Abstract
Purpose
The accurate and timely diagnosis of malignant pleural effusion (MPE) in lung cancer patients is important because MPE has a poor prognosis and is classified as stage IV disease. Molecular biomarkers for pleural effusion, such as circulating extracellular microRNAs (miRNAs) isolated from pleural fluid, may help in the diagnosis of MPE. The present study examined whether miRNAs that are deregulated in lung cancer (miR-134, miR-185, and miR-22) can serve as diagnostic markers for lung adenocarcinoma-associated MPE (LA-MPE).
Materials and Methods
Real-time reverse transcription quantitative polymerase chain reaction was used to measure the expression of the three miRNAs in samples from 87 patients with pleural effusion comprising 45 LA-MPEs and 42 benign pleural effusions (BPEs). The area under the receiver operating characteristic curve (AUC) was then used to evaluate the diagnostic performance of each of the three miRNAs and compare it with that of the common tumor marker, carcinoembryonic antigen (CEA).
Results
The expression of all three miRNAs was significantly lower in LA-MPE than in BPE (p <0.001). The AUCs for miR-134, miR-185, miR-22, and CEA were 0.721, 0.882, 0.832, and 0.898, respectively. Combining CEA with the three miRNAs increased the diagnostic performance, yielding an AUC of 0.942 (95% confidence interval, 0.864 to 0.982), with a sensitivity of 91.9% and a specificity of 92.5%.
Conclusion
The present study suggests that the expression levels of circulating extracellular miR-134, miR-185, and miR-22 in patients with pleural effusion may have diagnostic value when differentiating between LA-MPE and BPE.
Keywords: Adenocarcinoma, Lung, miR-134, miR-185, miR-22, Pleural effusion
Introduction
Approximately 15% of lung cancer patients present with a pleural effusion at the time of initial diagnosis, and 50% develop a pleural effusion later in their disease course [1,2]. Pleural effusions can occur in patients with lung carcinomas of all histological types; however, they are more common in those with adenocarcinoma (40% of all pleural effusion cases) [2]. Metastatic malignant pleural effusion (MPE) due to lung cancer has a poor prognosis; MPE has been reclassified as M1a disease because its outcome is equivalent to that of distant metastasis [3]. Therefore, a diagnosis of MPE is important for lung cancer patients. Lung cancer patients suspected of having a pleural effusion undergo a chest X-ray and contrast-enhanced computed tomography to confirm the presence of fluid in the pleural cavity and to evaluate the pleura for nodularity or thickening, which may suggest metastasis [4]. Image-guided aspiration of the pleural fluid is often performed; however, the diagnosis of MPE by pleural fluid cytology has a sensitivity of only 60% with repeated large-volume (>50 mL) thoracentesis [5]. Therefore, it would be useful to identify biomarkers in the pleural fluid that can be used to improve diagnostic sensitivity. A systemic review found that carcinoembryonic antigen (CEA) showed good specificity (94%) but poor sensitivity (54%) for the diagnosis of MPE [6]. In addition, several studies examined different combinations of biomarkers in an attempt to improve diagnostic accuracy. Thus, there is a need to identify alternative noninvasive diagnostic biomarkers to improve the diagnosis of pleural effusions.
MicroRNAs (miRNAs) are a family of endogenous small (approximately 22 nucleotides in length) non-coding and functional RNAs that regulate the expression of many genes [7]. Aberrant expression of miRNAs occurs in many types of cancer, and several miRNAs function as tumor suppressor genes or oncogenes [8]. Recently, circulating extracellular miRNAs (in serum, plasma, pleural fluid, ascites, and other body fluids) were identified as noninvasive biomarkers for the diagnosis, prognosis, and prediction of responses to cancer treatment [9,10]. The present study examined the expression levels of three circulating miRNAs (miR-134, miR-185, and miR-22) in pleural effusion fluid; these miRNAs are deregulated in lung cancer. miR-134 regulates cell proliferation, apoptosis, and migration, and inhibits epithelial to mesenchymal transition (EMT) in non-small cell lung cancer (NSCLC) [11,12]; miR-185 suppresses the proliferation in lung adenocarcinoma cell lines and induces cell cycle arrest at the G1 phase [13]; miR-22 inhibits the growth and invasion of lung cancer by downregulating the expression of ErbB3 [14]. Given the diagnostic and prognostic potential of miR-134, miR-185, and miR-22 in lung cancer, we investigated the diagnostic significance of these miRNAs in patients with benign pleural effusion (BPE) and lung adenocarcinoma-associated MPE (LA-MPE) and compared it with that of the most commonly used tumor marker, CEA.
