Abstract
The brain-specific immediate early gene Arc/Arg3.1 is induced in response to a variety of stimuli, including sensory and behavior-linked neural activity. Here we report the generation of transgenic mice, termed TgArc/Arg3.1-d4EGFP, expressing a 4-hour half-life form of enhanced green fluorescent protein (d4EGFP) under the control of the Arc/Arg3.1 promoter. We show that d4EGFP-mediated fluorescence faithfully reports Arc/Arg3.1 induction in response to physiological, pathological and pharmacological stimuli, and that this fluorescence permits electrical recording from activated neurons in the live mouse. Moreover, the fluorescent Arc/Arg3.1 indicator revealed activity changes in circumscribed brain areas in distinct modes of stress and in a mouse model of Alzheimer’s disease. These findings identify the TgArc/Arg3.1-d4EGFP mouse as a versatile tool to monitor Arc/Arg3.1 induction in neural circuits, both in vitro and in vivo.
Keywords: immediate early genes, Arc, Arg3.1, green fluorescent protein (GFP), transgenic mice, synaptic plasticity, neural circuits, stress
Introduction
The immediate early gene (IEG) Arc/Arg3.1 (activity-regulated cytoskeleton-associated protein or activity-regulated gene 3.1) was identified as a hippocampal transcript strongly induced by seizures and synaptic plasticity-inducing electrical stimulation in the rat hippocampus (Link et al., 1995; Lyford et al., 1995). Since the publication of the two original papers, Arc/Arg3.1 has become one of the best-studied brain-expressed IEGs, and a large amount of data has demonstrated Arc/Arg3.1 induction under physiological and pathological conditions.
Arc/Arg3.1 activation and function have been investigated most in the rodent hippocampus, where this immediate early gene plays an essential role in spatial processing, learning and memory. Hippocampal Arc/Arg3.1 transcripts are induced during exploration of a novel environment in a subset of context-activated pyramidal neurons (Guzowski et al., 2004; Guzowski et al., 1999; Vazdarjanova and Guzowski, 2004), and the levels of Arc/Arg3.1 expression correlate with learning in hippocampal-dependent spatial tasks (Guzowski et al., 2001). Furthermore, reduction or loss of Arc/Arg3.1 expression in the hippocampus, either by infusion of antisense oligodeoxynucleotides in the rat (Guzowski et al., 2000) or by genetic deletion in the mouse (Plath et al., 2006), interferes with synaptic plasticity and hippocampus-dependent learning and memory. Thus, although the detailed mechanisms through which Arc/Arg3.1 affects hippocampal functions are still under investigation, with recent evidence pointing to regulation of AMPA receptor endocytosis (Chowdhury et al., 2006; Rial Verde et al., 2006; Shepherd et al., 2006), mapping of Arc/Arg3.1 transcriptional induction provides a high-resolution approach to linking the activity of neural circuits to complex hippocampus-based behavior.
Arc/Arg3.1 induction has also been employed for mapping neural activity in several cortical and subcortical areas in the rodent brain. For example, Arc/Arg3.1 is induced by light exposure in context (orientation tuning) activated neural networks in the visual cortex (Wang et al., 2006), and its expression correlates with induction of synaptic plasticity during a critical postnatal developmental period, when cortical responses are shaped by sensory experience. Other brain regions where Arc/Arg3.1 was shown to be activated by physiological stimuli include the olfactory bulb (Guthrie et al., 2000) and piriform cortex (Zou and Buck, 2006) in response to odor application, the accessory olfactory bulb in mating (Matsuoka et al., 2002), the primary somatosensory cortex, parietal cortex and dorsal striatum after exploration of a novel environment (Ramirez-Amaya et al., 2005; Vazdarjanova et al., 2006), and the suprachiasmatic nucleus of the hypothalamus in response to light exposure (Nishimura et al., 2003).
Besides in studies of physiological brain activity, Arc/Arg3.1 induction has also been analyzed under conditions modeling human neuropathology in the rodent. Generalized seizures induce robust Arc/Arg3.1 induction in the hippocampus, especially in the dentate gyrus (DG) (Link et al., 1995; Lyford et al., 1995). Other human disease-related conditions that cause changes in Arc/Arg3.1 activation in specific brain regions include acute and chronic stress (Kozlovsky et al., 2008; Mikkelsen and Larsen, 2006; Ons et al., 2004; Trneckova et al., 2007), chronic neuroinflammation (Rosi et al., 2005), opiate withdrawal (Lucas et al., 2008), brain injury (Rickhag et al., 2007; Tan et al., 2007; Temple et al., 2003), and Alzheimer’s disease (Dickey et al., 2004; Dickey et al., 2003; Lacor et al., 2004; Palop et al., 2005). In addition, Arc/Arg3.1 gene activation can also serve to monitor and compare effects of different drugs, such as antidepressants regulating monoamine metabolism (Castro et al., 2003; Larsen et al., 2007; Pei et al., 2003; Tordera et al., 2003), the psychostimulant methylphenidate used to treat attention deficit hyperactivity disorder (Chase et al., 2007), or drugs of abuse, including morphine (Ziolkowska et al., 2005), cocaine and amphetamine (Tan et al., 2000).
Taken together, a large body of experimental evidence links the induction of Arc/Arg3.1 to different physiological and pathological as well as pharmacological neural circuit activity in the rodent brain. Methods of in situ hybridization and immunohistochemistry that are traditionally used for Arc/Arg3.1 detection, however, can only be applied to fixed brain sections, and hence do not allow analysis of live Arc/Arg3.1-expressing cells by, for example, electrophysiology in brain slices. In addition, in situ hybridization and immunohistochemistry are relatively labor intensive. We therefore generated a novel Arc/Arg3.1 “indicator” mouse, termed TgArc/Arg3.1-d4EGFP, which expresses a destabilized (4-hour half-life) version of the enhanced green fluorescent protein (d4EGFP) under the control of the Arc/Arg3.1 promoter on a BAC transgene. This mouse model can be used for the convenient visualization of Arc/Arg3.1-positive neural circuits in ex vivo microscopy and in vitro and in vivo electrophysiology. As a salient experimental example, the fluorescent Arc/Arg3.1 indicator gene proved sufficiently sensitive to detect changes in neural activity in a mouse model of Alzheimer’s disease, at an age well before any previously described amyloid-β (Aβ)-linked alterations.
Materials and Methods
Generation of BAC Arc/Arg3.1d4EGFP mice
A BAC clone containing the Arc/Arg3.1 gene (~64 kb upstream and ~63 kb downstream of the Arc/Arg3.1 ATG) was isolated by screening the CITB mouse BAC library (Research Genetics, Cat.No. 960050) and confirmed by direct sequencing of the BAC termini and the Arc/Arg.1 ORF. The targeting vector pSV1.RecA (Yang et al., 1997) used for the BAC modification contained: arm A (671bp fragment homologous to the promoter region upstream of the ATG), the ORF of a destabilized (4-hr half-life) EGFP (subcloned from pd4EGFP N1; BD Biosciences, Clontech), the intron/polyA signal of the rabbit β-globin gene, the kanamycin resistance gene flanked by FRT sites, and arm B (528 bp homologous to the sequence 3’ of the Arc/Arg3.1 polyA signal). After two rounds of homologous recombination (integration of the shuttle vector bearing the targeting construct and subsequent removal of the vector backbone; each step verified by Southern blotting) the kanamycin resistance gene was removed by transformation with a plasmid expressing Flp recombinase (pMAK-705FLP.amp) (Yang et al., 1997). The modified genomic fragment was released from pBeloBAC 11 with Not I endonuclease and purified on a Sepharose CL-4B gel column. Pronuclear injection of the modified BAC transgene in fertilized oocytes (B6D2F2 strain) resulted in the generation of 15 founders, 11 independent lines transmitted the transgene to progenitors; these mice were then bred into NMRI background and 9 lines were analyzed. PCR primers used for genotyping were: pair 1, gcagatgaacttcagggtcagc (GFP-5rev) and gcagagctcaagcgagttctc (arcRA-A200-5p), amplifying a PCR product of 351 bp; pair 2, cgtaatacgactcactatagggcg (T7-24) and cccctggaatatacccgacc (B5 start-rev), amplifying a PCR product of 271 bp. All studies reported here were performed with mice of line 3 or 11. The mice were raised on a 12-h light/dark cycle with water and food ad libitum.
Co-localization of intrinsic d4EGFP signal and Arc/Arg3.1-immunoreactivity in the brain of TgArc/Arg3.1-d4EGFP mice
Mice from nine independent founder lines were killed by overdose with isoflurane and rapidly perfused transcardially with 4% paraformaldehyde (PFA) in PBS (pH 7.4). Brains were removed and postfixed overnight at 4°C in the same fixative. 50 μm coronal or saggital sections of whole brains were prepared on a vibratome (Leica) and collected in PBS. Sections were then preincubated at room temperature (RT) in preblock solution (PBS, pH 7.4, 1% Triton X-100, 5% normal goat serum, NGS), followed by overnight incubation with antibodies against Arc/Arg 3.1 (polyclonal rabbit Arg3.1 antiserum, 1:1000) in PBS containing 0.5% TritonX-100 and 1% NGS. The next day, sections were washed with PBS (3 × 10 min) and incubated for 1h at RT with CY3-conjugated anti-rabbit secondary antibodies (Jackson Immuno Research Laboratories, Inc.), diluted 1:300 in PBS containing 0.5% TritonX-100 and 1% NGS. Sections were then washed in PBS (3 × 10 min), affixed to slides and coverslipped with Mowiol (Sigma). All images were acquired on a confocal microscope Leica TCS NT microscope; digitized images were analyzed using Adobe Photoshop (Adobe, Mountain View, CA).
