Abstract
Objective
It was previously suggested that the malic enzyme 2 (ME2) as the candidate gene for psychosis in fine mapping of chromosome 18q21. Chromosome 18q21 is also one of the possible regions that can contribute to addiction.
Methods
We performed a pilot study for discovering candidate gene of chromosome 18q21 in the methamphetamine abusers for elucidating the candidate gene for methamphetamine addiction leading to psychosis. We have selected 30 unrelated controls (16 males, 14 females; age=59.8±10.4) and 37 male methamphetamine abusers (age=43.3±7.8). We analyzed 20 single nucleotide polymorphisms (SNPs) of 7 neuronal genes in chromosome 18q21 for DNA samples that was checked for the data quality and genotype error. The association between the case-control status and each individual SNP was measured using multiple logistic regression models (adjusting for age and sex as covariates). And we controlled false discovery rate (FDR) to deal with multiple testing problem.
Results
We found 3 significant SNPs of 2 genes in chromosome 18q21 (p-value<0.05; adjusting for age as covariate) in methamphetamine abusers compared to controls. We also found 2 significant SNPs of 1 gene (p-value<0.05; adjusting for age and sex as covariates) (rs3794899, rs3794901:MAPK4). Two SNPs in MAPK4 gene were significant in both statistical groups.
Conclusion
MAPK4, the gene for mitogen-activated protein kinase 4, is one of the final 6 candidate genes including ME2 in 18q12-21 in our previous finemapping for psychosis. Our results suggest that MAPK4 can be a candidate gene that contribute to the methamphetamine addiction leading to psychosis.
Keywords: Substance-related disorders, Methamphetamine, Psychotic disorders, Malic enzyme 2, Mapk4, Comorbidity
INTRODUCTION
Psychotic disorders, including schizophrenia and bipolar disorder, are debilitating mental illnesses that lead to progressive deterioration in the social and occupational functioning of individuals,1,2) and increased economic burden of the society.3,4) Methamphetamine, a central nervous system stimulant, is the most commonly abused drug in Korea as well as other parts of East Asia.5,6,7) Methamphetamine abuse is a significant public health concern not only in Korea, but also in other parts of the world.5,8) In humans, abuse of methamphetamine leads to serious psychiatric conditions,9,10,11,12,13) including a variety of psychotic disorders.
Studies suggested that genetic factors contribute to the development of these psychotic disorders.14) Although bipolar disorder and schizophrenia were considered as distinct illnesses, recent studies have shown that these disorders are genetically overlapping.15,16,17,18,19,20,21,22) Linkage studies identified wide regions containing a number of genes, and required subsequent analyses to examine the association of specific genes to the disease phenotypes.23) Previous studies conducted in the Costa Rican population have shown evidence of linkage disequilibrium between markers within the 18q21 region of both these psychiatric phenotypes: severe bipolar disorder and schizophrenia.22,24) Further analyses revealed that psychotic symptomatology (hallucinations, delusions, disorganized thought, disorganized behavior) was the relevant phenotype showing association to the 18q21 region, as individuals, both with and without a history of mania, showed evidence of linkage disequilibrium to this region.22) Psychotic symptoms are a core feature of both schizophrenia and schizoaffective disorder, and are frequently observed during acute phases of bipolar disorder.
Development of new molecular genetic techniques has propelled advanced studies aimed at understanding the genetic basis of drug abuse. It is possible that personality factor mediates intermediately in the genetic contributions to drug abuse. Classical genetic studies documented the contributions of a highly complex set of genetic factors to the abuse of multiple addictive substances.25) Human psychopathology of psychosis emerges as anxiety, a state of mental disequilibrium. Men become depressed when their anxieties are not resolved. Psychopathologically, psychosis and mania are deviations from reality in which, depressed patients fall, and addiction is the final defense beyond psychosis, thereby leading to comorbidity. The comorbidity of psychosis and addiction is very clear from a psychopathologic perspective. Clinically, addictive symptoms are prevalent among patients with schizophrenia and bipolar disorder. The close interrelationship between psyche and soma has been studied.26) The comorbidity of psychosis and addiction needs to be analyzed genetically from the perspectives of several broad processes, including personality, memory, and cognition, for understanding the mechanisms underlying the psychiatric symptoms. The genetic regulation of these processes may explain the methamphetamine addiction leading to psychosis.
Genetics is commonly used for explaining the biological basis of psychiatric disorders. Fine mapping of chromosome 18 revealed that malic enzyme 2 (ME2) is a candidate gene for psychosis.27) In an effort to identify the genes within the 18q21 that may be contributing to the development of psychotic disorders in the Costa Rican population, we performed additional fine mapping with microsatellite markers by utilizing a sample that allowed us to establish the association between psychotic disorders and this region of the chromosome.22) Marker D18S474, located at 71.32 centimorgan (cM) from the pter (sex-averaged distance, Marshfield Genetic Map),28) had shown evidence of linkage disequilibrium (LD) with all the four psychotic phenotypes tested (psychotic disorders with and without a history of mania, Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) Schizophrenia, or Schizoaffective Disorder).29) For the follow-up study, we focused on the 5 cM region surrounding D18S474. This region (from D18S450 to D18S484) contains 21 known genes, many of which are expressed in the central nervous system. To determine if any of these genes were relevant to mania or psychosis, we analyzed mRNA expression patterns in bipolar versus control post-mortem brains, and reviewed previous studies that examined altered gene expression patterns in schizophrenia and bipolar disorder. Further, single nucleotide polymorphism (SNP) analyses were performed to test the association between a specific gene within this region and the phenotypes of psychosis and mania. Based on the results of our genetic mapping and gene expression, we report that ME2, a mitochondrial enzyme involved in energy metabolism in neurons,30) is associated with a spectrum of psychotic disorders, including schizophrenia, schizoaffective disorder, and bipolar disorder.
A number of studies suggested that chromosome 18 contains several regions that likely harbor candidate genes contributing to the susceptibility to develop addiction.31) However, data from genome-wide association studies in humans and mice suggested a larger role for "higher order" pharmacogenomics in the molecular genetics of addiction. For instance, it is possible that overlapping genetic vulnerabilities influences the probability for developing dependence on a variety of addictive substances. We performed a pilot study for fine mapping of chromosome 18q21 of methamphetamine abusers for identifying the candidate genes responsible for the methamphetamine addiction leading to psychosis.
