Background: Although glycosaminoglycan-induced GAPDH prefibrillar species accelerates α-synuclein aggregation, its role in toxicity remains unclear.
Results: The toxic effect exerted by α-synuclein oligomers on cell culture was abolished by GAPDH protofibril, which was identified and structurally characterized.
Conclusion: GAPDH protofibrils can efficiently sequester α-synuclein toxic oligomers.
Significance: GAPDH protofibrils may play an important role in neuronal proteostasis and could open a novel therapeutic strategy for synucleinopathies.
Keywords: α-Synuclein, Cell Permeabilization, Molecular Modeling, Parkinson Disease, X-ray Scattering, GAPDH, Protofibril Structure
Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a multifunctional enzyme that has been associated with neurodegenerative diseases. GAPDH colocalizes with α-synuclein in amyloid aggregates in post-mortem tissue of patients with sporadic Parkinson disease and promotes the formation of Lewy body-like inclusions in cell culture. In a previous work, we showed that glycosaminoglycan-induced GAPDH prefibrillar species accelerate the conversion of α-synuclein to fibrils. However, it remains to be determined whether the interplay among glycosaminoglycans, GAPDH, and α-synuclein has a role in pathological states. Here, we demonstrate that the toxic effect exerted by α-synuclein oligomers in dopaminergic cell culture is abolished in the presence of GAPDH prefibrillar species. Structural analysis of prefibrillar GAPDH performed by small angle x-ray scattering showed a particle compatible with a protofibril. This protofibril is shaped as a cylinder 22 nm long and a cross-section diameter of 12 nm. Using biocomputational techniques, we obtained the first all-atom model of the GAPDH protofibril, which was validated by cross-linking coupled to mass spectrometry experiments. Because GAPDH can be secreted outside the cell where glycosaminoglycans are present, it seems plausible that GAPDH protofibrils could be assembled in the extracellular space kidnapping α-synuclein toxic oligomers. Thus, the role of GAPDH protofibrils in neuronal proteostasis must be considered. The data reported here could open alternative ways in the development of therapeutic strategies against synucleinopathies like Parkinson disease.
Introduction
Neurodegenerative diseases like prion, Alzheimer, Huntington, and Parkinson diseases are classically characterized by brain proteinaceous aggregates. Lewy bodies and Lewy neurites, the neuropathological hallmarks of Parkinson disease and several neurological diseases, are mainly constituted by intracellular filamentous aggregates of the protein α-synuclein (α-SN)4 (1). In addition, recent studies have demonstrated the presence of misfolded or aggregated extracellular α-SN, suggesting that the pathogenic action of this protein might involve the transfer of α-SN from one cell to another through the extracellular space, with deadly consequences to the recipient cell (2–6). In vitro studies revealed that α-SN amyloid aggregation is a nucleation-dependent event that occurs in a process ranging from monomer via oligomers to fibrils (7, 8). Several studies conducted in vitro and in vivo showed that oligomeric α-SN intermediates are more toxic to cells than the monomeric or fibrillar forms of the protein (8–11). One of the main pathways of α-SN oligomer-induced pathogenicity is related to the impartment of biomembranes (12).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and glycosaminoglycans (GAGs) have been found to be associated with α-SN amyloid aggregates in Parkinson disease (11, 13, 14). On the one hand, GAPDH colocalizes with α-SN in amyloid aggregates in post-mortem tissue of patients with sporadic Parkinson disease and promotes the formation of Lewy body-like inclusions in cell culture. GAPDH is a homotetrameric enzyme largely expressed in cells and well known for its central role in energy production. However, recent data suggest that GADPH also possesses highly diverse nonglycolitic functions in the intra- or extracellular space (15) and has also been related to neurodegenerative diseases (16–22). Moreover, genomic analysis suggests that GAPDH has a protective effect on late-onset Alzheimer disease (23). On the other hand, GAGs are present in most, if not all, types of amyloids inside and outside of the cells (24, 25). In vitro, GAGs have proved to affect protein aggregation kinetics (26). We recently reported that sulfated GAGs, like heparin and heparan sulfates, are able to trigger GAPDH amyloid aggregation under pH and temperature physiological conditions (27). The heparin-induced GAPDH species formed during the early stages of the aggregation process (HI-GAPDHESS) are able to accelerate α-SN aggregation with a remarkable efficiency (27). In this study, we show for the first time that the interaction among GAGs, GAPDH, and α-SN exerts a protective role on dopaminergic cell survival. We have also performed a structural characterization of the HI-GAPDHESS by using small angle x-ray scattering (SAXS) combined with mass spectrometry, protein docking, and molecular dynamics simulations. Among the HI-GAPDHESS mixture, we were able to identify a native-like dimer as well as a protofibrillar species 22 nm long with a diameter of 12 nm. The experimental results strongly suggest that the protofibrils are the scavengers of the α-SN toxic oligomeric species. A protofibril all-atom model consistent with experimental constraints is herein presented. Upon secretion in the extracellular space, GAPDH might interact with GAGs leading to the formation of GAPDH protofibrils, which could improve neuron survival by sequestering toxic species of α-SN. In this context, our results could pave the way for a novel therapeutic strategy on neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Preparation of Oligomer-rich α-SN Samples
Expression and purification of recombinant human α-SN were performed as described previously (28). The purity of the protein was assessed by SDS-PAGE. Monomeric α-SN stock solutions were prepared in 20 mm HEPES, pH 7.4. Prior to measurements, protein solutions were filtered and centrifuged for 30 min at 12,000 × g. The protein concentration was determined by the measurement of absorbance at 275 nm using extinction coefficient ϵ275 = 5600 cm−1 m−1. The aggregation protocol was adapted from previous studies (29, 30). Monomeric α-SN solutions (140 μm) in 20 mm HEPES, pH 7.4, were incubated in a Thermomixer® comfort (Eppendorf) at 37 °C and 700 rpm. Aggregation was monitored with an ISS (Champaign, IL) PC1 spectrofluorometer using a thioflavin T (ThT) fluorescence assay on aliquots withdrawn from the incubation mixture at different times, according to LeVine (28). Oligomer-containing samples (α-SNoli) were harvested at 16 h of incubation, before the onset of the exponential fibril-growth phase.
Transmission Electron Microscopy
Aggregated α-SN (10 μl) was adsorbed onto Formvar-coated carbon grids (200 mesh), washed with Milli-Q water, and stained with 2% (w/v) uranyl acetate. The samples were imaged in a JEM-1200 Ex (Jeol) transmission electron microscope equipped with a GATAN camera, model 785.
Infrared Spectroscopy Measurements
Samples at 4 mg/ml 20 mm pD 7 D2O/HEPES buffer of α-SN were collected after 16 h of orbital incubation at 37 °C. Because monomeric α-SN was higher than the oligomeric species after 16 h of incubation, we partially separated it from the monomer using an Amicon Ultra-0.5 100-kDa cutoff filter and assembled it in a thermostatted cell between two CaF2 windows with a path length of 50 nm. The spectra were recorded in a Nicolet 5700 spectrometer equipped with a DTGS detector (Thermo Nicolet, Madison, WI) as described previously (31). The D2O contribution in the amide I′ region was eliminated by subtracting the buffer spectra from that of the solution at the same temperature to obtain a flat baseline between 2000 and 1700 cm−1. Fourier self- deconvolution and determination of band position of the original amide I′ band were performed as described (31).
Human Neuroblastoma Cell Culture
SH-SY5Y cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptavidin, at 37 °C and 5% CO2. For the assay, cells were seeded in 96-well plates at 15,000 cells/well and maintained in 100 μl of DMEM supplemented with 10% FBS and 1% penicillin/streptavidin for 24 h at 37 °C and treated as follows: control untreated cells received 25 μl of HEPES buffer (20 mm, pH 7.4); heparin-treated cells received 25 μl of a 75 μg/ml heparin solution; GAPDH-treated cells received 25 μl of a GAPDH (50 μm in HEPES) solution preincubated for 24 h at 37 °C; HI-GAPDHESS-treated cells received 25 μl of a GAPDH (50 μm) solution preincubated with 75 μg/ml heparin for 1 h and α-SNoli-treated cells received 25 μl of an α-SN (140 μm) solution preincubated for 16 h at 37 °C; GAPDH + α-SNoli-treated cells received 25 μl of a GAPDH (50 μm) + α-SNoli (140 μm); and HI-GAPDHESS + α-SNoli-treated cells received 25 μl of a HI-GAPDHESS (50 μm) + α-SNoli (140 μm) mixture. Mixtures containing GAPDH and α-SN were preincubated for 1 h at 37 °C under orbital agitation before addition to the cells. H2O2 (75 μm final in medium) was used for cell death control. Cell viability was determined using the colorimetric MTT metabolic activity assay (32). All experiments were performed in sextuplicate, and the relative cell viability (%) was expressed as a percentage relative to the untreated control cells.