BrdU protocol
Six three month-old male Arc-d4EGFP mice were housed two per cage, with free access to food and water. BrdU (Sigma) was diluted in water containing 1% sucrose, at a concentration of 1 mg/ml (Shimshek et al., 2006). Mice were treated with BrdU for 1 or 2 weeks and killed sequentially at 1, 2 or 4 weeks after start of the treatment (2 mice per time point). Mice were killed by isofluran, transcardially perfused with 4% paraformaldehyde (PFA) pH 7.4, and brains were postfixed in 4% PFA overnight. After sectioning of brains on a vibratome, the 50 μm coronal sections were incubated in 50% formamide and 50% 2x standard citrate saline (SSC, pH 7,0) for 2 hrs at 60 C. After washing in 2xSSC, sections were incubated in 2M HCl for 30 min at 37 °C and washed in Na-borate buffer (pH 9.0) for 10 min at RT, washed in PBS and incubated with 1% Triton X-100, 10% NGS solution in PBS for 2 hrs at RT. Next, sections were incubated with antibodies against GFP (rabbit anti-GFP antibody, Molecular Probes, Cat#A11122, dilution 1:10,000) and BrdU (rat monoclonal anti-BrdU antibodies, Accurate chemical and scientific corporation, ACCU, Cat# OBT0030G, dilution 1:400) in 0.5% Triton X100, 1% NGS in PBS O/N at RT. Sections were then washed in PBS and incubated with FITC-conjugated anti-rabbit and CY3-conjugated anti-rat secondary antibodies (Jackson ImmunoResearch Labs, dilution 1:250) in 0.5% Triton X-100, 1% NGS on PBS for 2 hrs at RT. After washing in PBS, sections were mounted on slides, dried and coverslipped with Mowiol.
Two-photon imaging and whole cell recording
In vivo two-photon targeted whole-cell recordings were obtained in three to four week-old mice, as described (Komai et al., 2006a; Komai et al., 2006b). Briefly, mice (postnatal day 43–51) were anesthetized with urethane (1.5–2 g/kg body weight, i.p.), a metal plate was attached to the skull, and a large craniotomy (about 2 mm diameter) was opened over the barrel cortex. The dura was removed while the chamber was superfused with external solution (mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 25 NaHCO3, 1.25 NaH2PO4, and 25 glucose. The exposed surface was covered with 1.5% agarose. Two-photon images were acquired with an Ultima laser-scanning microscope (Prairie Technologies, Middleton, WI), using a 40X objective (NA 0.8). The excitation source was a Coherent Ultra Ti:sapphire laser, tuned to a center wavelength of 950 nm. Green and red fluorescence were detected simultaneously with two photomultiplier tubes (Hamamatsu). The internal solution was (mM): 135 K-gluconate, 4 KCl, 10 HEPES, 4 Mg-ATP, 0.3 Na2-GTP, 10 phosphocreatine, 0.015 Alexa 594, and 0.3% biocytin (pH 7.3). Recordings were obtained by an Axoclamp-2A amplifier (Axon Instruments, Foster City, CA), filtered at 10 kHz, and digitized at 10–20 kHz. For whisker stimulation, mechanical stimuli (9.5° deflection angle) were delivered by a capillary attached to a piezoelectric apparatus (Piezo Systems, Cambridge, MA) for 200 ms at a frequency of 1 Hz. After recording, the mice were perfused transcardially with 4% PFA in PBS for morphological analyses.
Electroconvulsive shock treatment
Four-month-old male Arc/Arg3.1-d4EGFP mice (line 3 and 11) were anesthetized with pentobarbital (50 mg/kg body weight, ip) and received electric current through ear clips (60 Hz, sine wave, 80 V, 1 sec) as described (Yamagata and Obata, 1996). One or 3 hrs after the current delivery, mice were again anesthetized and perfused with 4% PFA in 0.1 M PB for 12 min at RT. The mice immediately perfused with the fixative after the first anesthetization served as control (naïve in the Fig. 2). Coronal brain slices (50 μm) were cut with a vibrating microslicer (VT-1000, Leica) and subjected for immunohistochemical analysis of the endogenous Arc/Arg3.1 protein and the d4EGFP expressions by the protocol mentioned above. Briefly, slices were blocked with 10% NGS containing 0.1% Triton X-100 antibody in PBS for 1 hr and reacted with either anti-Arc (Arg3.1 antiserum, provided by Dietmar Kuhl, dilution 1:1,500) or anti-GFP (Molecular Probes, Cat#A11122, dilution 1:2,000) rabbit antibody in 1% NGS containing PBS overnight at 4 C. After reaction with biotinylated anti-rabbit antibody (Vector laboratory, dilution 1:200), immunoreactions were visualized with the ABC elite kit (Vector laboratory) and DAB development procedure.
Figure 2. Induction of d4EGFP and Arc/Arg3.1 in the hippocampus following an electroconvulsive shock (ECS).
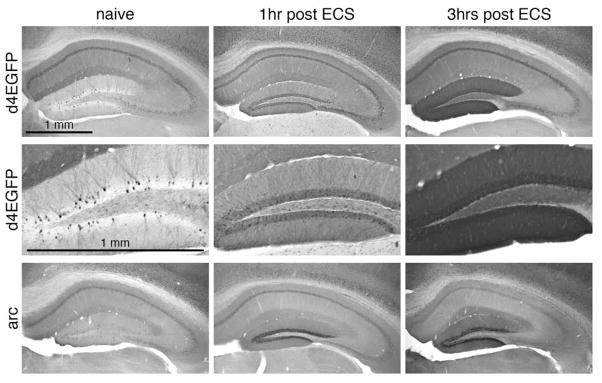
Immunohistochemistry with anti-GFP (top and mid panels) and anti-Arc/Arg3.1 antibodies revealed robust induction of both d4EGFP and Arc/Arg3.1 protein in the hippocampus, and particularly in the DG granule cells) at 1 and 3 hours post ECS. Naïve indicates non-treated control animal. Scale bar = 1 mm (top left panel).
p-MPPI protocol
Two month-old male TgArc/Arg3.1-d4EGFP mice (line 11; 21–24 g) received p-MPPI by i.p. injection at a dose 8.5 mg per kg between 8 and 9 a.m. (3 mice were used per experimental group); control mice were injected at the same time with saline. The mice were returned to their home cage, killed and perfused at 3, 6 and 12 hrs after p-MPPI injection (perfusant contained 4% PFA and 0.01% glutaraldehyde in phosphate buffer, pH 7.4). Coronal sections (40 μm) were cut on a Leica vibratome, collected every 120 μm along the rostrocaudal axis (approximately between +3.0 and −2.5 mm from Bregma), immunoreacted with rabbit polyclonal anti-GFP antibody (1:10,000, Molecular Probes) and visualized by use of the ABC kit (Vector Laboratories) and nickel-intensified DAB procedure. Sections were mounted onto SuperFrost slides, dehydrated and coverslipped. Mosaic images of sections were taken on a Leica DM6000 B microscope equipped with a motorized OASIS XY stage, Z focus control and Surveyor with Turboscan software (Objective Imaging, Ltd.), and monochrome camera QImaging Retiga EXi FAST 1394. Mosaic montage-images of the entire sections (taken with 10x objective) were analyzed manually by placing a 500×500 μm frame over brain regions of interest, including prefrontal cortex, dentate gyrus, lateral septum, caudate putamen and central nucleus of amygdala. Cell counts were made from 3 sections per region, bilaterally, for each mouse and performed by a person blind to the experimental treatment.
Stress protocols
Male TgArc/Arg3.1-d4EGFP mice (two to three months old) were habituated in individual cages with free access to water and chow for a week prior to the experiment. The mice were divided into five groups: 1) Home cage control (n= 4); 2) Single restraint (n=6; mice were placed in individual plastic cylinders with inner diameter of 3 cm for 1 hr and killed immediately or placed in their home cages and killed 2 hrs later); 3) Single intraperitoneal injection of bacterial endotoxin lipopolysaccharide, LPS (n=4; mice were injected with LPS from Escherichia coli serotype 0111:B4, L-3012, lot 36H4130, Sigma, St Louis, MO, USA, at a dose of 200 μg per 100 g of body weight (Grinevich et al., 2001), diluted in 300 μl of sterile saline and killed 0, 3, 6 and 24 hrs later); 4) Repeated restraint (n= 4; after 1 hr restraint, mice were returned to home cage for 2 hrs, again immobilized for 1 hr and killed immediately after the second restraint period); 5) Combination of restraint and LPS (n=6; mice were restrained for 1 hr, returned to home cage for 2 hrs, received LPS injection and killed 3 hrs later).
To perform the neurochemical characterization of d4EGFP-positive neurons in the PVN after combined stress, sections were stained with anti-GFP, CRH, vasopressin or oxytocin antibodies (tissue processing for CRH immunohistochemistry was done as described in (Asan et al., 2005). Sections containing the PVN were preincubated in preblock solution followed by overnight incubation (RT) with chicken anti-GFP antibody combined with polyclonal rabbit anti-CRH (Peninsula Lab, dilution 1:10.000) or monoclonal mouse anti-oxytocin (1:100) or monoclonal mouse anti-vasopressin antibody (1:100; oxytocin and vasopressin antibodies kindly provided by Dr. Harold Gainer, NIH; (Ben-Barak et al., 1985; Whitnall et al., 1985).