Materials and Methods
1. Patients and pleural fluid collection
We analyzed pleural fluid samples from 87 patients with pleural effusion who visited Chungbuk National University Hospital and Kangwon National University Hospital between February 2009 and April 2012. Pleural effusion was diagnosed as benign based on the clinical context and the absence of malignant cells in at least two separate samples from the same patient. BPE samples were obtained from patients showing no clinical or radiological evidence of malignancy prior to the study. LA-MPE was confirmed by pathological diagnosis based on the presence of adenocarcinoma cells. LA-MPE samples were obtained from patients showing no evidence of another malignant tumor and either 1) positive immunocytochemical staining for thyroid transcription factor-1 in MPE cell blocks, or 2) tumors that were histologically and/or clinically diagnosed as primary adenocarcinomas of the lung.
All effusion samples were collected before the start of cancer-directed therapy and were transported to the laboratory within 30 minutes of collection. The samples were then centrifuged at 11,300 ×g for 5 minutes. The supernatant and sediment from each sample were aliquoted into separate Eppendorf tubes and stored at -80℃ until use. All patients provided written informed consent to participate in the study, which was reviewed and approved by the Institutional Review Board of Chungbuk National University Hospital.
2. RNA extraction and real-time reverse transcription quantitative polymerase chain reaction
RNA was extracted from each sample (500 µL of sample) using a urine miRNA purification kit (Genolution Pharmaceuticals Inc., Seoul, Korea) according to the manufacturer's instructions. The amount of RNA extracted from each sample was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
The expression levels of miR-134, miR-185, and miR-22 in 42 BPE samples and 45 LA-MPE samples were measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Isolated miRNA (100 ng) was reverse transcribed using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol and using a specific miRNA primer that was provided with the TaqMan MicroRNA Assay kit (Applied Biosystems). Quantification of miRNA was performed by real-time RT-qPCR using an Applied Biosystems 7500 Fast Real-Time PCR System along with the TaqMan MicroRNA Assay, TaqMan Universal PCR Master Mix, and No AmpErase UNG (Applied Biosystems). All reactions were run in triplicate and Cq data were determined using default threshold settings. Relative miRNA expression levels were calculated using the 2-ΔΔCt method. The selection of suitable genes for the relative quantification of miRNA expression is essential to avoid inaccurate results and to improve the comparability of gene expression data. We previously examined the expression of the selected miRNAs, two small nuclear RNAs (RNU6B and U6 snRNA), and two small nucleolar RNAs (RNU44 and RNU48) to identify the most stably expressed genes for use as reference genes in pleural effusion samples [15]. U6 snRNA was identified as the most stable reference gene in the BPE and LA-MPE samples; thus, the expression levels of the three miRNAs were normalized to those of U6 snRNA.
3. Immunoassay for CEA
After performing the miRNA analyses, additional immunoassays were conducted to measure the levels of CEA expression in patients with pleural effusion. CEA levels were measured in an electrochemiluminescence immunoassay using a Cobas 8000 e602 analyzer (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions.
4. Statistical analysis
Differences in miR-134, miR-185, miR-22, and CEA expression between LA-MPE and BPE samples were assessed using the Mann-Whitney U test. A receiver operating characteristic (ROC) curve was plotted and the area under the curve (AUC) was calculated to evaluate the diagnostic performance of the three miRNAs and CEA. The best sensitivity/specificity pair was selected (with a specificity of least 0.6). Risk scores were assigned to all patients according to a linear combination of the expression level of the miRNAs and CEA, weighted according to the regression coefficient. Cox stepwise regression and stratification analyses were also conducted. All statistical analyses were performed using SPSS ver. 15.0 (SPSS Inc., Chicago, IL). The MedCalc ver. 10.4.7.0 (MedCalc, Mariakerke, Belgium) software was used to perform the ROC analysis. All p-values were two-sided and p<0.05 was considered statistically significant.
