Background: Interaction between PAT-4 (ILK) and UNC-112 (kindlin) is required for localization of each protein to integrin adhesions.
Results: PAT-4 missense mutations were identified that restore binding of a mutant UNC-112 protein and its proper localization.
Conclusion: Residues mutated in the suppressor mutants cluster in two regions on the surface of a homology model of PAT-4.
Significance: An interaction surface between PAT-4 and UNC-112 is suggested.
Keywords: Adhesion, C. elegans, Muscle, Protein Structure, Protein-Protein Interactions, Integrin-linked Kinase, Kindlin, Suppressor Genetics
Abstract
Caenorhabditis elegans striated muscle cells attach to basement membrane and transmit the force of muscle contraction through integrin adhesion complexes. The cytoplasmic tail of β-integrin (PAT-3) is associated with a conserved four-protein complex that includes UNC-112 (kindlin), PAT-4 (integrin-linked kinase), PAT-6 (α-parvin/actopaxin), and UNC-97 (PINCH). The proper localization of UNC-112 to muscle integrin adhesion sites requires PAT-4. A recent report (Qadota, H., Moerman, D. G., and Benian, G. M. (2012) A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (kindlin) to integrin adhesion sites. J. Biol. Chem. 287, 28537–28551) suggests a possible molecular mechanism for this requirement: that UNC-112 exists in closed inactive and open active conformations, and conversion to the open active form is promoted by binding to PAT-4 (ILK). Previously, we also reported identification of a single missense mutation in UNC-112, D382V, which abolishes both binding to PAT-4 and normal localization to integrin adhesion sites in vivo. In this report, we describe isolation and characterization of PAT-4 missense mutations that permit binding with UNC-112 D382V and place nine affected residues on a homology model of PAT-4. These nine residues cluster in two regions on the surface of PAT-4, do not overlap the likely binding surface for PAT-6 (α-parvin), and therefore may reside along the interaction surface of PAT-4 for UNC-112 (kindlin). We also show that one of these PAT-4 mutations restores the ability of UNC-112 D382V to localize to integrin adhesions and participate in complex formation.
Introduction
Integrin adhesion sites consist of the transmembrane heterodimeric protein integrin and >100 different associated proteins (2–4). These protein complexes have an important role in cell crawling; as a cell moves forward, new adhesion sites are assembled at the leading edge, and old adhesion sites are disassembled at the back end of the cell. Integrin adhesion sites are also important in transmitting the force of muscle contraction to the outside of a mammalian striated muscle cell. These cells contain many myofibrils mechanically liked by intermediate filaments, and at the periphery of these cells, myofibrils are attached to the muscle cell membrane via integrin adhesion complexes called costameres (5, 6).
In the model genetic organism Caenorhabditis elegans, the striated muscle contains a narrow zone of myofibrils adjacent to the cell membrane along the outer side of the muscle cell (7, 8). As in mammalian striated muscle, the thin filaments are attached to Z-disks (called dense bodies in nematodes), and the thick filaments are organized around M-lines. In addition, all of the dense bodies and M-lines are attached to the muscle cell membrane, and thus most of the force of muscle contraction is transmitted through these structures. The membrane-proximal regions of the dense bodies and M-lines consist of integrin and associated proteins (9–11). The cytoplasmic tail of β-integrin is associated with four conserved proteins: UNC-112 (kindlin), PAT-4 (integrin-linked kinase, ILK2), PAT-6 (actopaxin/α-parvin), and UNC-97 (PINCH). PAT-4 has been shown to interact with UNC-112, PAT-6, and UNC-97 (12–15). Each of these proteins is required for myofibril assembly in embryonic muscle (15–17).
In contrast to C. elegans, which has one kindlin (UNC-112), humans have three kindlins, each encoded by a separate gene (18). Human kindlin 1 is expressed primarily in epithelial cells such as keratinocytes and intestinal epithelial cells. Kindlin 2 is expressed everywhere except for hematopoietic cells and is the only kindlin expressed in heart muscle. Kindlin 3 is expressed in hematopoietic cells, with highest expression in megakaryocytes (which give rise to platelets). Inherited recessive mutations in kindlin 1 result in a serious skin disease (Kindler syndrome), and recessive mutations in kindlin 3 result in severe dysfunction of both platelets and leukocytes (leukocyte adhesion deficiency type III). Nematode UNC-112 is equally similar to human kindlin 1 and kindlin 2, each sharing 42% identical residues. UNC-112 is less homologous to kindlin 3; there is an internal span of about 130 residues in which there is little sequence similarity (supplemental Fig. 1).
In C. elegans, the relationship between UNC-112 and PAT-4 has been analyzed in several ways. The proper localization of UNC-112 to the base of M-lines and dense bodies requires PAT-4; the converse is also true. In addition, the protein kinase domain of PAT-4 interacts with the N-terminal half of UNC-112 (12, 13).
