Background: Although Musashi mediates target mRNA polyadenylation, the underlying molecular mechanism has not been elucidated.
Results: Germ line development defective-2, a poly(A) polymerase, associates with Musashi and is necessary and sufficient for Musashi-directed polyadenylation.
Conclusion: Germ line development defective-2 mediates Musashi-dependent mRNA translation.
Significance: Germ line development defective-2 couples Musashi to polyadenylation and translational activation of target mRNAs.
Keywords: Oocyte, Polyadenylation, RNA-binding Protein, Translation Control, Xenopus
Abstract
The mRNA-binding protein, Musashi, has been shown to regulate translation of select mRNAs and to control cellular identity in both stem cells and cancer cells. Within the mammalian cells, Musashi has traditionally been characterized as a repressor of translation. However, we have demonstrated that Musashi is an activator of translation in progesterone-stimulated oocytes of the frog Xenopus laevis, and recent evidence has revealed Musashi's capability to function as an activator of translation in mammalian systems. The molecular mechanism by which Musashi directs activation of target mRNAs has not been elucidated. Here, we report a specific association of Musashi with the noncanonical poly(A) polymerase germ line development defective-2 (GLD2) and map the association domain to 31 amino acids within the C-terminal domain of Musashi. We show that loss of GLD2 interaction through deletion of the binding domain or treatment with antisense oligonucleotides compromises Musashi function. Additionally, we demonstrate that overexpression of both Musashi and GLD2 significantly enhances Musashi function. Finally, we report a similar co-association also occurs between murine Musashi and GLD2 orthologs, suggesting that coupling of Musashi to the polyadenylation apparatus is a conserved mechanism to promote target mRNA translation.
Introduction
Growing evidence has revealed the cell's ability to control translation of specific subpopulations of mRNAs both spatially and temporally. A common theme of targeted translational control is regulation of 3′ poly(A) tail length, wherein a short poly(A) tail results in translational inactivation, and long poly(A) tail promotes translational activation (1–3). This form of translation control has been primarily studied in oocytes of the frog Xenopus laevis. Immature (Dumont stage VI) oocytes are arrested in late G2 and, in response to progesterone stimulation, resume the cell cycle and proceed into meiosis. Once stimulated, the oocyte nucleus (germinal vesicle) breaks down (GVBD),2 which is marked by the appearance of a white spot on the cell's animal pole, and the oocyte continues to metaphase of meiosis II, at which point it is mature and competent to be fertilized (4).
The oocyte maturation process (resumption of the cell cycle and progression through meiosis) involves a highly regulated signaling cascade, which is dependent upon synthesis of new proteins such as the Mos proto-oncogene, B-type cyclins, and Musashi (5–11). Because transcription is suppressed during maturation, the oocyte controls synthesis of critical proteins through selective translation of maternally derived mRNAs, in a strict temporal order (9, 11–18). This dependence on mRNA translation and lack of interference from transcriptional pathways make oocyte maturation an excellent model system to study the mechanisms of translational control (19).
Central to control of the signaling cascade that leads to GVBD are the translational regulators Pumilio, Musashi, and the cytoplasmic polyadenylation element-binding protein 1 (CPEB1). Activating these factors occurs through a sequential hierarchy and results in the translational activation of target mRNAs controlled by each respective factor in a temporally orchestrated manner (20). Pumilio mediates repression of Ringo until progesterone stimulation leads to dissociation of Pumilio from the Ringo mRNA, resulting in Ringo translation (21, 22). Ringo then activates free cyclin-dependent kinase subunits, triggering phosphorylation of several targets, including Musashi (23). Musashi phosphorylation triggers the cytoplasmic polyadenylation and translation of several target mRNAs, including those encoding Mos, cyclin B5, and Nrp1A/B (Musashi-1) prior to GVBD (10, 23–25). Subsequently, Mos-dependent activation of MAPK signaling primes CPEB1 for activation (26), leading to de-repression and translational activation of CPE-dependent mRNAs, including those encoding cyclin B1, Wee1, and CPEB4 (27–30). At GVBD, activation of the maturation-promoting factor (cyclin B/cyclin-dependent kinase) results in partial degradation of CPEB1 and functional substitution of CPEB1 with CPEB4 (30). CPEB1 activation appears to be mediated by several mechanisms depending upon the repression complex assembled on the mRNA (31–33). In immature stage VI oocytes, the atypical poly(A) polymerase GLD2 is found in a complex with CPEB1 and the deadenylase PARN. Progesterone-stimulated maturation triggers phosphorylation of CPEB1 and expulsion of PARN, resulting in the “unopposed” polyadenylation of CPE-containing mRNAs by GLD2 (34, 35).
Although a fairly detailed understanding of Pumilio and CPEB1 function has emerged, the mechanism(s) by which Musashi directs translational activation of target mRNAs in response to progesterone is unknown. Musashi does not appear to mediate repression of the Musashi-binding element (MBE-containing mRNAs in immature oocytes) but does mediate both cytoplasmic polyadenylation and translational activation in response to progesterone stimulation (24, 25, 36). In this study, we identify GLD2 as a Musashi-interacting factor that contributes to cytoplasmic polyadenylation of MBE target mRNAs prior to GVBD. GLD2 associates with Musashi-1 and Musashi-2 in both immature and progesterone-stimulated oocytes. The interaction of GLD2 with Musashi appears to be functional as assessed by several criteria. First, deletion of the region containing the GLD2 interaction domain compromises the ability of Musashi to mediate cell cycle progression. Second, inhibition of GLD2 synthesis with antisense oligonucleotides ablates cell cycle progression and attenuates Musashi-directed mRNA cytoplasmic polyadenylation. Finally, overexpression of GLD2 and Musashi exerts a synergistic acceleration of Mos mRNA polyadenylation and oocyte maturation. We also demonstrate interaction between the mammalian orthologs of GLD2 and Musashi-1, suggesting that GLD2 interaction with Musashi is a conserved mechanism to direct cytoplasmic polyadenylation and activation of Musashi target mRNA translation.
EXPERIMENTAL PROCEDURES
Oocyte Culture and Microinjections
Dumont Stage VI immature Xenopus laevis oocytes were isolated and cultured as described previously (37). Oocytes were micro-injected using a Nanoject II Auto-Nanoliter Injector (Drummond Scientific). mRNA for oocyte injection was made by linearization of the plasmid and in vitro transcription using SP6 (Promega) or T7 (Invitrogen) RNA polymerase as appropriate. Oocytes were stimulated to mature with 2 μg/ml progesterone. The appearance of a white spot on the animal pole was used to score the rate of GVBD. Where indicated, progesterone-stimulated oocytes were segregated when 50% of the oocytes completed GVBD (GVBD50) into those that had not (−) or had (+) completed GVBD. In the event of ambiguous morphology, oocytes were fixed for 10 min in ice cold 10% trichloroacetic acid and dissected for the presence or absence of a germinal vesicle. Animal protocols were approved by the UAMS Institutional Animal Care and Use committee, in accordance with Federal regulations.
Oocyte Lysis and Sample Preparation
Oocytes were lysed in 10 μl/oocyte of ice cold Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 2 mm EDTA, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm PMSF, 1× protease inhibitor (Thermo Scientific)). Yolk and cell debris were removed by centrifugation at 12,000+ rpm for 10 min in a refrigerated tabletop centrifuge. Where required, a portion of the lysate was transferred immediately following lysis to STAT-60 (Tel-Test, Inc) for RNA extraction using the manufactures protocol followed by a subsequent purification by precipitation in 4 m LiCl at −80 °C for 30 min and centrifugation at 12,000 rpm for 10 min in a refrigerated tabletop centrifuge.