Results
1. Patient characteristics
Eighty-seven pleural effusion samples (42 BPE and 45 LA-MPE) were analyzed by RT-qPCR. Table 1 shows the baseline characteristics of the patients in the BPE and LA-MPE groups. The median age of the BPE patients was 61 years (range, 22 to 92 years) and that of the LA-MPE patients was 68 years (range, 33 to 92 years). The majority of BPE patients were diagnosed with tuberculous pleurisy or parapneumonic effusion. Twenty-seven patients with LA-MPE had stage M1b disease at presentation.
Table 1.
Characteristics of the study populations
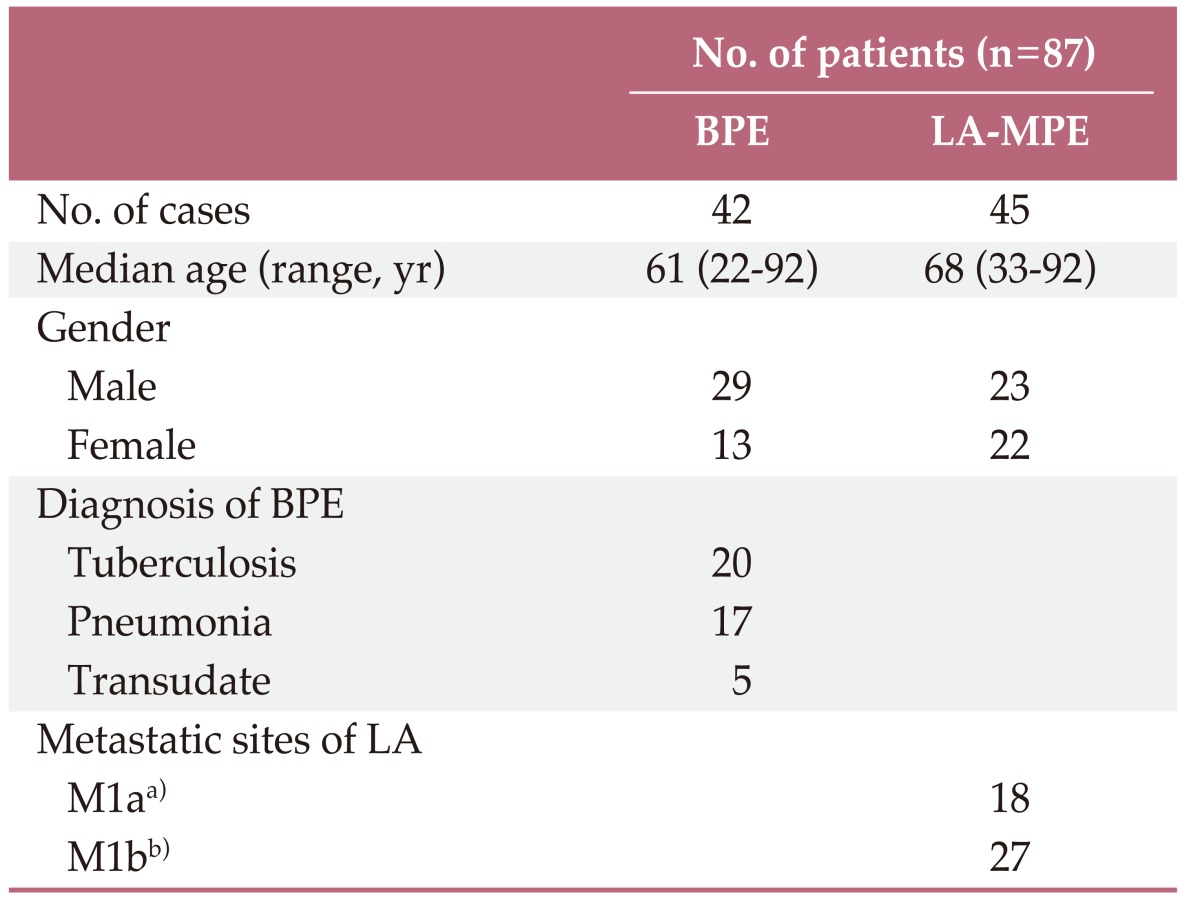
BPE, benign pleural effusion; LA-MPE, lung adenocarcinoma-associated malignant pleural effusion; LA, lung adenocarcinoma. a)M1a, separate tumor nodule(s) in a contralateral lobe, a tumor with pleural nodules, or malignant pleural (or pericardial) effusion, b)M1b, distant metastasis.
