Background: The significance of telomeric protein translocation to the spindle pore is unknown.
Results: TAP68 mediates TRF1 transit to the centrosome in mitotic cells.
Conclusion: Repression of TAP68 induces genomic instability possibly through inhibition of TRF1 and tankyrase recruitment to centrosomes.
Significance: Identifying TRF1-interacting proteins that regulate its localization is crucial for understanding the role of telomeric proteins in mitosis.
Keywords: Cell Biology, Centriole, Mitosis, Phosphorylation, Telomeres, Nek2A Kinase, TAP68, TRF1
Abstract
The telomere capping protein TRF1 is a component of the multiprotein complex “shelterin,” which organizes the telomere into a high order structure. Besides telomere maintenance, telomere-associated proteins also have nontelomeric functions. For example, tankyrase 1 and TRF1 are required for the maintenance of faithful mitotic progression. However, the functional relevance of their centrosomal localization has not been established. Here, we report the identification of a TRF1-binding protein, TAP68, that interacts with TRF1 in mitotic cells. TAP68 contains two coiled-coil domains and a structural maintenance of chromosome motifs and co-localizes with TRF1 to telomeres during interphase. Immediately after nuclear envelope breakdown, TAP68 translocates toward the spindle poles followed by TRF1. Dissociation of TAP68 from the telomere is concurrent with the Nek2A-dependent phosphorylation at Thr-221. Biochemical characterization demonstrated that the first coiled-coil domain of TAP68 binds and recruits TRF1 to the centrosome. Inhibition of TAP68 expression by siRNA blocked the localization of TRF1 and tankyrase 1 to the centrosome. Furthermore, siRNA-mediated depletion of TAP68 perturbed faithful chromosome segregation and genomic stability. These findings suggest that TAP68 functions in mediating TRF1-tankyrase 1 localization to the centrosome and in mitotic regulation.
Introduction
Telomeres, which are specialized nucleoprotein complexes at the ends of eukaryotic chromosomes that consist of tandem repeated DNA sequences bound by various telomeric proteins, prevent chromosomal termini from undergoing unregulated degradation, recombination, and fusion (1–3). From budding yeast to human cells, duplex telomeric DNA repeat-binding factors and their interacting proteins have been identified and characterized (4–6). Although significant divergence has occurred in these proteins across species, there are also extensive levels of conservation. For example, in fission yeast, duplex telomeric DNA repeats are bound by Taz1p, a homologue of the human telomeric repeat binding factor proteins (7, 8). Like Taz1p, human TRF1 directly binds duplex telomere repeats and interacts with multiple proteins, including TERF1-interacting nuclear factor 2 (TIN2), tankyrase, and PIN2/TERF1-interacting telomerase inhibitor 1 (PINX1) (6, 8–11). Interestingly, the chromatin association of TRF1 is regulated by key mitotic kinase PLK1 during mitosis (12, 13). Two related poly(ADP)-ribose polymerases that were originally identified as TRF1-interacting proteins, tankyrase 1 and 2, inhibit the binding of TRF1 to telomeres (14). Tankyrase 1 and 2 are nearly identical in amino acid sequence and form homo- and heterodimers, suggesting that they are functionally similar (15).
In addition to their telomeric functions, telomere-associated proteins also have nontelomeric functions. For example, TRF1 expression correlates with stem cell pluripotency (16). The functional involvement of TRF1 in mitosis is well established. The spindle pole localization of TRF1 has been reported more than 10 years ago (4, 17). Later, it was reported that TRF1 interacts with spindle assembly checkpoint protein Mad1 and key mitotic kinase NEK2A (NIMA-related kinase 2) (18, 19). Recently, accumulating evidence suggests that TRF1 is required for faithful mitotic progression (20–22). In addition, PinX1 phosphorylation has been associated with the regulation of faithful chromosome segregation (23, 24). Tankyrase localizes to mitotic spindle poles during mitosis and is required for normal spindle formation and sister chromatid segregation through as yet unclear mechanisms (25). In addition, poly(ADP-ribose) is required for spindle assembly and structure (26, 27). Similar to TRF1, the function of tankyrase 1 is also regulated by PLK1 (Polo-like kinase 1)-mediated phosphorylation (28).
Centrosome is the major microtubule organization center and is essential for the correct assembly of bipolar spindle in animal cells. Aberrations in the number and structure of the centrosome lead to genomic instability and have been characterized in a variety of solid tumors as well as leukemias and lymphomas (29, 30). Although the mitotic role of TRF1 is well established, the mechanism by which TRF1 and its complex is recruited and tethered to the centrosome in vertebrates is unknown. In this study, we aimed to assess the mechanism by which the telomere-associated proteins regulate chromosome stability using a proteomic analysis to isolate TRF1-interacting proteins during mitosis. A protein of unknown function was identified and was designated as TAP68 (TRF1-associated protein of 68 kDa). Biochemical analysis demonstrated the direct interaction between TRF1 and TAP68. These two proteins co-localize on telomere in interphase and at mitotic spindle poles in mitosis. The spindle pole localization of TRF1 depends on TAP68. We further demonstrated that phosphorylation of TAP68 at residues Thr-221 and Thr-457 by kinase NEK2A and PLK1 coordinates TRF1 translocation from telomere to spindle pole. Together, our data reveal TAP68 plays a regulatory role in TRF1 localization to centrosomes and uncover the novel mechanism of telomeric protein in the regulation of accurate mitotic progression.
EXPERIMENTAL PROCEDURES
Affinity Precipitation of the TRF1 Complex
Mitotic extracts from HeLa S3 cells (1 × 109) were prepared in lysis buffer (25 mm HEPES, pH 6.9, 150 mm NaCl, 2 mm EGTA, 5 mm MgCl2, 0.2% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml chymostatin, and 10 μg/ml pepstatin A) and incubated with 100 μl of protein A-Sepharose beads coupled with a rabbit anti-TRF1 antibody (Ab13792; Novus Biologicals, Inc.) using a previously described protocol (24). The column was washed four times with lysis buffer, and the bound proteins were eluted with 250 μg/ml antigen peptide, resolved by SDS-PAGE, and stained with Coomassie Blue. Specific bands were individually excised and digested with trypsin (Promega, Madison, WI), and the tryptic peptides were sequenced using a MALDI-TOF and LC-MS/MS mass spectrometry as described previously (25–28). The specificity of the TRF1-TAP68 interaction was validated using a different TRF1 antibody Ab1423 (Novus Biologicals, Inc) in high stringency lysis buffer containing 1% Nonidet P-40 and 0.5% deoxycholate.
Preparation of Samples for Mass Spectrometry
In-gel digestions were done essentially as described previously (31, 32). Briefly, the protein band, P1, was excised and subjected to digestion by modified porcine trypsin (50–100 ng/digestion; Promega). Peptides were recovered, co-crystallized with cyano-4-hydroxycinnamic acid, and analyzed using MALDI-TOF mass spectrometry (Voyager DE-STR, Perspective Biosystems) equipped with a nitrogen laser. All MALDI spectra were externally calibrated using a standard peptide mixture (Sigma). Some post-source decay spectra were acquired on a TofSpec SE MALDI-TOF mass spectrometer (Micromass, Manchester, UK). Database interrogations based on experimentally determined peptide masses were carried out using mass spectrometry (MS)-Fit, and post-source decay data interrogation was performed using MS-Tag as described previously (31).