For neurochemical characterization of d4EGFP-positive neurons in the hypothalamic arcuate nucleus we combined chicken polyclonal anti-GFP antibodies with rabbit polyclonal antibodies against β-endorphin (National Hormone and Peptide Program, Director Dr. A.F. Parlow, AFP-791579Rb, dilution 1:5.000) or against neuropeptide Y (NPY) (Phoenix Pharmaceuticals, Inc., dilution 1:5000).
GFP was visualized with FITC-conjugated anti-chicken IgG antibodies. CRH, β-endorphin, NPY, oxytocin, and vasopressin were detected either by CY3-conjugated anti-rabbit IgG antibodies or CY3-conjugated anti-mouse antibodies (Jackson Immuno-Research Laboratories, Inc) diluted 1:300. After staining, the sections were washed in PBS (3 × 10 min), affixed to slides and coverslipped with Mowiol (Sigma).
All images were acquired on a confocal Leica TCS NT microscope; digitized images were analyzed using Adobe Photoshop (Adobe, Mountain View, CA). Adobe Photoshop was also used for the PVN selection and overlays.
Monitoring of Arc/Arg3.1-d4EGFP activation in a mouse model of Alzheimer’s disease
TgArc/Arg3.1-d4EGFP mice were crossed with 5xFAD mice carrying two transgenes, expressing human amyloid precursor protein (APP) with three familial Alzheimer’s disease (FAD) mutations and presinilin (PS 1, PS2) with two FAD mutations (Oakley et al., 2006). Three transgenic TgArc/Arg3.1-d4EGFP/5xFAD mice (2 males, 1 female) and three control TgArc/Arg3.1-d4EGFP mice (2 males, 1 female) from the same litter (1 month old, males housed separate from females) were killed by isoflurane directly after removal from home cages to analyze baseline d4EGFP labeling. After perfusion with 4% PFA (pH 7.4) and overnight postfixation in the same fixative, brains were vibratome-sectioned coronally, and the 50 μm sections were stained with anti-GFP antibodies (Molecular Probes). d4EGFP was visualized either with FITC-conjugated or streptavidin-conjugated secondary antibody and DAB staining. Three sections, developed with DAB, from each mouse at the same coronal level of the dorsal hippocampus, were chosen for counting d4EGFP-immunoreactive cells. Total cell number in the dorsal and ventral blades of the dentate gyrus was estimated using the optical fractionator method (Oakley et al., 2006) and Stereo Investigator software (Microbrightfield, Inc): dorsal or ventral blades of left and right dentate gyrus were traced at low magnification (5x) using a live image; the 63x (oil immersion) objective was used to achieve optical sectioning during stereological analysis. The Stereo Investigator software placed dissector frames (sampling grids were 100×100 μm) using a systematic-random sampling design within each contour. The data were analyzed by Student’s t-test.
Institutional approvals for animal work
Experimental procedures involving stress induction and whisker trimming were performed at the Max Planck Institute for Medical Research, Heidelberg and approved by the Animal Ethics Committee (Karlsruhe, Germany) and the Max-Planck Society. ECS experiment was carried out at National Institute for Physiological Sciences, Japan, under the approval of the National Institute for Physiological Science’s Animal Care and Use Committee.
Results
Neural activity induces d4EGFP expression in TgArc/Arg3.1-d4EGFP mice
We generated transgenic mice expressing an EGFP form with shortened half-life (d4EGFP) under the Arc/Arg3.1 promoter, contained on a bacterial artificial chromosome (BAC) (Fig. 1A and Methods). All founder lines showed similar baseline expression of Arc/Arg3.1 and d4EGFP (Figure 1B–D; Supplemental Figure 1). As Arc/Arg3.1 is highly induced in the hippocampal dentate gyrus (DG) granule cells following generalized seizures (Link et al., 1995; Lyford et al., 1995), we compared in our transgenic mice (founder lines 3 and 11) the induction of d4EGFP and Arc/Arg3.1 protein at 1 and 3 hours after electroconvulsive shock (ECS) treatment. As shown in Figure 2, both d4EGFP and native Arc/Arg3.1 were robustly induced in DG granule cells. These data demonstrate correct activation of the transgene-contained Arc/Arg3.1 promoter by ictal activity.
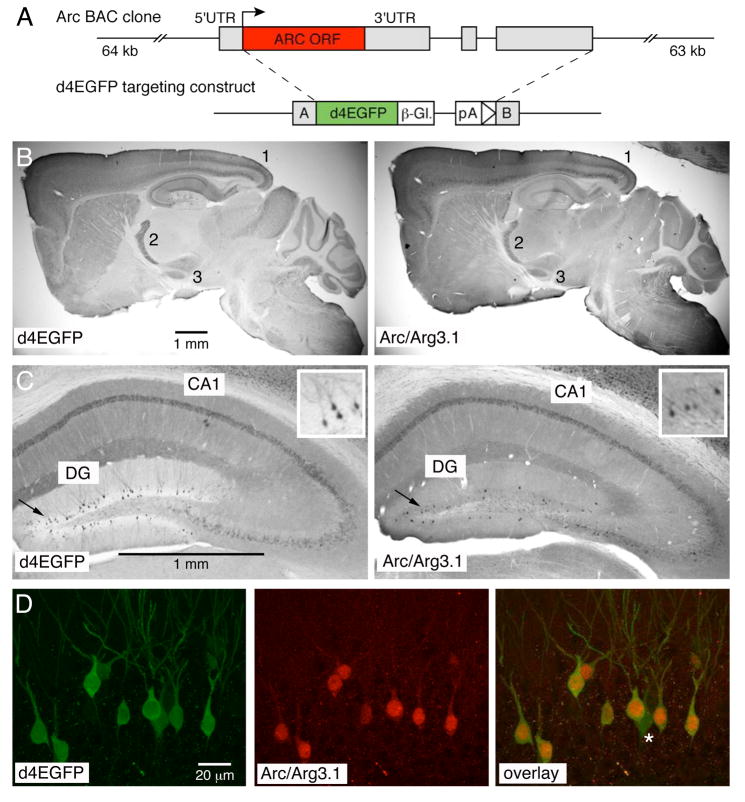
Figure 1. Comparison of d4EGFP and native Arc/Arg3.1 expression.
(A) Top: schema of the 127 kb BAC clone; Arc/Arg3.1 coding sequence is shown as a red box, UTR’s as gray boxes. Bottom: d4EGFP construct used to generate the BAC-derived TgArc/Arg3.1-d4EGFP transgene; d4EGFP coding sequence is shown as a green box, β-globin poly-A and intron as an interrupted white box, and the targeting Arms A and B as gray boxes. (B) Immunohistochemistry with anti-GFP (left panel) and anti-Arc/Arg3.1 antibodies (right panel) reveals overall comparable patter of expression, with prominent labeling in deep and superficial cortical layers (1), reticular thalamus (2) and subthalamic nucleus (3); the immunosignal was visualized with HRP-conjugated secondary antibodies and DAB reaction; see also Supplemental Figure 1 for comparison of all nine transgenic lines and Arc/Arg3.1 mRNA expression. (C) Higher magnification of anti-GFP (left panel) and anti-Arc/Arg3.1 immunosignal in the hippocampus. Note the prominent labeling of many cells in the CA1 and sparse labeling in the dentate gyrus (DG) regions, respectively. Insert in the right upper corner of each panel shows zoom-in view of the DG labeling. (D) Confocal microscopy of direct d4EGFP signal (left panel) and Cy3-visualized anti-Arc/Arg3.1 immunosignal (mid panel) reveals high level of colocalization in the DG region (overlay, right panel). See Supplemental Figure 2 for further examples of d4EGFP and Arc/Arg3.1 colocalization in the DG and cortex.
In a second set of experiments, we used sensory stimulation in the rodent somatosensory “barrel” cortex as a more physiological induction paradigm. The barrel cortex represents tactile information derived from the facial whiskers on the rodent’s snout, with an anatomical organization that matches sensory information from one facial whisker to one “primary” cortical column (Woolsey and Van der Loos, 1970). Trimming of all but two whiskers for 24 hours results in an upregulation of cortical responses evoked by deflection of the two spared whiskers (Diamond et al., 1993). As shown in Figure 3, this form of sensory stimulation caused a strong induction of d4EGFP expression in the D1 and D2 cortical columns representing the spared D1 and D2 whiskers. This demonstrates that our indicator mice can be used to map Arc/Arg3.1 induction in response to sensory-evoked neural circuit plasticity in the mouse cortex.
Figure 3. Induction of d4EGFP in the barrel cortex by sensory-stimulation.
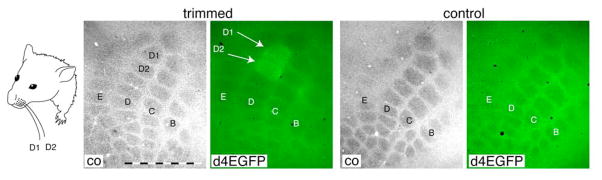
All but two D1 and D2 facial whiskers were trimmed, as shown in the schematic drawing, and cortical d4EGFP expression was analyzed 24 hours later. Anti-GFP immunostaining visualized with FITC-conjugated secondary antibodies revealed prominent induction of d4EGFP expression in the D1 and D2 columns in the trimmed, but not in the control untrimmed, animal. The scale bar = 1 mm. Cytochrome oxidase (co) staining (left in two panels) was used to visualize individual columns in the barrel cortex, which are organized in rows A though D; position of the D1 and D2 columns are marked in the image.