METHODS
We have selected 30 unrelated controls (16 males, 14 females; age=59.8±10.4 years) and 37 male methamphetamine abusers (age=43.3±7.8 years). We analyzed 56 SNPs of 18 neuronal genes in chromosome 18 for DNA samples that was checked for the data quality and genotype error. The association between the case-control status and each individual SNP was measured using multiple logistic regression models (adjusting for age and sex as covariates). And we controlled false discovery rate (FDR) to deal with multiple testing problem.
Sample Collection
All subjects were recruited in accordance with the principles of the Declaration of Helsinki and with approval from the Institutional Review Boards of the Bugok National Hospital in Korea. Each subject was interviewed by a psychiatrist who was blind to the previous history of the subject, using the Diagnostic Interview for Genetic Studies (DIGS).32) In addition, information on each subject was gathered through an interview with a family member, using the Family Interview for Genetic Studies (FIGS).32) Medical records (inpatient and outpatient) were also abstracted. All affected subjects (i.e., those given formal psychiatric diagnoses for the present study) were diagnosed using a best estimate diagnostic process, as described by Walss-Bass et al.22) The process arrives at a lifetime consensus diagnosis or diagnosis using the DSM-IV.
Design for reference data
A study was performed in 30 normal Korean subjects. The general inclusion criterion was that subjects should be normal and belong to the age groups specified in the sample. Subjects with conditions suspected of affecting any biases were excluded from the study. Subjects were also excluded if they had been receiving corticoid, estrogen, androgen, T3 (triiodothyronine)-T4 (thyroxine) therapy or diphenylhydantoin, vitamin D, bisphosphonate, calcitonin, fluoride, thiazide diuretics, or barbiturates for more than 6 months, as all these drugs affect any biases.33,34,35,36,37,38) Since it was impossible to obtain a population-based register for technical and legal reasons, the selection of control subjects meeting the inclusion/exclusion criteria was made from volunteers (students, hospital workers, patients' relatives, etc.) attending the hospital.
Experimental design
The subjects of the study included inpatients at the Addiction Inpatient Unit of the Bugok National Hospital in Korea, which has an established protocol for evaluation, treatment, and follow-up of patients with drug abuse. We identified 37 patients that fulfilled DSM-IV diagnostic criteria of methamphetamine abuse or dependence.39) The patients also satisfied the following criteria: (1) duration of methamphetamine abuse longer than 1 year, and its frequency more than 100 times and (2) absence of concomitant diseases that might affect any biases. Factors that may influence any biases were also recorded. We could not include female patients under environmental circumstances.
Genotyping
DNA from 30 control persons and 37 patients, which were affected with methamphetamine abuse, was genotyped using 56 SNPs of 18 neuronal genes in chromosome 18 (fluorescently labeled markers of the highest heterozygosity available within chromosome 18). These markers spanned the region of chromosome 18, functionally relevant to neuropsychiatric disorders. We have first searched for functionally relevant candidate genes to the comorbidity of psychosis and addiction. And then we finally selected 56 SNPs of 18 neuronal genes in chromosome 18. The genotyping process was carried out as described previously in Lee et al.27) by laboratory personnel who were blinded to phenotype data. Molecular genetic studies are represented as two major studies (linkage study and association study). Linkage study progress in the concept of linkage equilibrium between gene and so close marker (e.g., microsatellite) and association study progress in the concept of LD between gene and so very, very close marker (e.g., SNP). Earlier studies had included micro satellites in my previous fine mapping association analyses. But in this pilot study for discovering candidate genes rather than candidate regions, we chose to focus on SNPs, which are more effective markers than microsatellites for detecting association due to strong LD with disease causing genetic variants as Korean population may have diverged many generations previously.40)
Standard PCR was performed using the ABI 877 automated thermocycler (Applied Biosystems Inc., Foster City, CA, USA) or the PE 9700 PCR instrument (Applied Biosystems Inc.). Amplified fragments were analyzed on the 7900HT Sequence Detection System (Applied Biosystems Inc.) for SNPs. Genotypes were assigned using GeneScan, Genotyper, and SDS software (Applied Biosystems Inc.). Each genotype was scored separately by two individuals, who were blind to diagnosis of the subjects. Genotype scores were compared using a software program implemented in Microsoft Excel (Microsoft Inc., Redmond, WA, USA), discrepancies were discussed with review of the original gels, and final genotypes agreed upon. Genotypes were checked for violations of Mendelian inheritance by PEDSYS program INFER (Southwest Foundation for Biomedical Research, San Antonio, TX, USA). Genotypes for two SNPs were discarded from the statistical analyses due to recurrent Mendelian discrepancies.
Fine Mapping Analyses
In the fine mapping stage (where 20 SNPs markers were tested), we selected 20 SNPs of 7 neuronal genes in chromosome 18q21. We screened NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/projects/mapview/maps.cgi?taxid=9606&chr=18) for exploring candidate genes in chromosome 18q21 for the methamphetamine addiction leading to psychosis. Finally selected 7 neuronal genes were MBD1, MAPK4, MRO, ME2, ELAC1, SMAD4, MEX3C. They were chosen based on functional and positional relevance to neuropsychiatric disorders. Twenty SNPs were genotyped and analyzed from those 7 genes.
SNPs selection (Table 1)
Table 1.
