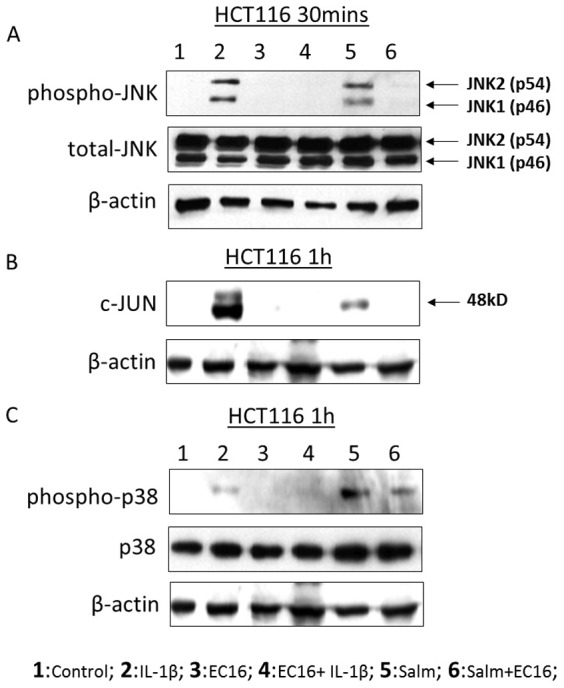
Cells were then lysed and proteins were tested using western blot. Three independent experiments were done.
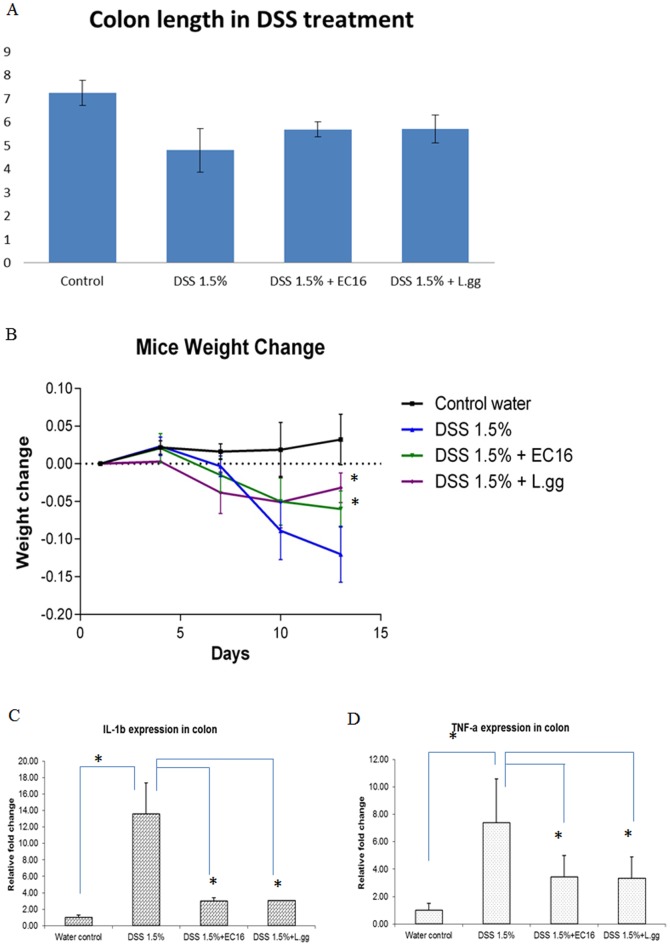
E.faecalis suppress IL-1β and TNF-α expression in DSS induced colitis mice model
To examine the immunomodulatory effects of E.faecalis in vivo, we induced colitis using Dextran Sulphate Sodium salt (DSS) in mice and then treated them with E.faecalis EC16 or Lactobacillus rhamnosus GG (L.gg), a well studied probiotic strain. Colon length, which is an indicator of inflammation, was shortened by DSS treatment. Consistent with an immunorepressive role, both E.faecalis and L.gg significantly alleviated this shortening (Figure 7A). Furthermore, E.faecalis as well as L.gg prevented DSS-induced weight loss in these animals (Figure 7B). IL-1β and TNF-α which were activated by DSS were significantly down regulated by EC16 and L.gg (Figure 7C,D)
Figure 7. E.faecalis treated DSS mild colitis model.