Pulldown and RNase Treatment
Oocytes were injected with 57.5 ng of each in vitro transcribed mRNA and incubated for 16 h at 18 °C. Lysates were prepared as described above. 300 μl of oocyte lysate was added to 450 μl ice cold Nonidet P-40 lysis buffer and incubated with 50 μl of 50% glutathione-Sepharose conjugated bead slurry (GE) at 4 °C for 6 h with gentle rotation. Beads were then gently pelleted by centrifugation at 500 rpm for 5 min; the supernatant was removed and replaced with 500 μl fresh Nonidet P-40 lysis buffer. This process was repeated 3 times. On the third wash, 200U of RNase1 (Ambion) was added and incubated for 15 min at 37 °C. Following final centrifugation, all Nonidet P-40 was removed and 50 μl of LDS sample loading buffer (Invitrogen) was added. Beads were incubated for 10 min at 70 °C, then crushed by centrifugation at 12,000 rpm for 10 min. Finally, 45 μl of the sample was loaded per each lane of a 10% NuPAGE gel (Invitrogen) and electrophoresed.
Western Blotting
For each lane, half-oocyte equivalents of lysate were prepared in NuPAGE LDS sample loading buffer and electrophoresed through a 10% NuPAGE gel then transferred to a 0.2 μm-pore-size nitrocellulose membrane (Protran; Midwest Scientific). The membrane was blocked with 5% nonfat dried milk in TBST (20 mm Tris, pH 7.5, 150 mm NaCl, 0.05% Tween 20) for 60 min at room temperature, or overnight at 4 °C. Following incubation with primary antibody, filters were washed three times for 10 min in TBST, incubated with horseradish peroxidase conjugated secondary antibody then washed 3 × 10 min in TBST. Blot were developed using enhanced chemiluminescence in a Fluorchem 8000 Advanced Imager (Alpha Innotech Corp.). Western blots were quantified using Fluorchem FC2 software (Alpha Innotech Corp.).
Antibodies
The HA antibody (Cell Signaling) was used at 1:1000. The GST (Santa Cruz Biotechnology) and GFP (Invitrogen) antibodies were used at 1:5000. The Tubulin antibody (Sigma) was used at 1:10,000. The Xenopus GLD2 antibody was a generous gift from Dr. Marvin Wickens and used at 1:1000. All working antibody preparations were made in TBST + 0.5% nonfat milk.
Polyadenylation Assays
cDNAs for polyadenylation assays were synthesized using RNA ligation-coupled PCR as described previously (38). The increase in PCR product length is specifically due to extension of the poly(A) tail (36, 38). The same reverse primer was used for all reactions and has the sequence: 5′-GCTTCAGATCAAGGTGACCTTTTT-3′. The Mos forward primer has the sequence: 5′-GCAAGGATATGAAAAAAAGATTTC-3′. The Cyclin B1 primer has the sequence: 5′-GTGGCATTCCAATTGTGTATTGTT-3′.
Antisense Oligodeoxynucleotide Injections and Rescue
Antisense oligodeoxynucleotide 5′-TCCCTCGTCGCTTCTCCTCTTTCTGT-3′ was designed to target endogenous XGLD2-a and XGLD2-b mRNAs. Antisense oligodeoxynucleotides targeting Xenopus Musashi-1 and Musashi-2 were previously described (24). Control oocytes were injected with randomized oligonucleotide with the sequence 5′-TAGAGAAGATAATCGTCATCTTA-3′ (12). A total of 100 ng of antisense oligonucleotides was injected for each condition and oocytes were incubated at 18 °C for 16 h. For Musashi rescue assays, antisense injected oocytes were subsequently injected with 23 ng RNA encoding wild-type Musashi-1 or a deletion mutant Musashi-1, as indicated. The oocytes were then incubated for 1 h at room temperature to allow expression of the protein, then stimulated to mature with progesterone.
Statistical Analysis
All quantitated data are presented as the mean ± S.E. Statistical significance was assessed by one way Analysis of Variance followed by the Bonferroni post hoc test or by Student's t test when only two groups were compared. A probability of p < 0.05 was adopted for statistical significance.
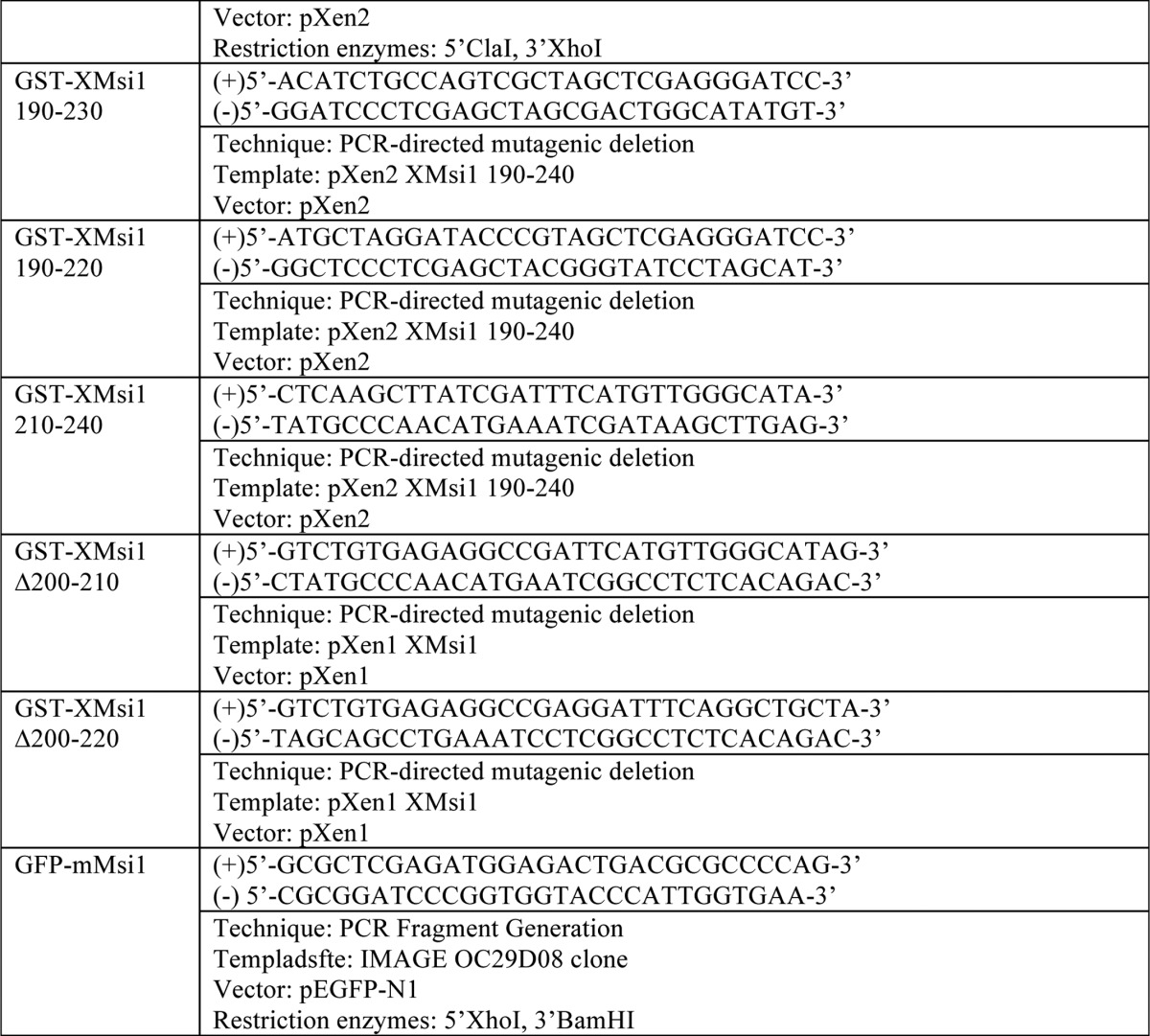
Plasmid Construction
See Table 1.