Selected 20 SNPs of 7 neuronal genes in chromosome 18q21 in fine mapping for methamphetamine abuse
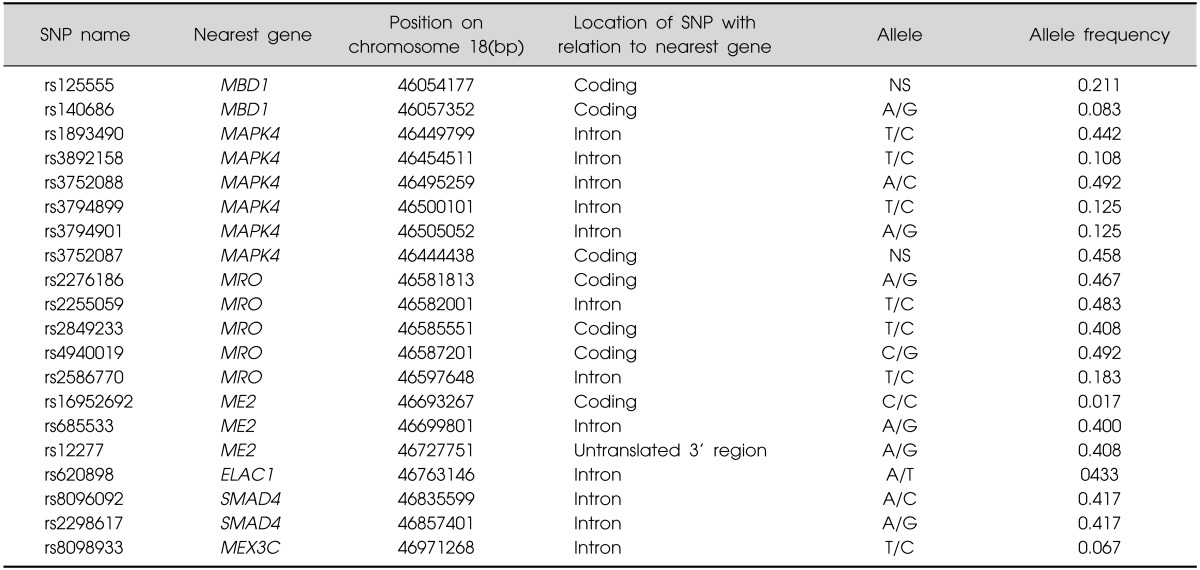
SNP, single nucleotide polymorphism; NS, not-specified.
We have first searched possible candidate genes functionally relevant to neuropsychiatric disorders in chromosome 18q21 from the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/projects/mapview/maps.cgi?taxid=9606&chr=18), which was referenced from positional relevance in my previous fine mapping analyses.22,27) Finally genotyped SNPs in selected neuronal genes were chosen from previous literatures and availability in Illumina Customized Sentrix Array Matrix genotyping_v6A with Standard Illumina procedures using Illumina BeadStation 500G array scanner in the NCBI ENTREZ SNP (http://www.ncbi.nlm.nih.gov/sites/entrez).
Case-control association tests
Case-control association tests were performed for the phenotype of "methamphetamine abuse". All other subjects were classified as unknown controls for purposes of the analyses. For testing of association, we performed analyses for all 56 SNPs, using the phenotype of methamphetamine abuse.
Additional analyses
We have set analyses into four forms. The first set was the original case-control analysis and the second set was the original case-control analysis adjusted by male sex because cases are composed of only males. Sequent analyses, adjusted by age, were performed because the age difference between two groups was significant. The third set was the original case-control analysis adjusted by age. Finally, the fourth set was the original case-control analysis adjusted by age and sex.
Statistical Analysis
We calculated genotype frequencies for each individual polymorphism and evaluated Hardy-Weinberg equilibrium to check the data quality and genotype error. Chi-square test statistic is used to compare the observed numbers of each genotype with those expected for the population following chi-square distribution with a one degree of freedom.41)
The association between the case-control status and each individual SNP, measured by the odds ratio and its corresponding 95% confidence interval using multiple logistic regression models (adjusting for sex and age as covariates). All analyses were performed assuming a dominant, recessive, additive, allelic effect for each polymorphism. In the dominant model, both the heterozygous variant and the rare homozygous variant were combined. In the recessive model, the variant was defined as only the rare homozygous genotype and in the additive model, each genotype variant has the same effect and in allele model, rare allele variant has effect. The likelihood ratio test was used to test the effect of each SNP at the 5% significant level.
And we controlled FDR to deal with multiple testing problem.42) We used the Benjamini and Hochberg method to control FDR.
All data were processed and analyzed by the R software, version 2.6.2. (Institute for Statistics and Mathematics, Wirtschaftsuniversitat Wien, Vienna, Austria; http://www.r-project.org).