Calcein Release Assay
The lipid mixture used for these experiments was extracted from brain membranes of Wistar white rats by Folch method (33). Rats were provided by the “Bioterio” Instituto de Química Biológica (Facultad de Bioquímica, Química y Farmacia, UNT, Tucumán, Argentina). The rats were obtained by exocria and maintained under controlled temperature and humidity under a 12-h light/dark cycle. The animals were maintained and treated in accordance with the criteria established in the “Guide for the Care and Use of Laboratory Animals,” published by the Institute of Laboratory Animal and National Research (1999). After extraction, lipids were stored in chloroform/methanol (2:1, v/v). For the preparation of large multilamellar vesicles, lipids were dried under nitrogen onto the wall of a Corex glass tube, placed in a vacuum oven to completely remove any remaining solvent, and then rehydrated in 25 mm Tris, 50 mm calcein, pH 7.4 buffer. To obtain small unilamellar vesicles (SUVs), multilamellar vesicles were sonicated with probe-type sonifier under nitrogen and controlled temperature. To remove titanium debris, the suspension was centrifuged for 15 min at 1100 × g (34). To separate calcein-loaded SUV from free dye, a Sephadex G-75 gel filtration medium (Pharmacia Biotech) was used. During incubation, changes in the fluorescence intensity of the different mixtures were monitored at λexc = 490 nm and λem = 510 nm (35) in an ISS (Champaign, IL) PC1 spectrofluorometer. GAPDH (50 μm) solution preincubated with 75 μg/ml heparin for 1 h and α-SNoli-treated SUVs received 25 μl of an α-SN (140 μm) solution preincubated for 16 h at 37 °C; GAPDH + α-SNoli-treated SUVs received 25 μl of a GAPDH (50 μm) + α-SNoli (140 μm); HI-GAPDHESS + α-SNoli-treated SUVs received 25 μl of an HI-GAPDHESS (50 μm) + α-SNoli (140 μm). Mixtures containing GAPDH and α-SN were preincubated for 1 h at 37 °C under orbital agitation before addition to 50 μm lipid vesicles. Total dye release was completed by the addition of 0.1 volume % Triton X-100. The percentage of probe release was calculated as shown in Equation 1,
where IF, IT, and IB are the fluorescence intensity of the dye released by the protein, total dye released, and control blank.
Small Angle X-ray Scattering Data
The scattering intensity of a noninteracting polydisperse system, which can be regarded as a mixture with p components, can be written as shown in Equation 2 (36),
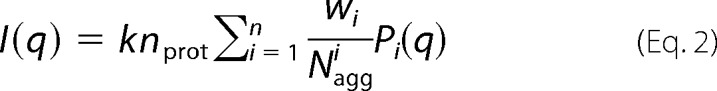 |
where k is a constant related to the experimental setup and should be the same for different SAXS curves collected within the same beamline setup; Naggi is the aggregation number of the i-th species, for instance, the values for the tetramer, dimer, and cylinder are 4, 2, and Nagg, respectively; Pi(q) is the form factor of the i-th species, and wi is weight in this model. It is important to consider that nprot wi is the concentration of the i-th species, or even the total amount of protein composed in each species.
The scattering curves of GAPDH in the presence of heparin were analyzed as a linear combination of three different species on solution (tetramers, dimers and an effective cylinder) in such a way that we get Equation 3,
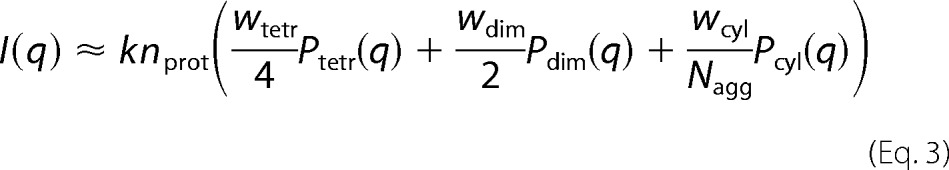 |
where wtetr, wdim, and wcyl are the weight of the tetramer, dimer, and an effective cylinder, respectively, and they respond to the following equation: wtetr + wdim + wcyl = 1. All SAXS data analysis was performed with GENFIT software (37, 38).
It should be remarked here that the choice of such a set of species in solution was based on the previous analysis of the distance distribution function p(r) (30), which is a model-independent procedure. The previous p(r) analysis thus showed that a rod-like protein aggregate evolved in the solution, coexisting with a native tetramer, reaching a maximum dimension of 25 nm with a cross-section radius of ∼6 nm up to 180 min of GAPDH-heparin incubation (30). However, as we mention below, the modeling fails to reproduce the experimental data at a high q range if smaller species as dimers are not included in the data analysis. The presence of monomers, instead of dimers in solution, was also probed. However, to get a good fitting to the experimental data, the modeled cylinders resulted in maximum dimensions on the order of 50 nm, which is not consistent with p(r) analysis.
Therefore, concerning the SAXS models employed here, both the tetramer and the dimer were calculated using the protein crystallographic structure (Protein Data Bank code 1J0X), through SASMOL methodology (37). Such a methodology assumes that the structures of the protein in the crystal and in solution are the same.
Regarding the form factor of an effective cylinder, it can be described as shown in Equation 4 (39),
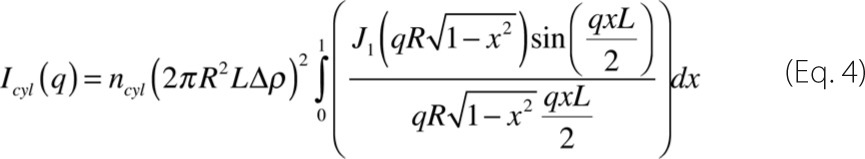 |
where R is the cylinder radius; L its length; Δρcyl = (ρcyl − ρsol) is the electron density contrast between the cylinder (ρcyl) and the solvent (ρsol) and ncyl the cylinder numeric density, which can be written as a function of the total protein concentration as: ncyl = nprotwcyl, with nprot the total protein concentration, and wcyl the weight of the cylinder in the SAXS curve.
Furthermore, it is possible to rewrite some of the cylinder's structural parameters as a function of others as shown in Equation 5,
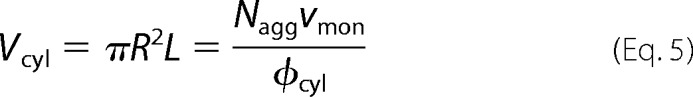 |
where Vcyl is the cylinder volume, and Nagg is the monomer-like aggregation number inside the cylinder. The volume of the GAPDH monomer is known from its crystallographic structure (νmon = 44,936 Å3). ϕcyl is the volume fraction of GAPDH inside the cylinder, i.e. ϕcyl = Naggν/Vcyl. Finally, both L and ρcyl can be now re-written as a function of ϕcyl as shown in Equations 6 and 7,
 |
and
ρprot is the theoretical electron density of the protein, calculated from its sequence (ρprot = 0.4235 e/Å3), and ρsol is the electron density of the solvent (= 0.333 e/Å3). After such rearrangement, the cylinder can be modeled using R, Nagg, and ϕcyl as fitting parameters and all the other variables concerning this model can be written as a function of these three variables.
Applying such methodology, the final set of fitting parameters are then R, Nagg, ϕcyl, and wtetr, wdim, and wcyl besides the experimental constant k.
Photoinduced Cross-linking of GAPDH in the Presence of Heparin
The mixture of GAPDH/heparin at a final concentration of 1 mg/ml, 0.5 mg/ml was preincubated at 37 °C under agitation for 0, 5, 30, 60, 90, and 120 min. For each mixture obtained at different time intervals, the photoreaction was carried out in a 10 × 10-mm quartz fluorescence cuvette with 1000 μl, following a similar procedure to the one previously described by Fancy et al. (40, 41). Concentration of the photosensitizer tris(bipyridine)ruthenium(II) dichloride was 10 μm, and a 20-fold molar excess of the electron acceptor ammonium persulfate was present with a time exposure to light of 1 min The samples were placed in a cuvette holder with a magnetic stirrer and irradiated with a Superlite SUV-DC illuminator (Lumatec, Germany) equipped with a 200-watt DC super pressure short arc mercury lamp. After the irradiation, 20-μl aliquots of the samples were diluted with 5 μl of SDS-loading buffer containing β-mercaptoethanol to quench the radical reaction. The samples were boiled for 10 min and then 20 μl of each sample were loaded per lane on 10% SDS acrylamide gels. Electrophoresis was carried out in a Bio-Rad Mini PROTEAN® system in slab gels using the buffer and fixing described by Weber and Osborn (42). The gel was stained with Coomassie Blue staining method. The protein molecular weight marker was Precision Plus ProteinTM Standards. Conversely, 200-μl aliquots of the samples were placed in ISS (Champaign, IL) PC1 spectrofluorometer where the emission spectrum of tyrosine and dityrosil was made. Emission spectra were registered setting λexc at 275 and 320 nm for selective excitation of Tyr or di-Tyr, respectively.