2. Expression levels of miR-134, miR-185, miR-22, and CEA in BPE and LA-MPE
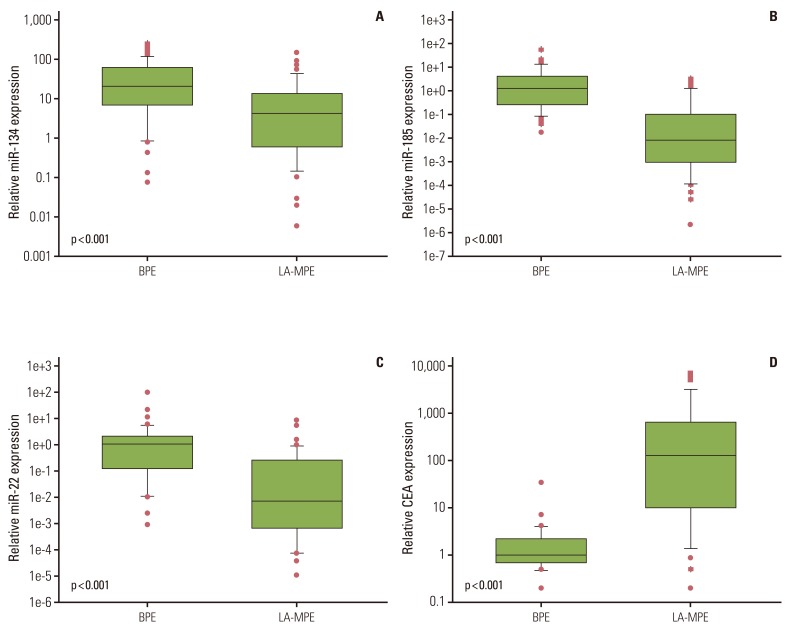
The expression levels of miR-134, miR-185, and miR-22 in LA-MPE samples were significantly lower than in BPE samples (all p<0.001, Mann-Whitney U test) (Fig. 1A-C). The median expression levels of miR-134, miR-185, and miR-22 were 19.758 (range, 0.069 to 243.601), 1.229 (range, 0.015 to 47.948), and 1.074 (range, 0.001 to 95.377), respectively, in BPE samples and 4.209 (range, 0.005 to 143.520), 0.008 (range, 0.000 to 2.852), and 0.008 (range, 0.000 to 8.562), respectively, in LA-MPE samples.
Fig. 1.
Comparison of miR-134 (A), miR-185 (B), miR-22 (C), and carcinoembryonic antigen (CEA) (D) expression levels in benign pleural effusion (BPE) and lung adenocarcinoma-associated malignant pleural effusion (LA-MPE) samples. Statistical significance was determined by the Mann-Whitney U test.
Of the 87 effusion samples, 77 were available for CEA testing. The median CEA expression level was significantly higher in the LA-MPE samples than in the BPE samples (127 ng/mL; range, 0.2 to 6,574 ng/mL vs. 1.0 ng/mL; range, 0.2 to 33.3 ng/mL) (p<0.001) (Fig. 1D).
3. Diagnostic performance of miR-134, miR-185, miR-22, and CEA for LA-MPE
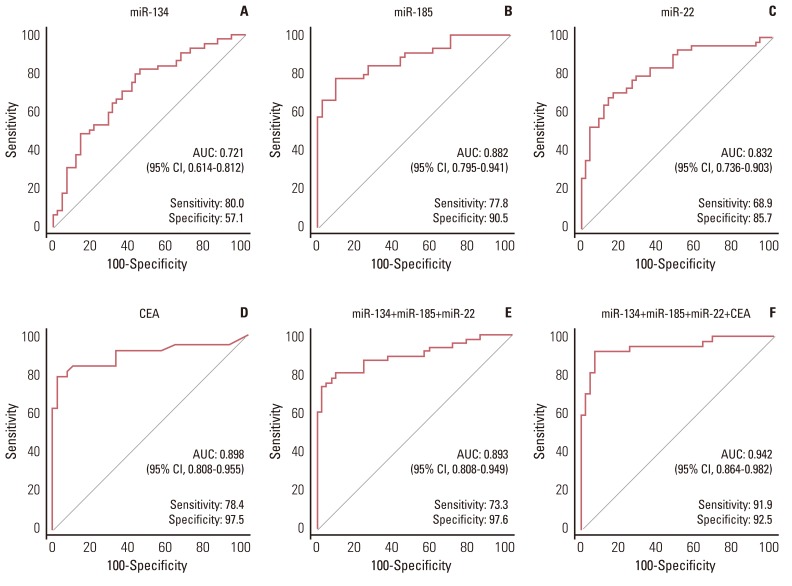
The diagnostic performance of miR-134, miR-185, miR-22, and CEA (and a combination of these markers) in differentiating BPE from LA-MPE was evaluated by ROC curve analysis (Table 2). Cut-off points were determined such that they maximized the sum of sensitivity and specificity. The cut-off points for miR-134, miR-185, miR-22, and CEA were 16.99, 0.092, 0.051, and 7.6 ng/mL, respectively. The diagnostic accuracy of miR-134, miR-185, and miR-22, as measured by the AUC, was 0.721, 0.882, and 0.832, respectively (Fig. 2A-C). The AUC for CEA was 0.898, with a sensitivity of 78.4% and a specificity of 97.5% (Fig. 2D). The predicted probability of being diagnosed with LA-MPE from the logit model based on the three miRNAs [logit=(-0.020×expression level of miR-134)+(-1.103×expression level of miR-185)+(-0.452×expression level of miR-22)] was used to