RESULTS
We found 3 significant SNPs of 2 genes in chromosome 18q21 (p-value<0.05; adjusting for age as covariate) in methamphetamine abusers compared to controls (rs3794899, rs3794901:MAPK4; rs2849233:MRO). We also found 2 significant SNPs of 1 gene (p-value<0.05; adjusting for age and sex as covariates) (rs3794899, rs3794901:MAPK4). Two SNPs in MAPK4 gene were significant in both statistical groups.
Demographic and Clinical Characteristics of Patients of Methamphetamine Abuse (Table 2)
Table 2.
Demographic and clinical characteristics in methamphetamine abusers and controls
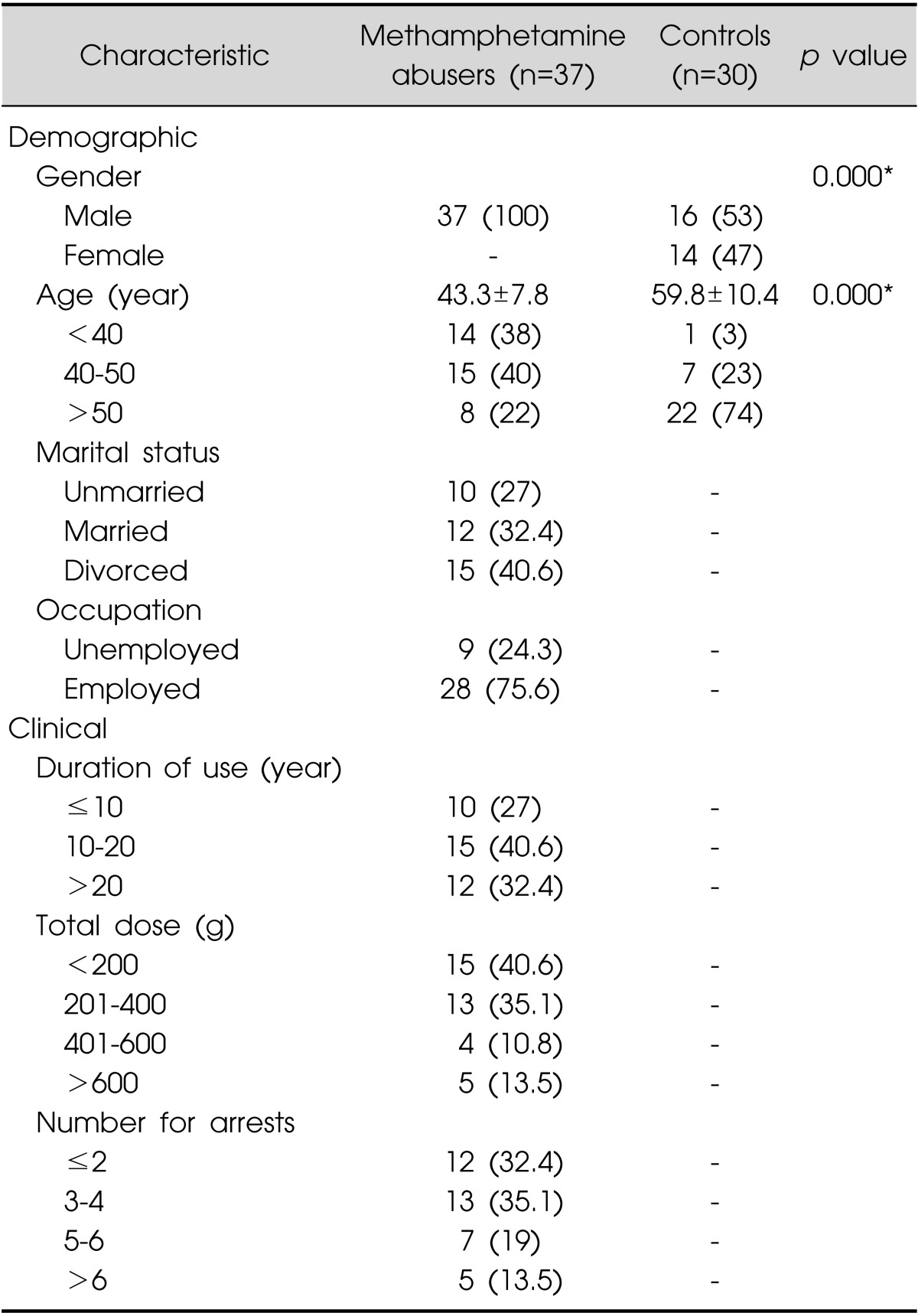
Values are presented as number (%) or mean±standard deviation.
*p<0.001
Demographic and clinical characteristics of the study patients are listed in Table 2. The duration and total dose of methamphetamine abuse varied widely among patients. The mean duration of methamphetamine abuse was 14.35±1.10 years (mean±standard error [SE]), and the mean total dose of methamphetamine that had been consumed during the period of drug abuse was 326.46±40.94 g (mean±SE), estimated solely on the basis of the patients' statement and uncorrected for purity of drug. We compared the height, weight, and body mass index of methamphetamine abusers with those of the Korean male population from data obtained from Korean Statistical Information Service (KOSIS, 2004).43) There were no significant differences between our study and control groups for these parameters.
SNP Fine Mapping Analyses
Set 1 (original case-control association): We found 1 significant SNP of one gene in chromosome 18q21 (p-value <0.05) in methamphetamine abusers compared to controls (rs2276186:MRO) (Table 3).
Table 3.
Significant SNPs results in the original case-control association (set 1)
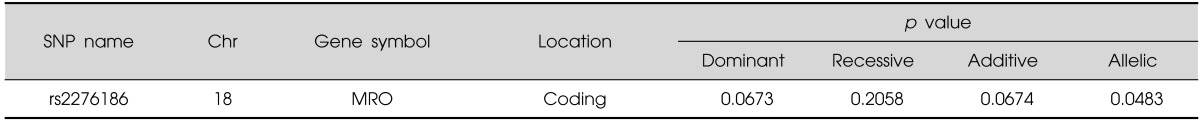
SNP, single nucleotide polymorphism.
Set 2 (original case-control association adjusted by sex): We also found 1 significant SNP of one gene (p-value<0.05; adjusting for sex as covariates) (rs3794899: MAPK4) (Table 4).
Table 4.
Significant SNPs results in the original case-control association adjusted by sex (set 2)

SNP, single nucleotide polymorphism.
Set 3 (original case-control association adjusted by age): We found 3 significant SNP of 2 genes in chromosome 18q21 (p-value<0.05; adjusting for age as covariate) in methamphetamine abusers compared to controls (rs3794899, rs3794901:MAPK4; rs2849233:MRO) (Table 5).
Table 5.
Significant SNPs results in the original case-control association adjusted by age (set 3)
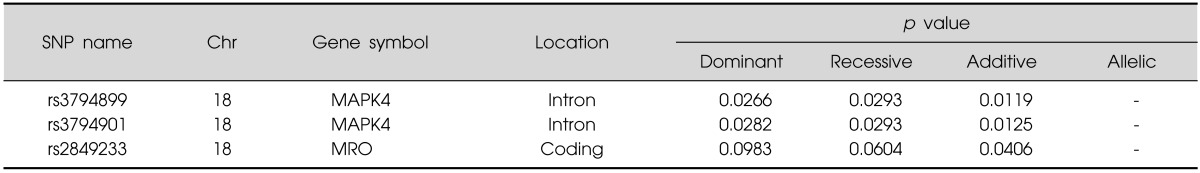
SNP, single nucleotide polymorphism.
Set 4 (original case-control association adjusted by age and sex): We also found 2 significant SNPs of one gene (p-value<0.05; adjusting for age and sex as covariates) (rs3794899, rs3794901:MAPK4) (Table 6).
Table 6.
Significant SNPs results in the original case-control association adjusted by age and sex (set 4)
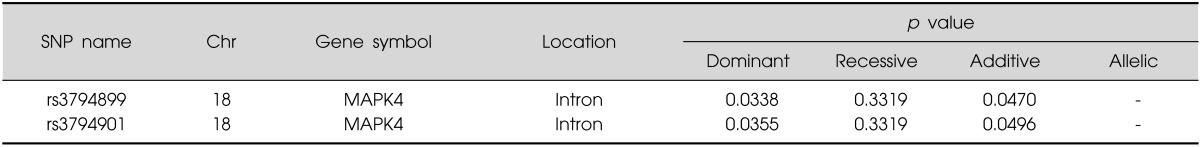
SNP, single nucleotide polymorphism.