Trypsin Fingerprinting and Mass Spectrometric Analysis
For determination of photoinduced cross-linking modifications, trypsin fingerprinting was applied. Aliquots of photoinduced cross-linked tetrameric and aggregated GAPDH samples were separated by SDS-PAGE analysis as described above, treated with dithiothreitol/iodoacetamide, and digested in-gel using 20 μl of trypsin solution (25 ng/μl). Extracts from each sample were desalted using C18 resin accommodated in a tip (μZip Tip Millipore). The peptides were extracted from the resin with an α-cyanohydroxycinnamic acid solution used as matrix. The masses of the digestion products were analyzed using reflex mode MALDI-TOF mass spectrometry (ABI Mass Spectrometer 4800 plus MALDI-TOF-TOF). The identity of the signals was determined using GPMAW 9.2.
Computational Modeling of the Protofibril
Protein-protein docking models were generated using SymmDock (43). The crystallographic structure of the asymmetric unit for rabbit muscle GAPDH (44) deposited in RCSB (Protein Data Bank code 1J0X, chains O and P) was used as the building block. Complex models were predicted using 2–18-fold cyclic, dihedral, cubic, and helical symmetry by geometric docking algorithms. To predict the complex, we have applied the SymmDock algorithm twice in a two-step fashion. First, from the dimer we predicted a hexamer with C3 rotational symmetry, which in turn served as the input to build the 36-mer structure. The top 100 solutions of each run were analyzed, and candidate models failing to match the geometrical features derived from SAXS studies were discarded.
To further relax the structure of the protofibril model, molecular dynamic simulations were performed using a multiscale approach. At first, coarse grained simulations of the system were performed using the MARTINI force field (45) and its extension to proteins (46) as implemented in the GROMACS package. Elastic network constraints were also applied to maintain the internal structure of individual subunits (47). The elastic bond strength and the upper cutoff were set to 500 kcal/mol and 0.8 nm, respectively. Based on a comparison with all atom simulations, these were the parameters that better conserve the native structure while preserving realistic dynamics of the system. It should be noted that constraints introduced by the elastic network apply only to the tertiary structure, whereas the quaternary structure is free to change. The systems were simulated for 4 μs (16 μs effective time) coupled to Nose-Hoover thermostat and Parrinello-Rahman barostat. In a second stage, all-atom detail was included in the model by using the reverse transformation protocol described by Rzepiela et al. (48).
Contact Maps Calculation
Contact between a given pair of residues at the interface is defined as shown in Equation 8 (49),
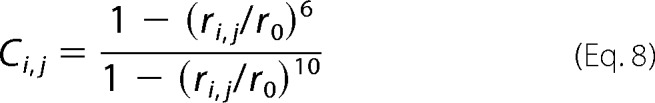 |
where r0 = 8.5 Å, and ri,j is the average distance between Cα atoms of residues i and j during the last microsecond of simulation. Through the use of this continuous function, we define a tight interaction between two residues as Ci,j > 0.8.
Spatial Aggregation Propensity
Regions prone to aggregation were predicted using the algorithm called Spatial Aggregation Propensity (50) and mapped on the protein's solvent accessible surface using VMD (51). This parameter is defined in Equation 9,
 |
where solvent-accessible areas of side chain atoms are computed at each simulation snapshot; solvent-accessible areas of fully exposed residues are calculated over a simulation of the tripeptide Ala-Xaa-Ala; and residue hydrophobicity is taken from the hydrophobicity scale of Black and Mold (52) and normalized so that glycine has hydrophobicity of zero. In this way, negative spatial aggregation propensity (SAP) values correspond to hydrophilic regions, whereas positive SAP values correspond to hydrophobic patches.
RESULTS
HI-GAPDHESS Modifies α-SNoli Toxicity and Membrane Stability
As we have shown previously, early stage species present in the heparin-induced GAPDH amyloid aggregation pathway (HI-GAPDHESS) can modulate α-SN aggregation kinetics (27). Here, we analyze whether HI-GAPDHESS can also modulate the toxicity of α-SNoli on a dopaminergic cell model using SH-SY5Y cell cultures (53).
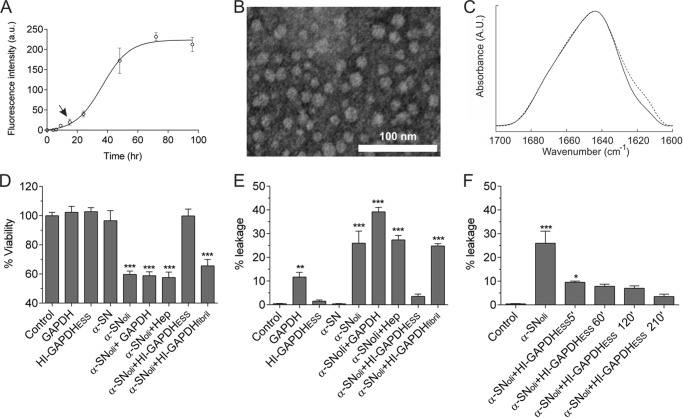
Several protocols have been devised to obtain oligomeric aggregates of α-SN, leading to different structural and functional arrangements of the protein Here, we used a protocol adapted from Refs, 29, 30. Fig. 1A shows the aggregation kinetics of α-SN as monitored by ThT fluorescent assay. The oligomers were harvested at 16 h, which corresponds to the lag phase. The fact that these species do not enhance ThT fluorescence suggests that they may lack the cross-β-structure characteristic of amyloid fibrils (54). In fact, the α-SN oligomers appeared as spheroidal and polydisperse species as shown by transmission electron microscopy (Fig. 1B). The FTIR analysis of the oligomeric state revealed an increase of β-structure compared with the monomeric state as judged by the shoulder at ∼1618 cm−1 (Fig. 1C). Taken together, these data suggest that the α-SNoli used in this work is compatible with the soluble on-pathway oligomers described previously (55).
FIGURE 1.
HI-GAPDHESS sequesters α-SNoli abolishing its deleterious effects. A, kinetics of 140 μm α-SN aggregation monitored by ThT fluorescence. The arrow shows the point where α-SNoli is harvested. B, uranyl acetate-stained transmission electron microscopy images of α-SNoli. a.u., arbitrary units. C, FTIR spectra of the deconvoluted amide I′ region of monomeric α-SN (solid line) and α-SNoli (dashed line). Deconvolution was performed using a Lorentzian line shape of 18 cm−1 and a resolution enhancement factor of 1.75. D, cell viability of SH-SY5Y after the addition of 20 mm HEPES, pH 7.40 (control), or 50 μm GAPDH. For HI-GAPDHESS, the enzyme (50 μm) was preincubated with heparin (75 μg/ml) for 2 h at 37 °C. α-SN and α-SNoli corresponds to α-synuclein (140 μm) preincubated for 0 and 16 h at 37 °C with orbital agitation, respectively. For α-SNoli + GAPDH, α-SNoli was preincubated for 1 h at 37 °C with GAPDH (50 μm). α-SNoli + HI-GAPDHESS corresponds to α-SNoli preincubated for 1 h at 37 °C with HI-GAPDHESS. MTT test was used to estimate cell viability. E, changes in liposomal membrane permeability induced by distinct aggregated species. The fluorescence signal was normalized with the signal observed after Triton X-100 addition, which induced complete rupture of the vesicles. α-SN monomer was employed as leakage negative control. HI-GAPDHESS, α-SNoli, α-SNoli + GAPDH, α-SNoli, and α-SNoli + HI-GAPDHESS were prepared as described above. F, effect of heparin/GAPDH incubation time on the production of species with the ability to protect membranes against content leakage induced by α-SNoli. The α-SNoli was preincubated with HI-GAPDHESS harvested after 5, 60, 120, and 210 min of GAPDH/heparin mixture under orbital agitation at 37 °C. Results were evaluated by analysis of variance (p = 0.001), and asterisks indicate significant differences versus the control (***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05).
The addition of α-SNoli prepared as done previously to SH-SY5Y cells culture induced approximately 40% of cell death (Fig. 1D). In contrast, preincubation of α-SNoli with HI-GAPDHESS during 60 min abolished α-SNoli toxicity. This protective effect was not observed when α-SNoli was preincubated with either native GAPDH or heparin, indicating that only intermediate species formed during the heparin-induced GAPDH fibrillation can efficiently protect cells against α-SNoli toxicity. Of note, the protective effect of GAPDH was lost once the fibrillar state was reached (Fig. 1D). In addition, neither native GAPDH nor HI-GAPDHESS affected cell viability on their own (Fig. 1D).
One of the proposed mechanisms by which α-SNoli exerts its toxic effect is by inducing a perturbation in the cell membrane integrity (56–58). To evaluate whether HI-GAPDHESS interferes with this mechanism, we studied the effect of preincubation of α-SNoli with HI-GAPDHESS on a membrane model system. To do so, we monitored the release of a fluorescent probe entrapped in rat brain lipid vesicles upon the addition of α-SNoli alone or preincubated with HI-GAPDHESS. The addition of α-SNoli induced the release of calcein from liposomes, confirming the ability of these species to affect membrane permeability. Noteworthy, preincubation of α-SNoli with HI-GAPDHESS significantly diminished such an effect (Fig. 1E). On the contrary, preincubation of α-SNoli with GAPDH in its native tetrameric or in its fibrillar state did not prevent calcein leakage indicating that inhibition of the α-SNoli-mediated membrane permeabilization should be associated with an unidentified intermediate species among HI-GAPDHESS, which does not affect the membrane integrity on its own. It is important to note that GAPDH in its native tetrameric state induces changes in membrane permeability as described previously (59). Nevertheless, this feature is lost when the enzyme is incubated with heparin (Fig. 1E). Therefore, we demonstrate that during the early stages of aggregation, the heparin/GAPDH incubation mixture is enriched in a particular subpopulation capable of sequestering the α-SNoli.