TABLE 1.
Plasmid construction
For PCR fragment generation, the template was subjected to PCR amplification using the indicated primers. Resulting PCR fragments were then purified using agarose gel electrophoresis followed by cleanup using a QIAquick gel extraction kit (Qiagen). Next, the fragments and destination vector were digested using the indicated restriction enzymes and again purified and cleaned up using gel electrophoresis and the QIAquick kit. The fragment and vector were then ligated using the T7 Quick Ligase (New England Biolabs). Finally, the ligated fragment/vector was used to transform competent DH5-α Escherichia coli. For PCR-directed mutagenic deletion, the template was subjected to PCR amplification of the entire plasmid. Primer sequence “looped out” the desired sequence for deletion. PCR-directed stop codon insertion is the same as the PCR-directed mutagenic deletion, except primers directed insertion of a stop codon rather than deletion.
RESULTS
Musashi Specifically Associates with the Poly(A) Polymerase, GLD2
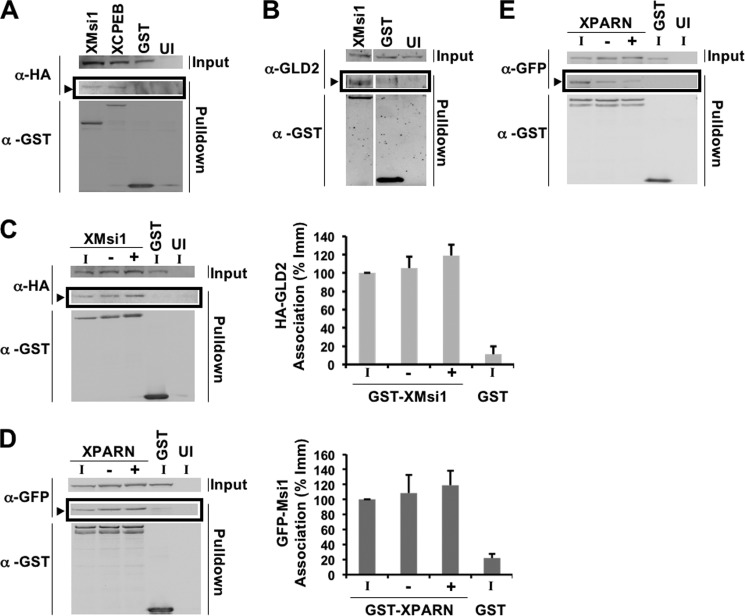
Musashi has been previously demonstrated to regulate the polyadenylation status of target mRNAs (10, 24, 25). However, Musashi itself is not a poly(A) polymerase. We therefore hypothesized that Musashi must recruit a poly(A) polymerase to the 3′ UTR of mRNAs containing a MBE. The noncanonical poly(A) polymerase GLD2, which does not bind target mRNAs directly (42), has been previously suggested to mediate CPE-directed cytoplasmic polyadenylation (34). Further, GLD2 has been reported to associate with the Mos mRNA (39), which we have previously shown to be under Musashi-directed control (25). We therefore hypothesized that GLD2 may mediate Musashi-directed polyadenylation. To test this idea, we used a co-association assay to determine whether Musashi-1 and GLD2 associate in oocytes. Briefly, immature Stage VI oocytes were co-injected with mRNA encoding GST-Musashi-1 and HA-GLD2. Oocytes were then allowed to express the proteins, lysed, subjected to GST pulldown in the presence of RNase1 (to eliminate proteins that simply co-associate by virtue of interacting with the same mRNA) and specific interacting proteins detected by Western blotting. A GLD2-specific band was identified as a co-associating protein in GST-Musashi-1-injected oocytes, but not in oocytes injected with the GST moiety alone (Fig. 1A, arrowhead). As a positive control, GLD2 association was similarly detected with GST-CPEB1 (Fig. 1A). We also injected the mRNA encoding GST-Musashi-1 or GST, and endogenous GLD2 interaction was detected using a GLD2 specific antibody (Fig. 1B, arrowhead). No GLD2 association was detected with the GST moiety alone.
FIGURE 1.
Musashi-1 associates specifically with the noncanonical poly(A) polymerase, GLD2. A, oocytes were co-injected with mRNA encoding HA-GLD2 and either GST-XMsi1, GST-CPEB1, or GST. The injected oocytes were incubated overnight to express the introduced proteins and then lysed. Lysates were then subjected to GST pulldown and treatment with RNase I. Associations were visualized by Western blotting. GST-XMsi1 and GST-CPEB1 associate with HA-GLD2 in an RNase I-independent manner, although the GST tag alone does not (arrowhead). UI, uninjected oocytes. B, oocytes were injected with GST-XMsi1 or GST and allowed to express the protein before lysis and pulldown. An antibody targeting endogenous GLD2 detects GLD2 associating with GST-XMsi1 but not GST (arrowhead). UI, uninjected oocytes. C, oocytes were co-injected with mRNA encoding HA-GLD2 and either GST-XMsi1 or GST. Following incubation, oocytes were stimulated to mature with progesterone. When 50% of oocytes reached GVBD, lysate was made using immature (I) and progesterone-stimulated oocytes pre-GVBD (−) and post-GVBD (+). HA-GLD2 associates with GST-XMsi1 in immature and progesterone-stimulated oocytes (arrowhead). UI, uninjected oocytes. A representative experiment is shown, and the composite results of three independent experiments are shown graphically (right panel). D, oocytes were injected with mRNA encoding GFP-XMsi1 and GST-XPARN or GST. Oocytes were then treated as in C. XMsi1 associates with PARN in immature and progesterone-stimulated oocytes (arrowhead). A representative experiment is shown, and the composite results of three independent experiments are shown graphically (right panel). E, oocytes were injected with mRNA encoding GFP-XCPEB1 and GST-XPARN or GST. Oocytes were then treated as in C and D. As described previously, cytoplasmic polyadenylation element-binding protein dissociates from PARN after progesterone addition. A representative experiment is shown.
To determine whether GLD2 and Musashi-1 remain associated during maturation, when Musashi is known to actively direct polyadenylation of target mRNAs, we co-injected oocytes with mRNA encoding GST-Musashi-1 and HA-GLD2 as before. Following incubation, the oocytes were either left untreated or stimulated with progesterone before lysis. When 50% of the oocyte population reached GVDB (GVBD50), they were sorted into those oocytes that had completed GVBD and those which had not. This sorting of oocytes was critical because Musashi has been shown to direct progesterone-stimulated polyadenylation prior to GVBD (24, 25). Following GST pulldown and visualization by Western blotting, GLD2 was found to associate with GST-Musashi-1 in progesterone-stimulated oocytes prior to GVBD (Fig. 1C). When normalized for the amount of GST-Musashi-1 recovered in each experiment, no significant difference in GLD2 association with Musashi was observed between immature and progesterone-stimulated oocytes (Fig. 1C, Graph). No GLD2 association was detected with the GST moiety alone. We conclude that GLD2 specifically interacts with Musashi-1 in both immature and progesterone-stimulated oocytes.