SNPs in 2 genes (MAPK4, MRO) of 7 genes were significantly associated with methamphetamine abuse in all four sets. Two SNPs in MAPK4 gene were significantly associated with methamphetamine abuse in final set 4 adjusted by age and sex. The MAPK4 gene lies between D18S473 and D18S474, finally concentrated region in our previous fine mapping analysis for psychosis and mania.27) Given the very basic mechanism for protein kinase of this gene, we focused on this gene.
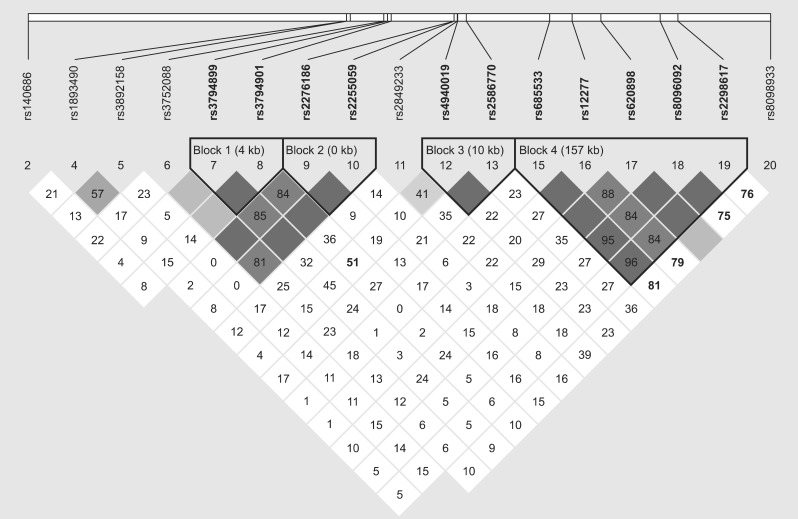
LD analyses of selected 7 positionally only relevant genes which was focused in 18q21 fine mapping research was performed. Twenty SNPs genotyped in our fine mapping for methamphetamine abusers and controls are also represented (Fig. 1).
Fig. 1.
Linkage disequilibrium (LD) analyses of selected 7 positionally only relevant genes which was focused in 18q21 fine mapping research. Twenty single nucleotide polymorphisms (SNPs) genotyped in our fine mapping for methamphetamine abusers and controls are also represented. Haplotype block pattern constructed by the Haploview program (Barrett et al. 2005) is shown. The number in each cell represents the LD. Parameter D' (×100), blank cells mean D'=1. Each cell is painted with graduated color relative to the strength of LD between markers, which is defined by both D' value and confidence bounds on D'. SNPs are indicated by a SNP ID number (rs number).
DISCUSSION
The results of fine mapping study presented here are in line with our earlier findings regarding psychosis and mania.22,27) Our findings suggest that MAPK4 gene could be associated with methamphetamine abuse. Therefore, this gene could have the possibility of being involved in the processes of methamphetamine abuse leading to psychosis in our samples of Korean population.
The MAPK4 gene that encodes mitogen-activated protein kinase 4 was one of the final 6 candidate genes in our previous fine mapping studies of 18q12-21 in our previous fine mapping for psychosis, although not directly associated.27) Our results suggest that MAPK4 is a candidate gene contributing to the methamphetamine addiction leading to psychosis.
Our previous findings27) (fine mapping of a chromosomal locus associated with psychosis and mania, and mRNA expression analyses in post-mortem brain) suggested that a specific mitochondrial enzyme, ME2, likely contributed to the expression of a spectrum of phenotypes ranging from schizophrenia to bipolar disorder. ME2 is a mitochondrial enzyme involved in neuronal glucose metabolism and in the synthesis of 2 key neurotransmitters, namely, GABA and glutamate.30) These findings are consistent with the reports that mitochondrial enzyme dysfunction plays a role in the pathophysiology of psychotic disorders.44,45,46,47,48,49) Previous studies conducted in the Central Valley of Costa Rica (CVCR; one using samples of severe bipolar disorder, the other using subjects primarily diagnosed with schizophrenia) showed evidences for the existence of LD between the phenotypes of schizophrenia and bipolar disorders and the 18q21,22,50) 2.41 cM region (2.7 Mb) spanning from D18S450 to D18S474. Additionally, three separate linkage studies have pointed to a genetic predisposition locus in the general region for SC,51) BP,52) or both BP and SC.53) Maziade et al.53) showed evidence of linkage in a combined sample of SC and BP at the marker D18S472, which is approximately the same position as the marker D18S474 used in the present study.
In our earlier report,27) we described the fine mapping analyses for the 18q21 locus in the CVCR. Microsatellite analyses narrowed the area of strongest association of the psychosis phenotype to the region from 45.74 Mb (D18S473) to 46.94 Mb (D18S474). There are 11 known genes within this 1.20-Mb region: MYO5B (45.60 Mb-45.98 Mb), FLJ32743 (46.01 Mb-46.05 Mb), MBD1 (46.05 Mb-46.06 Mb), CXXC1 (46.06 Mb-46.07 Mb), C18 or f24 (46.16 Mb-46.17 Mb), LOC390853 (46.24 Mb-46.34 Mb), MAPK4 (46.44 Mb-46.51 Mb), MRO (46.58 Mb-46.60 Mb), ME2 (46.66 Mb-46.73 Mb), ELAC1 (46.75 Mb-46.77 Mb), and SMAD4 (46.81 Mb-46.86 Mb). All of the genes in this region are of special interest because of their known functions and/or their relationship (association) with neuropsychiatric disorders. MBD1 is known to be associated with Rett syndrome and autism spectrum disorders.54,55) The ME2 gene is shown to be associated with idiopathic generalized epilepsy; the strongest association was seen with variants in the promoter region area of this gene.56) In the present study, the SMAD4 gene, which contains a single SNP, showed the strongest association to psychosis, and therefore is a candidate gene for schizophrenia and bipolar disorder.57) SMAD4 have been shown to be involved in neuronal proliferation and differentiation.58)
Genotyping evidence showed direct association of haplotypes containing SNPs within the ME2 and SMAD4 genes (hcv2752066-hcv7457521-hcv22274384) and the phenotype of psychosis. An individual SNP (hcv22274383) that is physically outside of the ME2 gene, but in strong LD with the ME2 gene, also showed significant association to the phenotype of psychosis. It is likely that a gene variant or mutation within either of the genes in LD with the ME2 gene (SMAD4 and ELAC1) is responsible for the association detected in this study. Further studies, including sequencing for possible novel mutations in the ME2 and SMAD4 genes, are necessary to definitively assess whether ME2, SMAD4, or ELAC1 contain specific variants that may directly cause the spectrum of psychotic illnesses seen in CVCR sample. Once the microsatellite analyses had highlighted the region from D18S473 to D18S474, and the mRNA analyses suggested ME2 as a gene of interest, the previous study focused only on SNPs that were in tight LD with the ME2 gene. Further analyses of the region spanning from D18S450 to D18S484 are required to test other genes for evidence of association to either mania or psychosis. Of note, all other SNPS (i.e., all those not in strong equilibrium with the ME2 and SMAD4 genes), including the two SNPs within the MBD1 gene, showed no evidence of an association with either psychosis or mania in the current sample. The evidence of association of the ME2 gene with the phenotype of psychosis in the CVCR sample, together with the evidence that the ME2 gene expression was low in the post-mortem brain samples suggest that this gene is involved in the development of psychotic and/or manic disorders.