Colon lengths (cm) (A) and Mice weight change (B) with 1.5% DSS with or without EC16 and L.GG treatment as described in Material and Method. Real time PCR on IL-1β (C) and TNF-α (D) expression in colon. Each group contains 6–8 mice. Data was expressed as mean value ±SD. Student's T-test was used for statistical analysis as described in Materials and Methods. * p<0.05, ** P<0.01.
Discussion
In this study, we describe the isolation and primary characterization of E.faecalis strains isolated from healthy newborns in Indonesia. E.faecalis represented the most frequently isolated bacteria from the children and selected isolates possessed ability to strongly inhibit inflammatory markers in IECs. The selected isolates with the most potent anti-inflammatory effect were characterized to define a potential mechanism of action and we found that MAPK pathways were regulated. Specifically, E. faecalis was able to mitigate activation of JNK and p38 coincident with reduced expression of IL-8 in vitro as well as IL-1β and TNF-α in vivo.
Enterococci have previously been reported among the common early colonizers in humans [6]. Thus our study population in Indonesia provides a surprising consistency of early colonizing bacteria. Since E.faecalis accounts for 90-95% commensal enterococci in adult human intestines, the finding of E.faecalis as more prominent than the next most prominent species, E.faecium, is not unexpected. In our study L.casei was the second most frequently isolated lactic acid bacteria in 3 day old infants. Although the study cohort is not big, to our knowledge this is the first report of lactobacillus among early colonizers, which might reflect environmental differences for this Indonesian cohort.
None of the lactobacilli isolates were strongly immunosuppressive as determined by IL-8 expression levels. Certain E.faecalis isolates, on the other hand, could strongly inhibit IL-8 in IECs. Our previous findings showed that E.faecalis can induce anti-inflammatory cytokine IL-10 in intestinal epithelial through PPAR-gamma [33]. Taken them together, this may explain lower abundance enterococcus colonization [34] and high rate of NEC in preterm infant[35]. Furthermore, infants who developed NEC carry less E.faecalis [13], [36]. Therefore, E.faecalis may possess the capacity to modulate and attenuate the inflammatory responses further to prevent inflammatory diseases such as NEC in infants. Interestingly, the behavior of the three IEC lines differed substantial in response to many bacteria, suggesting that, in contrast to standard analyses, the use of a single IEC for characterization may be inadequate. The anti-inflammatory effects were confirmed in an in vivo system. This finding is consistent with previous study showing that E.faecalis has a great protective effect in DSS-induced experimental colitis model in mice [37]. It is not clear, at this stage, why only certain isolates of E.faecalis in this study could reduce IL-8 production in vitro. It will be interesting to potentially examine these other isolates for their behavior in DSS-induced colitis model in vivo.
Interestingly, while E.faecalis isolates decreased IL-8 protein level inside and outside the cells, its mRNA level was upregulated. Recent researches also showed that MAPK can regulate eIF4E [38] and eIF4E overexpression is associated with increased IL-8 expression [39]. Therefore, inhibition of MAPK pathway might suppress eIF4E, which results in reduction in IL-8 translation. In the DSS-colitis model as well as patients with inflammatory bowel disease (IBD), p38 levels are increased in the muscularis propria of colonic tissue [20]. When treated with p38 inhibitor, mucosal IL-1β and TNF-α levels were reduced in DSS colitis model [21] consistent with what we found for E.faecalis treatment. Thus, the phenomena we see with E.faecalis may be exclusively a result of MAPK inhibition. Alternately, E.faecalis may be affecting other pathways, for example our previous findings showed LAB can suppress TLR3, TLR9 and TRAF6 mRNA levels [40]. Therefore E.faecalis could suppress TLR pathways and further suppress MAPK pathway as well as NFκB-mediated transcription.