Enrichment of HI-GAPDHESS in a Protofibrillar Species Revealed by SAXS
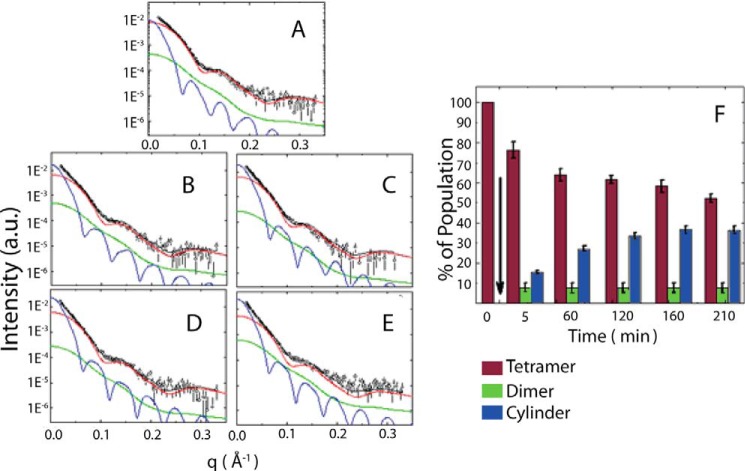
According to our previous studies, HI-GAPDHESS consists of prefibrillar species coexisting with native GAPDH tetramer (27). To further inquire into the nature of the putative GAPDH species capable of recruiting α-SNoli, we utilized SAXS because this technique is able to provide low resolution information on the structure of the species present in a polydisperse system as well as on their relative amounts (36, 60–63). In this way, we have previously shown that heparin triggers the formation of an elongated species with maximum dimension of ∼25 nm coexisting with the native tetramer through the analysis of the distance distribution function p(r) (27). Here, we performed a deeper characterization of the species present during the early steps of the heparin-induced GAPDH fibrillation process. We analyzed the SAXS curves obtained by GAPDH in the absence or in the presence of heparin after 5, 60, 120, 160, and 210 min of incubation (Fig. 2). In the absence of heparin, no significant changes were evidenced in the GAPDH SAXS curves indicating the stability of the protein (27). However, in the presence of heparin, we modeled the scattering curves as a mixture composed of the native tetrameric protein (Protein Data Bank code 1J0X) and an anisometric scattering particle represented by a homogeneous cylinder. As the modeling failed to reproduce the SAXS experimental data in the high q range, we evaluated the inclusion of additional smaller particles for a better fit (data not shown). Interestingly, the best fitting, performed with GENFIT software (37, 38), was achieved when dimers were included in the SAXS data analysis, resulting in fitting parameters for the protofibril consistent with p(r) analysis (30). The quality of the fittings was rather good, indicating that the modeling is compatible with the scattering data for the whole set of experiments. The fitting parameters correspond to the amount of protein in the tetrameric (wtetr), dimeric (wdim), and cylindrical (wcyl) forms (Table 1). Further parameters, the cylinder radius (R), the protein fraction inside the cylinder (ϕcyl), and the number of monomers composing the cylinder, which corresponds to the aggregation number (Nagg), can also be extracted from the SAXS data analysis.
FIGURE 2.
SAXS modeling of the heparin/GAPDH incubation mixture. Small angle x-ray scattering curves of GAPDH in the presence of heparin (open circles) at 5 min (A); 60 min (B); 120 min (C); 160 min (D); and 210 min (E). The solid black line represents the sum of three different models: GAPDH tetramer (red line), dimer (green line), and an effective cylinder (blue line). See text for details. F, relative population of GAPDH species present in the incubation mixture after the addition of heparin. The arrow indicates the moment when GAPDH and heparin were mixed. The bars represent the native-like tetramer (red vertical bar), the native-like dimer (green bar), and the protofibril (blue bar). a.u., arbitrary units.
TABLE 1.
Adjustment parameters from the cylinder-like aggregate along time
The parameters used are as follows: R, cylinder radius; L, length; ϕcyl, the cylinder fraction volume; Nagg, aggregation number (i.e. the number of GAPDH monomers inside the cylinder); ρcyl, electronic density; wcyl, weight of the cylinder in the SAXS curve. In the last two lines the average and the S.D. values are shown.
| Time (min) | k | wtetr | wdim | wcyl | R | L | ϕcyl | Nagg | ρcyl |
|---|---|---|---|---|---|---|---|---|---|
| 10−5 | % | % | % | Å | Å | e/Å3 | |||
| 2 | 5.08 | 76.5 | 7.96 | 15.56 | 59.6 | 200 | 0.7 | 35.0 | 0.3932 |
| 52 | 5.00 | 63.99 | 9.04 | 26.97 | 57.18 | 222 | 0.68 | 35.0 | 0.3915 |
| 106 | 5.00 | 61.5 | 4.94 | 33.56 | 59.07 | 210 | 0.65 | 33.0 | 0.3885 |
| 160 | 5.00 | 58.35 | 4.94 | 36.7 | 59.61 | 222 | 0.66 | 36.0 | 0.3891 |
| 212 | 5.12 | 52.41 | 11.11 | 36.41 | 60.02 | 219 | 0.69 | 38.0 | 0.3923 |
| Average | 5.04 | 7.6 | 59.10 | 215 | 0.68 | 35.4 | 0.391 | ||
| S.D. | 0.05 | 2.4 | 1.00 | 9 | 0.02 | 1.6 | 0.002 |
Taken together, these results suggest that in the early stages of heparin-induced fibrillation, GAPDH coexists in three different aggregation states in solution: native tetramers, native-like dimers, and higher order aggregates with a cylindrical shape. These cylinder-like species, structurally characterized for the first time in the present results, are on average 21.5 ± 0.9 nm long with 11.8 ± 0.2 nm diameter and are composed of 35 ± 2 monomers (Table 1). Following the nomenclature proposed by Kodali and Wetsel (64), we will refer to these species as protofibrils.
Time-evolution of different HI-GAPDHESS subpopulations was obtained through the SAXS data analysis (Fig. 2F). The native tetrameric species diminished as the protofibrillar species concentration increased during the first 210 min of incubation of GAPDH in the presence of heparin, whereas the dimer population remained unaltered during this time interval. Noteworthy, there is a good agreement between the increment of the protofibrillar species (Fig. 2F) in the heparin/GAPDH incubation mixture and its capability to protect the membrane against α-SNoli (Fig. 1F).
All-atom Model of GAPDH Protofibrillar Species
Structural characterization of transient protofibrillar species at the atomic level is not a trivial task using classical structural techniques; therefore, we used computational modeling. Briefly, we have analyzed the most probable arrangement of GAPDH subunits within the protofibril considering the following constraints obtained from the SAXS data analysis: (i) it has a cylindrical shape, a diameter, and long axis lengths of 11.8 and 21.5 nm respectively; (ii) the number of subunits to be fitted into the cylinder (Nagg) is 35 ± 2; (iii) the subunits maintain a native-like fold as observed on FTIR spectra recorded during the early stages of heparin- induced aggregation (27); (iv) a native-like dimer can be used as the building block because this species is also present in the early stages of the fibrillation process (Fig. 2).
Using SymmDock (43), we generated 100 docking models taking the GAPDH dimer as building block. Based on the particle size restrictions derived from SAXS, we narrowed the possibilities down to only one model, which was then submitted to relaxation protocols through molecular dynamic simulations techniques. Because of the time and length scales needed for this study, we used a multiscale approach. The first step used was the MARTINI coarse grain force field with elastic network constraints (47) to perform the simulations (45). In the second step, all-atom detail was introduced back into the model (Fig. 3) by using the reverse transformation protocol described by Rzepiela et al. (48). This approach could preserve the overall fold of each subunit, as evidenced by the root mean square deviation of the subunits from its x-ray structure, which remained under 0.28 nm throughout the whole simulation. The presence of improper protein-protein contacts in the proposed protofibril model was evaluated by following the evolution of the protein's solvent-accessible surface area, where an increment is indicative of repulsions at the interfaces. The assembly model that we propose for the protofibril remains stable within the microsecond time scales (data not shown), indicating that there is no repulsion in the interfaces.