Previous analyses of GLD2 interaction with CPEB1 have suggested that GLD2 function is opposed in immature oocytes by the presence of the deadenylase PARN within the CPEB1 complex. Progesterone stimulation resulted in phosphorylation of CPEB1 and expulsion of PARN, leading to the unopposed action of GLD2 on CPE-containing mRNAs (35). We therefore sought to determine whether PARN associated with Musashi-1 in immature oocytes and was similarly expelled in response to progesterone. We found that PARN can associate in an RNase-insensitive manner with Musashi-1 in immature oocytes and we do not observe dissociation of PARN from Musashi-1 complexes in response to progesterone stimulation (Fig. 1D). By contrast, significant dissociation of PARN from CPEB1 was observed after progesterone stimulation (Fig. 1E), consistent with an earlier study (35). These results indicate that PARN interaction with CPEB1 and Musashi-1 is differentially regulated in progesterone-stimulated oocytes.
Deletion Mapping of the GLD2 Binding Domain within the Musashi Protein
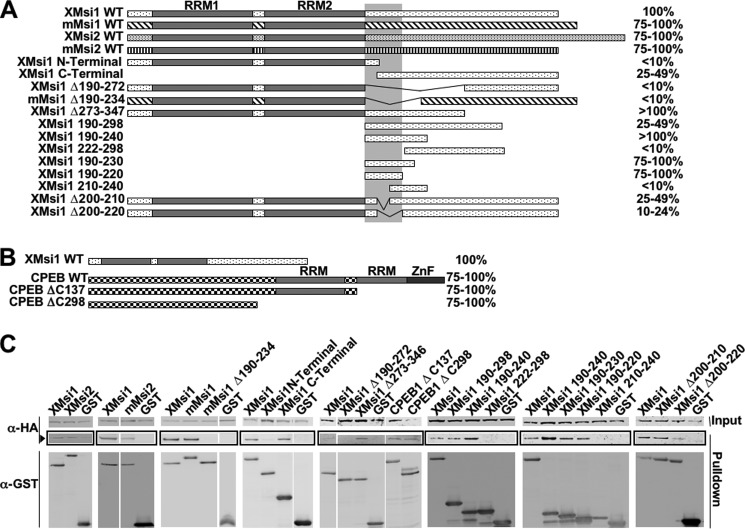
As an initial step toward testing the functional significance of the Musashi/GLD2 association, we sought to map domain(s) of Musashi-1 required for association with Xenopus GLD2 protein. A series of deletion mutants of the Musashi-1 (Fig. 2A) and CPEB-1 (Fig. 2B) proteins were generated and tested for GLD2 co-association in the GST-pulldown assay (Fig. 2C). The GLD2 minimal interaction domain appears to reside within amino acids 190–220, which lie C-terminal to the two RNA recognition motif domains. This 31-amino acid domain was sufficient for GLD2 to interact with Musashi-1 when fused to GST (Fig. 2, A and C, XMsi1 190–220). The Xenopus Musashi-2 protein was also able to co-associate with GLD2, as were the mammalian Musashi-1 and Musashi-2 proteins (Fig. 2, A and C). We conclude that GLD2 interacts within the C-terminal region of Musashi and that this interaction appears to be evolutionarily conserved for both the Musashi-1 and Musashi-2 isoforms.
FIGURE 2.
Musashi/GLD2 interaction domain lies within amino acids 190–220 of Musashi. A, oocytes were co-injected with mRNA encoding HA-GLD2 and one of the listed GST-Musashi constructs. Co-injection with HA-GLD2 and the GST moiety alone served as a negative control in all experiments. Following incubation to allow protein expression, the oocytes were lysed and subjected to GST pulldown and RNase I treatment, followed by Western blotting. Horizontal bars schematically represent the Musashi constructs, and % value denotes degree of GLD2 interaction as quantified by spot densitometry and normalized to wild-type Musashi-1 in each case. The vertical bar marks amino acids 190–220, the deduced GLD2 interaction domain. B, mapping of the domain of CPEB1 that associates with GLD2. Methodology was the same as described in A. C, representative experiments from A and B showing GLD2 association (arrowhead) with the indicated Musashi and cytoplasmic polyadenylation element-binding protein constructs.
Because the GLD2 interaction domain on CPEB1 has not been previously determined, deletions of the CPEB1 protein were tested for association with GLD2. We found that the GLD2 protein associates with the N-terminal half of the CPEB1 protein (Fig. 2, B and C). However, no obvious linear consensus sequence was found between this portion of CPEB1 and the 190–220 domain of Musashi-1.
Interaction of GLD2 with Musashi Promotes Target mRNA Polyadenylation and Cell Cycle Progression
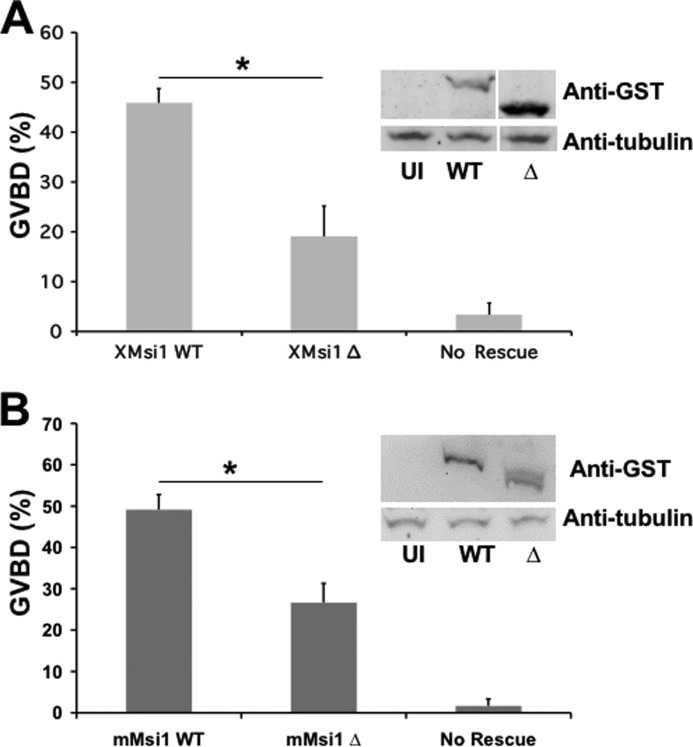
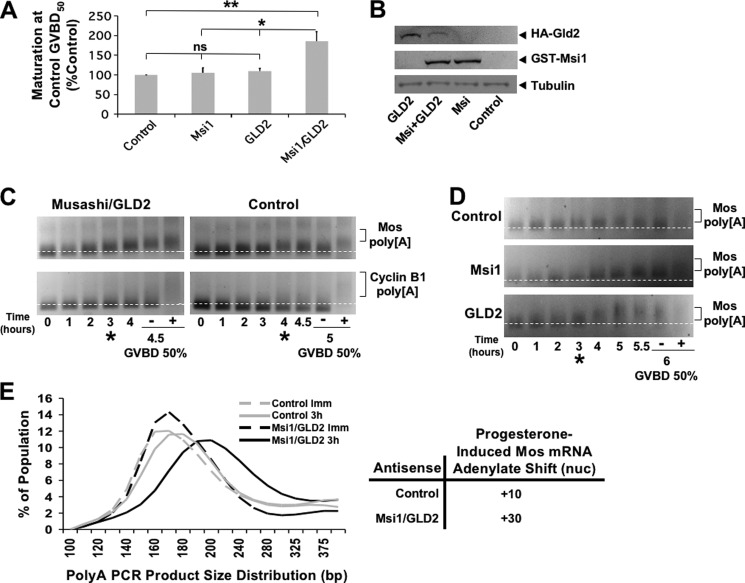
To test the functional significance of the Musashi:GLD2 interaction, we compared the ability of wild-type and GLD2 interaction-defective mutant Musashi proteins to mediate cell cycle progression in response to progesterone stimulation. In this assay, endogenous Musashi function is specifically abrogated by injection of antisense oligonucleotides targeting both endogenous Musashi-1 and Musashi-2 mRNAs but not by scrambled, control oligonucleotides (24). As expected, the Musashi antisense oligonucleotides inhibited progesterone-stimulated GVBD (Fig. 3, A and B, No Rescue). Re-injection of RNA encoding GST fused to wild-type Xenopus Musashi-1 (Fig. 3A) or murine Musashi-1 (Fig. 3B) rescued progesterone-stimulated oocyte maturation as expected (24). By contrast, injection of similar levels of deletion mutants lacking the GLD2 interaction domain in either the Xenopus Musashi-1 (Fig. 3A, XMsi1 Δ) or murine Musashi-1 (Fig. 3B, mMsi1 Δ) demonstrated a significantly reduced ability to mediate timely progression through oocyte maturation (assessed as % GVBD at the time the wild-type Musashi-1 rescue achieved GVBD50). Taken together, the data indicate that deletion of the GLD2 interaction domain attenuated Musashi-directed cell cycle progression.