E.faecalis, as one of the first colonizer, could suppress the inflammatory responses and shape the immune system. Infant intestine usually undergo acute inflammation when exposed to Gram-negative bacteria. The presence of E. faecalis may help the intestine maintain the immune balance in response to such challenges. Our finding that E.faecalis performed as good as recognized probiotics indicates their potential to serve as a probiotic. However, the therapeutic effects must be examined thoroughly as E.faecalis was also reported as an opportunistic pathogen in hospital infections. Since we found dead E.faecalis also have the ability to suppress IL-8 secretion, the use of dead E.faecalis could mitigate the risk of opportunistic E.faecalis infection.
Materials and Methods
Bacteria culture and Identification
All bacterial strains were isolated from 16 healthy infants in Indonesia. This study is reviewed and approved by the National University of Singapore Institutional Review Board (NUS IRB), approval no. NUS1469. A written consent form was obtained from participants guardians. The inclusion criteria included natural birth and breast feeding. The exclusion criteria included antibiotic intake 2 weeks before and within the study, received probiotics/culture milk 2 months prior and during the course of the study.
Those bacteria were first Gram stained followed by API 50 CH test strips for rod-shaped bacteria and rapid ID 32 STREP test for the coccal shaped strains. All bacteria strains were then cultured and extracted for DNA. 16S rDNA were directly sequenced using primer 1100 reverse 5V-GGGTTGCGCTCGTTG-3V to obtain partial sequence of the 16S rDNA [41]. Phylogenetic tree calculation was based on a sequence distance method and utilizes the Neighbor Joining (NJ) algorithm of Saitou and Nei [42].
Cell culture and Infection
Caco-2, HT-29 and HCT116 were obtained from American Type Culture Collection (Manassas, VA) and maintained in ATCC recommended medium. Before cell infection, 1×105 cells were cultured in sterile 24-well flat-bottom plates (Nalge Nunc International, USA) for 24 hours. Caco-2, HT-29 and HCT116 cells were incubated in fresh medium without (control) or with bacteria at a multiplicity of infection (MOI) of 100 for 6 hours. Supernatants were harvested for ELISA assay (BD bioscience, San Diego, CA). and proteins were harvested for Western blotting analysis.
Conditional Medium and Cell Inserts
The cell culture supernatants obtained from IECs, bacteria and coculture of bacteria and IECs were harvested sterilely and conditional media were then added to the cell cultures prepared. The supernatant were then collected for cytokines assay. For cell inserts test, HCT116 cells were plated in wells of Transwell (USA) multiple well plate. 100 µl of bacterial suspensions were added to each insert. To sonicate bacterial cells, protease inhibitors were added to the suspensions. The bacterial cells were then disrupted by sonication (SANYO, Japan) on ice for 10 cycles with 30 sec pulses and 1min rest [43]. Cell debris was pelleted down and added in the cell culture prepared as above. Another bacterial suspensions were exposed to UV light for 5 min [44] to make sure at least 99% bacteria were killed and then co-culture with cells.
TNF-α, IL-1β and S. typhimurium induced cytokine secretion and protein production
200 ng/ml TNF-α (Preproteck, INC, rocky hill, NJ), 0.2 ng/ml IL-1β (Preproteck, INC, rocky hill, NJ) and S. typhimurium at a MOI of 100 were added to Caco-2 and HCT116 cells. Cells were then infected with E. faecalis EC16 with or without TNF-α/IL-1β/S. typhimurium. Cells without any treatment were used as controls. The cells were then cultured at 37°C with 5% CO2 for 30 mins, 1 h, 2 h, 4 h and 6 h. Supernatants were collected and the concentration of IL-8 was determined by ELISA (BD bioscience, San Diego, CA). Other cytokines like INF-γ, TNF-α, IL-2, IL-5, IL-17 and ICAM-1 were determined using cytokine assay (Bio-Rad, USA). The proteins were harvested for Western blotting assay.
Microarray analysis
A total of 406 human immunology signaling pathway related cDNA clones were detected in this study (Superarray, USA) according to manufacture's instruction. Raw data are available at Gene Expression Omnibus with accession number of GSE56485. Signals were analyzed using the web-based GEArray Expression Analysis Suite (Supperarray, US). GeneSpring GX 7.3.1 and Ingenuity pathway analysis was also used to analyze the data. Data were excluded with bad or absent flags. In this experiment, 2-3 replicates were done for one treatment. Student t test was applied for controlling the false-positive rate (P<0.05 was considered significant), clustering analysis was generated by the software.