Our recent report (1) provides a possible molecular explanation for the PAT-4 dependence of UNC-112 localization. We demonstrated that the cytoplasmic tail of β-integrin (PAT-3) binds to UNC-112, but not to PAT-4, and that the N- and C-terminal halves of UNC-112 bind to each other. PAT-4 was shown to compete with the C-terminal half of UNC-112 for binding to the N-terminal half of UNC-112. From these protein interaction data we proposed the following model: UNC-112 exists in closed inactive and open active conformations, and conversion to the open active form is promoted by binding to PAT-4 (ILK). Support of this model came from analysis of two types of UNC-112 mutants; one type of mutant, UNC-112 D382V cannot bind to PAT-4; another type of mutant, represented by UNC-112 T346A or E349K, does not permit interaction of the N- and C-terminal halves of UNC-112. Whereas UNC-112 D382V does not localize to integrin adhesion sites (similar to deficiency of pat-4), UNC-112 double mutants (D382V,T346A or D382V,E349K) do localize properly, suggesting that detachment of the two halves of UNC-112, perhaps in an “open” conformation, favors interaction with β-integrin. In this report, we describe isolation and characterization of PAT-4 missense mutations that permit binding with UNC-112 D382V and place the affected residues on a homology model of PAT-4. We also demonstrate that one of these PAT-4 mutations restores the ability of UNC-112 D382V to localize to integrin adhesions and participate in complex formation.
EXPERIMENTAL PROCEDURES
Nematode Strains and Culture
Wild-type, strain N2, was used in these studies. Nematodes were grown at 20 °C on NGM agar plates with Escherichia coli strain OP50 (19).
Yeast Two-hybrid Assays and Plasmids
The general methods used for performing yeast two-hybrid assays were described previously (13, 20). To create UNC-112 with mutation D382V as a bait plasmid (pGBDU-UNC-112 (D382V)), an UNC-112 N-terminal cDNA fragment with the D382V mutation (digested with BamHI and BglII) was cloned into the BamHI and BglII sites of a pGBDU-UNC-112 C-terminal half plasmid. Other bait plasmids (pGBDU-UNC-97 and pGBDU-PAT-6) were described previously (13, 14).
Screening for PAT-4 Suppressor Mutations That Can Bind to UNC-112 (D382V)
We utilized random mutagenesis by PCR, as described previously (1). PAT-4 cDNAs with mutations were amplified with two primers (5′ in (GAA GAT ACC CCA CCA AAC) and 3′ in (AAA GAA GGC AAA ACG ATG)) and a PAT-4 cDNA plasmid (pDM#280) (13) as a template. An amplified cDNA and digested vector (pACT with XhoI) were co-transformed into PJ69-4A yeast strain (21) with pGBDU-UNC-112 with D382V (PAT-4 nonbinding mutation) and then transformants were spread onto −His plates; total screening number = 12,700. After 3 days incubation at 30 °C, His+ colonies were streaked onto −Ade plates. From 62 His+Ade+ colonies, clones were isolated and tested for binding to UNC-112 (D382V). Among 62 isolated clones, 40 clones retained their ability to bind to UNC-112 (D382V), and the DNA sequences of 37 of these clones were determined (Table 1).
TABLE 1.
Missense mutations in PAT-4 that permit binding to UNC-112 D382V
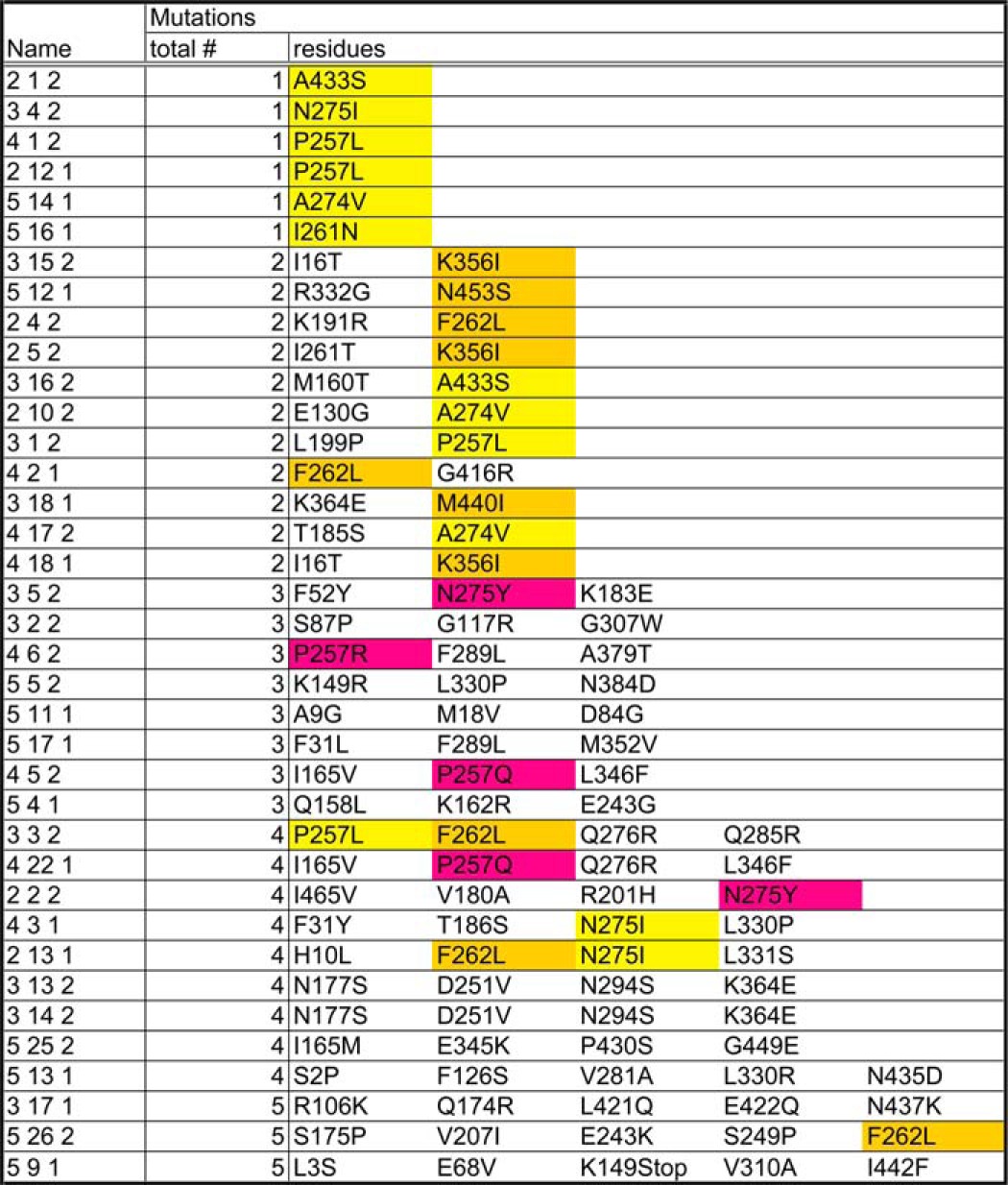
Yellow, amino acid residues affected in single PAT-4 mutants (from original screening); gold, amino acid residues affected in single PAT-4 mutants (from site-directed mutagenesis); magenta, the same amino acid residues affected in single mutants, but changed to different residues in multiply mutant PAT-4.