Rapid onset and decay of d4EGFP expression in response to single-dose pharmacological activation
We used the destabilized d4EGFP to monitor both the induction and decay of Arc/Arg3.1 expression. For this purpose, the TgArc/Arg3.1-d4EGFP mice were treated with a single dose of the 5-HT1A receptor antagonist p-MPPI (4-(2′-methoxy-phenyl)-1-(2′-(n-2″-pyridinyl)-p-iodobenzamido)-ethyl-piperazine), which was shown to induce Arc/Arg3.1 mRNA in the frontal and parietal cortices, hippocampus and caudate putamen (Pei et al., 2003; Tordera et al., 2003). Analysis of d4EGFP expression in mice injected intraperitoneally (i.p.) with p-MPPI (8.5 mg/kg) revealed robust induction of d4EGFP labeling 6 hours post injection in many cortical, hippocampal, striatal, septal, and thalamic regions relative to saline-injected controls (Figure 4A). The most prominent d4EGFP signals were observed in prefrontal cortex, caudate putamen, lateral septum and central amygdala (Figure 4B). Importantly, at 12 hours post p-MPPI injection, d4EGFP labeling had returned to baseline in all brain regions (Figure 4). This demonstrates that both onset and decay of Arc/Arg3.1 expression are faithfully monitored in TgArc/Arg3.1-d4EGFP mice.
Figure 4. Treatment with the 5-HT1A receptor antagonist p-MPPI causes a robust induction of d4EGFP.
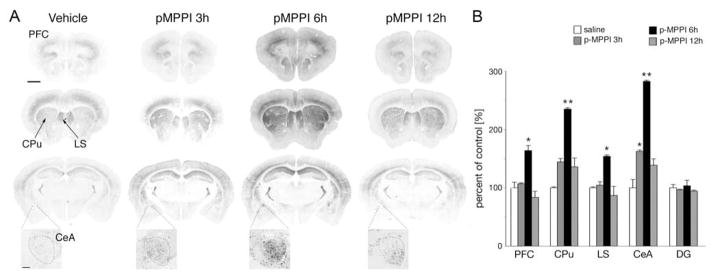
(A) Representative images of d4EGFP labeling in brain sections from control (vehicle) and p-MPPI treated animals reveal modest induction in the central nucleus of amygdala at 3 hours and robust induction in the prefrontal cortex, caudate putamen, lateral septum and the central nucleus of amygdala at 6 hours post p-MPPI injection. The level of d4EGFP expression returned to baseline at 12 hours. At each time point, three coronal sections taken approximately at +1.8, +0.9, and −1.58 mm from Bregma (order from top to bottom) are shown. Zoom-in regions show the central nucleus of amygdala (lateral subdivision of the central nucleus of amygdala is outlined). Scale bar for all large sections = 1 mm (upper left image); scale for enlarged CeA images = 100μm. (B) Quantification of d4EGFP-positive neurons, as percent change from control (vehicle treated) animals (% mean ± SD): PFC = 100 ± 9.6; 107.0 ± 1.7; 163.7 ± 8.8; 83.5 ± 10.5; CPu = 100 ± 14.1; 144.5 ± 7.5; 234.8 ± 0.2; 136.0 ± 5.7; LS =100 ± 1.6; 104.5 ± 5.8; 154.0 ± 2.3; 86.8 ± 15.2; CeA = 100 ± 14.3; 162.3 ± 2.4; 282.4 ± 2.0; 138.8 ± 10.9; DG = 100 ± 5.5; 96.5 ± 1.2; 103.5 ± 9.5; 94.3 ± 1.7. Abbreviations: CeA – central nucleus of amygdala. CPu – caudate putamen, LS – lateral septum, PFC – prefrontal cortex, DG – dentate gyrus. Statistical significance * p < 0,05, ** p<0.01 (One way ANOVA).
In vitro and in vivo electrophysiological analyses of d4EGFP-labeled neurons
Transgenic expression of fluorescent proteins, typically EGFP, has been used in many electrophysiological studies to identify neurons that differ from neighboring non-labeled cells by genetic manipulation. We thus tested whether d4EGFP-labeled neurons in TgArc/Arg3.1-d4EGFP mice show sufficient fluorescence for targeted electrophysiological analysis in vitro in brain slices and in vivo in anesthetized mice. Using standard wide-field fluorescence microscopy, it was relatively easy to visualize in brain slices d4EGFP-positive neurons in sparse labeled areas, such as the hippocampal DG. Labeled DG granule cells had mature intrinsic properties indistinguishable from non-labeled neighboring neurons (data not shown), suggesting that Arc/Arg3.1 expression is not linked to neurogenesis in the hippocampus. This was confirmed by BrdU labeling, as BrdU-positive and d4EGFP-positive DG granule cells showed no overlap in TgArc/Arg3.1-d4EGFP mice treated with BrdU in drinking water for one or two weeks and killed one, two or four weeks after the start of treatment (Supplemental Figure 3).
It proved more challenging, however, to distinguish d4EGFP-positive and -negative neurons by wide-field fluorescence microscopy in hippocampal CA1 and other brain regions with more subtle cell-to-cell differences in native Arc/Arg3.1 levels. Reliable targeting of d4EGFP-positive cells was possible when assisted by two-photon microscopy (Denk et al., 1990), which allows for high-resolution imaging at considerable tissue depth (data not shown). Indeed, use of two-photon microscopy allowed us to visualize in the somatosensory barrel cortex of anesthetized mice d4EGFP-positive neurons at a depth of several hundred micrometers and examine their receptive field properties by two-photon targeted patching (TPTP) (Margrie et al., 2003) (Figure 5).
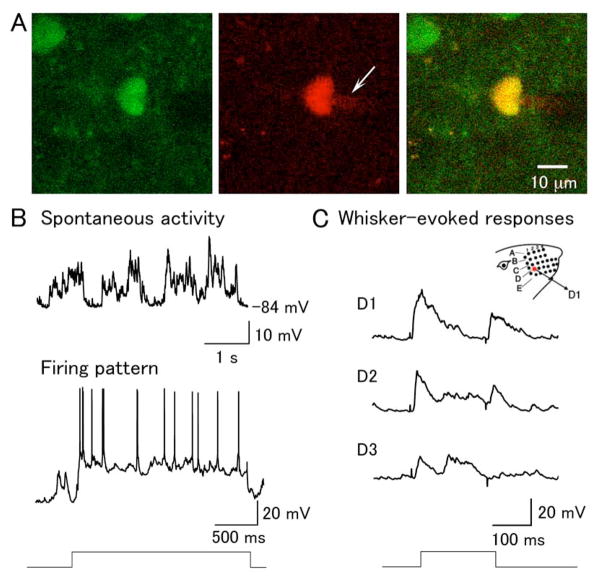
Figure 5. In vivo whole-cell recording of d4EGFP-positive neurons in the barrel cortex.
(A) An example of a d4EGFP-positive in vivo-patched layer 2/3 neuron. Left: d4EGFP signal; middle: Alexa 594 red signal in the internal recording solution, with a visable shadow of the patch pipette tip (white arrow); right overlay. (B) Spontaneous activity and firing pattern of d4EGFP-positive neuron. Top trace shows typical two-state membrane fluctuations (up and down states) observed in cortical neurons in anesthetized animals; bottom trace shows spiking in the same cell, elicited with DC current injection. (C) Deflection of the D1, D2, and D3 whiskers, as illustrated in the schema on the top right, evoked the corresponding responses shown below. The time course of the whisker deflection is shown at the bottom.
Arc/Arg3.1-d4EGFP labeling monitors neural activity in stress
Numerous IEGs, including c-fos, c-jun, and zif268 (egr1), have been used extensively for identification of brain circuits affected by stress (Kovacs, 1998; Martinez et al., 2002). Interestingly, previous studies reported a lack of Arc/Arg3.1 induction after a single restraint-based psychophysical stress (Mikkelsen and Larsen, 2006; Ons et al., 2004), suggesting that Arc/Arg3.1 is less responsive to stress-induced cell signaling. In agreement with the literature, a single-restraint stress paradigm did not induce expression of d4EGFP in the hypothalamic paraventricular nucleus (PVN), one of the main structures controlling neuroendocrine and autonomic responses to stress (Aguilera, 1994; Turnbull and Rivier, 1999) (Figure 6A–B). In contrast, repeated restraint stress as well as immune stress (i.p. injection of bacterial endotoxin lipopolysaccharide, LPS) induced d4EGFP labeling in PVN in the dorsal aspect of the lateral magnocellular subdivision. When combined, the two protocols had an additive effect, causing a prominent d4EGFP labeling in PVN neurons that were also positive for corticotropin-releasing hormone (CRH) (Figure 6C–I; note that oxytocin- or vasopressin-expressing PVN neurons were not d4EGFP-labeled). These data suggest that Arc/Arg3.1 contributes to stress-induced cellular adaptation in PVN CRH-positive neurons, in response to stronger stress stimuli compared to c-fos and several other IEGs.
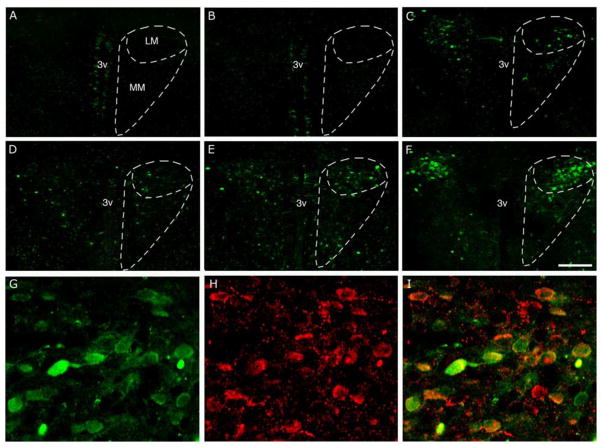
Figure 6. Induction of d4EGFP in the hypothalamic paraventricular nucleus (PVN) in different stress paradigms.