The mitogen-activated protein kinase 4 encoded by MAPK4 gene is a member of the mitogen-activated protein kinase family. Growth factor receptor tyrosine kinases activate mitogen-activated protein kinases, which then translocate into the nucleus where they phosphorylate nuclear targets. A wide variety of extracellular stimuli activates MAPK pathways leading to cell proliferation, differentiation, growth, and migration. A typical MAPK cascade involves a three-kinase module architecture by which a signal from an upstream kinase is transmitted to a downstream kinase by sequential phosphorylation. Four well-defined groups of MAPKs are known (ERK1/2, c-Jun N-terminal kinases, p38s, and ERK5), but additional members, including ERK3, ERK4 (p63 MAPK, ERK3-related, ERK3, MAPK4, Prkm4), and ERK8, have been identified. ERK4 (p63 MAPK) was described in humans,59) soon after ERK1, ERK2, and ERK3 were identified.60) Among MAPKs, ERK4 is most closely related to ERK3, displaying 62% overall amino acid sequence identity and 73% within the predicted kinase domain. Both ERK4 and ERK3 lack the highly conserved activation loop ("a-loop") motif TXY between the kinase subdomains VII and VIII, but possess a segmented sequence at this position. Even the APE motif of subdomain VIII, which is highly conserved in other MAPKs is replaced by an SPR motif in ERK4 and ERK3. ERK4 and ERK3 carry long C-terminal extensions. Human Erk4 was mapped on chromosome 18q12-21,61) and a cDNA for the rat homolog rMNK2 has been isolated.62) Activators and relevant substrates of ERK4 remain elusive, and the enzymatic activities of the atypical ERKs have not been defined. Initially, the MAPK-activated protein kinase MK5,63,64) also known as p38-regulated and -activated kinase (PRAK), was described as a member of the MK family and a downstream target of p38.65,66) Previous data suggested that MK5 is not a physiological substrate for p38 in vivo,67) because the stimuli that activate the p38 pathway fail to activate MK5, and the binding of endogenous p38 to MK5 is weaker than its interaction with other established substrates, namely, MK2 and MK3. Interestingly, it has been recently demonstrated that MK5 strongly interacts with and is activated by ERK3.67,68) Given this very basic mechanism for the cellular neurophysiological cascades, the MAPK4 gene could influence the intracellular signaling pathways leading to the comorbidity of psychosis and addiction.
Whole genome SNP association studies suggested that many candidate "cell adhesion" genes located on chromosome 18 are involved in the molecular genetics of addiction. The CHST9, OSBPL1A, IMPACT, DOK6, HRH4, ZNF407, KIAA1713, and CCDC102B genes were identified by the whole genome SNP association studies of 420 unrelated European-American substance abusers versus 320 control European-Americans, and 560 unrelated African-American substance abusers versus 360 African-American controls.58,69,70)
Our study has several limitations. One limitation of the current study is the relatively low statistical significance of association (global p values showing association of methamphetamine abuse to SNPs within or in strong LD with the MAPK4 gene were between 0.0496 and 0.0119). Therefore, our findings need to be confirmed using a larger sample from Korea or other population. The second limitation is that, the individual SNPs that were shown to be associated with psychosis in our previous fine mapping were within ME2 gene (not the MAPK4 gene). Third, the control subjects in our samples were never exposed to methamphetamine. It is very critical, but very difficult to recruit sufficient number of very age and sex matched control in case-control association analysis. Therefore, we additionally analyzed case-control association with adjustment by age and sex for these unmatched controls for the results to be negative (rs1990162) although not recruiting matched contol in this study due to the very many limitations. Also we are now designing for recruiting matched control. Meanwhile there are some opinions among researchers that it is sufficient to do case-control association study without age-sex adjustment in spite of possible population stratification by covariate or confounder. Finally, denser genotyping, sequencing, and functional studies are needed to confirm whether the Mapk4 gene or other genes in tight LD with ME2, contributes to the comorbidity of psychosis and addiction in the Korean and other populations, which could be very overinclusive with many limitations including very small number of unctrolled subjects.
Acknowledgments
This work was supported by Clinical Research Grant from Pusan National University Hospital 2014.