Homology Modeling
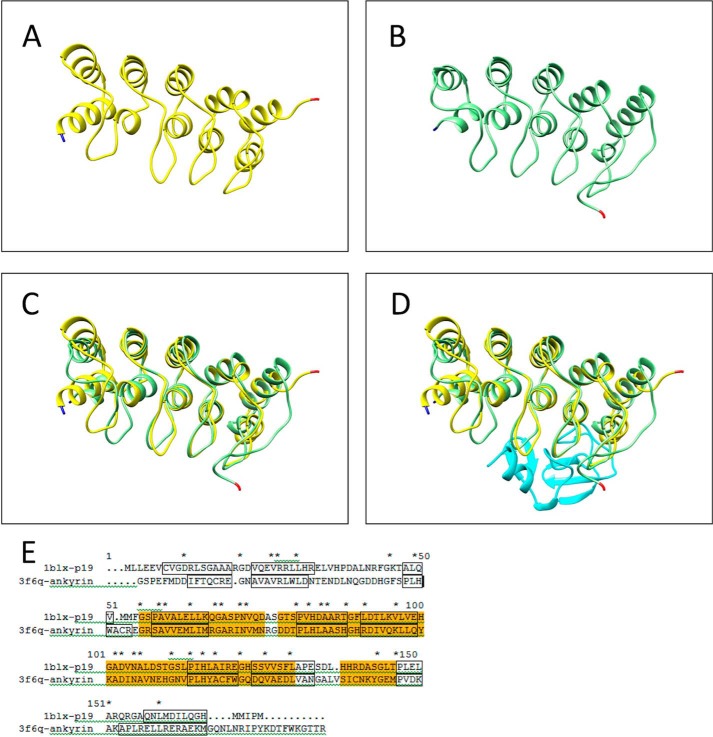
Domains within PAT-4 were defined by use of PFAM. Molecular modeling of C. elegans PAT-4 (ILK, protein kinase domain, residues 260–460) was completed using Modeler (version 9v7, University of California San Francisco (22)) for model construction and the UCSF Chimera package (version 1.6.2, University of California San Francisco (23)) for visualization, structure comparison, and analysis. The PAT-4 model was built and minimized based on the crystal structure of human ILK (Protein Data Bank ID code, 3KMW) chain A (24). Initial sequence alignment between human ILK (residues 185–452) and C. elegans PAT-4 (residues 199–464) was generated from NCBI Blast. The final alignment is shown in Fig. 1B. Overlay of the structures (C. elegans PAT-4 with α-parvin) was completed in Chimera using the Matchmaker-Structure Comparison tool (Tools for integrated sequence-structure analysis with UCSF Chimera (25)). Structural similarity of the ankyrin repeats of ILK (PAT-4) to p19INK4d was determined by the Dali Protein Structure 3D Alignment Server (26). Fig. 6 shows the structural and sequence alignment of ankyrin repeats and p19INK4d.
FIGURE 1.
Location of PAT-4 suppressor mutations, effects of these mutations on protein interactions, the sequence alignment of PAT-4 with ILK. A, on the left top is a schematic representation of PAT-4 with ankyrin repeats (ANK) in blue and pseudokinase domain (residues 210–461) in yellow, as defined by PFAM. Below are indicated the nine missense mutations with their approximate locations, all in the pseudokinase domain. Underlining indicates the 5 residues that are identical to those in human ILK. B, ClustalW alignment of the pseudokinase domains of PAT-4 with ILK used to create the homology model of PAT-4. The sequence represented for ILK is the sequence of the protein expressed and crystallized, as reported by Fukuda et al. (24). In the row between the two sequences, identical residues are indicated, and + indicates conservation. Over the span of 266 residues, 59% are identical (157 of 266). Yellow highlighting denotes the residues affected in the nine PAT-4 suppressor mutations.
FIGURE 6.