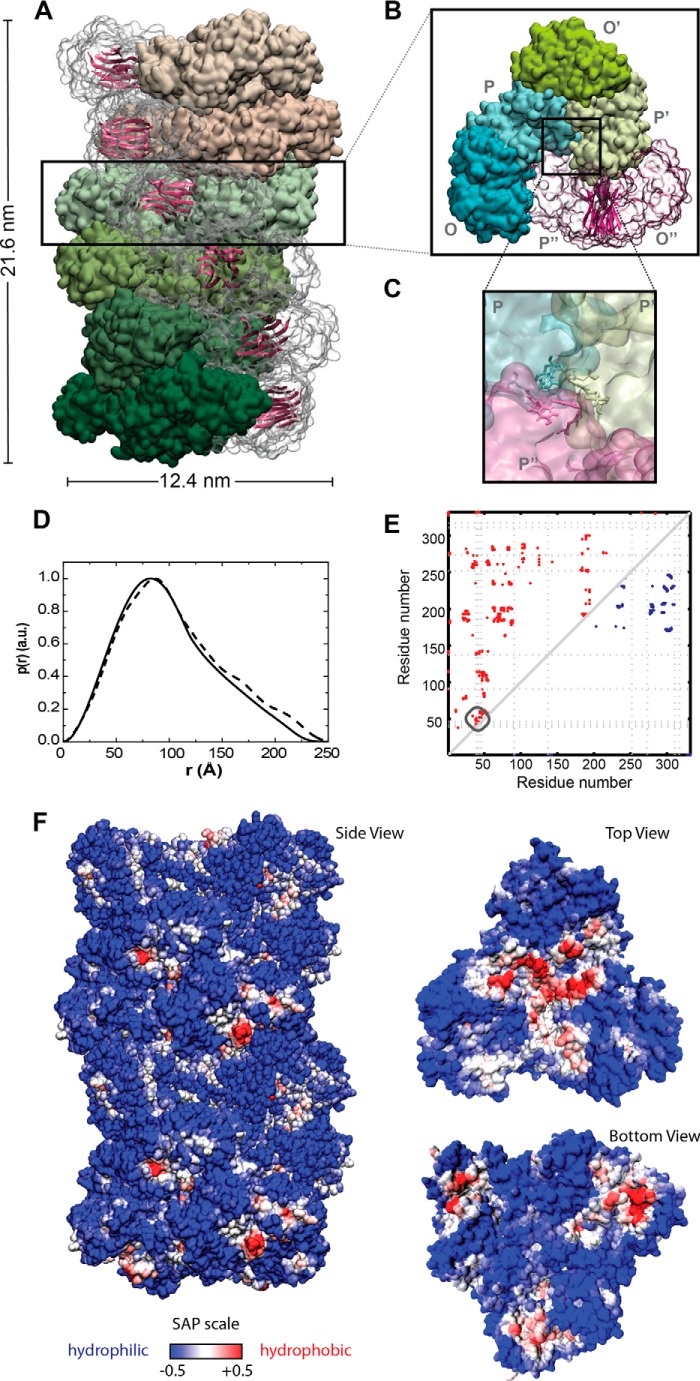
FIGURE 3.
All-atom model of the heparin-induced GAPDH protofibril. A, surface representation of the whole protofibril (side view), with each layer colored in a different shade of green or pink; B, surface representation for one of the repetitive layers along the elongation axis (top view). The dimer formed between subunits O and P is represented in a transparent surface shown in schematic representation the β-sheets forming the intersubunit interface and that might be involved in the cross-β structures in the mature fibril. C, surface representation of the contact between the dimeric building blocks at the core of the protofibril. Tyr residues 39, 42, and 46 are represented in Licorice. D, pair distance distribution functions, p(r), of the effective cylinder obtained from SAXS (solid line), along with the theoretical p(r) function of the model (dashed line). E, intersubunit contact map for the tetrameric (blue dots on upper right) versus putative protofibril (red dots on upper left) assembly of GAPDH. The dotted line represents the position of the tyrosine residues. F, spatial aggregation propensity at R = 10 mapped onto the protofibril solvent-accessible surface. The red regions indicate aggregation-prone sites with hydrophobic patches exposed.
The reliability of the model obtained is given by the good agreement between the experimental and the theoretical distance distribution p(r) function data as estimated with SASMOL software (Fig. 3) (37). According to our model, the protofibril has the shape of a cylinder with 21.6 nm height and 12.4 nm diameter. Within the protofibril, the dimers are arranged into layers of hexamers stacked along the long axis of the cylinder and presented a helical twist with a structure that repeats every 12 layers. It is important to note that in the model presented, the β-sheets within the catalytic domain of each subunit were oriented in tandem perpendicular to the elongation axis of the protofibril. Under this scheme, it seems to be possible that the flexible loops present in the interface between the layers might take another course, allowing the formation of a hydrogen bond network connecting these β-sheets, giving rise to the characteristic cross-β-structure of the amyloid fibrils.
Thus, according to the proposed model, heparin induces changes in the quaternary structure of GAPDH leading to the formation of new protein-protein interfaces in the protofibril. Changes at the interfaces could be visualized by plotting the contact maps, which depict the proximity between two residues at the interface in the protofibril compared with the native tetramer (Fig. 3E). An analysis of our model shows that Tyr-39, Tyr-42, and Tyr-46 are in close proximity to the interface between three subunits in the core of the protofibril (Fig. 3, C and E). In the tetramer, these residues are exposed to the solvent and do not participate in the formation of protein-protein interfaces.
To understand the molecular basis of the interaction between the protofibril and α-SNoli, we evaluated the exposure of hydrophobic patches on the protein surface. Hydrophobic interactions were shown to play an important role in protein aggregation, and thus several algorithms were developed in an attempt to identify these regions. Aggregation-prone regions were predicted through the calculation of the spatial aggregation propensity (50). Mapping this parameter on the protein's surface reveals the presence of a hydrophobic patch at the edge of the protofibril (Fig. 3F). This hydrophobic patch might act as a scaffold for recruiting additional subunits of GAPDH for the elongation of the protofibril, as well as for recruiting α-SNoli into a mixed fibril.
Mass Spectrometry Validates the GAPDH Protofibril Model
To experimentally validate the proposed GAPDH protofibrillar model, we resourced to covalent stabilization through photo-induced cross-linking of unmodified proteins (PICUP). This method allows the formation of Tyr–Tyr bonds when these residues are in close proximity (41). To do this, aliquots from an incubation mixture containing GAPDH and heparin were harvested at different times and photolyzed in the presence of tris(bipyridine)ruthenium(II) dichloride and ammonium persulfate. The formation of new di-tyrosine bonds was shown by the increase of the characteristic emission peak at 410 nm in the fluorescent spectra only in heparin-containing mixtures (Fig. 4A). The SDS-PAGE analysis (Fig. 4B) of each of these samples shows the presence of new covalently stabilized dimeric and trimeric GAPDH species. In the absence of heparin, no covalent cross-link was detected.
FIGURE 4.
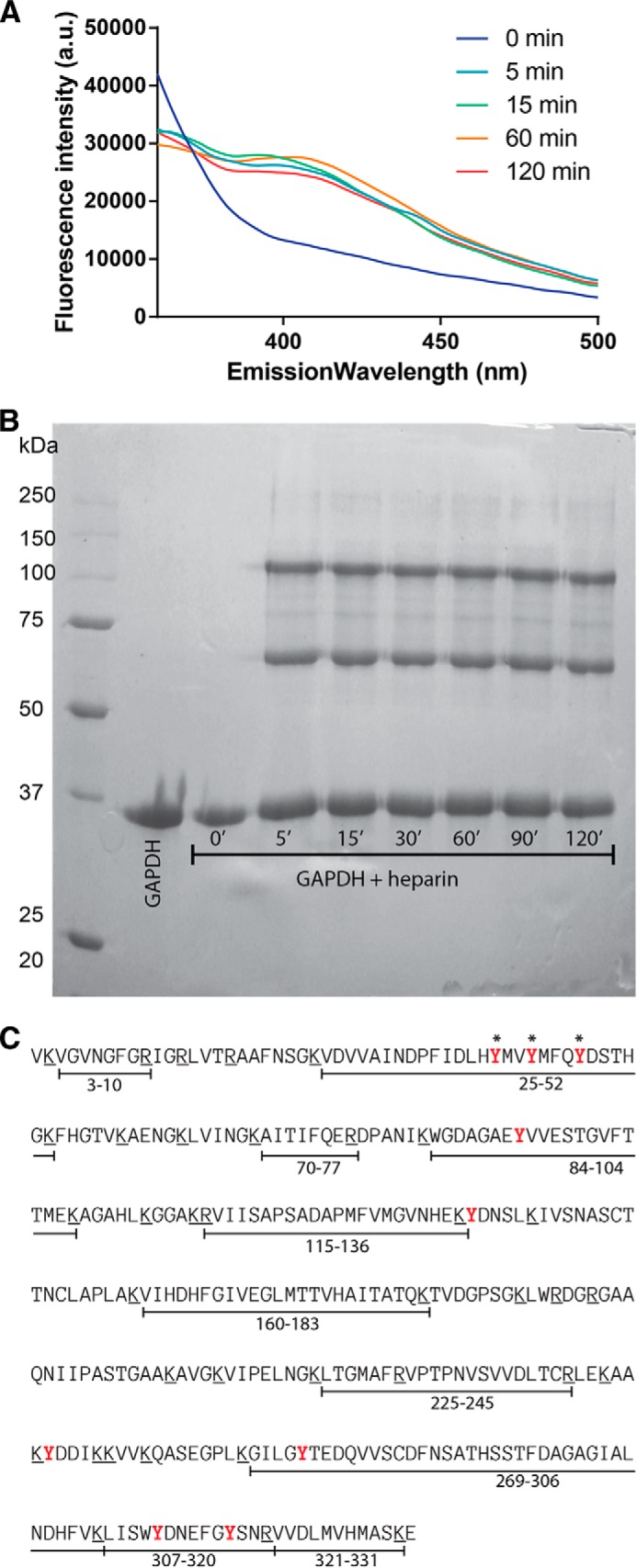
Characterization of new protein-protein interfaces in GAPDH protofibril. A, fluorescence emission spectral changes of the photolyzed solution of GAPDH incubated in the presence of heparin; excitation at 275 nm reflects the formation of Tyr–Tyr bonds. A.U., arbitrary units. B, electrophoresis gel of the GAPDH alone or after incubation with heparin at different time intervals treated with PICUP. C, amino acid sequence of GAPDH. Putative trypsin cleavage sites (Arg and Lys residues) are underlined. The peptide fragments detected on the MS/MS analysis of the monomer band are indicated below the sequence. Tyr residues available for cross-linking are depicted in red, and those forming Tyr-Tyr bridges in the PICUP-stabilized dimer and trimer are marked with asterisks.