Representative images of coronal hypothalamic sections are shown from (A) naïve control mouse, (B) after single restraint stress, (C) after repeated restraint stress, (D–E) after LPS administration at 3 and 6 hrs, respectively, and (F) after combined restrain and LPS treatment. Note the increase in the number of d4EGFP-positive cells after different repeated stress paradigms (C–F). White dashed lines outline medial and lateral magnocellular subdivisions of the PVN. The d4EGFP signal (G) overlaps with corticotropin-releasing hormone immunoreactivity (H) in the majority of d4EGFP-positive neurons as seen on a single-plane confocal image (I). Scale bars = 200μm (panels A–F), 40 μm (panels G–I). Abbreviations: 3v – third ventricle, LM and MM - lateral and medial magnocellular subdivisions of the PVN (Paxinos and Franklin, 2003).
The stress protocol by single LPS injection also led to a prominent d4EGFP induction in cells located in the ventral part of the hypothalamus, specifically in the lateral part of the arcuate nucleus and retrochiasmatic area (Fig. 7 A–I). The arcuate nucleus, a major feeding center of the brain, is mainly composed of two populations of neurons expressing either neuropetide Y (NPY) or proopiomelanocortin (POMC) (Elmquist, 2001). Double staining with antibodies against β-endorphine (a derivative of POMC) and NPY revealed colocalization of d4EGFP with β-endorphine in cell somas and processes (Fig. 7 J–L) but not with NPY (Fig 7 M–O). These observations, together with the finding of Arc/Arg3.1 induction in PVN CRH neurons, suggest a role of Arc/Arg3.1 in regulating POMC neurons for a metabolic adaptation to immune challenge.
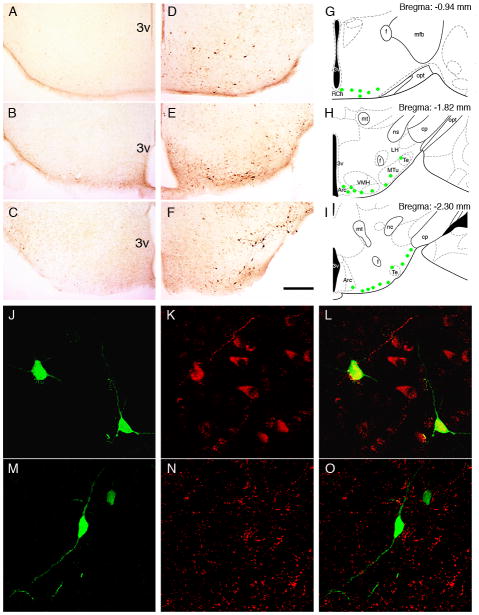
Figure 7. Induction of d4EGFP in the arcuate nucleus and retrochiasmatic area in immune stress.
Immunohistochemistry with anti-GFP antibody revealed no signal in control mice (A–C) and a profound upregulation of d4EGFP expression 6 hrs after a single LPS injection in the ventral hypothalamus (D–F). d4EGFP-positive neurons were mainly located in the lateral part of the arcuate nucleus and retrochiasmatic nucleus (extending to the lateral tuberal region), as represented by green circles in the schematic drawings in G–I (Paxinos, Franklin, 2003). Co-staining with antibodies against EGFP (J, M), beta-endorphin (K) or neuropeptide Y (N) revealed overlap of d4EGFP expression with β-endorphin (L) but not NPY (O). Scale bars = 200 μm (A–F), 40 μm (J–O). Abbreviations: 3v - third ventricle, Arc - arcuate nucleus, cp - cerebral peduncle, f - fornix, LH - lateral hypothalamus; mfb - medial forebrain bundle, mt - mammillothalamic tract, MTu - medial tuberal nucleus, ns - nigrostriatal bundle, opt - optic tract, RCh - retrochiasmatic area, Te - Terete hypothalamic nucleus, VMH - ventromedial hypothalamic nucleus.
Arc/Arg3.1-mediated d4EGFP induction can monitor neural activity in rodent model of Alzheimer’s disease
In a final set of experiments, we examined Arc/Arg3.1 activation in a mouse model of Alzheimer’s disease (AD). In a recent study, acute application of soluble β amyloid (Aβ) oligomers, proposed to cause cognitive dysfunctions in early AD (Klein et al., 2004; Walsh and Selkoe, 2007), was shown to bind to dendritic spines and induce Arc/Arg3.1 expression in vitro in cultured hippocampal neurons (Lacor et al., 2004). To find out whether Arc/Arg3.1 induction can be detected in young AD transgenic mice, we crossed our TgArc/Arg3.1-d4EGFP indicator mice with AD mice expressing human amyloid precursor protein (APP) and presenilin 1 (PS1) transgenes (5XFAD mouse model; (Oakley et al., 2006). As shown in Figure 8, prominent induction of d4EGFP labeling was detected in CA1 (Figure 8B) and DG granule cells (Figure 8B,D), as well as a moderate induction in deep cortical layers (Figure 8B) in one-month-old transgenic Tg5XFAD/TgArc/Arg3.1-d4EGFP mice, but not in littermates lacking the AD transgenes. These data support the hypothesis that early Aβ expression enhances the induction of Arc/Arg3.1 in affected neural circuits. They further indicate that the TgArc/Arg3.1-d4EGFP mice should prove useful in monitoring early neural activity changes in mouse models of human neuropathologies.
Figure 8.
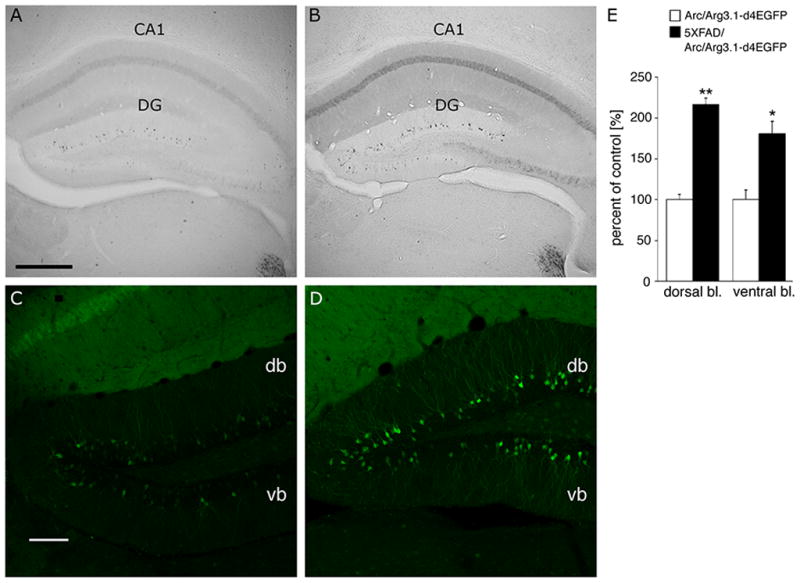
Increased d4EGFP expression in a mouse model of Alzheimer’s disease Representative images of DG hippocampus (coronal sections) from one month-old TgArc/Arg3.1-d4EGFP and Tg5XFAD/TgArc/Arg3.1-d4EGFP littermate mice are shown after visualization of d4EGFP by DAB (A, B) or immunofluorescence (C,D). Note the increased number of and labeling intensity of d4EGFP-positive neurons in CA1 (B) and DG granule cells (B, D) in the Tg5XFAD/TgArc/Arg3.1-d4EGFP mice. Brain sections stained with anti-GFP antibody and visualized by biotinylated (DAB method) or FITC-conjugated secondary antibodies. The fluorescent images are single confocal scans. Abbreviations: db – dorsal blade of DG, vb – ventral blade of DG; scale bars = 250 μm (A,B) and 100 μm (C,D). (E) Count of d4EGFP-positive neurons in Tg5XFAD/TgArc/Arg3.1-d4EGFP mice, as percent change from control TgArc/Arg3.1-d4EGFP littermates, in the DG: (% mean ± SD): 216.3 ± 7.7 vs. 100 ± 6.4 (**p < 0.002) in the dorsal blade, and 180.4 ± 15.3 vs. 100 ± 11.6 (*p < 0.03) in the ventral blade.
Discussion
Arc/Arg3.1 is a well-recognized IEG, which can be induced in diverse structures of the rodent brain in response to a wide range of behavioral, sensory, pharmacological and pathophysiological challenges (for reviews see (Clayton, 2000; Tzingounis and Nicoll, 2006). Most studies employed “ex vivo” measurements of Arc/Arg3.1 in postmortem brains. Here we report the generation of transgenic BAC mice faithfully expressing a destabilized form of EGFP in Arc/Arg3.1-expressing neurons. These TgArc/Arg3.1-d4EGFP mice allowed us to perform in vitro and in vivo recordings of Arc/Arg3.1 expressing neurons. In the course of our electrophysiological studies we demonstrated a “normal” range of responses of d4EGFP-positive layer 2/3 neurons of barrel cortex, suggesting that d4EGFP does not alter the electrophysiological properties, which opens many possibilities for functional studies. For example, one might employ two-photon targeted patching in fluorescent Arc/Arg3.1 neurons to test whether Arc/Arg3.1 induction may play a role in synaptic plasticity in somatosensory cortical neurons in response to different sensory stimulation protocols (Fox, 2002).