construct a ROC curve. The AUC for the combined miRNAs was 0.893 (95% confidence interval [CI], 0.808 to 0.949), with a sensitivity of 73.3% and a specificity of 97.6% (Fig. 2E). The predicted probability of being diagnosed with LA-MPE from the logit model based on a combination of the three miRNAs plus CEA [logit=(-0.017×expression level of miR-134)+(-1.148×expression level of miR-185)+(0.332×expression level of miR-22)+(0.127×expression level of CEA)] was also used to construct a ROC curve. The AUC for a combination of all four markers was 0.942 (95% CI, 0.864 to 0.982), with a sensitivity of 91.9% and a specificity of 92.5% (Fig. 2F).
Table 2.
Sensitivity, specificity, and areas under the curves for miR-134, miR-185, miR-22, CEA, and combinations of these markers, in lung adenocarcinoma-associated malignant pleural effusion samples
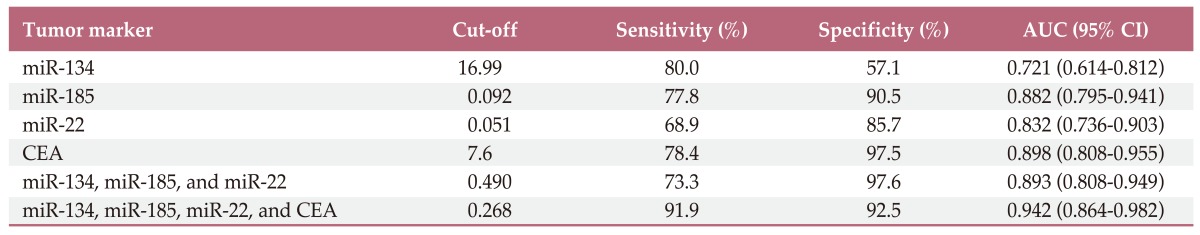
CEA, carcinoembryonic antigen; AUC, area under the curve; CI, confidence interval.
Fig. 2.
Receiver operating characteristic (ROC) curve analysis. The ROC plots for miR-134 (A), miR-185 (B), miR-22 (C), carcinoembryonic antigen (CEA) (D), a combination of three miRNAs (E), and a combination of three miRNAs plus CEA (F) were used to differentiate lung adenocarcinoma-associated malignant pleural effusion from benign pleural effusion. AUC, area under the receiver operating characteristic curve; CI, confidence interval.
Discussion
The present study demonstrated that LA-MPE samples showed significantly lower expression of miR-134, miR-185, and miR-22 than BPE samples. This suggests that all three miRNAs may be useful diagnostic biomarkers for differentiating between BPE and LA-MPE. In addition, the diagnostic performance of the three combined miRNAs (AUC, 0.893) was comparable with that of CEA (AUC, 0.898). CEA is a useful diagnostic biomarker for patients with pleural effusion; however, CEA is found to confer good specificity but relatively poor sensitivity for the diagnosis of LA-MPE [6]. The combination panel comprising miR-134, miR-185, miR-22, and CEA improved the diagnostic value (AUC, 0.942); in particular, the sensitivity increased to 91.9%.