To identify the Tyr involved in new covalent bonds stabilizing the dimeric and trimeric species detected in the SDS-PAGE, we performed trypsin digestion of the isolated bands, followed by comparative analysis of the generated peptides using mass spectrometry. Most of the expected peptides were detected in the sample corresponding to the monomer (Fig. 4C). It is important to note the presence of a signal compatible with fragment 25–52 in the monomer sample, bearing Tyr-39, Tyr-42, and Tyr-46 which could be involved in the formation of Tyr–Tyr bonds stabilizing the dimers or trimers (Table 2). Nevertheless, the signal compatible with the segment peptide 25–52 was no longer detected in dimer or trimer samples. On the contrary, the dimer and trimer digestion samples show the presence of additional signals, such as the one compatible with ∼9657 Da, which could be assigned to a new fragment formed by cross-linking Tyr residues between segment peptides 9–58 and 25–58.
TABLE 2.
GAPDH peptides found in digested monomer, dimer, and trimer samples
Putative dipeptides bearing a di-tyrosine link are shown at the bottom of the table. NS indicates no signal.
| Peptide fragment | Mass H+ (theoretical) | Mass H+ (experimental) |
||
|---|---|---|---|---|
| Monomer | Dimer | Trimer | ||
| 3–10 | 805.4315 | 805.40 | 805.406799 | 805.399353 |
| 25–52 | 3317.5646 | 3317.72 | NS | NS |
| 70–77 | 977.5415 | 977.472778 | 977.497131 | 977.500793 |
| 84–104 | 2277.038 | 2277.04248 | 2276.95728 | 2276.073 |
| 115–136 | 2370.2283 | 2369.23364 | 2369.13135 | 2369.22168 |
| 160–183 | 2618.3759 | 2618.44653 | 2618.27832 | 2618.31665 |
| 269–306 | 4087.8614 | 4087.98267 | 4087.95044 | 4088.08081 |
| 307–320 | 1763.8024 | 1763.79981 | 1763.80115 | 1763.74927 |
| 321–331 | 1229.6381 | 1229.55957 | 1229.64111 | 1229.55627 |
| 9–42 and 25–39 | 5536.8957 | NS | 5534.09 | |
| 14–46 and 25–44 18–52 and 35–52 | 6238.0784 6238.60 | NS | 6238.61 | |
| 14–63 and 25–52 | 8942.3633 | NS | 8943.81 | |
| 9–58 and 25–58 | 9657.8132 | NS | 9657.75 | |
The difference observed in the cross-linking properties of GAPDH might be understood in terms of the differences in the Tyr-Tyr interactions in the interface in the protofibril compared with the native tetramer (Fig. 3E). Accordingly, in the crystallographic structure of the native tetrameric protein, the region corresponding to fragment 25–52 is exposed to the solvent, although on the protofibril this region is confined to the subunit-subunit interfaces, just at the core of each layer (Fig. 3E) allowing Tyr-39, Tyr-42, and Tyr-46 in close proximity for cross-linking. When the protofibril is exposed to the denaturing conditions of SDS-PAGE, all the subunits disassemble except those with covalent stabilization, giving rise to the dimeric and trimeric species, along with the monomeric species (Fig. 4B). Altogether, these results give experimental support to the structural model proposed for the GAPDH protofibril.
DISCUSSION
We have previously shown that some intermediates present in the early stages of heparin-induced GAPDH amyloid aggregation (HI-GAPDHESS) are able to accelerate the kinetics of α-SN aggregation. Here, we demonstrate that these HI-GAPDHESS species are able to abolish α-SNoli deleterious activity at the cell membrane level possibly by sequestering SNoli. Interestingly, the capacity of HI-GAPDHESS to reduce membrane damage (Fig. 1F) increases with the amount of protofibril species present in the solution (Fig. 2F). The protofibril revealed by the SAXS analysis composes a proteic cylinder-like envelope 22 nm long and a 6-nm radius. The all-atom model for such a protofibril, obtained using rigid body docking and molecular dynamics simulations, shows that GAPDH subunits are arranged into layers of hexamers stacked along the cylinder long axis with a helical twist that gives rise to a repeating structure every 12 layers. Also, the β-sheets within the catalytic domain of each subunit are oriented in tandem perpendicular to the elongation axis of the protofibril. We propose that the flexible loops present in the interface between the layers might take a new direction, allowing the formation of a hydrogen bond network connecting these β-sheets, giving rise to the characteristic cross-β-structure of the amyloid fibrils as observed by ThT fluorescence and FTIR spectroscopy (27). Cross-linking experiments coupled to mass spectrometry gives additional support to the model, validating the hypothesis that Tyr-39, Tyr-42, and Tyr-46 intervene in the formation of new protein-protein interfaces in the protofibril.
The structural characterization of the protofibril acquires relevance in view of its beneficial features. Using a dopaminergic cell model, we demonstrated the ability of heparin-induced GAPDH protofibrillar species to protect cells against exogenous addition of α-SNoli. This protection seems to be mediated by abolishing the membrane perturbation induced by α-SNoli, as indicated by fluorescence assays on a simplified cellular membrane model. It is tempting to speculate that heparin-induced GAPDH protofibrils behave as an efficient polymerization seed for α-SN, acting as scavengers for α-SNoli toxic species, pushing them to reach the fibrillar nontoxic state, and preventing them from reaching the cytoplasmic membrane. In a previous work, we demonstrated the formation of a mixed fibril when GAPDH protofibrils are incubated in the presence of α-SNoli (27). The lack of an all atom model for α-SNoli species prevented us from applying the computational tool such as docking studies for inquiring on the specific nature of the protein-protein interaction. Still, several works have evidenced the relevance of hydrophobic interactions in protein aggregation. In particular, the algorithm SAP, which is based on the detection of hydrophobic patches on the protein surface, has proved useful for predicting putative aggregation-prone regions in proteins (50). An analysis of GAPDH protofibrils revealed a hydrophobic patch at the protofibril edge (Fig. 3), which could account for the recruitment of α-SNoli into a mixed protofibril as depicted in Fig. 5.
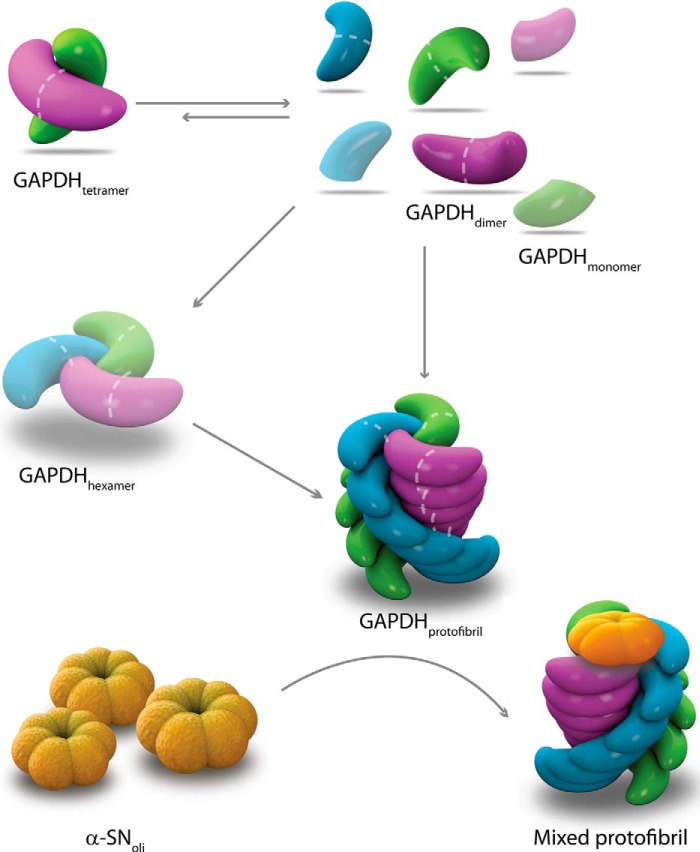
FIGURE 5.