Molecular Cloning and Plasmid Construction
The full-length TRF1 mRNA was amplified from the human tonsil tissue by retrotranscriptase (RT)-PCR with the following primers: sense, 5′-CGGGTACCTTAACATGGCGGAGGATGTTTCCT-3′, and antisense, 5′-CCGGAATTCAATACTTAACTGTCCTTTCATCAA-3′. The thermo-cycling conditions were as follows: 1 cycle at 95 °C for 3 min; 30 cycles at 94 °C for 45 s, 52 °C for 30 s, and 72 °C for 90 s; and 1 cycle at 72 °C for 10 min. The resulting product was introduced into the pCR-II TOPO vector for DNA sequencing. After confirmation by sequencing, the coding sequence of TRF1 was amplified by PCR with the EcoRI-containing forward primer 5′-CGGAATTCCGGGCGGAGGATGTTTCC-3′ and the BamHI-containing reverse primer 5′-CGGGATCCGCGGTCTTCGCTGTCTGA-3′, using the pCR-II TOPO/TRF1 construct as template. The thermo-cycling conditions were as follows: 1 cycle at 95 °C for 3 min; 30 cycles at 94 °C for 45 s, 50 °C for 30 s, and 72 °C for 90 s; and 1 cycle at 72 °C for 10 min. The PCR product was then subcloned into the following vectors for in vivo and in vitro expression: pEGFP-C2, p3×FLAG-myc-CMV-24 (Sigma), and pET28a (Novagen). The coding region of TAP68 was isolated from a human testis cDNA library (BD Biosciences) using temperature-gradient PCR with the following primers: sense, 5′-CCCGATCTGCTCAACTTCAA-3′, and antisense, 5′-CTACTCAGCCAGGCTGTTGC-3′. The thermo-cycling conditions were as follows: 1 cycle at 95 °C for 3 min; 30 cycles at 94 °C for 45 s, 50–62 °C for 30 s, and 72 °C for 90 s; and 1 cycle at 72 °C for 10 min. The candidate fragment was cloned into the pMD18-T vector (Takara Biotechnology, Dalian, China) and completely sequenced. The clone with the correct insert was utilized as the template for PCR amplification with the high fidelity DNA polymerase, Pyrobest (Takara Biotechnology), and the target fragment was inserted into pEGFP-C2 by blunt-end cloning (BD Biosciences). All plasmid constructs were sequenced for verification. Site-directed mutagenesis was carried using a standard PCR technique. Both TRF1 and TAP68 coding regions were found to be identical to the sequences detected in NCBI GenBankTM (accession numbers U40705 and NM_007032, respectively).
Antibody Preparation
The anti-TAP68 antibody was generated after immunization of two rabbits with purified His-tagged TAP68 protein. The rabbit IgG fraction was purified using affinity beads coupled with full-length TAP68 protein as described previously (33). In some cases, mouse IgG from TAP68-immunized mice was used for immunofluorescence and Western blot analyses. The specificity of the TAP68 antibodies was verified using Western blot analysis of HeLa cell lysates. The other antibodies used in this study were as follows: rabbit polyclonal anti-TRF1 antibodies (Novus Biologicals, Inc.); Cy5-conjugated NuMA antibody (Novus Biologicals, Inc.); mouse monoclonal anti-γ-tubulin (TUB2.1; Cy5-conjugated; Sigma); β-tubulin GTU-88 antibodies (Sigma); mouse monoclonal anti-GFP antibody (BD Biosciences); anti-tankyrase 1 antibody (BD Biosciences); anti-FLAG M2 gel and mouse monoclonal anti-FLAG antibody conjugated with horseradish peroxidase (HRP; Sigma); HRP-conjugated goat anti-rabbit IgG antibody and Texas Red-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA); FITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch); rhodamine-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch).
In Vitro Kinase Assay
Both the His-tagged wild-type and phosphomutant TAP68 proteins were expressed in Escherichia coli BL21(DE3)pLys and purified as described previously (34). The fusion proteins bound to nickel beads were suspended in phosphorylation buffer (25 mm HEPES, pH 7.5, 5 mm MgSO4, 50 mm NaCl, 2 mm EGTA) prior to use. To verify whether Thr-221 is a substrate for NEK2A or Thr-457 is a substrate for PLK1, 2 μg of purified His-TAP68 fusion proteins, both wild-type and mutants, were incubated with 1 unit of active Nek2A or PLK1 (Upstate Biotechnology, Lake Placid, NY) in kinase buffer containing 20 mm HEPES, pH 7.5, 10 mm MgCl2, 5 mm EGTA, 1 mm dithiothreitol, 10 μm ATP, and 5 μCi of [32P]ATP (PerkinElmer Life Sciences) in a total volume of 30 μl. Kinase reactions were carried out at 30 °C for 30 min and terminated by the addition of 10 μl of 4× SDS-PAGE sample buffer and separated by 6–16% gradient SDS-PAGE. The gel was stained with Coomassie Brilliant Blue, dried, and quantified using a PhosphorImager (GE Healthcare).
Immunofluorescence Microscopy
For immunofluorescence, HeLa cells were grown onto sterile, acid-treated 18-mm coverslips in 24-well plates (Corning Glass, Corning, NY) and maintained in DMEM with 10% FBS until nearly 80% confluence. Cells were rinsed for 1 min with PHEM buffer (100 mm PIPES, 20 mm HEPES, pH 6.9, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol) and permeabilized for 1 min with PHEM plus 0.2% Triton X-100 as described previously (33, 35). Extracted cells were then fixed in freshly prepared 4% paraformaldehyde plus 0.05% glutaraldehyde in PHEM and rinsed three times in PBS. After the cells were blocked with 0.05% Tween 20 in PBS (TPBS) containing 1% BSA, they were incubated with various primary antibodies in a humidified chamber for 1 h followed by three washes to remove unbound antibody. Monoclonal antibodies bound to γ-tubulin and β-tubulin were visualized using rhodamine-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI, Sigma). Slides were examined with an Axiovert-200 fluorescence microscope (Carl Zeiss, Göttingen, Germany) with a ×63 1.40 NA PlanApo objective, and the images were captured by AxioVision 3.0 software and analyzed with Image-5 (Carl Zeiss).
Co-immunoprecipitation Assay
For co-immunoprecipitation experiments, the FLAG-tagged TRF1 construct was co-transfected with GFP-tagged TAP68 constructs or GFP vector alone (20 μg/dish) into human 293T cells using calcium phosphate. After 4 h, fresh medium was added. The cells were detached from the dishes by flushing with cold PBS 36 h after transfection, harvested by centrifugation, and lysed in pre-cooled lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100, 1 mm dithiothreitol, 2 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin). After incubation for 30 min on a shaker at 4 °C, detergent-insoluble debris was removed by centrifugation at 14,000 × g at 4 °C for 15 min. Cell lysates were further treated with λ-phosphatase (New England Biolabs) for 30 min at 30 °C after incubation and centrifugation at 14,000 rpm at 4 °C for 15 min. Cleared lysates were transferred to a new tube and incubated on a rocking platform at 4 °C for 4 h with either 10 μl of FLAG-M2-coupled agarose beads or 10 μl of protein A/G coupled with 10 μg of anti-GFP antibody. After the beads were washed three times with lysis buffer and two times with PBS, proteins were eluted with 50 μl of Laemmli loading buffer and resolved by 10% SDS-PAGE. Separated proteins were subsequently transferred onto nitrocellulose membrane for Western blot analysis with either GFP or FLAG monoclonal antibodies.
TAP68 Knockdown
The siRNA sequence used for silencing TAP68 corresponds to the coding region 336–354 (relative to the start codon). As a control, either a duplex targeting cyclophilin or scrambled sequence was used (36). The 21-mer oligonucleotide RNA duplexes were synthesized by Dharmacon Research, Inc. (Boulder, CO). Double thymidine synchronized cells were transfected with siRNA oligonucleotides using Oligofectamine (Invitrogen). The efficiency of the siRNA oligonucleotide on TAP68 protein knockdown was assessed by Western blot analysis after 48–72 h.
Subcellular Fractionation
2 × 106 interphase HeLa cells were used and incubated with 10× v/v of microtubule-stabilizing buffer (PEM buffer) solutions, which contained 5 mm PIPES, pH 7.2, 0.5 mm EDTA, 5 mm MgCl2, 5 mm NaCl, 1 mm PMSF, 2 mg/ml aprotinin, 2 mg/ml pepstatin A, 2 mg/ml leupeptin, at room temperature for 5 min and centrifuged at 2500 × g for 2 min to collect the cells. Cell pellets were re-dissolved in pre-cold 4 °C hypotonic solutions (PEM buffer plus 0.1% digitonin) (Sigma) and sonicated for four bursts at setting level 2 for 5 s each in a 4 °C ice bath with a BILON92 sonicator (Shanghai Bilon Instruments Co. Ltd., Shanghai, China) and sonicated on ice for four bursts of 5 s each by using a probe-tip sonicator BILON92. The homogenates were further centrifuged at 4 °C, 15,000 × g for 10 min. The supernatants (cytoplasm portion) and cell pellets (nuclei portion) were transferred separately to pre-cooling Eppendorf tubes and ready for further Western blot assay.