The ILK ankyrin-PINCH LIM1 complex is similar in size and shape to the cell cycle inhibitor p19INK4d. Structural similarity of the ankyrin repeats of human ILK (PAT-4) to the cell cycle inhibitor p19INK4d determined by the Dali Protein Structure 3D Alignment Server. This structural alignment provides a possible binding site for the ankyrin repeats of ILK to the protein kinase domain of ILK (PAT-4). A, structure of p19INK4d (yellow) as bound to CDK6, from crystal structure 1BLX. B, structure of the ankyrin repeats of human ILK (PAT-4) bound to the first LIM domain of PINCH (UNC-97) (light green) from crystal structure 3F6Q. C, p19INK4d and ankyrin repeats superimposed (Chimera, Matchmaker) (25). D, p19INK4d and ankyrin repeats with LIM1 domain (cyan) of PINCH (UNC-97) bound to the ankyrin repeats as in 1BLX. The N terminus and C terminus of each protein is marked with blue and red, respectively. E, sequence alignment between p19INK4d and ankyrin repeats. Helices from the structures are outlined in black, identical residues are marked with an asterisk (*). Region of high conservation is highlighted in orange (∼28% identity).
Expression of HA-tagged UNC-112 and myc-tagged PAT-4 in C. elegans Using a Heat Shock Promoter
pPD49.78/83-HA-UNC-112 (D382V) was described previously (1). XhoI fragments of PAT-4 cDNA of either wild type or from pACT clones were cloned into the SalI site of pGBDU-C2, resulting in pGBDU-PAT-4 (wild type or P257L). The SmaI-BglII fragment of pGBDU-PAT-4 (wild type or P257L) was cloned into pBS-myc (kindly provided from Dr. Kozo Kaibuchi, Nagoya University) via the SmaI and BamHI sites, resulting in pBS-myc-PAT-4 (wild type or P257L). To construct pPD49.78- or pPD49.83-myc-PAT-4 plasmids, the SpeI fragment of pBS-myc-PAT-4 (wild type or P257L) was cloned into pPD49.78 or pPD49.83 (gifts from Dr. Andy Fire, Stanford University, Palo Alto, CA) via a NheI site. These vectors contain two different heat shock promoters.
pPD49.78/83-HA-UNC-112 (D382V) and pPD49.78/83-myc-PAT-4 (WT or P257L) were mixed with pTG96 (SUR-5::NLS::GFP) as a transformation marker (27) and injected into wild-type N2 worms. Transgenic lines with extrachromosomal arrays containing pPD49.78/83-HA-UNC-112 (D382V)/pPD49.78/83-myc-PAT-4 (WT) (called sfEx47) or pPD49.78/83-HA-UNC-112 (D382V)/pPD49.78/83-myc-PAT-4 (P257L) (called sfEx50) and pTG96 were established by picking up GFP-positive worms using a GFP dissection microscope. Expression of HA-tagged UNC-112 (D382V) and myc-tagged PAT-4 (WT, P257L) proteins was induced by incubation of the transgenic worms at 30 °C for 2 h (heat shock). We prepared Laemmli-soluble protein extracts (30) from these transgenic nematodes with or without heat shock and verified the expression of HA-tagged UNC-112 and myc-tagged PAT-4 proteins by Western blotting, reacting with anti-HA (Sigma-Aldrich H3663, 1/200 dilution) and anti-myc (Sigma-Aldrich H5546, 1/200 dilution).
Immunostaining
Nematodes were fixed using the method described previously (28, 29). Antibody staining with anti-HA (Sigma Aldrich H3663, 1/200 dilution), anti-GFP (Invitrogen A11122, 1/200 dilution) and anti-UNC-95 (1/100 dilution) were performed as described previously (30). Secondary antibodies were anti-rabbit Alexa Fluor 488 (Invitrogen) for anti-UNC-95 and anti-GFP, and anti-mouse Alexa Fluor 594 (Invitrogen) for anti-HA, each used at 1/200 dilution. Stained samples were mounted on a glass slide with a coverslip containing mounting solution (20 mm Tris (pH 8.0), 0.2 m DABCO, and 90% glycerol). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat ×63/1.4 oil objective, in 2.5× zoom mode. The color balances of the images were adjusted by using Adobe Photoshop.
Integration of Extrachromosomal Arrays
The extrachromosomal arrays containing pPD49.78/83-HA-UNC-112 (D382V), pPD49.78/83-myc-PAT-4 (WT or P257L), and pTG96 were integrated into the genome by UV irradiation (31) with some modifications.3 The resulting integrated nematode lines are called sfIs8, for HA-UNC-112 (D382V) and myc-PAT-4 (WT); sfIs10, for HA-UNC-112 (D382V) and myc-PAT-4 (P257L).
Detection of Complex Formation from Worm Lysates
Nematode-integrated transgenic lines sfIs3 and sfIs5 (1) were induced to express HA-tagged UNC-112 wild type or UNC-112 D382V, respectively, by heat shocking mostly adult animals by exposure to 30 °C for 3 h, and worm lysates were prepared as described by Qadota et al. (1). To 500 μl of each worm lysate was added 50 μl of a 1:1 slurry of monoclonal anti-HA-agarose beads (Sigma-Aldrich A2095) and mixing at 4 °C for 2 h. The beads were pelleted and washed three times with lysis buffer, transferred to new tubes, pelleted, and as much liquid was removed as possible. The proteins from these beads were eluted with 33 μl of 2× Laemmli sample buffer, and portions were separated on 10% SDS-polyacrylamide gels, and blotted. Separate lanes were reacted with the following antibodies: rabbit anti-HA (Sigma-Aldrich H6908) at 1/200 dilution; rabbit anti-PAT-4 (1), affinity-purified, at 1/100; rat anti-PAT-6 (32), affinity-purified, at 1/200 dilution; and rabbit anti-UNC-97 (20), affinity-purified, at 1/100. Reactions were detected by ECL.