Schematic diagram of the heparin-induced GAPDH fibrillation pathway and its interaction with α-SNoli. Heparin interacts with GAPDH inducing dissociation of tetramer into dimers, which then reassemble into the growing protofibril. The existence of additional potential intermediates (shadowed monomer and hexamer) is also proposed. The model of interaction between GAPDH protofibrils and α-SNoli is based on the hydrophobic patch present at the edge of the protofibril allowing the recruitment α-SNoli into a mixed protofibril.
In this scenario, although α-SN is mainly found in the cytosol, identification of this protein outside the cell (65, 66) suggests that α-SN amyloid pathology may spread during disease progression (3, 4, 67) possibly by a prion-like mechanism, i.e. by a self-templating change in protein structure (68). The GAPDH protofibril could play an important role sequestering α-SNoli species spreading through the extracellular space. Here, we used heparin to trigger the formation of GAPDH protofibrils, and although this GAG is not considered to exist inside the brain, it is representative of highly sulfated heparan sulfate domains classically found in the extracellular matrix of any tissue (69). In this scenario, a role for GAPDH protofibrils in cellular proteostasis seems possible because heparan sulfates from brain extracellular matrix can colocalize and thus interact with secreted GAPDH inducing its dissociation and reassociation into new species capable of scavenging α-SNoli. According to our hypothesis, the dissociation of the tetramer into dimers represents a key step into the formation of species involved in proteostasis of α-SNoli (Fig. 5). We cannot deny the presence of monomers as well as additional structural species in solution which, if present, they might be in very small amounts and below the limits of detection of the methods used herein. Interestingly, drugs used for the treatment of Parkinson disease increase the stability of GAPDH as a dimer (70). This mechanism could be involved in neuroprotection and represents a new strategy for therapeutic design in Parkinson disease.
Acknowledgments
We thank the National Laboratory of Synchrotron Light (Campinas, SP, Brazil) for the use of SAXS beam line facilities and Centro Nacional de Supercomputaçao-Universidad Federal do Rio Grande do Sul (Porto Alegre, Brazil) for the computational clusters facilities. We also thank Profs. Paolo Mariani and Francesco Spinozzi, both from Universitá Politecnica delle Marche, Ancona, Italy, who provided us with the GENFIT software.
This work was supported in part by grants from Florencio Fiorini Foundation; Ecos-Sud Grant A12S02, and CIUNT Grant D439-1.
- α-SN
- α-synuclein
- GAG
- glycosaminoglycan
- SAXS
- small angle x-ray scattering
- SUV
- small unilamellar vesicle
- PICUP
- photo-induced cross-linking of unmodified protein
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ThT
- thioflavin T
- SAP
- spatial aggregation propensity.
REFERENCES
- 1. Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee H. J., Patel S., Lee S. J. (2005) Intravesicular localization and exocytosis of α-synuclein and its aggregates. J. Neurosci. 25, 6016–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee H. J., Suk J. E., Bae E. J., Lee S. J. (2008) Clearance and deposition of extracellular α-synuclein aggregates in microglia. Biochem. Biophys. Res. Commun. 372, 423–428 [DOI] [PubMed] [Google Scholar]
- 4. Lee S. J. (2008) Origins and effects of extracellular α-synuclein: implications in Parkinson's disease. J. Mol. Neurosci. 34, 17–22 [DOI] [PubMed] [Google Scholar]
- 5. Chai Y. J., Kim D., Park J., Zhao H., Lee S. J., Chang S. (2013) The secreted oligomeric form of α-synuclein affects multiple steps of membrane trafficking. FEBS Lett. 587, 452–459 [DOI] [PubMed] [Google Scholar]
- 6. Steiner J. A., Angot E., Brundin P. (2011) A deadly spread: cellular mechanisms of α-synuclein transfer. Cell Death Differ. 18, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wood S. J., Wypych J., Steavenson S., Louis J. C., Citron M., Biere A. L. (1999) α-Synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson's disease. J. Biol. Chem. 274, 19509–19512 [DOI] [PubMed] [Google Scholar]
- 8. Conway K. A., Harper J. D., Lansbury P. T., Jr. (2000) Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry 39, 2552–2563 [DOI] [PubMed] [Google Scholar]
- 9. Chiti F., Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 10. Caughey B., Lansbury P. T. (2003) Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 11. Cremades N., Cohen S. I., Deas E., Abramov A. Y., Chen A. Y., Orte A., Sandal M., Clarke R. W., Dunne P., Aprile F. A., Bertoncini C. W., Wood N. W., Knowles T. P., Dobson C. M., Klenerman D. (2012) Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butterfield S. M., Lashuel H. A. (2010) Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew Chem. Int. Ed. Engl. 49, 5628–5654 [DOI] [PubMed] [Google Scholar]
- 13. Tsuchiya K., Tajima H., Kuwae T., Takeshima T., Nakano T., Tanaka M., Sunaga K., Fukuhara Y., Nakashima K., Ohama E., Mochizuki H., Mizuno Y., Katsube N., Ishitani R. (2005) Pro-apoptotic protein glyceraldehyde-3-phosphate dehydrogenase promotes the formation of Lewy body-like inclusions. Eur. J. Neurosci. 21, 317–326 [DOI] [PubMed] [Google Scholar]
- 14. Leverenz J. B., Umar I., Wang Q., Montine T. J., McMillan P. J., Tsuang D. W., Jin J., Pan C., Shin J., Zhu D., Zhang J. (2007) Proteomic identification of novel proteins in cortical Lewy bodies. Brain Pathol. 17, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sirover M. A. (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 1810, 741–751 [DOI] [PubMed] [Google Scholar]
- 16. Zheng L., Roeder R. G., Luo Y. (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- 17. Engel M., Seifert M., Theisinger B., Seyfert U., Welter C. (1998) Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A. Two old enzymes combine for the novel Nm23 protein phosphotransferase function. J. Biol. Chem. 273, 20058–20065 [DOI] [PubMed] [Google Scholar]
- 18. Kumagai H., Sakai H. (1983) A porcine brain protein (35 K protein) which bundles microtubules and its identification as glyceraldehyde 3-phosphate dehydrogenase. J. Biochem. 93, 1259–1269 [DOI] [PubMed] [Google Scholar]
- 19. Xu K. Y., Becker L. C. (1998) Ultrastructural localization of glycolytic enzymes on sarcoplasmic reticulum vesicles. J. Histochem. Cytochem. 46, 419–427 [DOI] [PubMed] [Google Scholar]
- 20. Lin H., Zhu Y. J., Lal R. (1999) Amyloid β protein(1–40) forms calcium-permeable, Zn2+-sensitive channel in reconstituted lipid vesicles. Biochemistry 38, 11189–11196 [DOI] [PubMed] [Google Scholar]
- 21. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 22. Butterfield D. A., Hardas S. S., Lange M. L. (2010) Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J. Alzheimers Dis. 20, 369–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen M., Cox C., Belbin O., Ma L., Bisceglio G. D., Wilcox S. L., Howell C. C., Hunter T. A., Culley O., Walker L. P., Carrasquillo M. M., Dickson D. W., Petersen R. C., Graff-Radford N. R., Younkin S. G., Ertekin-Taner N. (2010) Association and heterogeneity at the GAPDH locus in Alzheimer's disease. Neurobiol. Aging 33, 203 e225–e233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Díaz-Nido J., Wandosell F., Avila J. (2002) Glycosaminoglycans and β-amyloid, prion and tau peptides in neurodegenerative diseases. Peptides 23, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 25. Holmes B. B., DeVos S. L., Kfoury N., Li M., Jacks R., Yanamandra K., Ouidja M. O., Brodsky F. M., Marasa J., Bagchi D. P., Kotzbauer P. T., Miller T. M., Papy-Garcia D., Diamond M. I. (2013) Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U.S.A. 110, E3138–E3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motamedi-Shad N., Monsellier E., Torrassa S., Relini A., Chiti F. (2009) Kinetic analysis of amyloid formation in the presence of heparan sulfate: faster unfolding and change of pathway. J. Biol. Chem. 284, 29921–29934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Torres-Bugeau C. M., Ávila C. L., Raisman-Vozari R., Papy-Garcia D., Itri R., Barbosa L. R., Cortez L. M., Sim V. L., Chehín R. N. (2012) Characterization of heparin-induced glyceraldehyde-3-phosphate dehydrogenase early amyloid-like oligomers and their implication in α-synuclein aggregation. J. Biol. Chem. 287, 2398–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoyer W., Antony T., Cherny D., Heim G., Jovin T. M., Subramaniam V. (2002) Dependence of α-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 322, 383–393 [DOI] [PubMed] [Google Scholar]