Immunoelectron Microscopy
For immunoelectron microscopy, HeLa cells were grown onto sterile, acid-treated 18-mm coverslips in 6-well plates (Corning Glass, Corning, NY) and maintained in DMEM with 10% FBS until nearly 75% confluent. After removing the medium, cells were washed for 1 min with PHEM buffer (100 mm PIPES, 20 mm HEPES, pH 6.9, 5 mm EGTA, 2 mm MgCl2, 4 m glycerol) and then incubated with PHEM buffer containing 1% digitonin to perforate the cell membrane for 5 min at room temperature. The extracted cells were pre-fixed in a freshly prepared 4% paraformaldehyde and 0.05% glutaraldehyde PHEM buffer for 10 min and rinsed with PBS three times for 5 min each. After blocking with PBST (0.05% Tween 20 in phosphate saline buffer, PBS) containing 1% BSA, cells were incubated with primary anti-TAP68 antibody at room temperature for 1 h. After washing three times with PBST for 3 min each, cells were incubated with colloidal gold particles conjugated with secondary antibody at room temperature for 1 h. Cells were fixed with 1% glutaraldehyde in PBS for 20 min and washed three times again with PBS. These cells were then treated with 2% osmium tetroxide for 30 min and dehydrated through graded ethanols (10, 20, 40, 60, and 80% and two times at 100%, 5 min in each) and 100% acetone (three times) over 10 min. Cells were then detached from the coverslips and embedded in acetone-Epon 812 (Sigma) overnight at 30 °C. After ultrathin sections were prepared, they were stained with lead acetate uranium and examined and photographed using a transmission electron microscope (JEOL 1200, Peabody, MA).
Statistical Analysis
All experiments were performed at least three times or more. The percentages of mitotic cells in bipolar, tripolar, and tetrapolar spindles were presented as means ± S.D. The independent t test was performed to compare the differences of percentage of mitotic cells in bipolar, tripolar, and tetrapolar spindles between the cells treated with TAP68 siRNA and control siRNA. Statistical analysis was considered significant when the two-sided p value was <0.05, using SPSS 15.0 statistics software (SPSS Inc., Chicago).
RESULTS
Identification of a Novel TRF1-binding Protein, TAP68
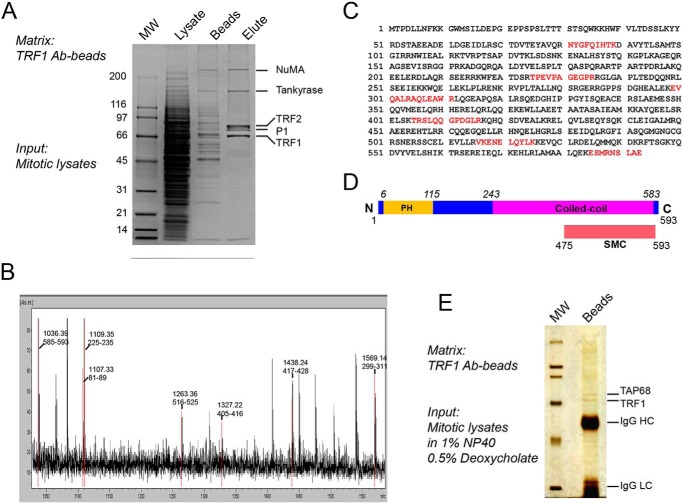
To further understand the role of TRF1 in mitotic regulation, soluble TRF1 and its potential binding partners were immunoprecipitated with an anti-TRF1 antibody from the lysates of mitotic HeLa cells because the TRF1-tankyrase 1 complex undergoes dynamic redistribution from the telomere to the centrosome upon the onset of mitosis (17). Mass spectrometric analysis of the immunoprecipitates showed that it contained several known telomere regulators, including tankyrase 1, TRF2, as well as a polypeptide (estimated mass of 68,000 Da) marked as P1 (Fig. 1A). The protein band of P1 was extracted from the acrylamide gel and digested with trypsin, and the resulting peptide fragments were subjected to mass spectrometric analysis (Fig. 1B). As shown in Fig. 1C, the six peptide sequences obtained matched the previously characterized open reading frame, AAH03618, that encodes a 593-amino acid protein with a predicted molecular mass of 68,041 Da. Computational analysis showed that AAH03618 contains one putative pleckstrin homology domain in its N terminus with two extended coiled-coil domains in its C terminus (Fig. 1D). In addition, AAH03618 contains a structural maintenance of a chromosome-like domain found in chromosome segregation-related ATPases (NCBI, www.ncbi.nlm.nih.gov) (26). Because the AAH03618 sequence distinguishes it from other TRF1-binding proteins, such as EB1 that localizes to the microtubule plus-end, we therefore referred to this protein as TAP68. The specific TRF1-TAP68 interaction was confirmed by using another anti-TRF1 antibody in high stringency lysis buffer containing 1% Nonidet P-40 and 0.5% deoxycholate. Under this condition, TAP68 remained firmly bound to TRF1, and the majority of other TRF1-binding proteins, such as TRF2 and tankyrase, were absent from the immunoprecipitate (Fig. 1E).
FIGURE 1.
Identification of a novel TRF1-binding protein, TAP68. A, mitotic HeLa cell extracts were applied to a TRF1 peptide antibody affinity column. After binding, columns were extensively washed, and bound proteins were eluted and separated by SDS-PAGE. The indicated proteins were extracted from the gel and digested with trypsin, and the amino acid sequence of the peptides was determined by MALDI-TOF and LC-MS/MS mass spectrometry. P1 is TAP68. B, protein band of P1 was extracted from the acrylamide gel and digested with trypsin, and the resulting peptide fragments were subjected to mass spectrometric analysis. C, P1 peptides match a previously uncharacterized protein with a 593-amino acid open reading frame of unknown function (AAH03618). D, schematic representation of the TAP68 domain organization predicted by NCBI's on-line conserved domain search tool. E, TRF1-TAP68 interaction was confirmed by immunoprecipitation assay. The anti-TRF1 antibody binding on protein A/G beads was incubated with mitotic cell lysate prepared in high stringency lysis buffer containing 1% Nonidet P-40 and 0.5% deoxycholate. Under this condition, TAP68 remained firmly bound to TRF1, whereas the majority of other TRF1-binding proteins, such as TRF2 and tankyrase, were absent from the co-immunoprecipitates.
TAP68 Binds to TRF1 via Its Coiled-coil Domains
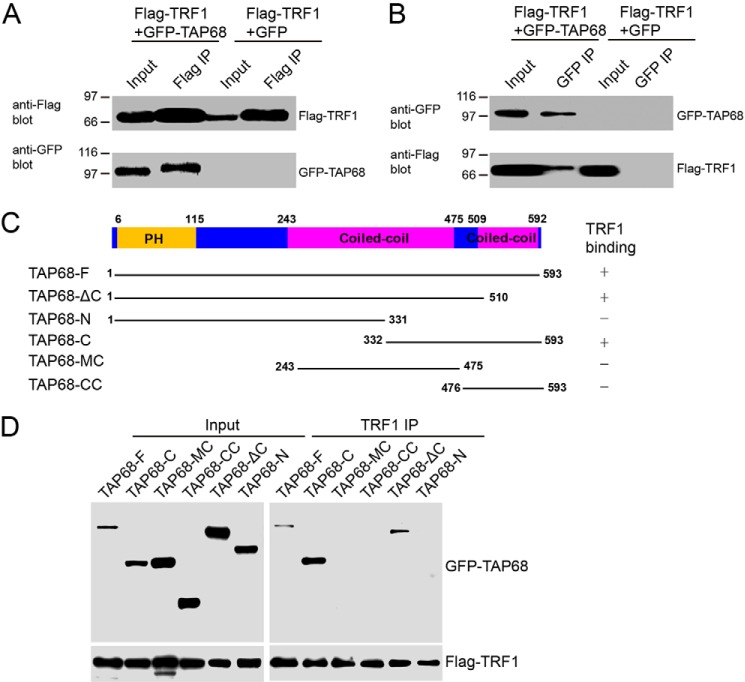
To further validate the interaction between TRF1 and TAP68, reciprocal co-immunoprecipitation was performed. First, we used anti-FLAG antibody to immunoprecipitate soluble TRF1 and its binding proteins from the lysates of mitotic HeLa cells transiently transfected to express FLAG-tagged TRF1 and GFP-tagged TAP68. Western blot analysis with anti-GFP antibody validated that TAP68 co-precipitated with TRF1 (Fig. 2A). No GFP was recovered in the FLAG immunoprecipitates, suggesting that the interaction between TRF1 and TAP68 is specific and independent of the GFP tag. Immunoprecipitation of TAP68 using an anti-GFP antibody confirmed the interaction between TAP68 and TRF1 (Fig. 2B).
FIGURE 2.