The expression of Arc/Arg3.1 has so far been preferentially evaluated by classical histological methods, such as in situ hybridization and immunohistochemistry. Despite good sensitivity, these methods have limitations for the detection of Arc/Arg3.1 mRNA or protein within the cell soma, due to the rapid transport of both mRNA and protein to dendritic compartments, which renders the identification of neuronal cell types cumbersome. The employment of TgArc/Arg3.1-d4EGFP mice helped overcome this problem due to the accumulation of d4EGFP in neuronal cell bodies and processes, even in the basal activity state. Similar labeling was seen in a previously published Arc/Arg3.1 knock-in d2EGFP mouse, in which the coding sequence for d2EGFP replaces that of Arc/Arg3.1 (Wang et al., 2006). In this mouse, however, the study of Arc/Arg3.1 functions can only be done in homozygous Arc/Arg3.1 knockout or heterozygous Arc/Arg3.1/d2EGFP mice. Our new TgArc/Arg3.1-d4EGFP lines thus represent a first mouse model that allows the study of wild-type Arc/Arg3.1-positive neurons. Among brain structures, the highest basal expression of d4EGFP in adult mice occurs in individual DG granule cells with d4EGFP-filled ramified dendrites, suggesting that these neurons are highly differentiated. This proposition was directly supported by results of BrdU treatment, which documented a lack of BrdU incorporation into the nuclei of d4EGFP-positive DG granule cells in adult TgArc/Arg3.1-d4EGFP mice.
The reliable activation of d4EGFP expression in TgArc/Arg3.1-d4EGFP mice has been confirmed by experimental paradigms previously applied by others. First, we showed that a generalized seizure (maximal electroconvulsive shock) induced simultaneous upregulation of Arc/Arg3.1 (Link et al., 1995; Lyford et al., 1995) and d4EGFP expression in the DG. Moreover, the application of the 5-HT1A receptor antagonist p-MPPI induced d4EGFP expression in various brain structures (prefrontal cortex, caudate putamen, lateral septum) previously seen activated by the same treatment (Toleda et al., 2003; Pei et al., 2003). In addition to these structures, the induction of d4EGFP was also found in the central nucleus of the amygdala, which has long been known to be responsive to 5-HT action (Middlemiss, 1984; Tork, 1988). Furthermore, the onset (3 hrs after p-MPPI injection), maximum (6 hrs), and decline (12–24 hrs) of d4EGFP expression in all analyzed brain structures reflect the advantage of the destabilized form of EGFP, as it allows monitoring the time-course of neural activity in different brain structures.
In addition to confirming previous results, we provide new results on the activation of Arc/Arg3.1 expression after stress and in a model of Alzheimer’s disease. With respect to stress, we showed an induction of d4EGFP expression in the hypothalamic PVN, which controls neuroendocrine and autonomic components of the stress response (Aguilera, 1994; Croiset et al., 2000; Turnbull and Rivier, 1999). In contrast to other IEGs, which are usually rapidly and profoundly induced in the PVN after a single psychophysical stress, induction of Arc/Arg3.1 required a repeated restraint protocol. However, in analogy to the induction of other IEGs, a single immune stress (LPS injection) led to d4EGFP induction in the PVN, which was further increased after combining both types of stressors. The induction was restricted to a distinct subset of CRH-positive neurons located in the PVN dorsal aspect of the lateral magnocellular subdivision. CRH is the central neurohormone mediating stress response and contributes to the regulation of the hypothalamic-pituitary-adrenal axis (Aguilera, 1994; Turnbull and Rivier, 1999) and autonomic CNS centers (Croiset et al., 2000). Although the cytoarchitectonic composition of the mouse PVN is not well defined, the region comprising d4EGFP/CRH-positive neurons may be similar to the dorsal aspect of the rat PVN which projects to autonomic centers of the brainstem and spinal cord (Sawchenko, 1987; Sawchenko and Swanson, 1985). Our preliminary experiments with retrograde tracing support the existence of direct connections between the dorsal-caudal aspect of the PVN lateral magnocellular subdivision and the autonomic center of the spinal cord (intermediolateral cell column) (data not shown). The exact connectivity of this subset of PVN neurons as well as the role of Arc/Arg3.1 in their activation upon stress will be the subject of further research.
Immune stress by LPS also induced d4EGFP expression in the arcuate nucleus and the retrochiasmatic hypothalamic region. These d4EGFP-positive neurons expressed POMC, a key anorexic peptide in the brain (Coll and Loraine Tung, 2009), which is upregulated in the arcuate nucleus after LPS injection (Sergeyev et al., 2001). This suggests that Arc/Arg3.1 induction in these neurons contributes to LPS-induced anorexia (Rorato et al., 2009). In addition, these neurons are known targets for leptin (Scott et al., 2009). LPS increases plasma levels of circulating leptin (Francis et al., 1999; Grunfeld et al., 1996; Sarraf et al., 1997), which in turn may augment POMC expression along with other factors, such as proinflammatory cytokines (Faggioni et al., 1997; Finck et al., 1998). Furthermore, neurons of the arcuate nucleus and retrochiasmatic area innervate sympathetic preganglionic neurons in the thoracic spinal cord, suggesting that this pathway may contribute to increased energy expenditure and decreased body weight following leptin administration (Elias et al., 1998). In line with this proposition, our preliminary results suggest that leptin induces d4EGFP expression in neurons of ventral hypothalamus (data not shown). In summary, our data suggest a role Arc/Arg3.1 in hypothalamic control of metabolism: Arc/Arg3.1-positive neurons in the arcuate nucleus and retrochiasmatic area may partake in the control of food intake and energy balance; the stimulatory effect of LPS on these neurons, likely mediated by leptin, may contribute to its anorexic action.
In the final set of experiments, the TgArc/Arg3.1-d4EGFP mice were used to report Arc/Arg3.1 expression in the 5XFAD mouse model of AD (Oakley et al., 2006). An induction of d4EGFP was observed in the DG and deep cortical layers in one month-old transgenic Tg5XFAD/TgArc/Arg3.1-d4EGFP mice; this finding provides evidence for pathology in neural activity that precedes plaque formation typically starting at three months of age in this mouse model (Oakley et al., 2006). This may suggest that Aβ oligomers alter Arc/Arg.1 expression, in a similar way as was observed in cultured neurons (Lacor et al., 2004). Interestingly, we did not detect changes in d4EGFP expression in three to four month old Tg5XFAD/TgArc/Arg3.1-d4EGFP mice (data not shown). Together with an earlier study demonstrating decreased Arc/Arg3.1 mRNA in six months and older APP+PS1 transgenic mice (Dickey et al., 2003), these findings suggest a complex disregulations of Arc/Arg3.1 function in Alzheimer’s disease. The impact of Arc/Arg3.1 elevation in early AD stages may contribute to deficits in long-term memory formation, as overexpression of Arc/Arg3.1 was shown to lead to a reduction of synaptic AMPA receptor levels (Chowdhury et al., 2006; Rial Verde et al., 2006).
In conclusion, we report the generation of new transgenic BAC mice that express d4EGFP under the control of the Arc/Arg3.1 promoter. Due to faithful d4EGFP expression in Arc/Arg3.1 neurons and the short d4EGFP half-life this mouse is a convenient model for in vitro, in vivo and ex vivo studies of Arc/Arg 3.1 functions as well as for the mapping of activated brain circuits in different physiological and pathophysiological conditions.
Supplementary Material
Comparison of d4EGFP expression in nine BAC transgenic lines
Top left panel shows detection of Arc/Arg3.1 mRNA by in situ hybridization (Allen Brain Atlas–Mouse Brain: http://www.brain-map.org; (Lein et al., 2007), with prominent expression in deep and superficial layers of somatosensory cortex (S1; short arrows), CA1 hippocampus, and reticular thalamus (Rt), and sparse expression in the DG hippocampus. All other panels show comparable d4EGFP protein expression in the nine mouse lines with transgene expression (lines 1, 2, 3, 6, 7, 11, 12, 14, and 15). Scale bar = 1 mm.
Colocalization of d4EGFP and Arc/Arg3.1 protein in the hippocampus and cortex
Confocal microscopy of direct d4EGFP signal (left panel) and Cy3-visualized anti-Arc/Arg3.1 immunosignal (mid panel) reveals high level of colocalization in the DG region (top and mid row panels) as well as in the cortex (bottom row panels). Overlay of the two signals is shown in panels on the right.
Lack of overlap between BrdU-positive and d4EGFP-positive cells the DG hippocampus
Mice were treated with BrdU in drinking water (1mg/ml) and killed 1, 2 or 4 weeks after the start of the treatment, as indicated above the panels (see Methods for more detail). The top panel row shows d4EGFP labeling, mid row shows BrdU staining and bottom row is an overlay of the two. There was no overlap between d4EGFP and BrdU signals at any time point. The images are maximal projections of 5–7 confocal z-sections (z step 1 μm) of coronal sections stained with primary anti-GFP and secondary anti-rabbit-FITC antibodies to visualize all d4EGFP-positive cells, and with primary anti-BrdU and secondary anti-mouse-Cy3 antibodies to visualize BrdU-positive cells. Scale bar = 100 μm.
Acknowledgments
We thank Dr. Harold Gainer (NINDS, NIH, Bethesda, USA) for monoclonal antibodies PS 41 and PS 38 against vasopressin and oxytocin; Dr. Albert F. Parlow (National Hormone and Peptide Program, Harbor-UCLA Medical Center, Torrance, USA) for polyclonal antibody against β-endorphin; and Dr. Andrei Rozov (University of Heidelberg, Germany) for initial electrophysiological recordings in acute hippocampal slices of Arc-d4EGFP mice. This work was supported by the Max-Planck Society, Human Frontier Science Program grant (RGY0079/2005-C) to P.O. and Alzheimer Association grant (NIRG-05-14907) to P.O.