TaqMan Low Density Array (TLDA)
The RNA was harvested using the RNA extraction kit (Roche, Switzerland). A 48-well format Taqman low density array was designed for a subset of genes that were differentially expressed in the array experiments including two endogenous controls 18 s and β-actin (Applied Biosystems, USA). Genes and ABI assay IDs are listed in Table S1. 0.5 µg total RNA was converted to cDNA using High-Capacity cDNA archive kit (Applied Biosystems, USA) and 10 ng cDNA in 100 µl TaqMan universal PCR master mix (Applied Biosystems, USA) was used for each port and run on an ABI 7900 system (Applied Biosystems, USA). Data was analyzed using the SDS2.2 software where baseline and threshold settings were automatically adjusted.
Sodium-Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and immuno blotting
Sodium-dodecyle sulfate-polyacrylamide gel electrophoresis was performed on a 10% gel by using 20 µg per lane of whole cell lysate. Electrophoresis was carried out using consistent voltage of 75 volts for approximately one and a half hour and then transferred onto a 0.22 µm Nitrocellulose membrane (Biorad, USA) at 85 volts for 2 hours in cold room. The membrane was then blocked in Tris buffer saline-Tween (TBST) containing 5% skim milk for at least 1 h. Specific primary antibodies were then diluted and the membrane was incubated overnight at 4 °C on an orbital shaker (Bellco, USA). After washing, appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies diluted in TBST containing 5% skim milk were added into the membrane and incubated for 1 hour at room temperature on an orbital shaker (Bellco, USA). Visualization of the immunolabeled bands was then carried out using ECL Plus Western Blotting Detection Reagents or ECL Advance Western Blotting Detection Kit (GE Healthcare, UK) according to manufacturer's instruction. The signals were exposed on X-ray film (Koda, USA).
Animals
Male C57BL/6 mice (8 to 10 weeks of age) were kept in SingHealth Experimental Medicine Centre and housed in collective cages at 22±1°C under a 12-h light/dark cycle (lights on at 07:00 h) with free access to laboratory chow and autoclaved tap water. Experiments were performed during the light phase of the cycle. The experimental procedures were previously approved by SingHealth Institutional Animal Care and Use Committee (IACUC) on the Ethical Use of Animals, where the study was carried out, and were conducted in accordance with Singapore regulations on animal welfare.
DSS induced mild colitis model in mice
Male C57BL/6 mice (n = 6–8 per group) were provided with a solution of filtered water containing 1.5% dextran sodium sulfate (DSS) (TdB Consultancy AB, Uppsala, Sweden) over a 12-day period. Every other day, the total of 200 ml of 1.5% DSS solution was replenished. The total volume of DSS solution consumed per mouse was proximally 4.4 ml/day. No differences between experimental groups were observed. On the third day of 1.5% DSS treatment, treatment group were fed with 10∧7 CFU/100 ml EC16 or 10∧7 CFU/100 ml L.GG using a gavage needle respectively. Control group were fed with PBS. The animals were provided with 1.5% DSS till day 12. On day 12, the animals were euthanized, colon were removed for length measurement and frozen in liquid nitrogen for future analysis.
Statistical analysis
Data are expressed as means ± SD. Significance of differences was determined using the Student's T-test and analysis of variance. P values<0.05 were considered to be statistically significant.
Supporting Information
IL-8 secretions in Caco-2 (A), HT-29 (B) and HCT116 (C) with the treatment of Lactobacillus and apoptosis assay in HCT116 (D,E). A total number of 107 cfu/ml bacteria were added into the cells for 6 h. Supernatants were harvested for cytokine assay as described in Materials and Methods. Three independent experiments were compiled to produce the data shown. Data were expressed as mean value ±SD. D. Apoptosis assay in HCT116 control. E. Apoptosis assay in HCT116 with EC16 treatment. One representative assay was shown from three independent experiments.