Research on the prevention and early diagnosis of cancer has focused on identifying biomarkers that can help to detect tumors using noninvasive diagnostic methods. Recently studies suggest that the profile of circulating miRNAs, as well as tissue miRNAs, could be used as a diagnostic biomarker for cancer [9,10]. miRNAs are found in all body fluids and show distinct compositions in different fluid types [10]. The stability of miRNAs in body fluids may be explained, at least in part, by the finding that they are present in extracellular vesicular structures, called exosomes, which protect them from degradation [10]. miRNA-containing exosomes are found not only in blood, but also in other types of body fluid. Circulating exosomes and tumor cells from patients with lung adenocarcinoma harbor similar miRNA signatures, and there appears to be a significant difference in exosomal miRNA levels between cancer patients and controls [16]. Therefore, it is possible that pleural fluid obtained from patients that are difficult to diagnose using clinical and radiological evaluation could also be assessed for the presence of these molecular biomarkers. Recently, Xie et al. [17] studied the expression levels of 22 miRNAs that were aberrantly expressed in the serum of lung cancer patients (82 patients with MPE and 28 patients with BPE). They reported that MPE showed higher expression levels of cell-free miR-24 and miR-30d than BPE. However, they included pleural effusion samples from heterogenous lung cancer patients and also examined ascites from gastric cancer patients.
In a previous study, we examined the differential expression of circulating miRNAs in BPE and LA-MPE samples using a peptide nucleic acid-based microarray [15]. The miRNA expression signatures were confirmed by RT-qPCR and cross-validated using an independent sample set. The results showed that the expression level of miR-198 was significantly lower in LA-MPE than in BPE. For the present study, we selected only three miRNAs (miR-134, miR-185, and miR-22) that are deregulated in lung cancer and examined their expression in plural effusion samples obtained from patients with BPE and LA-MPE by RT-qPCR. We found that the expression of miR-134, miR-185, and miR-22, was lower in LA-MPE than in BPE. Downregulation of certain miRNAs in cancer suggests that these miRNAs may have tumor suppressor characteristics. These three miRNAs are also reported to function as tumor suppressors in various cancers. The expression of miR-134 correlates with the invasive potential and EMT phenotype of NSCLC cells [11,12]. Functional assays showed that miR-134 inhibits EMT in NSCLC cells, and Forkhead box protein M1 (FOXM1), a potential metastasis promoter, was a direct and functional target of miR-134 [12]. Also, low expression of cell-free miR-134 in MPE is associated with poor survival in NSCLC patients [18]. miR-185 suppresses cell proliferation and induces G1 arrest in lung cancer cell lines [13]. The extent of the growth suppression induced by these miRNAs is similar to that induced by the tumor suppressor miRNA, let-7 [19]. However, miR-185 has a stronger effect on cell cycle progression than let-7. In contrast to its tumor suppressor role, miR-185 plays an oncogenic role in both renal cell carcinoma and colorectal cancers [20,21]. The implied involvement of miR-22 in the p53 tumor suppressor network suggests that it is a strong candidate tumor suppressor gene in human colon and liver cancers [22,23]. Moreover, miR-22 restores the cellular senescence program in cancer cells and acts as a tumor suppressor [24]. It also shows anti-lung cancer activity both in vitro and in vivo, possibly via the post-transcriptional regulation of ErbB3 [14]. However, miR-22 is also oncogenic in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7,8-diol-9,10-epoxide [25]. These discordant findings suggest that miRNAs may target different genes in different cell types, thereby contributing to distinct biological processes.
The present study has some limitations. First, patients with BPE as the control group were not matched with the LA-MPE patients with respect to age and gender, which may have affected the results. The second limitation of the study was that all of three miRNAs expression were downregulated in LA-MPE compared with BPE, and the cut-off points for miR-185 and miR-22 were very low. Therefore, the diagnostic performance of these miRNAs that have tumor suppressor characteristics could be worse than that of oncogenic miRNAs.
Conclusion
The present study showed that the circulating extracellular miR-134, miR-185, and miR-22 levels in pleural effusions can be used to differentiate between LA-MPE and BPE. Furthermore, a panel comprising all three miRNAs showed a diagnostic performance comparable with that of CEA. The combination of all three miRNAs plus CEA showed considerable clinical value for diagnosing LA-MPE, with much better sensitivity than CEA alone. Thus, we propose that miR-134, miR-185, and miR-22 could be used to develop a minimally invasive screening tool for the diagnostic evaluation of pleural effusions.