- 29. Kaylor J., Bodner N., Edridge S., Yamin G., Hong D. P., Fink A. L. (2005) Characterization of oligomeric intermediates in α-synuclein fibrillation: FRET studies of Y125W/Y133F/Y136F α-synuclein. J. Mol. Biol. 353, 357–372 [DOI] [PubMed] [Google Scholar]
- 30. Danzer K. M., Haasen D., Karow A. R., Moussaud S., Habeck M., Giese A., Kretzschmar H., Hengerer B., Kostka M. (2007) Different species of α-synuclein oligomers induce calcium influx and seeding. J. Neurosci. 27, 9220–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arrondo J. L., Castresana J., Valpuesta J. M., Goñi F. M. (1994) Structure and thermal denaturation of crystalline and noncrystalline cytochrome oxidase as studied by infrared spectroscopy. Biochemistry 33, 11650–11655 [DOI] [PubMed] [Google Scholar]
- 32. Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 33. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 34. Finer E. G., Flook A. G., Hauser H. (1972) Mechanism of sonication of aqueous egg yolk lecithin dispersions and nature of the resultant particles. Biochim. Biophys. Acta 260, 49–58 [DOI] [PubMed] [Google Scholar]
- 35. Kendall D. A., MacDonald R. C. (1982) A fluorescence assay to monitor vesicle fusion and lysis. J. Biol. Chem. 257, 13892–13895 [PubMed] [Google Scholar]
- 36. Barbosa L. R., Spinozzi F., Mariani P., Itri R. (2013) in Proteins in Solution and at Interfaces (Ruso J. M., Piñeiro A., eds) pp. 49–72, John Wiley & Sons, Inc., New York [Google Scholar]
- 37. Ortore M. G., Spinozzi F., Mariani P., Paciaroni A., Barbosa L. R., Amenitsch H., Steinhart M., Ollivier J., Russo D. (2009) Combining structure and dynamics: nondenaturing high-pressure effect on lysozyme in solution. J. R. Soc. Interface 6, S619–S634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barbosa L. R., Ortore M. G., Spinozzi F., Mariani P., Bernstorff S., Itri R. (2010) The importance of protein-protein interactions on the pH-induced conformational changes of bovine serum albumin: a small-angle x-ray scattering study. Biophys. J. 98, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fournet G., Guinier A. (1955) Small Angle Scattering of X-rays (translated by Walker C. B., Yudowitch K. L.) pp. 7–78, John Wiley & Sons, Inc., New York [Google Scholar]
- 40. Fancy D. A., Denison C., Kim K., Xie Y., Holdeman T., Amini F., Kodadek T. (2000) Scope, limitations and mechanistic aspects of the photo-induced cross-linking of proteins by water-soluble metal complexes. Chem. Biol. 7, 697–708 [DOI] [PubMed] [Google Scholar]
- 41. Fancy D. A., Kodadek T. (1999) Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. U.S.A. 96, 6020–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weber K., Osborn M. (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244, 4406–4412 [PubMed] [Google Scholar]
- 43. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) Geometry-based flexible and symmetric protein docking. Proteins 60, 224–231 [DOI] [PubMed] [Google Scholar]
- 44. Cowan-Jacob S. W., Kaufmann M., Anselmo A. N., Stark W., Grütter M. G. (2003) Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr. D. Biol. Crystallogr. 59, 2218–2227 [DOI] [PubMed] [Google Scholar]
- 45. Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H. (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 111, 7812–7824 [DOI] [PubMed] [Google Scholar]
- 46. Monticelli L., Kandasamy S. K., Periole X., Larson R. G., Tieleman D. P., Marrink S.-J. (2008) The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 4, 819–834 [DOI] [PubMed] [Google Scholar]
- 47. Periole X., Cavalli M., Marrink S.-J., Ceruso M. A. (2009) Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5, 2531–2543 [DOI] [PubMed] [Google Scholar]
- 48. Rzepiela A. J., Schäfer L. V., Goga N., Risselada H. J., De Vries A. H., Marrink S. J. (2010) Reconstruction of atomistic details from coarse-grained structures. J. Comput. Chem. 31, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 49. Vendruscolo M., Najmanovich R., Domany E. (1999) Protein folding in contact map space. Phys. Rev. Lett. 82, 656–659 [Google Scholar]
- 50. Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B. L. (2009) Design of therapeutic proteins with enhanced stability. Proc. Natl. Acad. Sci. U.S.A. 106, 11937–11942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 52. Black S. D., Mould D. R. (1991) Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 193, 72–82 [DOI] [PubMed] [Google Scholar]
- 53. Yang W. H., Yang C., Xue Y. Q., Lu T., Reiser J., Zhao L. R., Duan W. M. (2013) Regulated expression of lentivirus-mediated GDNF in human bone marrow-derived mesenchymal stem cells and its neuroprotection on dopaminergic cells in vitro. PLoS One 8, e64389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim H. Y., Cho M. K., Kumar A., Maier E., Siebenhaar C., Becker S., Fernandez C. O., Lashuel H. A., Benz R., Lange A., Zweckstetter M. (2009) Structural properties of pore-forming oligomers of α-synuclein. J. Am. Chem. Soc. 131, 17482–17489 [DOI] [PubMed] [Google Scholar]
- 55. Glabe C. G. (2008) Structural classification of toxic amyloid oligomers. J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 57. Volles M. J., Lee S. J., Rochet J. C., Shtilerman M. D., Ding T. T., Kessler J. C., Lansbury P. T., Jr. (2001) Vesicle permeabilization by protofibrillar α-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry 40, 7812–7819 [DOI] [PubMed] [Google Scholar]
- 58. van Rooijen B. D., Claessens M. M., Subramaniam V. (2010) Membrane permeabilization by oligomeric α-synuclein: in search of the mechanism. PLoS One 5, e14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morero R. D., Viñals A. L., Bloj B., Farías R. N. (1985) Fusion of phospholipid vesicles induced by muscle glyceraldehyde-3-phosphate dehydrogenase in the absence of calcium. Biochemistry 24, 1904–1909 [DOI] [PubMed] [Google Scholar]
- 60. Oliveira C. L., Behrens M. A., Pedersen J. S., Erlacher K., Otzen D. (2009) A SAXS study of glucagon fibrillation. J. Mol. Biol. 387, 147–161 [DOI] [PubMed] [Google Scholar]
- 61. Giehm L., Svergun D. I., Otzen D. E., Vestergaard B. (2011) Low-resolution structure of a vesicle disrupting α-synuclein oligomer that accumulates during fibrillation. Proc. Natl. Acad. Sci. U.S.A. 108, 3246–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Spinozzi F., Mariani P., Saturni L., Carsughi F., Bernstorff S., Cinelli S., Onori G. (2007) Met-myoglobin association in dilute solution during pressure-induced denaturation: an analysis at pH 4.5 by high-pressure small-angle x-ray scattering. J. Phys. Chem. B. 111, 3822–3830 [DOI] [PubMed] [Google Scholar]
- 63. Ortore M. G., Spinozzi F., Vilasi S., Sirangelo I., Irace G., Shukla A., Narayanan T., Sinibaldi R., Mariani P. (2011) Time-resolved small-angle x-ray scattering study of the early stage of amyloid formation of an apomyoglobin mutant. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 84, 061904 [DOI] [PubMed] [Google Scholar]
- 64. Kodali R., Wetzel R. (2007) Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 17, 48–57 [DOI] [PubMed] [Google Scholar]
- 65. El-Agnaf O. M., Salem S. A., Paleologou K. E., Cooper L. J., Fullwood N. J., Gibson M. J., Curran M. D., Court J. A., Mann D. M., Ikeda S., Cookson M. R., Hardy J., Allsop D. (2003) α-Synuclein implicated in Parkinson's disease is present in extracellular biological fluids, including human plasma. FASEB J. 17, 1945–1947 [DOI] [PubMed] [Google Scholar]
- 66. El-Agnaf O. M., Salem S. A., Paleologou K. E., Curran M. D., Gibson M. J., Court J. A., Schlossmacher M. G., Allsop D. (2006) Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson's disease. FASEB J. 20, 419–425 [DOI] [PubMed] [Google Scholar]
- 67. Luk K. C., Song C., O'Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M. (2009) Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sacino A. N., Giasson B. I. (2012) Does a prion-like mechanism play a major role in the apparent spread of α-synuclein pathology? Alzheimers Res. Ther. 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huynh M. B., Morin C., Carpentier G., Garcia-Filipe S., Talhas-Perret S., Barbier-Chassefière V., van Kuppevelt T. H., Martelly I., Albanese P., Papy-Garcia D. (2012) Age-related changes in rat myocardium involve altered capacities of glycosaminoglycans to potentiate growth factor functions and heparan sulfate-altered sulfation. J. Biol. Chem. 287, 11363–11373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Carlile G. W., Chalmers-Redman R. M., Tatton N. A., Pong A., Borden K. E., Tatton W. G. (2000) Reduced apoptosis after nerve growth factor and serum withdrawal: conversion of tetrameric glyceraldehyde-3-phosphate dehydrogenase to a dimer. Mol. Pharmacol. 57, 2–12 [PubMed] [Google Scholar]