FIGURE 3.
Deletion of the GLD2 binding domain in Musashi-1 compromises Musashi-mediated oocyte maturation. A, oocytes were injected with antisense oligonucleotides targeting endogenous Musashi-1 and Musashi-2. Following incubation, the oocytes were left untreated or re-injected with mRNA encoding wild-type (WT) or deletion mutant Δ190–272 of Xenopus Musashi-1 and allowed to express the protein prior to being stimulated to mature with progesterone. The Δ190–272 Musashi mutant protein is significantly compromised for rescue. The data represent three experiments. The Western panel inset demonstrates expression of the introduced Musashi rescue proteins. UI, uninjected, no rescue protein. * indicates p < 0.05. B, oocytes were injected with antisense oligonucleotides as described in A and subsequently re-injected with WT or Δ190–234 mutant mouse Musashi-1. The deletion mutant protein is significantly compromised for rescue of GVBD. * indicates p < 0.05.
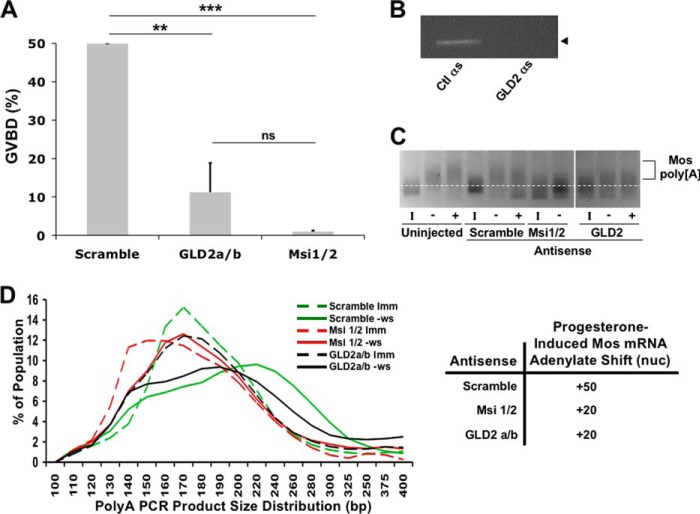
To complement the Musashi deletion analyses, we sought to inhibit endogenous GLD2 function through the use of antisense oligonucleotide injection. In contrast to control oligonucleotides, injection of GLD2 antisense oligonucleotides targeting both the GLD2a and GLD2b isoforms significantly attenuated progesterone-stimulated oocyte maturation (Fig. 4A). The Musashi-1/2 antisense oligonucleotides were used as a positive control for inhibition of GVBD. These findings compliment an earlier report where depletion of GLD2 using neutralizing antisera also blocked cytoplasmic polyadenylation (34). As expected, the GLD2 antisense oligonucleotides, but not control scrambled oligonucleotides, led to the selective cleavage of the endogenous GLD2 mRNA (Fig. 4B). Next, the polyadenylation status of the Musashi target mRNA, Mos, was examined. Treatment with Musashi-1/2 antisense caused a deadenylation of 20 nucleotides in immature oocytes, which was restored (but not subsequently extended) after progesterone addition (Fig. 4, C and D). GLD2 antisense-treated oocytes did not show any deadenylation but did show significantly attenuated polyadenylation (by ∼30 adenylate residues) compared with control oocytes after progesterone treatment (Fig. 4, C and D). Indeed, even in the few oocytes that were able to complete GVBD, GLD2 antisense-treated oocytes showed a dramatic attenuation of the poly(A) tail extension. We conclude that inhibition of endogenous GLD2 synthesis dramatically attenuates oocyte maturation and Musashi-dependent cytoplasmic polyadenylation of target mRNAs.
FIGURE 4.
GLD2 is necessary for Musashi-directed polyadenylation. A, oocytes were injected with antisense oligonucleotides targeting either GLD2a/b, Musashi-1/2, or a scrambled control oligonucleotide. Following incubation, the oocytes were stimulated to mature with progesterone treatment. Maturation was scored relative to control antisense-treated oocytes. GLD2 antisense severely attenuates oocytes maturation. The combined data of four independent experiments is shown. **, indicates p < 0.01; ***, indicates p < 0.001. B, representative PCR for endogenous GLD2 mRNA showing loss of intact GLD2 mRNA. C, representative poly(A) length assay for the Musashi target mRNA, Mos. A robust shift in PCR product mobility, indicative of polyadenylation, is seen in both uninjected and control (scrambled oligonucleotide)-injected oocytes. Oocytes injected with GLD2 antisense show dramatically attenuated polyadenylation of the Mos mRNA pool, similar to that seen with Musashi antisense injected oocytes. I, indicates immature oocytes; − indicates progesterone-stimulated pre-GVBD; + indicates progesterone-stimulated post-GVBD. D, graphic representation of the mobility shifts seen in C. The dashed lines are the distribution of MosPCR product in immature oocytes (Imm); solid lines are the distribution of MosPCR products in progesterone-stimulated oocytes pre-GVBD (−ws). The green lines are scrambled-control oligonucleotide-injected oocytes. Red lines represent Msi1/2 antisense-injected oocytes. Black lines are GLD2a/b antisense-injected oocytes. The line peaks indicate the median of the population of mRNA lengths. The shift of the peak between immature (dashed line) and progesterone-stimulated (solid line) indicates the extent of polyadenylation. Scrambled, control oligonucleotide-injected oocytes show a polyadenylation shift of 50 adenylate residues, although both Msi1/2 and GLD2a/b oligonucleotide-injected oocytes show a polyadenylation shift of only 20 adenylates. ns, not significant.
Further support for a functional interaction between Musashi-1 and GLD2 was obtained from overexpression studies. Injection of RNA encoding GLD2 or GST-Musashi-1 alone did not affect the rate of progesterone-stimulated oocyte maturation (Fig. 5A). However, co-injection of GLD2 with GST-Musashi-1 resulted in a 1.83 ± 0.25-fold increase in the rate of maturation (Fig. 5A). Protein expression from the injected RNAs was verified by Western blotting (Fig. 5B). When the polyadenylation status of the endogenous Mos mRNA was assessed, co-expression of GLD2 and GST-Musashi-1 resulted in an acceleration of Mos mRNA polyadenylation by ∼1 h (Fig. 5, C and E), compared with water-injected controls. Importantly, the polyadenylation of the late class cyclin B1 mRNA still occurred after completion of GVBD in oocytes co-expressing Musashi-1 and GLD2, so the relative temporal order of maternal mRNA translational activation was maintained. Additionally, the absolute length of the Mos mRNA poly(A) tail was not significantly altered, rather the time to initiation of polyadenylation was advanced. The acceleration in onset of polyadenylation of the Mos mRNA was not seen when Musashi-1 or GLD2 were overexpressed separately (Fig. 5D). We conclude that co-expression of GLD2 and Musashi-1 accelerates the time of initiation of polyadenylation of Musashi target mRNAs.