Acknowledgments
This research was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (2007-0054930).
All effusion samples were provided by Chungbuk National University Hospital and Kangwon National University Hospital, members of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs. All samples derived from the National Biobank of Korea were obtained with patient informed consent using institutional review board-approved protocols. We appreciate the experimental assistance of the cancer research team at the Bioevaluation Center (Korea Research Institute of Bioscience and Biotechnology, Republic of Korea).
Footnotes
Conflict of interest relevant to this article was not reported.
References
- 1.Memon A, Zawadzki ZA. Malignant effusions: diagnostic evaluation and therapeutic strategy. Curr Probl Cancer. 1981;5:1–30. doi: 10.1016/s0147-0272(81)80012-8. [DOI] [PubMed] [Google Scholar]
- 2.Johnston WW. The malignant pleural effusion: a review of cytopathologic diagnoses of 584 specimens from 472 consecutive patients. Cancer. 1985;56:905–909. doi: 10.1002/1097-0142(19850815)56:4<905::aid-cncr2820560435>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 4.See KC, Lee P. Advances in the diagnosis of pleural disease in lung cancer. Ther Adv Respir Dis. 2011;5:409–418. doi: 10.1177/1753465811408637. [DOI] [PubMed] [Google Scholar]
- 5.Nance KV, Shermer RW, Askin FB. Diagnostic efficacy of pleural biopsy as compared with that of pleural fluid examination. Mod Pathol. 1991;4:320–324. [PubMed] [Google Scholar]
- 6.Gu P, Huang G, Chen Y, Zhu C, Yuan J, Sheng S. Diagnostic utility of pleural fluid carcinoembryonic antigen and CYFRA 21-1 in patients with pleural effusion: a systematic review and meta-analysis. J Clin Lab Anal. 2007;21:398–405. doi: 10.1002/jcla.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 8.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Zhao YP, Liu WJ, Dong J, Chen WY, Zhang TP, et al. Circulating microRNAs in cancer: diagnostic and prognostic significance. Expert Rev Anticancer Ther. 2012;12:283–288. doi: 10.1586/era.11.197. [DOI] [PubMed] [Google Scholar]
- 10.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids: the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Wang H, Zhang S, Song J, Zhang Y, Wei X, et al. MiR-134 functions as a regulator of cell proliferation, apoptosis, and migration involving lung septation. In Vitro Cell Dev Biol Anim. 2012;48:131–136. doi: 10.1007/s11626-012-9482-3. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586:3761–3765. doi: 10.1016/j.febslet.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J Cancer Res Clin Oncol. 2012;138:1355–1361. doi: 10.1007/s00432-012-1194-2. [DOI] [PubMed] [Google Scholar]
- 15.Han HS, Yun J, Lim SN, Han JH, Lee KH, Kim ST, et al. Downregulation of cell-free miR-198 as a diagnostic biomarker for lung adenocarcinoma-associated malignant pleural effusion. Int J Cancer. 2013;133:645–652. doi: 10.1002/ijc.28054. [DOI] [PubMed] [Google Scholar]
- 16.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 17.Xie L, Wang T, Yu S, Chen X, Wang L, Qian X, et al. Cell-free miR-24 and miR-30d, potential diagnostic biomarkers in malignant effusions. Clin Biochem. 2011;44:216–220. doi: 10.1016/j.clinbiochem.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Lv M, Shen S, Zhou S, Wang P, Chen Y, et al. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One. 2012;7:e43268. doi: 10.1371/journal.pone.0043268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Brannon AR, Reddy AR, Alexe G, Seiler MW, Arreola A, et al. Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell renal cell carcinoma. BMC Syst Biol. 2010;4:51. doi: 10.1186/1752-0509-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, et al. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311–318. doi: 10.3892/ijo.2011.1043. [DOI] [PubMed] [Google Scholar]
- 22.Yamakuchi M, Yagi S, Ito T, Lowenstein CJ. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One. 2011;6:e20291. doi: 10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Jiang Y, Zhang H, Greenlee AR, Yu R, Yang Q. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7, 8-diol-9,10-epoxide. Toxicol In Vitro. 2010;24:1168–1175. doi: 10.1016/j.tiv.2010.02.016. [DOI] [PubMed] [Google Scholar]