Co-immunoprecipitation of HA-UNC-112 and myc-PAT-4
Transgenic lines carrying integrated arrays described above were induced to express HA-tagged UNC-112 D382V and either myc-tagged PAT-4 WT (sfIs8) or P257L (sfIs10) by heat shock, and worm lysates were prepared as described above. Immunoprecipitation using anti-HA beads and gel analysis was carried out as described in the previous section. Separate lanes were reacted with rabbit anti-HA (Sigma-Aldrich H6908) at 1/200 dilution, and rabbit anti-myc (Sigma-Aldrich H4439) at 1/200 dilution. Reactions were detected by ECL.
Calculation of Molar Ratios of PAT-4 to UNC-112 in Co-IP Experiments
Varying amounts (in nanograms) of recombinant GST-3×HA-myc (GST fused to three copies of HA and one copy of myc) on Western blots were reacted with either anti-HA or anti-myc, using the same concentration of antibodies and ECL exposure times as used in the immunoprecipitation experiments. Similarly, varying amounts of recombinant MBP-PAT-4 (1) on Western blots were reacted with anti-PAT-4. The intensity of ECL bands obtained from immunoprecipitations was matched with intensity of ECL bands obtained with known quantities of recombinant proteins. Molar ratios of PAT-4 to UNC-112 in the immunoprecipitation experiments were then calculated using these estimates of quantities of proteins in nanograms and their known molecular masses.
RESULTS
To obtain further information about the interaction between UNC-112 and PAT-4, we isolated PAT-4 mutants that could bind to UNC-112 D382V. Previously, we demonstrated that although wild-type UNC-112 binds to PAT-4, UNC-112 D382V fails to bind to PAT-4 (1). Using random PCR mutagenesis and the yeast two-hybrid system, we identified 37 PAT-4 mutants that can interact with UNC-112 D382V (Table 1). DNA sequencing revealed that 6 of the 37 PAT-4 mutants contained single amino acid changes, with one of them appearing twice independently (Table 1 and Fig. 1). An additional set of 4 single amino acid changes was found to be responsible for suppression by testing individual mutated residues from clones that had 2 mutations. Interestingly, although we mutagenized the entire pat-4 cDNA, all 9 single amino acid changes reside in the protein kinase domain of PAT-4, the region of PAT-4 demonstrated previously to be sufficient for interaction with UNC-112 (13). All 9 of these PAT-4 mutants also bind to wild-type UNC-112, suggesting that the structural changes in these mutants are minimal. PAT-4 also interacts with UNC-97 and PAT-6 (13, 14). Significantly, the mutations do not affect binding of PAT-4 to UNC-97 or PAT-6 (data not shown), further indicating the unique effect the mutations have on UNC-112 D382V binding, suggesting localization on the UNC-112 binding surface.
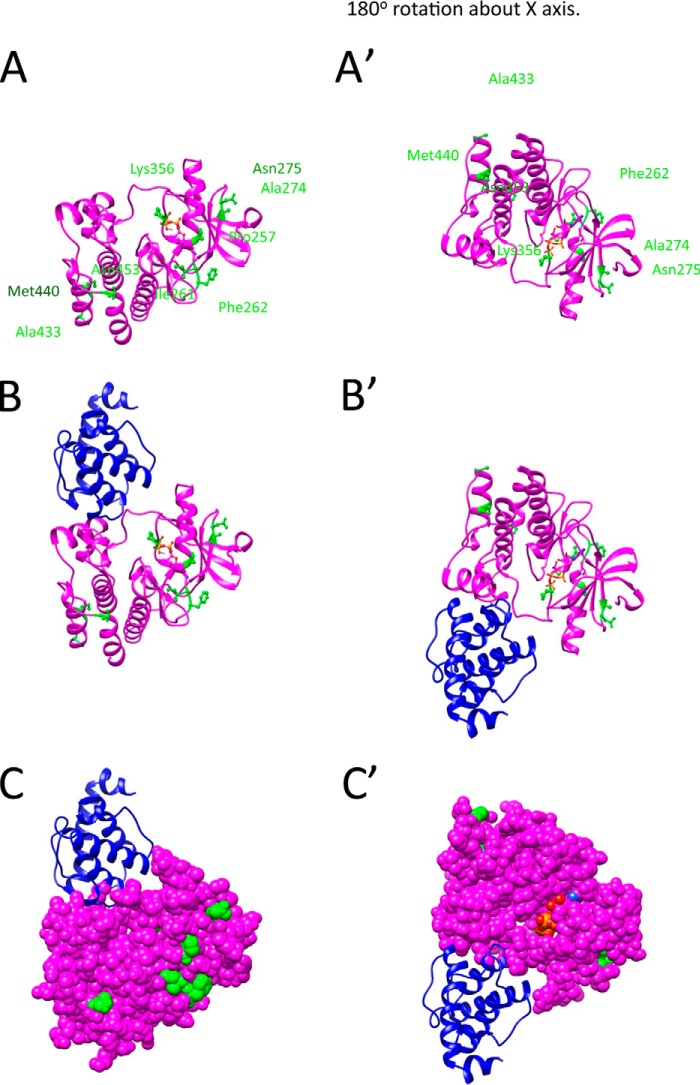
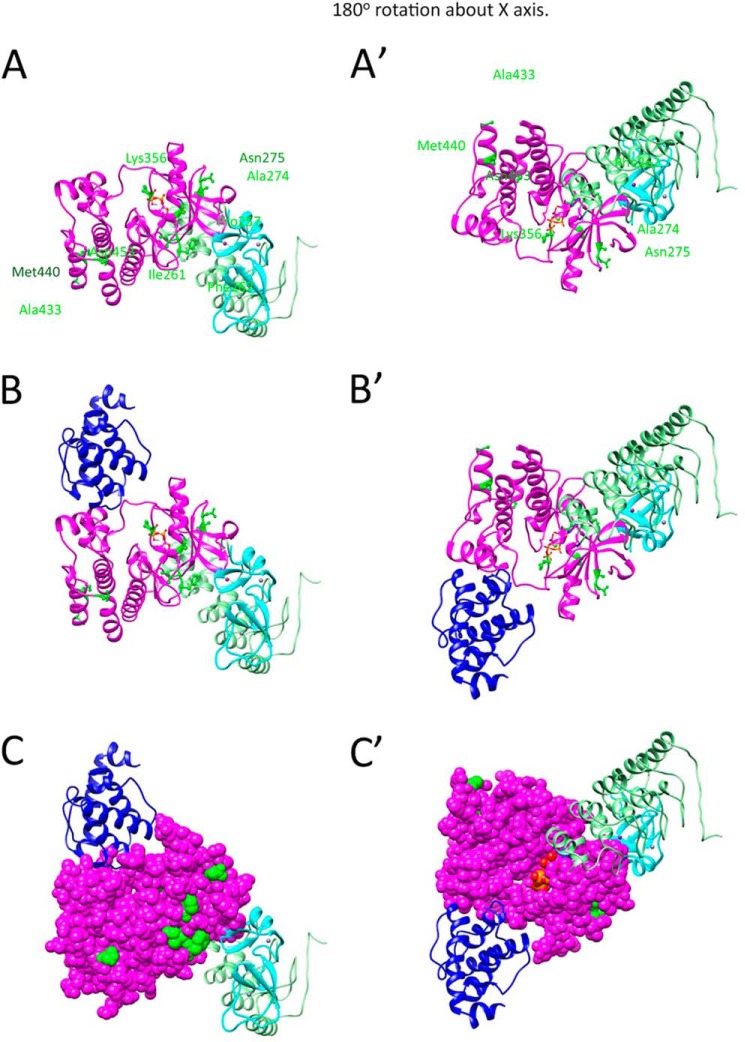