FIGURE 5.
GLD2 is sufficient of promote Musashi-mediated polyadenylation and oocyte maturation. A, oocytes were injected with nuclease-free water (control) or mRNA mixtures containing GST-Musashi-1, HA-GLD2a, or GST-Musashi-1 and HA-GLD2a. The oocytes were allowed to express the injected RNA and were then stimulated to mature with progesterone. The maturation rate relative to control in five independent experiments is shown graphically. *, indicates p < 0.05; **, indicates p < 0.01. ns, not significant. B, representative Western blot showing protein expression from an experiment in A. C, representative poly(A) assay comparing polyadenylation of two endogenous mRNAs from Musashi/GLD2 co-injected oocytes relative to water-injected (control) oocytes. The Mos mRNA is under Musashi control and therefore polyadenylated early (prior to GVBD), whereas the cyclin B1 mRNA is under cytoplasmic polyadenylation element-binding protein control and polyadenylated late (at or after GVBD). The earliest signs of polyadenylation (*) appear at 3 h for all 3 conditions. − indicates pre-GVBD; + indicates post-GVBD samples. D, poly(A) length assay comparing polyadenylation of Mos mRNA from Musashi, GLD2, or water-injected (control) oocytes. The earliest signs of polyadenylation appear at 3 h for all three conditions. This indicates that the acceleration in onset of polyadenylation seen in C requires overexpression of both Musashi-1 and GLD2 together. − indicates pre-GVBD; + indicates post-GVBD. E, graphic representation of the poly(A) assay (C) for the Mos mRNA at the 3rd h. The dashed line is immature (Imm) oocytes (0 h), and the solid line is at the 3-h time point. Gray lines represent water (control) injection, and the black lines are Musashi/GLD2 co-injected oocytes. At the 3rd h, Musashi/GLD2 co-injection has promoted a polyadenylation of ∼30 adenylate residues, whereas control injected oocytes have only extended their poly(A) tails by ∼10 adenylates.
Mammalian Orthologs of Musashi and GLD2 Co-associate
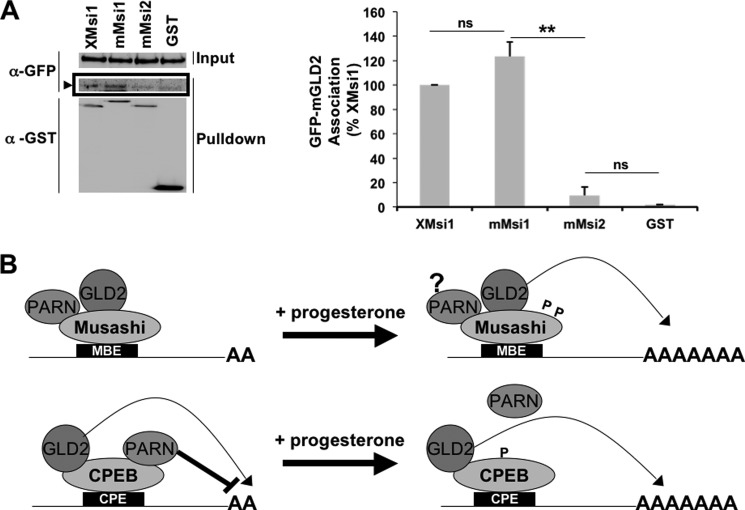
We finally asked if the murine Musashi and murine GLD2 proteins interact. We co-injected oocytes with GFP-tagged mouse GLD2 and GST-tagged mouse Musashi-1 or Musashi-2 or the GST moiety alone. Although GLD2 association with Musashi-1 (Fig. 6A) was detected, we did not see significant association between Musashi-2 and GLD2 (Fig. 6A). This finding indicates that mammalian Musashi-1 and GLD2 are capable of associating and that coupling of Musashi-1 with polyadenylation machinery may be an evolutionarily conserved mechanism of targeted polyadenylation and translational activation.
FIGURE 6.
Conservation of GLD2 and Musashi-1 co-association. A, oocytes were injected with GFP-tagged murine GLD2 (mGLD2) and GST-tagged Xenopus Musashi-1 (XMsi1), murine Musashi-1 (mMsi1), Musashi-2 (mMsi2), or the GST moiety alone. Co-associated proteins were assessed by Western blotting with GFP. A GFP-mGLD2-specific band was detected in both Xenopus and mouse GST-Musashi-1 pulldowns (arrowhead) but not with GST-mMusashi-2 or GST alone. A representative experiment is shown, as well as the average of three independent experiments (graph, right panel). **, indicates p < 0.01. ns, not significant. B, proposed model of Musashi regulation of polyadenylation of target mRNAs (upper panel). Our data indicate GLD2 mediates polyadenylation of Musashi target mRNAs following progesterone addition. The role and regulation of PARN within the Musashi complex are unclear (?) at the juncture. The cytoplasmic polyadenylation element-binding protein model, showing progesterone-dependent PARN expulsion, is shown for comparison (lower panel).
DISCUSSION
In this study, we provide the first evidence that Musashi can interact with the GLD2 poly(A) polymerase. The GLD2 enzyme is an atypical poly(A) polymerase in that it lacks inherent RNA binding function and is recruited to target mRNAs by association with sequence-specific RNA-binding proteins (34, 39, 42, 43). Interaction of GLD2 with Musashi serves to selectively recruit GLD2 to a subset of cellular mRNAs that possess Musashi-binding elements. GLD2 interaction with Musashi is RNase-insensitive, indicating that the interaction does not occur by simple co-occupancy of GLD2 and Musashi proteins on the same mRNA. Rather, the interaction occurs via protein-protein association in an mRNA-independent manner. As such, our findings link Musashi directly with the 3′ end processing machinery that controls modification of the poly(A) tail in the cytoplasm to stimulate translational activation of select mRNA species in response to extracellular stimulation.
Our findings complement prior reports linking GLD2 to CPE-dependent mRNA polyadenylation via recruitment through interaction with CPEB1 (34, 35). Both GLD2 and PARN are present in CPEB1 complexes in immature oocytes. In this context, PARN is thought to exert a dominant effect and thereby override GLD2 activity to maintain short poly(A) tails and translational dormancy. Upon progesterone stimulation, CPEB1 undergoes phosphorylation of Ser-174, which triggers expulsion of PARN from the complex and leads to the unopposed action of GLD2 and consequent extension of poly(A) tails of CPE-regulated mRNAs (23). We see a similar co-association of Musashi with both GLD2 and PARN in immature oocytes. However, upon progesterone stimulation and phosphorylation of Musashi on Ser-297 and Ser-322, which are critical for translational activation of Musashi target mRNAs (23), we do not observe expulsion of PARN from the Musashi complex (Fig. 1D). We do observe PARN dissociation from CPEB1 (Fig. 1E) implying that the mechanism of MBE-dependent mRNA polyadenylation and translational activation differs from that proposed for CPEB1 and does not appear to require PARN expulsion. At this juncture, it is not clear how the complex is remodeled to allow Musashi-directed mRNA cytoplasmic polyadenylation in response to progesterone stimulation. Presumably, PARN function is in some way attenuated within the Musashi complex and/or GLD2 activity is promoted, resulting in a net gain in poly(A) tail length. Interestingly, the continued presence of PARN in Musashi complexes in progesterone-stimulated oocytes may explain the deadenylation of certain Musashi target mRNAs that lack CPE sequences after GVBD (25, 36). Future studies will be required to directly assess the regulation of GLD2 and PARN within Musashi complexes assembled upon MBE-controlled mRNAs.