(TIF)
Cytokine productions in IECs. IL-8 production inside HCT116 cells (A, B), IL-8 secretion (C) and ICAM1, IL-2, IL-5, IL-17, IFN-γ and TNF-α in Caco-2 cells (D) with the treatment of 2 ng/ml of IL-1β and 107 cfu/ml of S. typhimurium and E. faecalis (EC16 and EC2) as described in Materials and Methods. Three replicates were done. Data was expressed as mean value ±SD.
(TIF)
Gene expression in Caco-2 cells tested using cDNA microarray. (A) The original photo of EC16 after exposure. (B) Volcano plot of EC16 obtained using Gene Spring software. Red dots represent the significantly changed genes. Yellow dots represent the genes not be regulated significantly. Only one of five sets of EC16 data were used as a representation.
(TIF)
Real-time PCR on E.faecalis treated HCT116 (dot) and Caco2 (black) cells. Experiments were done on 3-4 biological replicates and 2 technical replicates. Folds changes >1.5 and T-test with a P<0.05 was considered significant change.
(TIF)
ERK expression in HCT116 cells at 30 mins with the treatment of 2 ng/ml of IL-1β and S. typhimurium with and without E. faecalis EC16 at a MOI of 100. Total protein was harvested and protein production was analyzed using Western blotting as described in Materials and Methods. Experiments were repeated three times.
(TIF)
Gene ID and Taqman Primers ID.
(DOCX)
Acknowledgments
We also thank Dr. Patrick Reilly from National Cancer Center Singapore for critically reviewing the manuscript and his valuable suggestions. We also thank Dr. Sebastian Pott for the discussions during the study.
Funding Statement
The work is supported by National Research Foundation EWI Incentive for Research and Innovation Scheme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pinholt M, Ostergaard C, Arpi M, Bruun NE, Schonheyder HC, et al.. (2013) Incidence, clinical characteristics and 30-day mortality of enterococcal bacteraemia in Denmark 2006–2009: a population-based cohort study. Clin Microbiol Infect. [DOI] [PubMed]
- 2. Robyn J, Rasschaert G, Messens W, Pasmans F, Heyndrickx M (2012) Screening for lactic acid bacteria capable of inhibiting Campylobacter jejuni in in vitro simulations of the broiler chicken caecal environment. Benef Microbes 3: 299–308. [DOI] [PubMed] [Google Scholar]
- 3. Nueno-Palop C, Narbad A (2011) Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int J Food Microbiol 145: 390–394. [DOI] [PubMed] [Google Scholar]
- 4. Xu J, Gordon JI (2003) Honor thy symbionts. Proc Natl Acad Sci U S A 100: 10452–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Madan JC, Farzan SF, Hibberd PL, Karagas MR (2012) Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr Opin Pediatr 24: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orrhage K, Nord CE (1999) Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr Suppl 88: 47–57. [DOI] [PubMed]
- 7.Fanaro S, Chierici R, Guerrini P, Vigi V (2003) Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 91: 48–55. [DOI] [PubMed]
- 8. Iseki K (1987) [Development of intestinal flora in neonates]. Hokkaido Igaku Zasshi 62: 895–906. [PubMed] [Google Scholar]
- 9. Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M (2001) Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 108: 516–520. [DOI] [PubMed] [Google Scholar]
- 10. Schwiertz A, Gruhl B, Lobnitz M, Michel P, Radke M, et al. (2003) Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res 54: 393–399. [DOI] [PubMed] [Google Scholar]
- 11. Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P (1999) Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed 80: F167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hufnagel M, Liese C, Loescher C, Kunze M, Proempeler H, et al. (2007) Enterococcal colonization of infants in a neonatal intensive care unit: associated predictors, risk factors and seasonal patterns. BMC Infect Dis 7: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Normann E, Fahlen A, Engstrand L, Lilja HE (2013) Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta Paediatr 102: 129–136. [DOI] [PubMed] [Google Scholar]