Fukuda et al. (24) reported the crystal structure of human ILK kinase domain complexed with α-parvin. As shown in Fig. 1B, the pseudokinase domains of human ILK and C. elegans PAT-4 have extensive similarity, with 59% identical residues (157 of 266). Of the 9 PAT-4 suppressor mutations (indicated by yellow in Fig. 1B), 5 reside at conserved residues. (These conserved residues are underlined in Fig. 1A.) We used the crystal structure of human ILK to create a homology model of nematode PAT-4 kinase domain, and located the 9 residues mutated in our PAT-4 mutations on this structure. As shown in Fig. 2A, the suppressor mutations cluster in two regions on the surface of the PAT-4 homology model. Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, and Lys-356 cluster near the edge of the β-sheet and helix 1 in the N-terminal subdomain of the kinase, and Ala-433, Met-440, and Asn-453 cluster in the C-terminal helices of the C-terminal subdomain of the kinase. Fig. 2B shows the modeled PAT-4 together with the attached human α-parvin (blue). Note that α-parvin (PAT-6 ortholog) does not overlap or cover either cluster of suppressor residues on PAT-4. This is even more apparent when PAT-4 is viewed in space filling mode (Fig. 2C). The residues mutated in the suppressors are available to form a binding surface for another protein, perhaps UNC-112. That UNC-112 and PAT-6 bind along different surfaces of PAT-4 is consistent with the previously reported ternary complex of UNC-112, PAT-4, and PAT-6 (14).
FIGURE 2.
Homology model of PAT-4 with location of suppressor mutations. A, model of PAT-4 based on the crystal structure of human ILK is shown as a magenta ribbon. The positions of PAT-4 suppressor mutations are shown in green balls and sticks. ATP is shown colored by atom color in stick form. The view on the right was created by rotating the view on the left by 180° around the x axis. Note that the suppressor mutations cluster in two regions on the surface of PAT-4. Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, and Lys-356 cluster near the edge of the β-sheet and helix 1 in the N-terminal domain. Ala-433, Met-440, and Asn-453 cluster at the C-terminal helices. B, two views show the homology model of PAT-4 kinase with α-parvin (PAT-6 in nematodes). PAT-4 was substituted for human ILK in the human ILK·α-parvin complex (Protein Data Bank ID code 3KMW) (24). PAT-4 is shown in magenta, and α-parvin (PAT-6) is shown in dark blue. Note that α-parvin does not overlap or cover either cluster of the suppressor mutations. They are available to form another binding surface. C, two views show of the same complex as in B with PAT-4 in space fill mode. The suppressor mutations are shown in green. Note that a majority of the suppressor mutations (Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, Lys-356, and Asn-453) are on the far surface from α-parvin (PAT-6).