Biochemical characterization of TRF1-TAP68 interaction. A, exogenous TRF1 interacts with TAP68 in HeLa cells transfected with FLAG-tagged TRF1 and either GFP vector or GFP-tagged TAP68. After 36 h, HeLa cells were then extracted with 1% Nonidet P-40 plus 0.5% deoxycholate, and FLAG-tagged TRF1 protein and its binding proteins were immunoprecipitated (IP) with 20 μl of FLAG-M2 antibody-conjugated beads. Co-precipitated proteins were separated by SDS-PAGE and then immunoblotted for the presence of FLAG-TRF1 (upper panel) or GFP-TAP68 proteins (lower panel). B, exogenous TAP68 interacts with TRF1 in HeLa cells transfected with FLAG-tagged TRF1 and either GFP or GFP-tagged TAP68. After 36 h, HeLa cells were then extracted with 1% Nonidet P-40 plus 0.5% deoxycholate, and GFP-tagged TAP68 protein and its associated proteins were precipitated with 20 μl of GFP antibody-conjugated beads. Co-precipitated proteins were then detected by immunoblot for the presence of GFP-TAP68 (upper panel) or FLAG-TRF1 proteins (lower panel). C, schematic representation of different TAP68 truncation constructs. D, HeLa cells were transfected with full-length or different truncations of GFP-tagged TAP68 along with FLAG-tagged TRF1. After 36 h, HeLa cells were then extracted with 1% Nonidet P-40 plus 0.5% deoxycholate, and FLAG-tagged TRF1 and its accessory proteins were precipitated with 20 μl of rabbit TRF1 antibody-conjugated beads. Co-precipitated proteins were then immunoblotted for the presence of GFP-TAP68 proteins (upper panel) or FLAG-TRF1 (lower panel).
To map the region(s) of TAP68, which binds to TRF1, we immunoprecipitated soluble TRF1 and its binding proteins from lysates of mitotic HeLa cells exogenously expressing full-length and different deletion mutants of GFP-tagged TAP68 as shown in Fig. 2C. Western blot analysis with an anti-GFP antibody validated that the C-terminal 261 amino acids of TAP68 (TAP68-C) co-precipitated with TRF1 (Fig. 2D). But the TAP68ΔC that lacks the second coiled-coil domain can also bind TRF1. Thus, we concluded that both coiled-coil domains located on the C terminus of TAP68 were required for TRF1 binding. However, with assistance of the N-terminal domain, TAP68ΔC without the second coiled coil was able to bind TRF1.
TAP68 Co-localizes with TRF1 at the Telomeres in Interphase and Translocates to the Centrosome during Mitosis
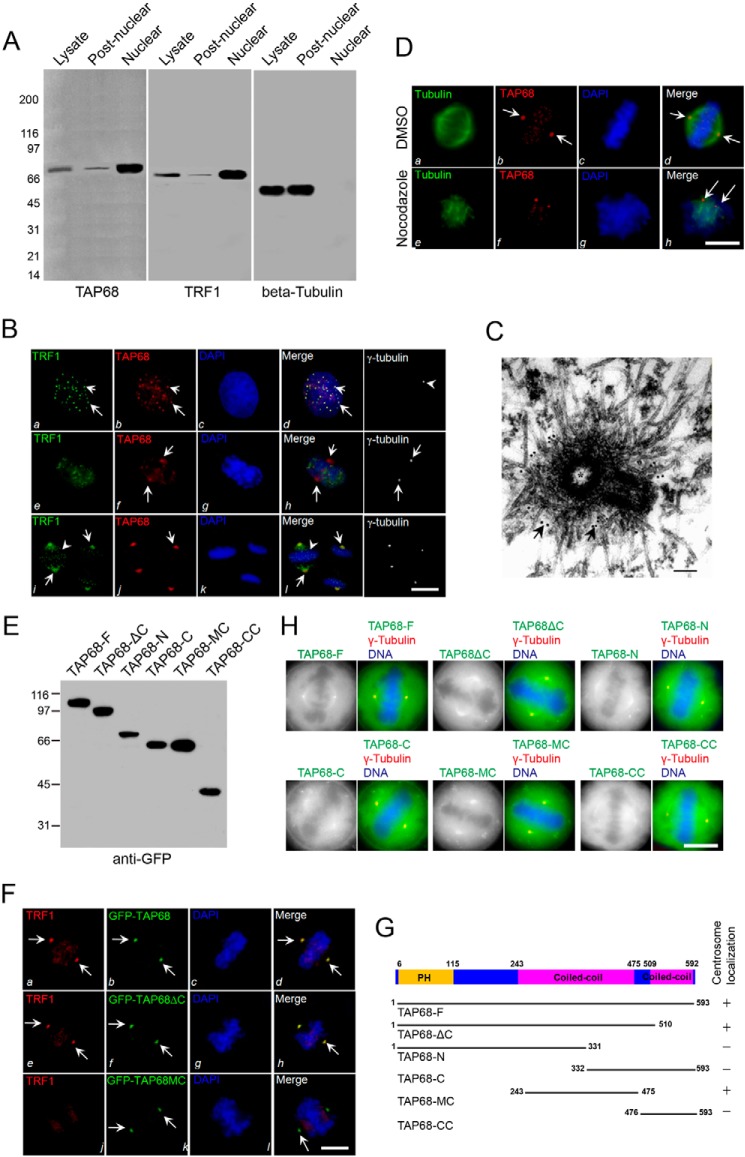
TRF1 is localized within telomeric nuclear speckles during interphase and translocates to the mitotic spindle during mitosis (17). To examine whether TAP68 is a nuclear protein, interphase HeLa cells were homogenized and separated into nuclear and post-nuclear fractions, and subcellular fractions were analyzed by Western blot using an anti-TAP68 mouse antibody. As shown in Fig. 3A (left and middle panels), both TAP68 and TRF1 were highly enriched in the nuclear fraction.
FIGURE 3.
TAP68 mediates TRF1 localization to the centrosome. A, TAP68 and TRF1 share similar subcellular localization. Nuclear and post-nuclear fractions were generated from interphase HeLa cell homogenates. Equal amounts of proteins (35 μg) from various fractions were separated by 5–15% gradient SDS-PAGE and detected by Western blot analyses using antibodies against TAP68, TRF1, and β-tubulin, respectively. B, representative images of interphase (top panels), late prophase/early prometaphase (middle panels), and metaphase (bottom panels) HeLa cells stained for TRF1 (green), TAP68 (red), DAPI (blue), and γ-tubulin (white). In interphase cells, TRF1 (panel a) and TAP68 (panel b) are readily seen in the nucleus as speckles and dots in the merge panel (panel d). In late prophase or the earliest prometaphase cells marked by nuclear membrane fragmentation, TAP68 appears at the centrosome (panel f, arrows), and TRF1 remains as speckles in the partially condensed chromosome (panel e, arrows). Both TAP68 and TRF1 become co-localized to the centrosome as the chromosomes align or after sister chromatid separation (panels i and j, arrows). A merged image shows the co-localization of TRF1 and TAP68 to the centrosome of mitotic cells (panel l). Bars, 10 μm. C, immunoelectron microscopy of late prophase cells indicated that TAP68 localizes to the pericentrioles. D, representative immunofluorescence images of HeLa cells treated with nocodazole or DMSO. 10 h after drug treatment, the cells were fixed and stained for tubulin (green), TAP68 (red), and DAPI (blue). TAP68 remained centrosome-associated in the absence of microtubules (panel h, arrows). E, ectopic expression of different GFP-tagged TAP68 proteins. HeLa cells were transfected GFP-tagged full-length TAP68 or its deletion mutants. After 36 h, HeLa cells were then harvested for Western blot analysis using a GFP antibody. F, representative images collected from metaphase HeLa cells transiently transfected with GFP-tagged TAP68 and its deletion mutants and stained for TRF1 (red), TAP68 (green), and DAPI (blue). In cells expressing full-length TAP68, both TAP68 and TRF1 are co-localized to the centrosome (panels a, b, and d, arrows). A similar co-distribution of TRF1 with TAP68 was observed in TAP68ΔC mutant-expressing cells (panels e, f, and h) but not TAP68MC-expressing cells (panels i, j, and l), demonstrating that the centrosomal localization region of TAP68 is independent of its TRF1-binding domain. Bars, 10 μm. G, schematic representation of the regions of TAP68, which specifies its centrosomal localization. H, representative images of HeLa cells transfected with different GFP-tagged TAP68 constructs. 24 h after transfection, cells were fix and co-stained for γ-tubulin (red) and DNA (blue). Bars, 10 μm.