- 14. Edelson MB, Bagwell CE, Rozycki HJ (1999) Circulating pro- and counterinflammatory cytokine levels and severity in necrotizing enterocolitis. Pediatrics 103: 766–771. [DOI] [PubMed] [Google Scholar]
- 15. Baggiolini M, Walz A, Kunkel SL (1989) Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Invest 84: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Djeu JY, Matsushima K, Oppenheim JJ, Shiotsuki K, Blanchard DK (1990) Functional activation of human neutrophils by recombinant monocyte-derived neutrophil chemotactic factor/IL-8. J Immunol 144: 2205–2210. [PubMed] [Google Scholar]
- 17. Huber AR, Kunkel SL, Todd RF 3rd, Weiss SJ (1991) Regulation of transendothelial neutrophil migration by endogenous interleukin-8. Science 254: 99–102. [DOI] [PubMed] [Google Scholar]
- 18. Otte JM, Cario E, Podolsky DK (2004) Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology 126: 1054–1070. [DOI] [PubMed] [Google Scholar]
- 19. Jijon HB, Buret A, Hirota CL, Hollenberg MD, Beck PL (2012) The EGF receptor and HER2 participate in TNF-alpha-dependent MAPK activation and IL-8 secretion in intestinal epithelial cells. Mediators Inflamm 2012: 207398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ihara E, Beck PL, Chappellaz M, Wong J, Medlicott SA, et al. (2009) Mitogen-activated protein kinase pathways contribute to hypercontractility and increased Ca2+ sensitization in murine experimental colitis. Mol Pharmacol 75: 1031–1041. [DOI] [PubMed] [Google Scholar]
- 21. Hollenbach E, Neumann M, Vieth M, Roessner A, Malfertheiner P, et al. (2004) Inhibition of p38 MAP kinase- and RICK/NF-kappaB-signaling suppresses inflammatory bowel disease. FASEB J 18: 1550–1552. [DOI] [PubMed] [Google Scholar]
- 22. Rafferty BJ, Unger BL, Perey AC, Tammariello SP, Pavlides S, et al. (2012) A novel role for the Rho-associated kinase, ROCK, in IL-1-stimulated intestinal epithelial cell responses. Cell Immunol 280: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jijon HB, Panenka WJ, Madsen KL, Parsons HG (2002) MAP kinases contribute to IL-8 secretion by intestinal epithelial cells via a posttranscriptional mechanism. Am J Physiol Cell Physiol 283: C31–41. [DOI] [PubMed] [Google Scholar]
- 24. Perez-Cano FJ, Dong H, Yaqoob P (2010) In vitro immunomodulatory activity of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713: two probiotic strains isolated from human breast milk. Immunobiology 215: 996–1004. [DOI] [PubMed] [Google Scholar]
- 25. Sjogren YM, Tomicic S, Lundberg A, Bottcher MF, Bjorksten B, et al. (2009) Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy 39: 1842–1851. [DOI] [PubMed] [Google Scholar]
- 26. Kalliomaki M, Satokari R, Lahteenoja H, Vahamiko S, Gronlund J, et al. (2012) Expression of microbiota, Toll-like receptors, and their regulators in the small intestinal mucosa in celiac disease. J Pediatr Gastroenterol Nutr 54: 727–732. [DOI] [PubMed] [Google Scholar]
- 27. Carey CM, Kostrzynska M (2013) Lactic acid bacteria and bifidobacteria attenuate the proinflammatory response in intestinal epithelial cells induced by Salmonella enterica serovar Typhimurium. Can J Microbiol 59: 9–17. [DOI] [PubMed] [Google Scholar]
- 28. Murch SH, Braegger CP, Walker-Smith JA, MacDonald TT (1993) Location of tumour necrosis factor alpha by immunohistochemistry in chronic inflammatory bowel disease. Gut 34: 1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Breese EJ, Michie CA, Nicholls SW, Murch SH, Williams CB, et al. (1994) Tumor necrosis factor alpha-producing cells in the intestinal mucosa of children with inflammatory bowel disease. Gastroenterology 106: 1455–1466. [DOI] [PubMed] [Google Scholar]
- 30. Braegger CP, Nicholls S, Murch SH, Stephens S, MacDonald TT (1992) Tumour necrosis factor alpha in stool as a marker of intestinal inflammation. Lancet 339: 89–91. [DOI] [PubMed] [Google Scholar]