References
- Aguilera G. Regulation of pituitary ACTH secretion during chronic stress. Frontiers in neuroendocrinology. 1994;15:321–50. doi: 10.1006/frne.1994.1013. [DOI] [PubMed] [Google Scholar]
- Asan E, Yilmazer-Hanke DM, Eliava M, Hantsch M, Lesch KP, Schmitt A. The corticotropin-releasing factor (CRF)-system and monoaminergic afferents in the central amygdala: investigations in different mouse strains and comparison with the rat. Neuroscience. 2005;131:953–67. doi: 10.1016/j.neuroscience.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Ben-Barak Y, Russell JT, Whitnall MH, Ozato K, Gainer H. Neurophysin in the hypothalamo-neurohypophysial system. I. Production and characterization of monoclonal antibodies. J Neurosci. 1985;5:81–97. doi: 10.1523/JNEUROSCI.05-01-00081.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Tordera RM, Hughes ZA, Pei Q, Sharp T. Use of Arc expression as a molecular marker of increased postsynaptic 5-HT function after SSRI/5-HT1A receptor antagonist co-administration. J Neurochem. 2003;85:1480–7. doi: 10.1046/j.1471-4159.2003.01782.x. [DOI] [PubMed] [Google Scholar]
- Chase T, Carrey N, Soo E, Wilkinson M. Methylphenidate regulates activity regulated cytoskeletal associated but not brain-derived neurotrophic factor gene expression in the developing rat striatum. Neuroscience. 2007;144:969–84. doi: 10.1016/j.neuroscience.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3. 1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–59. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DF. The genomic action potential. Neurobiology of learning and memory. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- Coll AP, Loraine Tung YC. Pro-opiomelanocortin (POMC)-derived peptides and the regulation of energy homeostasis. Molecular and cellular endocrinology. 2009;300:147–51. doi: 10.1016/j.mce.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Croiset G, Nijsen MJ, Kamphuis PJ. Role of corticotropin-releasing factor, vasopressin and the autonomic nervous system in learning and memory. European journal of pharmacology. 2000;405:225–34. doi: 10.1016/s0014-2999(00)00556-2. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–6. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Diamond ME, Armstrong-James M, Ebner FF. Experience-dependent plasticity in adult rat barrel cortex. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2082–6. doi: 10.1073/pnas.90.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Gordon MN, Mason JE, Wilson NJ, Diamond DM, Guzowski JF, Morgan D. Amyloid suppresses induction of genes critical for memory consolidation in APP + PS1 transgenic mice. J Neurochem. 2004;88:434–42. doi: 10.1111/j.1471-4159.2004.02185.x. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Loring JF, Montgomery J, Gordon MN, Eastman PS, Morgan D. Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein + presenilin-1 transgenic mice. J Neurosci. 2003;23:5219–26. doi: 10.1523/JNEUROSCI.23-12-05219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias CF, Lee C, Kelly J, Aschkenasi C, Ahima RS, Couceyro PR, Kuhar MJ, Saper CB, Elmquist JK. Leptin activates hypothalamic CART neurons projecting to the spinal cord. Neuron. 1998;21:1375–85. doi: 10.1016/s0896-6273(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Elmquist JK. Hypothalamic pathways underlying the endocrine, autonomic, and behavioral effects of leptin. Physiology & behavior. 2001;74:703–8. doi: 10.1016/s0031-9384(01)00613-8. [DOI] [PubMed] [Google Scholar]
- Faggioni R, Fuller J, Moser A, Feingold KR, Grunfeld C. LPS-induced anorexia in leptin-deficient (ob/ob) and leptin receptor-deficient (db/db) mice. The American journal of physiology. 1997;273:R181–6. doi: 10.1152/ajpregu.1997.273.1.R181. [DOI] [PubMed] [Google Scholar]
- Finck BN, Kelley KW, Dantzer R, Johnson RW. In vivo and in vitro evidence for the involvement of tumor necrosis factor-alpha in the induction of leptin by lipopolysaccharide. Endocrinology. 1998;139:2278–83. doi: 10.1210/endo.139.5.6012. [DOI] [PubMed] [Google Scholar]
- Fox K. Anatomical pathways and molecular mechanisms for plasticity in the barrel cortex. Neuroscience. 2002;111:799–814. doi: 10.1016/s0306-4522(02)00027-1. [DOI] [PubMed] [Google Scholar]
- Francis J, MohanKumar PS, MohanKumar SM, Quadri SK. Systemic administration of lipopolysaccharide increases plasma leptin levels: blockade by soluble interleukin-1 receptor. Endocrine. 1999;10:291–5. doi: 10.1007/BF02738628. [DOI] [PubMed] [Google Scholar]
- Grinevich V, Ma XM, Herman JP, Jezova D, Akmayev I, Aguilera G. Effect of repeated lipopolysaccharide administration on tissue cytokine expression and hypothalamic-pituitary-adrenal axis activity in rats. Journal of neuroendocrinology. 2001;13:711–23. doi: 10.1046/j.1365-2826.2001.00684.x. [DOI] [PubMed] [Google Scholar]
- Grunfeld C, Zhao C, Fuller J, Pollack A, Moser A, Friedman J, Feingold KR. Endotoxin and cytokines induce expression of leptin, the ob gene product, in hamsters. The Journal of clinical investigation. 1996;97:2152–7. doi: 10.1172/JCI118653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie K, Rayhanabad J, Kuhl D, Gall C. Odors regulate Arc expression in neuronal ensembles engaged in odor processing. Neuroreport. 2000;11:1809–13. doi: 10.1097/00001756-200006260-00003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44:581–4. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature neuroscience. 1999;2:1120–4. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–98. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein WL, Stine WB, Jr, Teplow DB. Small assemblies of unmodified amyloid beta-protein are the proximate neurotoxin in Alzheimer’s disease. Neurobiol Aging. 2004;25:569–80. doi: 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Komai S, Denk W, Osten P, Brecht M, Margrie TW. Two-photon targeted patching (TPTP) in vivo. Nat Protoc. 2006a;1:647–52. doi: 10.1038/nprot.2006.100. [DOI] [PubMed] [Google Scholar]
- Komai S, Licznerski P, Cetin A, Waters J, Denk W, Brecht M, Osten P. Postsynaptic excitability is necessary for strengthening of cortical sensory responses during experience-dependent development. Nature neuroscience. 2006b;9:1125–33. doi: 10.1038/nn1752. [DOI] [PubMed] [Google Scholar]
- Kovacs KJ. c-Fos as a transcription factor: a stressful (re)view from a functional map. Neurochemistry international. 1998;33:287–97. doi: 10.1016/s0197-0186(98)00023-0. [DOI] [PubMed] [Google Scholar]
- Kozlovsky N, Matar MA, Kaplan Z, Kotler M, Zohar J, Cohen H. The immediate early gene Arc is associated with behavioral resilience to stress exposure in an animal model of posttraumatic stress disorder. Eur Neuropsychopharmacol. 2008;18:107–16. doi: 10.1016/j.euroneuro.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Lacor PN, Buniel MC, Chang L, Fernandez SJ, Gong Y, Viola KL, Lambert MP, Velasco PT, Bigio EH, Finch CE, Krafft GA, Klein WL. Synaptic targeting by Alzheimer’s-related amyloid beta oligomers. J Neurosci. 2004;24:10191–200. doi: 10.1523/JNEUROSCI.3432-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MH, Rosenbrock H, Sams-Dodd F, Mikkelsen JD. Expression of brain derived neurotrophic factor, activity-regulated cytoskeleton protein mRNA, and enhancement of adult hippocampal neurogenesis in rats after sub-chronic and chronic treatment with the triple monoamine re-uptake inhibitor tesofensine. European journal of pharmacology. 2007;555:115–21. doi: 10.1016/j.ejphar.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5734–8. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Frenois F, Vouillac C, Stinus L, Cador M, Le Moine C. Reactivity and plasticity in the amygdala nuclei during opiate withdrawal conditioning: Differential expression of c-fos and arc immediate early genes. Neuroscience. 2008;154:1021–33. doi: 10.1016/j.neuroscience.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Meyer AH, Caputi A, Monyer H, Hasan MT, Schaefer AT, Denk W, Brecht M. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–8. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Martinez M, Calvo-Torrent A, Herbert J. Mapping brain response to social stress in rodents with c-fos expression: a review. Stress (Amsterdam, Netherlands) 2002;5:3–13. doi: 10.1080/102538902900012369. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Yamagata K, Sugiura H, Yoshida-Matsuoka J, Norita M, Ichikawa M. Expression and regulation of the immediate-early gene product Arc in the accessory olfactory bulb after mating in male rat. Neuroscience. 2002;111:251–8. doi: 10.1016/s0306-4522(01)00620-0. [DOI] [PubMed] [Google Scholar]
- Middlemiss DN. Stereoselective blockade at [3H]5-HT binding sites and at the 5-HT autoreceptor by propranolol. European journal of pharmacology. 1984;101:289–93. doi: 10.1016/0014-2999(84)90173-0. [DOI] [PubMed] [Google Scholar]
- Mikkelsen JD, Larsen MH. Effects of stress and adrenalectomy on activity-regulated cytoskeleton protein (Arc) gene expression. Neuroscience letters. 2006;403:239–43. doi: 10.1016/j.neulet.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Yamagata K, Sugiura H, Okamura H. The activity-regulated cytoskeleton-associated (Arc) gene is a new light-inducible early gene in the mouse suprachiasmatic nucleus. Neuroscience. 2003;116:1141–7. doi: 10.1016/s0306-4522(02)00786-8. [DOI] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, Berry R, Vassar R. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ons S, Marti O, Armario A. Stress-induced activation of the immediate early gene Arc (activity-regulated cytoskeleton-associated protein) is restricted to telencephalic areas in the rat brain: relationship to c-fos mRNA. J Neurochem. 2004;89:1111–8. doi: 10.1111/j.1471-4159.2004.02396.x. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Bien-Ly N, Massaro C, Yeung BZ, Yu GQ, Mucke L. Vulnerability of dentate granule cells to disruption of arc expression in human amyloid precursor protein transgenic mice. J Neurosci. 2005;25:9686–93. doi: 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates: Compact. 2. Elsevier; New York: 2003. [Google Scholar]