To ascertain the subcellular localization of TAP68 relative to TRF1, HeLa cells were next stained with a TAP68 mouse antibody followed by a rhodamine-conjugated secondary antibody (red), and TRF1 was labeled with a rabbit antibody followed by a fluorescein-conjugated secondary antibody (Fig. 3B, green). In addition, a Cy5-conjugated γ-tubulin mouse antibody was used to mark the centrosome (Fig. 3B, white, arrowhead; right panels). A pre-extraction procedure allows the effective labeling of the kinetochore and centrosomal proteins while preserving the fine cyto-structure of mitotic cells was also employed (33, 35). In the interphase cells shown in Fig. 3B (panels a–d), TAP68 staining appeared as 21 clearly resolved dots and speckles in the nucleus (red and arrows), whereas TRF1 staining showed typical telomeric nuclear speckles (green and arrows). Merged images confirmed that TAP68 is a putative telomeric protein at interphase.
In late prophase/early pro-metaphase, TAP68 staining was found emigrating toward the centrosome, which was confirmed with γ-tubulin staining (Fig. 3B, right panel). Immunoelectron microscopy of late prophase cells indicated that TAP68 localizes to the pericentrioles (Fig. 3C). However, TRF1 remained associated with condensing chromosomes without apparent centrosomal localization (Fig. 3B, panel h).
During metaphase and anaphase, TAP68 was fully relocated to the centrosome (Fig. 3B, panel j). Labeling with a TRF1 rabbit antibody revealed typical spindle pole staining in metaphase and anaphase cells, which was consistent with a previous report (4). The centrosomal staining of TAP68 was identical to the TRF1 labeling, which becomes apparent when the two images are merged (Fig. 3B, panel l, arrowhead). The centrosomal localization of TAP68 in nocodazole-treated cells indicated that its localization at the centrosome was independent of microtubules (Fig. 3D). These results indicate that TAP68 and TRF1 form a complex at the telomeres in interphase, and they migrate to centrosomes during mitosis.
The apparent temporal order of assembly of TAP68 and TRF1 to the centrosomes in mitosis led us to hypothesize that TAP68 may be important for TRF1 centrosomal localization. To test the role of TAP68 in TRF1 targeting, the localization of different segments of TAP68 tagged with GFP was examined. Western blot analysis showed that all the constructs, including the full-length GFP-TAP68, expressed proteins of the predicted size (Fig. 3E). To verify that the GFP tag would not interfere with localization of TAP68, the full-length TAP68 tagged with GFP was first tested for its targeting. In metaphase cells, GFP-TAP68 localized to centrosomes where the majority of TRF1 was also observed (Fig. 3F, panels a, b, and d, arrows). Thus, the GFP tag did not alter exogenous TAP68 localization. We then assessed the centrosomal localization of TAP68 deletion mutants relative to TRF1 in transfected HeLa cells. A schematic representation of TAP68 deletion mutants was shown in Fig. 3G. All TAP68 mutants containing the first coiled-coil domain (amino acids 243–475) were clearly localized to the spindle poles (Fig. 3F, panel f). In contrast, the C terminus of TAP68 containing the second coiled-coil domain and TRF1-binding region (amino acids 332–593) failed to localize to the spindle poles (Fig. 3G). Thus, the centrosomal localization domain of TAP68 resided within the first coiled-coil domain (Fig. 3F, panels f and j, arrows), which is sufficient for its centrosomal localization. However, the centrosomal localization of TRF1 was lost with the TAP68MC mutant, suggesting that TAP68 was required for TRF1 recruitment to the centrosome in mitosis (Fig. 3F, bottom panel).
TAP68 Specifies the Centrosomal Localization of TRF1 during Mitosis
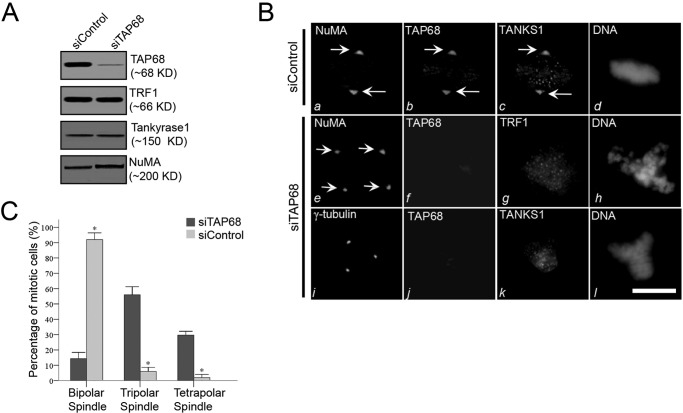
Targeted deletion of murine TRF1 resulted in an embryonic growth defect that was independent of telomere length regulations (37). Additional evidence has implicated TRF1 in the regulation of the mitotic spindle by interacting with a microtubule plus end-binding protein (EB1) (17). If TAP68 was required for the localization of TRF1 to the centrosome, depletion of TAP68 would abolish the centrosomal localization of TRF1. To examine the functional role of endogenous TAP68 in TRF1 localization to the centrosome, synchronized HeLa cells were depleted of TAP68 by transfection with small interfering RNA (siRNA) duplexes specific for TAP68. Western blot analysis confirmed the efficiency of TAP68 knockdown by its specific siRNA duplex but not by a scrambled siRNA. In addition, TAP68 siRNA does not alter TRF1, tankyrase 1, and NuMA protein levels (Fig. 4A). The observed 8-fold inhibition of TAP68 protein expression represented a more than 90% depletion of TAP68 in 91 ± 2% of the successfully transfected cells.
FIGURE 4.
TAP68 mediates TRF1 translocation to the centrosome and mitotic regulation. A, efficient siRNA-mediated suppression of TAP68. HeLa cells were transfected with a siRNA duplex specific for TAP68 and a scrambled control siRNA duplex. After 48 h, HeLa cells were then harvested for Western blot analysis using antibodies specific for TAP68, TRF1, tankyrase 1 (TANKS1), and NuMA. The analyses indicate the specificity and efficiency of the targeted protein suppression. B, representative immunofluorescence images of HeLa cells transfected with control or TAP68-specific siRNA duplex. 48 h post-transfection, cells were stained for the indicated antibodies and DNA, respectively. In the control duplex-transfected cells, NuMA, TAP68, and tankyrase 1 are all co-localized to the centrosome (panels a–c). In TAP68 siRNA duplex-treated cells, TAP68 protein expression was suppressed (panels f and j), and localization of TRF1 to the centrosome was diminished, although TRF1 distribution on chromosome remained (panel g). The distribution of tankyrase 1 to the centrosome was also eliminated in TAP68-depleted cells (panel k). However, the spindle pole localization of NuMA was not altered by the loss of TAP68 (panel e). Tetrapolar spindle in TAP68-depleted cells was revealed by NuMA staining (panel e). Bars, 10 μm. C, quantitative analyses of the mitotic spindle after TAP68 knockdown. HeLa cells were transfected with TAP68 siRNA oligonucleotide and control oligonucleotide for 36–48 h followed by fixation and staining for DNA, TAP68, and NuMA. After examination under a fluorescence microscope, the centrosome instability phenotypes were categorized as bipolar, tripolar, or tetrapolar spindles. Data represent the percentage of total mitotic cells evaluated. An average of 100 cells from three separate experiments was counted. * indicates a significant difference between the TAP68 siRNA- and control siRNA-transfected cells.
Next, we examined the requirement of TAP68 for targeting TRF1 to the centrosome. In TAP68-depleted cells, the centrosomal localization of TRF1 was reduced; however, no changes in NuMA association with the centrosome were noted (Fig. 4B), indicating the dependence of TRF1 on TAP68 to locate to the centrosome. Notably, depletion of TAP68 resulted in multiple spindle poles judged by the NuMA labeling (Fig. 4B, panel e) and γ-tubulin staining (Fig. 4B, panel i), suggesting that TAP68 functions in bipolar spindle formation and/or centrosomal stability. Further examination of tankyrase 1, a TRF1-binding protein, in TAP68-depleted cells revealed that very little tankyrase 1 was associated with the centrosomes with most remaining associated with chromosomes. Thus, stable localization of TRF1 and tankyrase 1 to the centrosome was dependent on TAP68.