Our data suggest a functional requirement for Musashi interaction with GLD2. Expression of wild-type Musashi-1 can rescue cell cycle progression in Musashi antisense-injected oocytes (24), whereas expression of Musashi mutant proteins encoding a deletion spanning the GLD2 interaction domain were significantly attenuated in their functional capability to rescue cell cycle progression. Furthermore, injection of antisense oligonucleotides targeting GLD2 specifically attenuated both oocyte maturation and polyadenylation of early class mRNAs in response to progesterone.
Although a role for GLD2 in Musashi function is clearly indicated, Musashi activity is not eliminated in the GLD2 interaction mutant. The residual Musashi-1 function may explained by several nonexclusive possibilities that include a second weak GLD2 interaction domain elsewhere on Musashi-1 that is not detected in our pulldown experiments, recruitment of an alternative poly(A) polymerase during maturation, or involvement of a potential polyadenylation-independent mechanism of Musashi-mediated translational activation (25).
Although our results are consistent with a requirement for Musashi to interact with GLD2 to mediate maternal mRNA translational activation, we cannot exclude the possibility that additional proteins may also interact with the same domain on the Musashi protein and thereby contribute to translational activation. For example, we note that a 44-amino acid domain of the mammalian Musashi-1 protein, including the GLD2 interaction region identified here (Fig. 2), has been reported to be necessary for repression of target mRNAs and association with the poly(A)-binding protein in rabbit reticulocyte lysate and HEK293T cells (44). However, the embryonic poly(A)-binding protein is the predominant poly(A)-binding protein in oocytes (45–47), and the function of the embryonic poly(A)-binding protein, if any, in Musashi-directed translational control is unclear. At this juncture, our data do not distinguish between indirect or direct interaction between Musashi and GLD2. Nonetheless, co-injection of both Musashi-1 and GLD2 acts synergistically to promote target mRNA translation and acceleration of progesterone-stimulated maturation, suggesting that GLD2 is a critical component of the Musashi translation promoting complex.
Deletion mapping localized the interaction domain to a 31-amino region within the C-terminal half of the protein (amino acids 190–220). This region lies outside of the two N-terminal RNA recognition motifs. Expression of the minimal region identified by deletion mapping fused to GST was sufficient to mediate interaction with GLD2 when expressed in oocytes (Fig. 2). The GLD2 interaction was also detected with the related Xenopus Musashi-2 isoform. In addition, both mammalian Musashi-1 and Musashi-2 demonstrated interaction with Xenopus GLD2 when ectopically expressed in the Xenopus oocyte. The 31-amino acid region is >73% identical between Xenopus Musashi-1 and Musashi-2 isoforms. CPEB1 interaction with GLD2 occurs via the N-terminal CPEB1 domain (Fig. 2, B and C); however, alignment between the conserved 31-amino acid region of Xenopus Musashi-1 and Musashi-2 with CPEB1 failed to identify an obvious region of homology. We suspect that the GLD2 interaction domain on target proteins may involve structural folding rather than a simple dependence upon a linear amino acid sequence. The region within GLD2 necessary for interaction with Caenorhabditis elegans GLD3, an RNA-binding protein, has been determined (42), but it is unclear if this domain mediates interaction with RNA-binding proteins in vertebrate cells. Further studies will be required to definitively map the GLD2 interaction domains within CPEB1 and Musashi to better define a consensus GLD2 target interaction motif.
Consistent with a conserved interaction of Musashi proteins and GLD2, we observe co-association of the mouse orthologs of GLD2 and Musashi-1. This finding suggests that mammalian Musashi-1 may employ a similar strategy as seen in oocytes to promote target mRNA translation. We did not, however, observe any significant interaction between mouse Musashi-2 and GLD2 (Fig. 6A), suggesting a Musashi isoform-specific interaction. Indeed, precedent for Musashi-1- and Musashi-2-specific protein associations has been suggested previously (48, 49). It is possible that mammalian Musashi-2 may not mediate polyadenylation. However, our recent work indicates that both mouse and human Musashi-2, like the Xenopus Musashi-2, do promote polyadenylation in Musashi antisense-treated Xenopus oocytes.3 Thus, an alternative possibility is that Musashi-2-directed polyadenylation of target mRNAs in mammalian cells may involve a different poly(A) polymerase.
Although Musashi proteins have been primarily implicated in promoting mammalian stem and progenitor cell self-renewal and proliferation via repression of target mRNAs (e.g. m-Numb and p21) (50–53), recent findings have emerged demonstrating that Musashi switches from a repressor to an activator of target mRNA translation in response to mammalian neural stem cell differentiation cues (54). A role for Musashi-mediated activation has also been proposed for Robo3/Rig-1 mRNA translation during midline crossing of precerebellar neurons (55) and activation of m-Numb mRNA translation in the gastric mucosa of mice (56), although the underlying mechanisms have not been elucidated. Our findings of Musashi association with GLD2 and target mRNA polyadenylation during Xenopus oocyte maturation may serve as a paradigm to explain Musashi-mediated activation of target mRNA translation in mammalian systems. Given the role of Musashi repression in promoting physiological and pathological stem cell self-renewal, our mechanistic insights into the bipartite function of Musashi may provide a foundation for future therapeutic control of stem cell function.
Acknowledgments
We thank Drs. M. Wickens and M. Rebbert for XGLD2 and Xrp-1 plasmids, respectively, and Linda Hardy for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HD35688 and Grants UL1TR000039 and KL2TR000063 from NCRR. This work was also supported by the Arkansas Breast Cancer Research Program, the Sturgis Charitable Trust, intramural funding support from the University of Arkansas for Medical Sciences College of Medicine Research Council, the University of Arkansas for Medical Sciences Translational Research Institute and the Arkansas BioSciences Institute.
M. C. MacNicol, C. E. Cragle, and A. M. MacNicol, manuscript in preparation.
- GVBD
- germinal vesicle breakdown
- MBE
- Musashi-binding element
- CPE
- cytoplasmic polyadenylation element
- PARN
- poly(A) ribonuclease.