- Pei Q, Zetterstrom TS, Sprakes M, Tordera R, Sharp T. Antidepressant drug treatment induces Arc gene expression in the rat brain. Neuroscience. 2003;121:975–82. doi: 10.1016/s0306-4522(03)00504-9. [DOI] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3. 1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–44. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 2005;25:1761–8. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3. 1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–74. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickhag M, Teilum M, Wieloch T. Rapid and long-term induction of effector immediate early genes (BDNF, Neuritin and Arc) in peri-infarct cortex and dentate gyrus after ischemic injury in rat brain. Brain research. 2007;1151:203–10. doi: 10.1016/j.brainres.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Rorato R, Menezes AM, Giusti-Paiva A, de Castro M, Antunes-Rodrigues J, Elias LL. Prostaglandin mediates endotoxaemia-induced hypophagia by activation of pro-opiomelanocortin and corticotrophin-releasing factor neurons in rats. Experimental physiology. 2009;94:371–9. doi: 10.1113/expphysiol.2008.045435. [DOI] [PubMed] [Google Scholar]
- Rosi S, Ramirez-Amaya V, Vazdarjanova A, Worley PF, Barnes CA, Wenk GL. Neuroinflammation alters the hippocampal pattern of behaviorally induced Arc expression. J Neurosci. 2005;25:723–31. doi: 10.1523/JNEUROSCI.4469-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf P, Frederich RC, Turner EM, Ma G, Jaskowiak NT, Rivet DJ, 3rd, Flier JS, Lowell BB, Fraker DL, Alexander HR. Multiple cytokines and acute inflammation raise mouse leptin levels: potential role in inflammatory anorexia. The Journal of experimental medicine. 1997;185:171–5. doi: 10.1084/jem.185.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchenko PE. Evidence for differential regulation of corticotropin-releasing factor and vasopressin immunoreactivities in parvocellular neurosecretory and autonomic-related projections of the paraventricular nucleus. Brain research. 1987;437:253–63. doi: 10.1016/0006-8993(87)91641-6. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Swanson LW. Localization, colocalization, and plasticity of corticotropin-releasing factor immunoreactivity in rat brain. Federation proceedings. 1985;44:221–7. [PubMed] [Google Scholar]
- Scott MM, Lachey JL, Sternson SM, Lee CE, Elias CF, Friedman JM, Elmquist JK. Leptin targets in the mouse brain. The Journal of comparative neurology. 2009;514:518–32. doi: 10.1002/cne.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeyev V, Broberger C, Hokfelt T. Effect of LPS administration on the expression of POMC, NPY, galanin, CART and MCH mRNAs in the rat hypothalamus. Brain Res Mol Brain Res. 2001;90:93–100. doi: 10.1016/s0169-328x(01)00088-2. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3. 1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–84. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshek DR, Jensen V, Celikel T, Geng Y, Schupp B, Bus T, Mack V, Marx V, Hvalby O, Seeburg PH, Sprengel R. Forebrain-specific glutamate receptor B deletion impairs spatial memory but not hippocampal field long-term potentiation. J Neurosci. 2006;26:8428–40. doi: 10.1523/JNEUROSCI.5410-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A, Moratalla R, Lyford GL, Worley P, Graybiel AM. The activity-regulated cytoskeletal-associated protein arc is expressed in different striosome-matrix patterns following exposure to amphetamine and cocaine. J Neurochem. 2000;74:2074–8. doi: 10.1046/j.1471-4159.2000.0742074.x. [DOI] [PubMed] [Google Scholar]
- Tan J, Ruttiger L, Panford-Walsh R, Singer W, Schulze H, Kilian SB, Hadjab S, Zimmermann U, Kopschall I, Rohbock K, Knipper M. Tinnitus behavior and hearing function correlate with the reciprocal expression patterns of BDNF and Arg3. 1/arc in auditory neurons following acoustic trauma. Neuroscience. 2007;145:715–26. doi: 10.1016/j.neuroscience.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Temple MD, Worley PF, Steward O. Visualizing changes in circuit activity resulting from denervation and reinnervation using immediate early gene expression. J Neurosci. 2003;23:2779–88. doi: 10.1523/JNEUROSCI.23-07-02779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordera R, Pei Q, Newson M, Gray K, Sprakes M, Sharp T. Effect of different 5-HT1A receptor antagonists in combination with paroxetine on expression of the immediate-early gene Arc in rat brain. Neuropharmacology. 2003;44:893–902. doi: 10.1016/s0028-3908(03)00096-0. [DOI] [PubMed] [Google Scholar]
- Tork J. Anatomy of the serotoninergic system. Ann NY Acad Sci. 1988;600:9–35. doi: 10.1111/j.1749-6632.1990.tb16870.x. [DOI] [PubMed] [Google Scholar]
- Trneckova L, Rotllant D, Klenerova V, Hynie S, Armario A. Dynamics of immediate early gene and neuropeptide gene response to prolonged immobilization stress: evidence against a critical role of the termination of exposure to the stressor. J Neurochem. 2007;100:905–14. doi: 10.1111/j.1471-4159.2006.04278.x. [DOI] [PubMed] [Google Scholar]
- Turnbull AV, Rivier CL. Regulation of the hypothalamic-pituitary-adrenal axis by cytokines: actions and mechanisms of action. Physiological reviews. 1999;79:1–71. doi: 10.1152/physrev.1999.79.1.1. [DOI] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3. 1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–7. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci. 2004;24:6489–96. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, Mikhael D, Worley PF, Guzowski JF, Barnes CA. Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. The Journal of comparative neurology. 2006;498:317–29. doi: 10.1002/cne.21003. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ. Abeta Oligomers - a decade of discovery. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Whitnall MH, Key S, Ben-Barak Y, Ozato K, Gainer H. Neurophysin in the hypothalamo-neurohypophysial system. II. Immunocytochemical studies of the ontogeny of oxytocinergic and vasopressinergic neurons. J Neurosci. 1985;5:98–109. doi: 10.1523/JNEUROSCI.05-01-00098.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey TA, Van der Loos H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain research. 1970;17:205–42. doi: 10.1016/0006-8993(70)90079-x. [DOI] [PubMed] [Google Scholar]
- Yamagata Y, Obata K. Ca2+/calmodulin-dependent protein kinase II in septally kindled rat brains: changes in protein level, activity and subcellular distribution in hippocampus and cerebral cortex. Neuroscience letters. 1996;211:109–12. doi: 10.1016/0304-3940(96)12724-5. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature biotechnology. 1997;15:859–65. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Ziolkowska B, Urbanski MJ, Wawrzczak-Bargiela A, Bilecki W, Przewlocki R. Morphine activates Arc expression in the mouse striatum and in mouse neuroblastoma Neuro2A MOR1A cells expressing mu-opioid receptors. Journal of neuroscience research. 2005;82:563–70. doi: 10.1002/jnr.20661. [DOI] [PubMed] [Google Scholar]
- Zou Z, Buck LB. Combinatorial effects of odorant mixes in olfactory cortex. Science. 2006;311:1477–81. doi: 10.1126/science.1124755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of d4EGFP expression in nine BAC transgenic lines
Top left panel shows detection of Arc/Arg3.1 mRNA by in situ hybridization (Allen Brain Atlas–Mouse Brain: http://www.brain-map.org; (Lein et al., 2007), with prominent expression in deep and superficial layers of somatosensory cortex (S1; short arrows), CA1 hippocampus, and reticular thalamus (Rt), and sparse expression in the DG hippocampus. All other panels show comparable d4EGFP protein expression in the nine mouse lines with transgene expression (lines 1, 2, 3, 6, 7, 11, 12, 14, and 15). Scale bar = 1 mm.
Colocalization of d4EGFP and Arc/Arg3.1 protein in the hippocampus and cortex
Confocal microscopy of direct d4EGFP signal (left panel) and Cy3-visualized anti-Arc/Arg3.1 immunosignal (mid panel) reveals high level of colocalization in the DG region (top and mid row panels) as well as in the cortex (bottom row panels). Overlay of the two signals is shown in panels on the right.
Lack of overlap between BrdU-positive and d4EGFP-positive cells the DG hippocampus
Mice were treated with BrdU in drinking water (1mg/ml) and killed 1, 2 or 4 weeks after the start of the treatment, as indicated above the panels (see Methods for more detail). The top panel row shows d4EGFP labeling, mid row shows BrdU staining and bottom row is an overlay of the two. There was no overlap between d4EGFP and BrdU signals at any time point. The images are maximal projections of 5–7 confocal z-sections (z step 1 μm) of coronal sections stained with primary anti-GFP and secondary anti-rabbit-FITC antibodies to visualize all d4EGFP-positive cells, and with primary anti-BrdU and secondary anti-mouse-Cy3 antibodies to visualize BrdU-positive cells. Scale bar = 100 μm.