The average proportion of mitotic cells in bipolar, tripolar, and tetrapolar spindles in HeLa cells treated with TAP68 siRNA oligonucleotides was 14.3, 56.0, and 29.7%, respectively (Fig. 4C). In contrast, the average percentages of mitotic cells in bipolar, tripolar, and tetrapolar spindles in HeLa cells treated with control siRNA oligonucleotides were 96.0, 6.0, and 2.0%, respectively. Thus, the proportion of mitotic cells with bipolar spindles was significantly decreased in TAP68 siRNA-transfected cells (p < 0.001). In contrast, tripolar and tetrapolar spindles were significantly increased in TAP68 siRNA-expressing cells (both p < 0.001; Fig. 4C).
TAP68 Residues Thr-221 and Thr-457 Are Phosphorylated by NEK2A and PLK1 during Mitosis
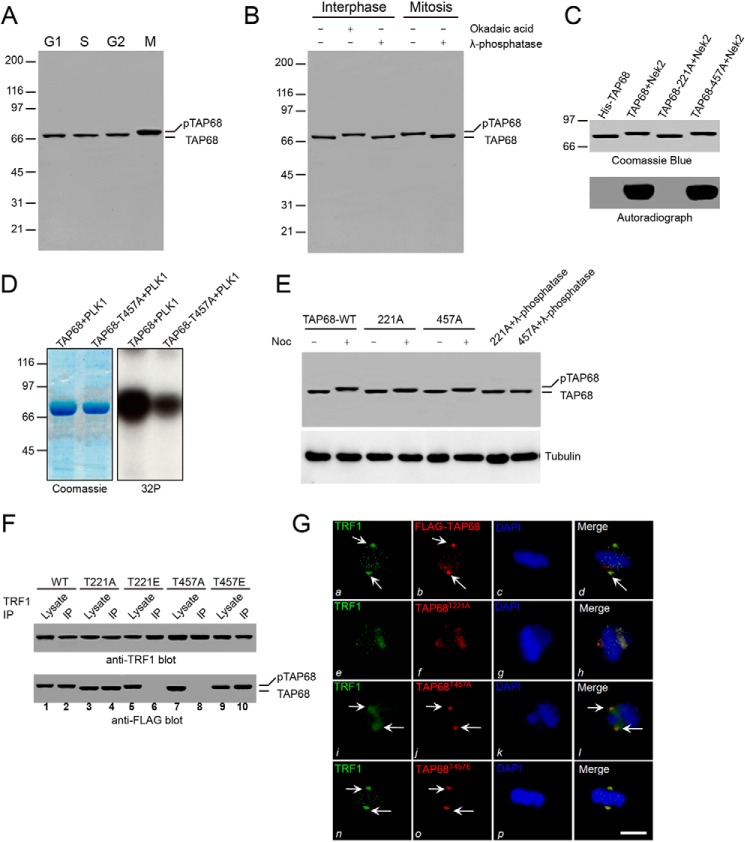
We next tested whether the changes in TAP68 localization during mitosis were induced by its phosphorylation. TAP68 was analyzed in lysates of HeLa cell that were synchronized at G1, S, G2, and M phases. In mitotic cells, a slower mobility TAP68 protein band was apparent compared with that of interphase cells (Fig. 5A). λ-Phosphatase treatment of the mitotic lysate induced a shift in TAP68 to the faster migrating form, although treatment of interphase cells with the serine/threonine phosphatase inhibitor, okadaic acid, resulted in a shift to the slower migrating form (Fig. 5B). These findings provide evidence that TAP68 is phosphorylated during mitosis.
FIGURE 5.
Phospho-regulation of TRF1-TAP68 interaction and centrosomal localization during mitosis. A, immunoblot analysis of TAP68 in HeLa cells synchronized at the indicated cell cycle phases. Note the mobility shift (pTAP68) from mitotic cell lysates. B, HeLa cell lysates (35 μg) from synchronized interphase and mitotic cells were treated with λ-phosphatase or the phosphatase inhibitor, okadaic acid, followed by Western blotting with an anti-TAP68 antibody. C, bacterially expressed histidine-tagged TAP68 fusion proteins, both wild-type and mutant (TAP68T221A and TAP68T457A), were phosphorylated in vitro in the presence of [32P]ATP and NEK2A kinase. Samples were separated by 6–16% gradient SDS-PAGE. The gel was dried and subsequently incubated with x-ray film. Note that in the presence of NEK2A, there was a dramatic incorporation of 32P into the wild-type TAP68 and TAP68T457A mutant, but not the TAP68T221A mutant. D, purified His-tagged TAP68-WT and TAP68-T457A were subjected to in vitro phosphorylation by recombinant PLK1 kinase. The left panel shows Coomassie Blue staining of the gel. The right panel shows the result of autoradiography. E, HeLa cell lysates from synchronized G1 and nocodazole-treated mitotic cells ectopically expressing wild-type or the indicated FLAG-TAP68 mutants were immunoblotted with an anti-FLAG and anti-α-tubulin antibodies, respectively. F, lysates from mitotic HeLa cells ectopically expressing FLAG-tagged TAP68-WT and the indicated mutants, immunoprecipitated with an anti-TRF1 antibody, and then immunoblotted with anti-TRF1 and anti-FLAG antibodies. TAP68T221A, which cannot be phosphorylated by NEK2A, was co-precipitated with TRF1 (lane 4), and its phospho-mimicking mutant TAP68T221E did not interact with TRF1 (lane 6). In contrast, the mutant with mimics Thr-457 phosphorylation by PLK1, TAP68T457E, interacted with TRF1, whereas the TAP68T457A mutant did not (lanes 10 and 8, respectively). Thus, phosphorylation of TAP68 at Thr-221 by NEK2A releases it from TRF1, whereas phosphorylation at Thr-457 by PLK1 is required for the TRF1-TAP68 interaction. G, representative images of HeLa cells ectopically expressing wild-type and mutant FLAG-TAP68. Cells were fixed and stained for TAP68 (red), TRF1 (green), and DNA (blue). Bars, 10 μm. Both wild-type FLAG-TAP68 and TRF1 are co-localized to the centrosome at metaphase (panels a and b, arrows). A merged image shows the co-localization of TRF1 and wild-type FLAG-TAP68 to the centrosome of mitotic cells (panel d). However, in HeLa cells ectopically expressing TAP68T221A, which cannot be phosphorylated by NEK2A, TAP68T221A remains associated with TRF1 at the telomere in late prometaphase cells (panel f) as evident in the merged image (panel h). In HeLa cells ectopically expressing TAP68T457A, which cannot be phosphorylated by PLK1, TAP68T457A localizes to the centrosome (panel j) with little TRF1 (panel i) as evidenced in the merged image (panel l, arrows), illustrating that PLK1-mediated phosphorylation is essential for TAP68-TRF1 association at the centrosome. Indeed, TRF1 co-localizes with TAP68 in HeLa cells ectopically expressing the phospho-mimicking mutant, TAP68T457E.
To define the region and specific residues of TAP68, which are phosphorylated during mitosis, we used anti-TAP68 antibodies to immunoprecipitate the TAP68 protein from mitotic HeLa cell lysates. Mass spectrometric analysis of the TAP68 protein isolated from mitotic cells suggested that both Thr-221 and Thr-457 were phosphorylated (data not shown). Computational analysis predicted that Thr-221 was a substrate of NEK2A, and Thr-457 was a substrate of PLK1 (38, 39). To test whether Thr-221 was a substrate of NEK2A, we performed in vitro phosphorylation on recombinant histidine-tagged fusion proteins, including both wild-type TAP68 and two mutant TAP68 proteins in which threonine 221 and 457 were replaced by alanine (T221A and T457A, respectively). The histidine-tagged wild-type and mutant TAP68 (T221A and T457A) migrated at about 70 kDa as shown in Fig. 5C. Incubation of the fusion proteins with [32P]ATP and the recombinant NEK2A kinase resulted in the incorporation of 32P into the TAP68-WT and TAP68-T457A proteins but not TAP68-T221A mutant (Fig. 5C). NEK2A-mediated phosphorylation was specific, because incubation of TAP68 with [32P]ATP in the absence of the kinase resulted in no detectable incorporation of radioactivity into the wild-type protein (Fig. 5C, 1st lane). Next, we performed in vitro kinase assay using TAP68-WT and TAP68–457A as substrates, respectively. Recombinant PLK1 kinase phosphorylated TAP68-WT strongly. In contrast, its phosphorylation toward TAP68–457A is very weak (Fig. 5D). Together, these studies confirmed the mass spectrometric analysis, and indicated that Thr-221 of TAP68 was a substrate of NEK2A, whereas Thr-457 was a substrate of PLK1. Mutation of Thr-221 and Thr-457 to alanine partially reduced FLAG-TAP68 phosphorylation in mitotic cells as determined by the mobility shift that was abolished with λ-phosphatase treatment (Fig. 5E), suggesting that NEK2A and PLK1 act cooperatively to phosphorylate TAP68 during mitosis. Thus, we concluded that both Thr-221 and Thr-457 are phosphorylated during mitosis.