References
- 1.McGuffin P, Asherson P, Owen M, Farmer A. The strength of the genetic effect. Is there room for an environmental influence in the aetiology of schizophrenia? Br J Psychiatry. 1994;164:593–599. doi: 10.1192/bjp.164.5.593. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann RF, Schloesser RJ, Gould TD, Manji HK. Mood stabilizers target cellular plasticity and resilience cascades: implications for the development of novel therapeutics. Mol Neurobiol. 2005;32:173–202. doi: 10.1385/MN:32:2:173. [DOI] [PubMed] [Google Scholar]
- 3.Bauer M, Unützer J, Pincus HA, Lawson WB NIMH Affective Disorders Workgroup. Bipolar disorder. Ment Health Serv Res. 2002;4:225–229. doi: 10.1023/a:1020968616616. [DOI] [PubMed] [Google Scholar]
- 4.Bartels SJ, Clark RE, Peacock WJ, Dums AR, Pratt SI. Medicare and medicaid costs for schizophrenia patients by age cohort compared with costs for depression, dementia, and medically ill patients. Am J Geriatr Psychiatry. 2003;11:648–657. doi: 10.1176/appi.ajgp.11.6.648. [DOI] [PubMed] [Google Scholar]
- 5.Chung H, Park M, Hahn E, Choi H, Choi H, Lim M. Recent trends of drug abuse and drug-associated deaths in Korea. Ann N Y Acad Sci. 2004;1025:458–464. doi: 10.1196/annals.1316.056. [DOI] [PubMed] [Google Scholar]
- 6.McKetin R, Kozel N, Douglas J, Ali R, Vicknasingam B, Lund J, et al. The rise of methamphetamine in Southeast and East Asia. Drug Alcohol Rev. 2008;27:220–228. doi: 10.1080/09595230801923710. [DOI] [PubMed] [Google Scholar]
- 7.Kulsudjarit K. Drug problem in southeast and southwest Asia. Ann N Y Acad Sci. 2004;1025:446–457. doi: 10.1196/annals.1316.055. [DOI] [PubMed] [Google Scholar]
- 8.Rawson RA, Condon TP. Why do we need an Addiction supplement focused on methamphetamine? Addiction. 2007;102(Suppl 1):1–4. doi: 10.1111/j.1360-0443.2006.01781.x. [DOI] [PubMed] [Google Scholar]
- 9.Lapworth K, Dawe S, Davis P, Kavanagh D, Young R, Saunders J. Impulsivity and positive psychotic symptoms influence hostility in methamphetamine users. Addict Behav. 2009;34:380–385. doi: 10.1016/j.addbeh.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Dore G, Sweeting M. Drug-induced psychosis associated with crystalline methamphetamine. Australas Psychiatry. 2006;14:86–89. doi: 10.1080/j.1440-1665.2006.02252.x. [DOI] [PubMed] [Google Scholar]
- 11.Zweben JE, Cohen JB, Christian D, Galloway GP, Salinardi M, Parent D, et al. Methamphetamine Treatment Project. Psychiatric symptoms in methamphetamine users. Am J Addict. 2004;13:181–190. doi: 10.1080/10550490490436055. [DOI] [PubMed] [Google Scholar]
- 12.Buffenstein A, Heaster J, Ko P. Chronic psychotic illness from methamphetamine. Am J Psychiatry. 1999;156:662. doi: 10.1176/ajp.156.4.662. [DOI] [PubMed] [Google Scholar]
- 13.Murray JB. Psychophysiological aspects of amphetamine-methamphetamine abuse. J Psychol. 1998;132:227–237. doi: 10.1080/00223989809599162. [DOI] [PubMed] [Google Scholar]
- 14.Shih RA, Belmonte PL, Zandi PP. A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int Rev Psychiatry. 2004;16:260–283. doi: 10.1080/09540260400014401. [DOI] [PubMed] [Google Scholar]
- 15.Van Broeckhoven C, Verheyen G. Report of the chromosome 18 workshop. Am J Med Genet. 1999;88:263–270. [PubMed] [Google Scholar]
- 16.Pulver AE, Mulle J, Nestadt G, Swartz KL, Blouin JL, Dombroski B, et al. Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol Psychiatry. 2000;5:650–653. doi: 10.1038/sj.mp.4000814. [DOI] [PubMed] [Google Scholar]
- 17.Siris SG. Depression in schizophrenia: perspective in the era of "Atypical" antipsychotic agents. Am J Psychiatry. 2000;157:1379–1389. doi: 10.1176/appi.ajp.157.9.1379. [DOI] [PubMed] [Google Scholar]
- 18.Bailer U, Leisch F, Meszaros K, Lenzinger E, Willinger U, Strobl R, et al. Genome scan for susceptibility loci for schizophrenia and bipolar disorder. Biol Psychiatry. 2002;52:40–52. doi: 10.1016/s0006-3223(02)01320-3. [DOI] [PubMed] [Google Scholar]
- 19.Cardno AG, Rijsdijk FV, Sham PC, Murray RM, McGuffin P. A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry. 2002;159:539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- 20.Potash JB, Zandi PP, Willour VL, Lan TH, Huo Y, Avramopoulos D, et al. Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am J Psychiatry. 2003;160:680–686. doi: 10.1176/appi.ajp.160.4.680. [DOI] [PubMed] [Google Scholar]
- 21.Berrettini W. Bipolar disorder and schizophrenia: convergent molecular data. Neuromolecular Med. 2004;5:109–117. doi: 10.1385/NMM:5:1:109. [DOI] [PubMed] [Google Scholar]
- 22.Walss-Bass C, Escamilla MA, Raventos H, Montero AP, Armas R, Dassori A, et al. Evidence of genetic overlap of schizophrenia and bipolar disorder: linkage disequilibrium analysis of chromosome 18 in the Costa Rican population. Am J Med Genet B Neuropsychiatr Genet. 2005;139B:54–60. doi: 10.1002/ajmg.b.30207. [DOI] [PubMed] [Google Scholar]
- 23.Escamilla M, Lee BD, Ontiveros A, Raventos H, Nicolini H, Mendoza R, et al. The epsin 4 gene is associated with psychotic disorders in families of Latin American origin. Schizophr Res. 2008;106:253–257. doi: 10.1016/j.schres.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Escamilla MA. Population isolates: their special value for locating genes for bipolar disorder. Bipolar Disord. 2001;3:299–317. doi: 10.1034/j.1399-5618.2001.30605.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Kim EY, Kwon do H, Lee BD, Kim YT, Ahn YB, Yoon KY, et al. Frequency of osteoporosis in 46 men with methamphetamine abuse hospitalized in a National Hospital. Forensic Sci Int. 2009;188:75–80. doi: 10.1016/j.forsciint.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Lee BD, Walss-Bass C, Thompson PM, Dassori A, Montero PA, Medina R, et al. Malic enzyme 2 and susceptibility to psychosis and mania. Psychiatry Res. 2007;150:1–11. doi: 10.1016/j.psychres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- 30.Hassel B. Pyruvate carboxylation in neurons. J Neurosci Res. 2001;66:755–762. doi: 10.1002/jnr.10044. [DOI] [PubMed] [Google Scholar]
- 31.Uhl GR, Drgon T, Johnson C, Fatusin OO, Liu QR, Contoreggi C, et al. "Higher order" addiction molecular genetics: convergent data from genome-wide association in humans and mice. Biochem Pharmacol. 2008;75:98–111. doi: 10.1016/j.bcp.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-864. [DOI] [PubMed] [Google Scholar]