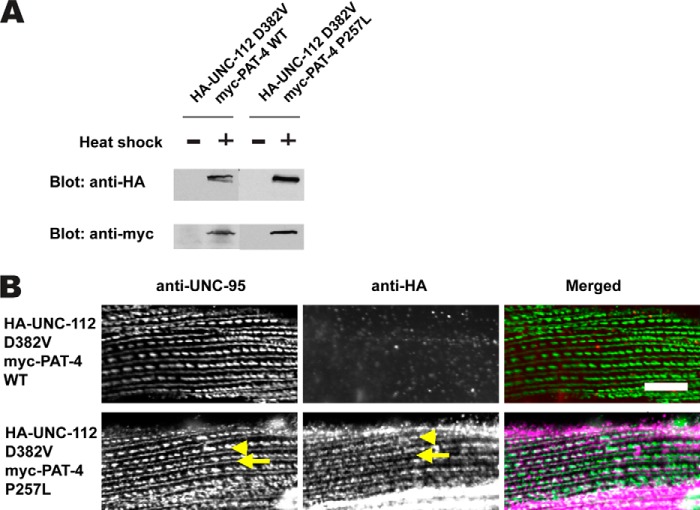
To examine the in vivo significance of the ability of mutant PAT-4 proteins to interact with UNC-112 D382V, we created transgenic lines expressing HA-tagged UNC-112 D382V and either myc-tagged wild-type PAT-4 or myc-tagged P257L mutant PAT-4. P257L was chosen because, as noted above, it is one of the five mutations that reside on a potential binding surface for UNC-112 and is also conserved in human PAT-4. The transgenic lines were designed for expression of the two tagged proteins by a heat shock promoter in a wild-type background. As shown in Fig. 3A, for each line, both the HA-tagged UNC-112 and myc-tagged PAT-4 are expressed only upon heat shock. Previously, we have shown that expression of HA-UNC-112 D382V in a wild-type background does not allow localization of the tagged protein to muscle integrin adhesion sites (dense bodies and M-lines) (1). Consistent with this result, even in the presence of ectopic myc-tagged wild-type PAT-4, HA-UNC-112 D382V fails to localize to these sites (Fig. 3B, upper row). However, co-expression of myc-tagged PAT-4 P257L allows localization to these structures (Fig. 3B, lower row). These results indicate that PAT-4 P257L binds to UNC-112 D382V, and this is consistent with our model that PAT-4 binding to UNC-112 is required for the localization of UNC-112 to integrin adhesion sites (Ref. 1 and see Fig. 5).
FIGURE 3.
Co-expression of HA-UNC-112 D382V and myc-PAT-4 P257L restores ability of UNC-112 D382V to localize to integrin adhesion sites. A, Western blot of lysates from transgenic worms expressing from a heat shock promoter HA-tagged UNC-112 D382V and either myc-tagged PAT-4 wild-type (WT) or PAT-4 P257L. Lysates were prepared after heat shock (+) or in the absence of heat shock (−) and reacted with anti-HA or anti-myc antibodies. Expression of the HA- and myc-tagged proteins depends on heat shock. B, localization of heat shock-expressed HA-tagged UNC-112 D382V in the presence of co-expressed myc-tagged PAT-4 wild type, or myc-tagged PAT-4 P257L. Adult worms were immunostained with anti-HA to detect transgenic UNC-112 and with anti-UNC-95 to visualize the optical plane of body wall muscle cells that contain dense bodies and M-lines (integrin adhesion sites). HA-UNC-112 D382V fails to localize to these structures in the presence of myc-PAT-4 WT but does localize to these structures in the presence of myc-PAT-4 P257L. Yellow arrowheads mark dense bodies, whereas yellow arrows mark M-lines.
FIGURE 5.
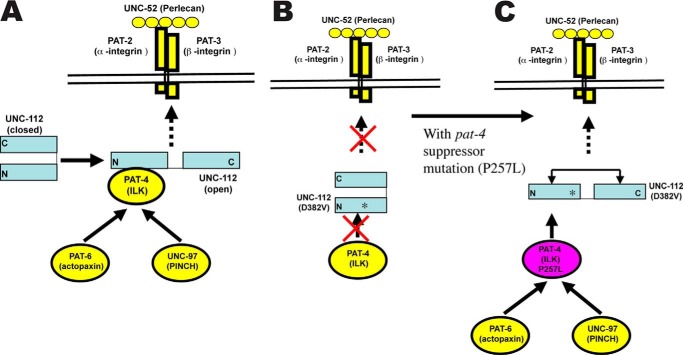
Model of interactions. A, as suggested in previous report (1), UNC-112 exists in closed inactive and open active conformations in its ability to bind to the cytoplasmic tail of PAT-3 (β-integrin). Transition to the open state is promoted by binding to PAT-4. As shown here, UNC-112 exists in a complex together with PAT-4, PAT-6, and UNC-97. B, a mutant form of UNC-112, D382V, which has reduced ability to interact with PAT-4, fails to localize to the base of integrin adhesion sites. C, compensating mutations in PAT-4 (such as P257L) permit binding between mutant PAT-4 and UNC-112 D382V and permit UNC-112 D382V to localize to the base of integrin adhesion sites. It is speculated that the interaction of PAT-4 P257L causes an opening up of UNC-112 D382V and thus binding to the cytoplasmic tail of PAT-3 (β-integrin).