Phosphorylation of TAP68 at Thr-457 Is Essential for the Association of TRF1 with TAP68 at the Centrosomes
TAP68 translocated to the spindle poles at the onset of prophase or early prometaphase (Fig. 3B), and this correlated with its phosphorylation (Fig. 5A), suggesting that phosphorylation of TAP68 may induce its dissociation from the telomere at the G2/M boundary. To evaluate the phospho-regulation of TAP68-TRF1 interaction, we used anti-TRF1 antibodies to immunoprecipitate TRF1, and bound TAP68 proteins from lysates of mitotic arrested HeLa cells exogenously express FLAG-tagged wild-type and mutant TAP68. Western blot analysis of the immunoprecipitate with anti-FLAG antibodies showed that both wild-type and the TAP68-T221A co-precipitate with TRF1 (Fig. 5F, 2nd and 4th lanes), although this association was abolished with the phosphorylation-mimicking mutant, TAP68-T221E (Fig. 5F, 8th lane). Interestingly, phosphorylation of Thr-457 was essential for TAP68-TRF1 interaction as TAP68-T457E was bound to TRF1 (Fig. 5F, 10th lane). Dephosphorylation of Thr-457 by phosphatase abolished the association of TAP68 with TRF1 (Fig. 5F, 6th lane), confirming the importance of Thr-457 in mediating TRF1-TAP68 interaction.
To correlate TAP68 phosphorylation with its centrosomal localization, both wild-type FLAG-TAP68 and phosphorylation mutant constructs were transiently transfected in HeLa cells. Both wild-type and the PLK-dependent phospho-mimicking mutant TAP68-T457E were localized to the centrosome (Fig. 5G, panels b and n, arrows) with a co-distribution of TRF1, indicating that phosphorylation of TAP68 at Thr-457 by PLK1 is essential for its association with TRF1 and for targeting TRF1 to the centrosome. Indeed, inhibition of Thr-457 phosphorylation (TAP68-T457A) prevented the localization of TRF1 to the centrosome (Fig. 5G, panel i, arrows). In contrast, the NEK2A-dependent phosphorylation-deficient mutant, TAP68-T221A, remained associated with TRF1 at the telomeres as the cells progressed toward metaphase (Fig. 5G, panels e–h), indicating that phosphorylation at Thr-221 was required for its dissociation from telomeric TRF at prophase or early prometaphase. This was consistent with the fact that TRF1 failed to precipitate TAP68-T221A from mitotic cell lysate. Thus, we concluded that NEK2A and PLK1 orchestrate TRF1-TAP68 interaction during mitosis.
DISCUSSION
Telomeres are essential for genome stability in all eukaryotes; dysfunctions in their assembly are associated with the chromosomal instability characteristic of cancer and aging (1, 3). Human telomere function requires two telomere-specific DNA TTAGGG repeat-binding proteins, TRF1 and TRF2, which interact with an array of proteins, including tankyrase, TIN2, PINX1, and protection of telomeres 1 (POT1) (1, 3). TRF1 and its binding proteins, such as tankyrase, relocate to the centrosome at the onset of mitosis (4–6). However, the molecular mechanisms underlying telomere dynamics and function in mitosis are less understood. In this study, we identified a novel centrosomal structural maintenance of chromosome domain protein, TAP68, which interacts with TRF1 and specifies its centrosomal localization in mitotic cells, providing insight into the function of the TRF1 protein complex in mitotic regulation. Immunoelectron microscopy confirmed that TAP68 localizes to the pericentrioles of mitotic cells. Moreover, depletion of TAP68 was sufficient to prevent TRF1 centrosomal translocalization and induced genomic instability. Interestingly, TAP68 is phosphorylated by NEK2A and PLK1, which regulate its transit from the telomere to the centrosome. Collectively, our results support a model in which TAP68 provides a functional link between telomeres and centrosomes in human cells. It would be of great interest to establish the structure-function relationship of TAP68 using structure-based analyses (40), which will enable us to consolidate the phospho-regulation into dynamic spindle regulation.
Oscillations in mitotic kinase activity drive progression through mitosis. A key mitotic kinase, NEK2A, exhibits a dynamic pattern of distribution during mitosis, which is important for faithful chromosome segregation during mitosis (36, 41–44). NEK2A is associated with the centrosome in interphase cells and becomes associated with spindle pole upon the breakdown of the nuclear envelope. Our finding that NEK2A phosphorylates TAP68 at Thr-221 in early prometaphase is consistent with the spatiotemporal dynamics of NEK2A during mitosis (36). In fact, TRF1/Pin2 was identified previously as both a protein (TRF1) that binds to telomeric DNA repeats (45) and as a protein (Pin2) that associates with the NIMA kinase (46), a NEK2A homologue in Aspergillus nidulans essential for entry into mitosis. We envision a model in which TAP68 is phosphorylated by NEK2A at the telomere at early prometaphase, which releases TAP68 for its trafficking to the centrosome where it is then phosphorylated by PLK1. Further studies will employ phospho-amino acid-specific antibodies to elucidate the spatiotemporal profile of phospho-TAP68 during mitosis. Given its close association with centrosome apparatus, it would be also important to ascertain if TAP68 exhibits microtubule binding activity and whether its association with microtubule is regulated by PLK1/Nek2A (47).
Abnormal centrosomal structure is frequently observed in cancer tissues and may be responsible for promoting disease progression (29, 48). As shown in this study, depletion of TAP68 in HeLa cells produced chromosome instability phenotypes similar to those seen in fission yeast in which aberrant expression of exogenous Pcp1 caused mitotic spindle defects, chromosome segregation abnormality, and supernumerary centrosomal structures (49). In fission yeast, disruption of TAZ1-Pcp1 interaction resulted in an anaphase bridge phenotype due mainly to the inability to resolve end-to-end chromosome fusions. In human cells, sister chromatid separation at the centromere and arms, but not at the telomeres, was associated with suppression of tankyrase 1 (50). In addition, a role for tankyrase 1 in bipolar spindle assembly has been reported (25, 26). Thus, conserved centrosome-telomere linkage events may play a role in mitotic chromosome segregation. Further studies will evaluate TAP68 levels in cancer cells with known chromosome instability to assess its possible role in disease progression.
Tankyrase is a poly(ADP-ribose) polymerase that transfers ADP-ribose to glutamic acid residues of its substrates (1). The localization of poly(ADP-ribose) to the centrosome is very similar to that of tankyrase (5). Perturbation of poly(ADP-ribose) by inhibitory antibody neutralization or poly(ADP-ribose) glycohydrolase treatment results in multipolar spindle assembly (26), which is similar to that seen in TAP68-depleted cells (Fig. 4B). The loss of tankyrase 1 at centrosomes correlated with the spindle multipolarity observed in TAP68-deficient cells, suggesting that poly(ADP-ribosylation) of centrosomal spindle proteins may be required for bipolar spindle formation and function in mitosis, consistent with Chang et al. (26).
NuMA is a spindle protein required for spindle pole assembly; it interacts directly with tankyrase 1 (51, 52). However, PARsylation of NuMA is not required for spindle function, which is consistent with our observation that centrosomal localization of NuMA is independent of tankyrase 1 (52). It would be interesting to elucidate the function and regulation of tankyrase 1 at the centrosome as well as its contribution to mitotic stability by identifying its substrates at the spindle pole. Proteomics-based strategies, such as those used in this study, that employ a combination of affinity isolation and mass spectrometry with biochemical manipulation of tankyrase 1 level at the spindle pole will allow us to unravel the molecular components whose functions are regulated by tankyrase 1-mediated protein PARsylation and mechanisms underlying normal centrosomal and telomeric function during mitosis.
This study is limited in that the function and clinical relevance of TAP68 remain unknown. Further studies will evaluate the effects of TAP68 silencing on cell proliferation as greater cell proliferation was observed with TRF1lox/lox mice (16). TAP68 suppression may also influence cell cycle progression. Knockdown of the TRF1-interacting protein, PinX1, delayed entry into mitosis, due to the loss of stabilization of TRF1 by PinX1 (53). Therefore, further studies will analyze the effects of TAP68 knockdown and TAP68 mutant expression on cell cycle progression. Alternatively, TAP68 may have a role in telomere maintenance in a similar fashion to Pin1 (54), which will be assessed in future studies.