REFERENCES
- 1. Dworkin M. B., Shrutkowski A., Dworkin-Rastl E. (1985) Mobilization of specific maternal RNA species into polysomes after fertilization in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 82, 7636–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosenthal E. T., Tansey T. R., Ruderman J. V. (1983) Sequence-specific adenylations and deadenylations accompany changes in the translation of maternal messenger RNA after fertilization of Spisula oocytes. J. Mol. Biol. 166, 309–327 [DOI] [PubMed] [Google Scholar]
- 3. Vassalli J. D., Huarte J., Belin D., Gubler P., Vassalli A., O'Connell M. L., Parton L. A., Rickles R. J., Strickland S. (1989) Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 3, 2163–2171 [DOI] [PubMed] [Google Scholar]
- 4. Ferrell J. E., Jr. (1999) Xenopus oocyte maturation: new lessons from a good egg. BioEssays 21, 833–842 [DOI] [PubMed] [Google Scholar]
- 5. Barkoff A., Ballantyne S., Wickens M. (1998) Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J. 17, 3168–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barkoff A. F., Dickson K. S., Gray N. K., Wickens M. (2000) Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 220, 97–109 [DOI] [PubMed] [Google Scholar]
- 7. Sagata N., Daar I., Oskarsson M., Showalter S. D., Vande Woude G. F. (1989) The product of the mos proto-oncogene as a candidate initiator for oocyte maturation. Science 245, 643–646 [DOI] [PubMed] [Google Scholar]
- 8. Sagata N., Oskarsson M., Copeland T., Brumbaugh J., Vande Woude G. F. (1988) Function of c-mos proto-oncogene product in meiotic maturation in Xenopus oocytes. Nature 335, 519–525 [DOI] [PubMed] [Google Scholar]
- 9. Sheets M. D., Wu M., Wickens M. (1995) Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature 374, 511–516 [DOI] [PubMed] [Google Scholar]
- 10. Arumugam K., Macnicol M. C., Macnicol A. M. (2012) Autoregulation of Musashi1 mRNA translation during Xenopus oocyte maturation. Mol. Reprod. Dev. 79, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freeman R. S., Ballantyne S. M., Donoghue D. J. (1991) Meiotic induction by Xenopus cyclin B is accelerated by coexpression with mosXe. Mol. Cell. Biol. 11, 1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferby I., Blazquez M., Palmer A., Eritja R., Nebreda A. R. (1999) A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 13, 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Howard E. L., Charlesworth A., Welk J., MacNicol A. M. (1999) The mitogen-activated protein kinase signaling pathway stimulates mos mRNA cytoplasmic polyadenylation during Xenopus oocyte maturation. Mol. Cell. Biol. 19, 1990–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murakami M. S., Vande Woude G. F. (1998) Analysis of the early embryonic cell cycles of Xenopus; regulation of cell cycle length by Xe-wee1 and Mos. Development 125, 237–248 [DOI] [PubMed] [Google Scholar]
- 15. Nakajo N., Yoshitome S., Iwashita J., Iida M., Uto K., Ueno S., Okamoto K., Sagata N. (2000) Absence of wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev. 14, 328–338 [PMC free article] [PubMed] [Google Scholar]
- 16. Roy L. M., Swenson K. I., Walker D. H., Gabrielli B. G., Li R. S., Piwnica-Worms H., Maller J. L. (1991) Activation of p34cdc2 kinase by cyclin A. J. Cell Biol. 113, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ballantyne S., Daniel D. L., Jr., Wickens M. (1997) A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell 8, 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Moor C. H., Richter J. D. (1997) The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 17, 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cragle C. E., MacNicol A. M. (2014) in Xenopus Development (Kloc M., Kubiak J. Z., eds) Chapter 3, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 20. MacNicol M. C., MacNicol A. M. (2010) Developmental timing of mRNA translation-integration of distinct regulatory elements. Mol. Reprod. Dev. 77, 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao Q., Padmanabhan K., Richter J. D. (2010) Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 16, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padmanabhan K., Richter J. D. (2006) Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 20, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arumugam K., MacNicol M. C., Wang Y., Cragle C. E., Tackett A. J., Hardy L. L., MacNicol A. M. (2012) Ringo/cyclin-dependent kinase and mitogen-activated protein kinase signaling pathways regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J. Biol. Chem. 287, 10639–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arumugam K., Wang Y., Hardy L. L., MacNicol M. C., MacNicol A. M. (2010) Enforcing temporal control of maternal mRNA translation during oocyte cell cycle progression. EMBO J. 29, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charlesworth A., Wilczynska A., Thampi P., Cox L. L., MacNicol A. M. (2006) Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 25, 2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keady B. T., Kuo P., Martínez S. E., Yuan L., Hake L. E. (2007) MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J. Cell Sci. 120, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 27. Cao Q., Richter J. D. (2002) Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21, 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Charlesworth A., Welk J., MacNicol A. (2000) The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′ untranslated region. Dev. Biol. 227, 706–719 [DOI] [PubMed] [Google Scholar]
- 29. de Moor C. H., Richter J. D. (1999) Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 18, 2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Igea A., Méndez R. (2010) Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 29, 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Charlesworth A., Meijer H. A., de Moor C. H. (2013) Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev. RNA 4, 437–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Radford H. E., Meijer H. A., de Moor C. H. (2008) Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 1779, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szostak E., Gebauer F. (2013) Translational control by 3′-UTR-binding proteins. Brief Funct. Genomics 12, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnard D. C., Ryan K., Manley J. L., Richter J. D. (2004) Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119, 641–651 [DOI] [PubMed] [Google Scholar]
- 35. Kim J. H., Richter J. D. (2006) Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24, 173–183 [DOI] [PubMed] [Google Scholar]
- 36. Charlesworth A., Ridge J. A., King L. A., MacNicol M. C., MacNicol A. M. (2002) A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 21, 2798–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machaca K., Haun S. (2002) Induction of maturation-promoting factor during Xenopus oocyte maturation uncouples Ca(2+) store depletion from store-operated Ca(2+) entry. J. Cell Biol. 156, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charlesworth A., Cox L. L., MacNicol A. M. (2004) Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in xenopus oocytes. J. Biol. Chem. 279, 17650–17659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rouhana L., Wang L., Buter N., Kwak J. E., Schiltz C. A., Gonzalez T., Kelley A. E., Landry C. F., Wickens M. (2005) Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11, 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacNicol M. C., Pot D., MacNicol A. M. (1997) pXen, a utility vector for the expression of GST-fusion proteins in Xenopus laevis oocytes and embryos. Gene 196, 25–29 [DOI] [PubMed] [Google Scholar]
- 41. Good P. J., Rebbert M. L., Dawid I. B. (1993) Three new members of the RNP protein family in Xenopus. Nucleic Acids Res. 21, 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J. (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316 [DOI] [PubMed] [Google Scholar]
- 43. Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J. (2006) The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. U.S.A. 103, 15108–15112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawahara H., Imai T., Imataka H., Tsujimoto M., Matsumoto K., Okano H. (2008) Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 181, 639–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cosson B., Couturier A., Le Guellec R., Moreau J., Chabelskaya S., Zhouravleva G., Philippe M. (2002) Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol. Cell 94, 217–231 [DOI] [PubMed] [Google Scholar]
- 46. Gorgoni B., Richardson W. A., Burgess H. M., Anderson R. C., Wilkie G. S., Gautier P., Martins J. P., Brook M., Sheets M. D., Gray N. K. (2011) Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 108, 7844–7849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voeltz G. K., Ongkasuwan J., Standart N., Steitz J. A. (2001) A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 15, 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawahara H., Okada Y., Imai T., Iwanami A., Mischel P. S., Okano H. (2011) Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 286, 16121–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. (2012) Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 40, 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Battelli C., Nikopoulos G. N., Mitchell J. G., Verdi J. M. (2006) The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21(WAF-1). Mol. Cell. Neurosci. 31, 85–96 [DOI] [PubMed] [Google Scholar]
- 51. Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. (2001) The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ito T., Kwon H. Y., Zimdahl B., Congdon K. L., Blum J., Lento W. E., Zhao C., Lagoo A., Gerrard G., Foroni L., Goldman J., Goh H., Kim S. H., Kim D. W., Chuah C., Oehler V. G., Radich J. P., Jordan C. T., Reya T. (2010) Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kharas M. G., Lengner C. J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., Gozo M., Einhorn W., Lane S. W., Scholl C., Fröhling S., Fleming M., Ebert B. L., Gilliland D. G., Jaenisch R., Daley G. Q. (2010) Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. MacNicol M. C., Cragle C. E., MacNicol A. M. (2011) Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle 10, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuwako K., Kakumoto K., Imai T., Igarashi M., Hamakubo T., Sakakibara S., Tessier-Lavigne M., Okano H. J., Okano H. (2010) Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron 67, 407–421 [DOI] [PubMed] [Google Scholar]
- 56. Takahashi T., Suzuki H., Imai T., Shibata S., Tabuchi Y., Tsuchimoto K., Okano H., Hibi T. (2013) Musashi-1 post-transcriptionally enhances phosphotyrosine-binding domain-containing m-Numb protein expression in regenerating gastric mucosa. PLoS One 8, e53540. [DOI] [PMC free article] [PubMed] [Google Scholar]