- 33.Tuck SP, Datta HK. Osteoporosis in the aging male: treatment options. Clin Interv Aging. 2007;2:521–536. doi: 10.2147/cia.s820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krum SA, Miranda-Carboni GA, Hauschka PV, Carroll JS, Lane TF, Freedman LP, et al. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J. 2008;27:535–545. doi: 10.1038/sj.emboj.7601984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaegashi Y, Onoda T, Tanno K, Kuribayashi T, Sakata K, Orimo H. Association of hip fracture incidence and intake of calcium, magnesium, vitamin D, and vitamin K. Eur J Epidemiol. 2008;23:219–225. doi: 10.1007/s10654-008-9225-7. [DOI] [PubMed] [Google Scholar]
- 36.Mazziotti G, Giustina A, Canalis E, Bilezikian JP. Glucocorticoid-induced osteoporosis: clinical and therapeutic aspects. Arq Bras Endocrinol Metabol. 2007;51:1404–1412. doi: 10.1590/s0004-27302007000800028. [DOI] [PubMed] [Google Scholar]
- 37.Blahos J. Treatment and prevention of osteoporosis. Wien Med Wochenschr. 2007;157:589–592. doi: 10.1007/s10354-007-0486-7. [DOI] [PubMed] [Google Scholar]
- 38.Migliaccio S, Brama M, Fornari R, Greco EA, Spera G, Malavolta N. Glucocorticoid-induced osteoporosis: an osteoblastic disease. Aging Clin Exp Res. 2007;19(3 Suppl):5–10. [PubMed] [Google Scholar]
- 39.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th ed (text revision) Washington DC: American Psychiatric Association; 1996. [Google Scholar]
- 40.Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 41.Weir BS. Genetic Data Analysis II: Methods for discrete population genetic data. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 42.Draăghici S. Data analysis tools for DNA microarrays. Boca Raton: Chapman & Hall/CRC; 2003. [Google Scholar]
- 43.Korean Statistical Information Service (KOSIS) Census of the Korean population by administrative distinct, sex and age [Internet] Office KNS; 2004. Available from http://kosis.kr/ [Google Scholar]
- 44.Maurer I, Zierz S, Müller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophr Res. 2001;48:125–136. doi: 10.1016/s0920-9964(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 45.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bubber P, Tang J, Haroutunian V, Xu H, Davis KL, Blass JP, et al. Mitochondrial enzymes in schizophrenia. J Mol Neurosci. 2004;24:315–321. doi: 10.1385/JMN:24:2:315. [DOI] [PubMed] [Google Scholar]
- 47.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 48.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 49.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 50.Escamilla MA, McInnes LA, Service SK, Spesny M, Reus VI, Molina J, et al. Genome screening for linkage disequilibrium in a Costa Rican sample of patients with bipolar-I disorder: a follow-up study on chromosome 18. Am J Med Genet. 2001;105:207–213. doi: 10.1002/ajmg.1205. [DOI] [PubMed] [Google Scholar]
- 51.Williams NM, Rees MI, Holmans P, Norton N, Cardno AG, Jones LA, et al. A two-stage genome scan for schizophrenia susceptibility genes in 196 affected sibling pairs. Hum Mol Genet. 1999;8:1729–1739. doi: 10.1093/hmg/8.9.1729. [DOI] [PubMed] [Google Scholar]
- 52.Ewald H, Mors O, Koed K, Eiberg H, Kruse TA. Susceptibility loci for bipolar affective disorder on chromosome 18? A review and a study of Danish families. Psychiatr Genet. 1997;7:1–12. doi: 10.1097/00041444-199700710-00001. [DOI] [PubMed] [Google Scholar]
- 53.Maziade M, Roy MA, Rouillard E, Bissonnette L, Fournier JP, Roy A, et al. A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol Psychiatry. 2001;6:684–693. doi: 10.1038/sj.mp.4000915. [DOI] [PubMed] [Google Scholar]
- 54.Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, et al. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 55.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 56.Greenberg DA, Cayanis E, Strug L, Marathe S, Durner M, Pal DK, et al. Malic enzyme 2 may underlie susceptibility to adolescent-onset idiopathic generalized epilepsy. Am J Hum Genet. 2005;76:139–146. doi: 10.1086/426735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itoh S, Itoh F, Goumans MJ, Ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- 58.Lu J, Wu Y, Sousa N, Almeida OF. SMAD pathway mediation of BDNF and TGF beta 2 regulation of proliferation and differentiation of hippocampal granule neurons. Development. 2005;132:3231–3242. doi: 10.1242/dev.01893. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez FA, Raden DL, Rigby MR, Davis RJ. Heterogeneous expression of four MAP kinase isoforms in human tissues. FEBS Lett. 1992;304:170–178. doi: 10.1016/0014-5793(92)80612-k. [DOI] [PubMed] [Google Scholar]
- 60.Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 61.Li L, Wysk M, Gonzalez FA, Davis RJ. Genomic loci of human mitogen-activated protein kinases. Oncogene. 1994;9:647–649. [PubMed] [Google Scholar]
- 62.García JI, Zalba G, Detera-Wadleigh SD, de Miguel C. Isolation of a cDNA encoding the rat MAP-kinase homolog of human p63mapk. Mamm Genome. 1996;7:810–814. doi: 10.1007/s003359900242. [DOI] [PubMed] [Google Scholar]
- 63.New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood LJ, et al. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni H, Wang XS, Diener K, Yao Z. MAPKAPK5, a novel mitogen-activated protein kinase (MAPK)-activated protein kinase, is a substrate of the extracellular-regulated kinase (ERK) and p38 kinase. Biochem Biophys Res Commun. 1998;243:492–496. doi: 10.1006/bbrc.1998.8135. [DOI] [PubMed] [Google Scholar]
- 65.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaestel M. MAPKAP kinases-MKs-two's company, three's a crowd. Nat Rev Mol Cell Biol. 2006;7:120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 67.Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, et al. Scaffolding by ERK3 regulates MK5 in development. EMBO J. 2004;23:4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, et al. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 2004;23:4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many "cell adhesion" genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- 70.Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4:129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]