- 31. Wolf A, Beuerlein K, Eckart C, Weiser H, Dickkopf B, et al. (2011) Identification and functional characterization of novel phosphorylation sites in TAK1-binding protein (TAB) 1. PLoS One 6: e29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang SH, Yu KN, Lee YS, An GH, Beck GR Jr, et al. (2006) Elevated inorganic phosphate stimulates Akt-ERK1/2-Mnk1 signaling in human lung cells. Am J Respir Cell Mol Biol 35: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Are A, Aronsson L, Wang S, Greicius G, Lee YK, et al. (2008) Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL-10 levels in colonic epithelial cells. Proc Natl Acad Sci U S A 105: 1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, et al. (2011) Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A 108: 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin HY, Chang JH, Chung MY, Lin HC (2013) Prevention of necrotizing enterocolitis in preterm very low birth weight infants: Is it feasible? J Formos Med Assoc. [DOI] [PubMed]
- 36. Stewart CJ, Marrs EC, Magorrian S, Nelson A, Lanyon C, et al. (2012) The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr 101: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 37. Chen LL, Wang XH, Cui Y, Lian GH, Zhang J, et al. (2009) Therapeutic effects of four strains of probiotics on experimental colitis in mice. World J Gastroenterol 15: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shveygert M, Kaiser C, Bradrick SS, Gromeier M (2010) Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol Cell Biol 30: 5160–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou S, Wang GP, Liu C, Zhou M (2006) Eukaryotic initiation factor 4E (eIF4E) and angiogenesis: prognostic markers for breast cancer. BMC Cancer 6: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang S, Ng LH, Chow WL, Lee YK (2008) Infant intestinal Enterococcus faecalis down-regulates inflammatory responses in human intestinal cell lines. World J Gastroenterol 14: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 43. Shon W, Lim S, Bae KS, Baek S, Lee W (2005) The expression of alpha4 integrins by human polymorphonuclear neutrophils in response to sonicated extracts of Enterococcus faecalis. J Endod 31: 369–372. [DOI] [PubMed] [Google Scholar]
- 44. Conner-Kerr TA, Sullivan PK, Gaillard J, Franklin ME, Jones RM (1998) The effects of ultraviolet radiation on antibiotic-resistant bacteria in vitro. Ostomy Wound Manage 44: 50–56. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IL-8 secretions in Caco-2 (A), HT-29 (B) and HCT116 (C) with the treatment of Lactobacillus and apoptosis assay in HCT116 (D,E). A total number of 107 cfu/ml bacteria were added into the cells for 6 h. Supernatants were harvested for cytokine assay as described in Materials and Methods. Three independent experiments were compiled to produce the data shown. Data were expressed as mean value ±SD. D. Apoptosis assay in HCT116 control. E. Apoptosis assay in HCT116 with EC16 treatment. One representative assay was shown from three independent experiments.
(TIF)
Cytokine productions in IECs. IL-8 production inside HCT116 cells (A, B), IL-8 secretion (C) and ICAM1, IL-2, IL-5, IL-17, IFN-γ and TNF-α in Caco-2 cells (D) with the treatment of 2 ng/ml of IL-1β and 107 cfu/ml of S. typhimurium and E. faecalis (EC16 and EC2) as described in Materials and Methods. Three replicates were done. Data was expressed as mean value ±SD.
(TIF)
Gene expression in Caco-2 cells tested using cDNA microarray. (A) The original photo of EC16 after exposure. (B) Volcano plot of EC16 obtained using Gene Spring software. Red dots represent the significantly changed genes. Yellow dots represent the genes not be regulated significantly. Only one of five sets of EC16 data were used as a representation.
(TIF)
Real-time PCR on E.faecalis treated HCT116 (dot) and Caco2 (black) cells. Experiments were done on 3-4 biological replicates and 2 technical replicates. Folds changes >1.5 and T-test with a P<0.05 was considered significant change.
(TIF)
ERK expression in HCT116 cells at 30 mins with the treatment of 2 ng/ml of IL-1β and S. typhimurium with and without E. faecalis EC16 at a MOI of 100. Total protein was harvested and protein production was analyzed using Western blotting as described in Materials and Methods. Experiments were repeated three times.
(TIF)
Gene ID and Taqman Primers ID.
(DOCX)