In summary, we describe a new TRF1-interacting protein, TAP68, which localizes to telomeres in interphase, migrating to centrosomes following nuclear envelope breakdown. At the centrosomes, TAP68 recruits TRF1. Repression of TAP68 results in genomic instability, which may be mediated by inhibition of TRF1 and tankyrase recruitment to centrosomes.
Acknowledgments
We thank the members of our laboratory for insightful discussions.
Footnotes
This work was supported by Chinese 973 Project Grants 2002CB713704, 2012CB917204, and 2013CB911203, National Natural Science Foundation of China Grants 30871109, 30770919, 30900640, 81000896, 81300418, and 31371363, and Zhejiang Provincial Natural Science Foundation of China Grant Y2090885.
REFERENCES
- 1. Smogorzewska A., de Lange T. (2004) Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 2. Armanios M., Blackburn E. H. (2012) The telomere syndromes. Nat. Rev. Genet. 13, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blackburn E. H. (2001) Switching and signaling at the telomere. Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 4. Nakamura M., Zhou X. Z., Kishi S., Kosugi I., Tsutsui Y., Lu K. P. (2001) A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr. Biol. 11, 1512–1516 [DOI] [PubMed] [Google Scholar]
- 5. Smith S., de Lange T. (1999) Cell cycle-dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112, 3649–3656 [DOI] [PubMed] [Google Scholar]
- 6. Smith S., Giriat I., Schmitt A., de Lange T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- 7. Cooper J. P., Nimmo E. R., Allshire R. C., Cech T. R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385, 744–747 [DOI] [PubMed] [Google Scholar]
- 8. Tomaska L., Willcox S., Slezakova J., Nosek J., Griffith J. D. (2004) Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J. Biol. Chem. 279, 50764–50772 [DOI] [PubMed] [Google Scholar]
- 9. Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 [DOI] [PubMed] [Google Scholar]
- 10. Kim S. H., Kaminker P., Campisi J. (1999) TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou X. Z., Lu K. P. (2001) The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107, 347–359 [DOI] [PubMed] [Google Scholar]
- 12. Nishiyama A., Muraki K., Saito M., Ohsumi K., Kishimoto T., Ishikawa F. (2006) Cell-cycle-dependent Xenopus TRF1 recruitment to telomere chromatin regulated by Polo-like kinase. EMBO J. 25, 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Z. Q., Yang X., Weber G., Liu X. (2008) Plk1 phosphorylation of TRF1 is essential for its binding to telomeres. J. Biol. Chem. 283, 25503–25513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith S., de Lange T. (2000) Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302 [DOI] [PubMed] [Google Scholar]
- 15. Riffell J. L., Lord C. J., Ashworth A. (2012) Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat. Rev. Drug. Discov. 11, 923–936 [DOI] [PubMed] [Google Scholar]
- 16. Schneider R. P., Garrobo I., Foronda M., Palacios J. A., Marión R. M., Flores I., Ortega S., Blasco M. A. (2013) TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 4, 1946. [DOI] [PubMed] [Google Scholar]
- 17. Nakamura M., Zhou X. Z., Kishi S., Lu K. P. (2002) Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 514, 193–198 [DOI] [PubMed] [Google Scholar]
- 18. Prime G., Markie D. (2005) The telomere repeat binding protein Trf1 interacts with the spindle checkpoint protein Mad1 and Nek2 mitotic kinase. Cell Cycle 4, 121–124 [DOI] [PubMed] [Google Scholar]
- 19. Fry A. M., Schultz S. J., Bartek J., Nigg E. A. (1995) Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 270, 12899–12905 [DOI] [PubMed] [Google Scholar]
- 20. Muñoz P., Blanco R., de Carcer G., Schoeftner S., Benetti R., Flores J. M., Malumbres M., Blasco M. A. (2009) TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 29, 1608–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., Tsai R. Y. (2009) GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 185, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohishi T., Hirota T., Tsuruo T., Seimiya H. (2010) TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 70, 2041–2052 [DOI] [PubMed] [Google Scholar]
- 23. Wang C., Yu J., Yuan K., Lan J., Jin C., Huang H. (2010) Plk1-mediated mitotic phosphorylation of PinX1 regulates its stability. Eur. J. Cell Biol. 89, 748–756 [DOI] [PubMed] [Google Scholar]
- 24. Yuan K., Li N., Jiang K., Zhu T., Huo Y., Wang C., Lu J., Shaw A., Thomas K., Zhang J., Mann D., Liao J., Jin C., Yao X. (2009) PinX1 is a novel microtubule-binding protein essential for accurate chromosome segregation. J. Biol. Chem. 284, 23072–23082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang P., Coughlin M., Mitchison T. J. (2005) Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 26. Chang P., Jacobson M. K., Mitchison T. J. (2004) Poly(ADP-ribose) is required for spindle assembly and structure. Nature 432, 645–649 [DOI] [PubMed] [Google Scholar]
- 27. Chang P., Coughlin M., Mitchison T. J. (2009) Interaction between poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell 20, 4575–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ha G. H., Kim H. S., Go H., Lee H., Seimiya H., Chung D. H., Lee C. W. (2012) Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ. 19, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nigg E. A., Raff J. W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 [DOI] [PubMed] [Google Scholar]
- 30. Krämer A. (2005) Centrosome aberrations–hen or egg in cancer initiation and progression? Leukemia 19, 1142–1144 [DOI] [PubMed] [Google Scholar]
- 31. Fang Z., Miao Y., Ding X., Deng H., Liu S., Wang F., Zhou R., Watson C., Fu C., Hu Q., Lillard J. W., Jr., Powell M., Chen Y., Forte J. G., Yao X. (2006) Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4. Mol. Cell. Proteomics 5, 1437–1449 [DOI] [PubMed] [Google Scholar]
- 32. Cheng Z., Ke Y., Ding X., Wang F., Wang H., Wang W., Ahmed K., Liu Z., Xu Y., Aikhionbare F., Yan H., Liu J., Xue Y., Yu J., Powell M., Liang S., Wu Q., Reddy S. E., Hu R., Huang H., Jin C., Yao X. (2008) Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene 27, 931–941 [DOI] [PubMed] [Google Scholar]
- 33. Yao X., Anderson K. L., Cleveland D. W. (1997) The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou R., Guo Z., Watson C., Chen E., Kong R., Wang W., Yao X. (2003) Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell 14, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu Y., Yao P. Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., Yao X. (2011) Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell. Biol. 3, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 37. Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. (2003) Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23, 6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fu G., Ding X., Yuan K., Aikhionbare F., Yao J., Cai X., Jiang K., Yao X. (2007) Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 17, 608–618 [DOI] [PubMed] [Google Scholar]
- 39. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M. (2012) The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat. Commun. 3, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Faragher A. J., Fry A. M. (2003) Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 14, 2876–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Du J., Cai X., Yao J., Ding X., Wu Q., Pei S., Jiang K., Zhang Y., Wang W., Shi Y., Lai Y., Shen J., Teng M., Huang H., Fei Q., Reddy E. S., Zhu J., Jin C., Yao X. (2008) The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene 27, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mardin B. R., Lange C., Baxter J. E., Hardy T., Scholz S. R., Fry A. M., Schiebel E. (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei R., Ngo B., Wu G., Lee W. H. (2011) Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol. Biol. Cell 22, 3584–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Steensel B., de Lange T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 [DOI] [PubMed] [Google Scholar]
- 46. Lu K. P., Hanes S. D., Hunter T. (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 47. Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G., Ding X., Yao X. (2011) PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286, 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nigg E. A. (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825 [DOI] [PubMed] [Google Scholar]
- 49. Flory M. R., Carson A. R., Muller E. G., Aebersold R. (2004) An SMC-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol. Cell 16, 619–630 [DOI] [PubMed] [Google Scholar]
- 50. Dynek J. N., Smith S. (2004) Resolution of sister telomere association is required for progression through mitosis. Science 304, 97–100 [DOI] [PubMed] [Google Scholar]
- 51. Silk A. D., Holland A. J., Cleveland D. W. (2009) Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang W., Dynek J. N., Smith S. (2005) NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 391, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yonekawa T., Yang S., Counter C. M. (2012) PinX1 localizes to telomeres and stabilizes TRF1 at mitosis. Mol. Cell. Biol. 32, 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee T. H., Tun-Kyi A., Shi R., Lim J., Soohoo C., Finn G., Balastik M., Pastorino L., Wulf G., Zhou X. Z., Lu K. P. (2009) Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat. Cell Biol. 11, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]