An additional function of PAT-4 is to recruit PAT-6 (α-parvin) and UNC-97 (PINCH) to integrin adhesion sites. Previously, it was reported that UNC-112 binds to PAT-4 (13), that PAT-4 binds to PAT-6 (14), and that PAT-4 binds to UNC-97 (13, 15). In addition, yeast three-hybrid assays suggest ternary complexes in the following combinations: UNC-112, PAT-4, PAT-6 (14); UNC-112, PAT-4, UNC-97 (15); and UNC-97, PAT-4, PAT-6 (15). From these data, we hypothesized the existence of a four-protein complex (Refs. 10, 30 and see Fig. 5). To obtain in vivo evidence for this four-protein complex, we used a co-immunoprecipitation approach from lysates of worms expressing HA-tagged UNC-112. Anti-HA beads were used to immunoprecipitate HA-UNC-112, and Western blotting was used to detect the presence of endogenous PAT-4, PAT-6, and UNC-97. When the experiment was conducted with HA-UNC-112 WT, PAT-4, PAT-6, and UNC-97 could be detected in the precipitate (Fig. 4A, left). However, when HA-UNC-112 D382V was precipitated, less PAT-4 was detected, and neither PAT-6 nor UNC-97 was detected (Fig. 4A, right). The difference in the amounts of endogenous PAT-4 co-immunoprecipitated by WT versus D382V UNC-112 can be compared by calculating the molar ratios using a method described under “Experimental Procedures.” As indicated on Fig. 4A, there is a 10-fold difference in these molar ratios. These results are consistent with the model that in nematode muscle, UNC-112 binds to PAT-4, and that PAT-4 binds to PAT-6 and UNC-97, and forms a stable four-protein complex (Fig. 5A).
FIGURE 4.
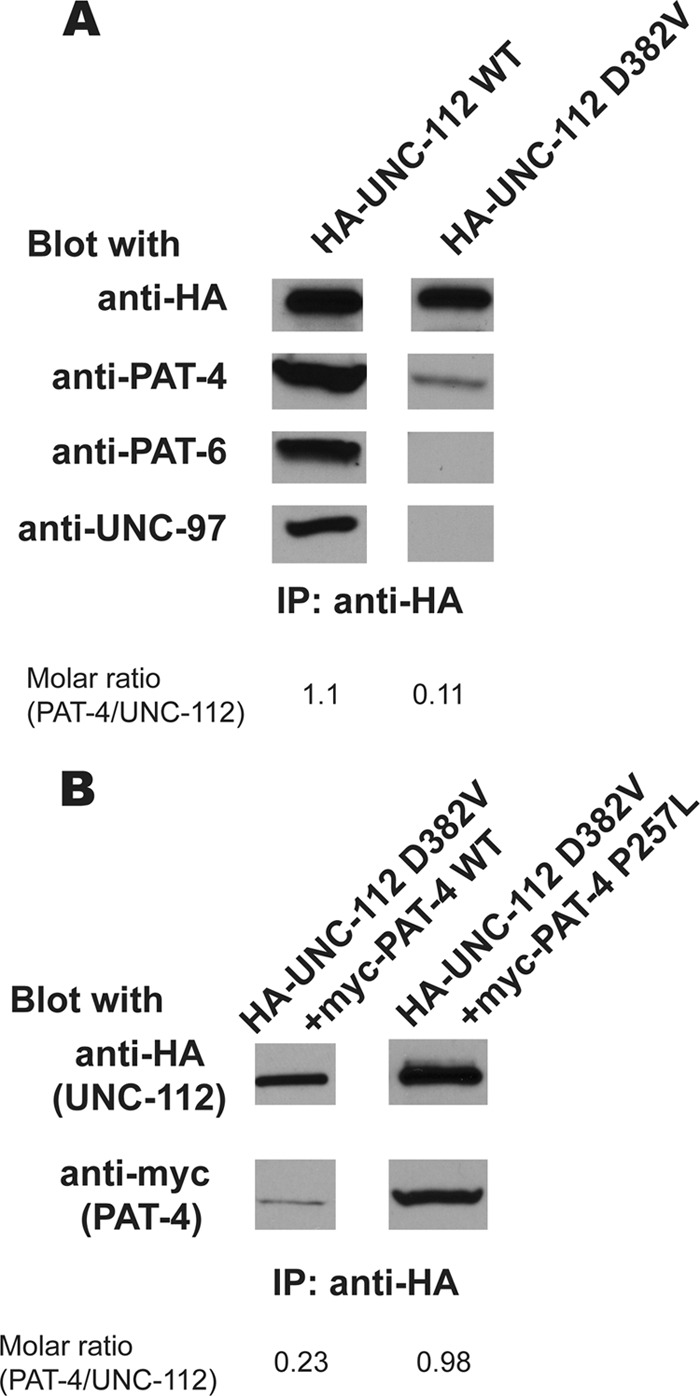
Demonstration of an UNC-112·PAT-4 complex, its reduction by UNC-112 D382V, and its restoration by PAT-4 P257L. A, demonstration of an UNC-112·PAT-4·PAT-6·UNC-97 complex. Its formation depends on the interaction of UNC-112 with PAT-4. HA-tagged UNC-112 wild-type (WT) or HA-tagged UNC-112 D382V proteins were immunoprecipitated (IP) from transgenic worms using anti-HA-coupled beads. After washing, eluted proteins were tested by immunoblotting for the presence of HA-UNC-112 and endogenous PAT-4, PAT-6, and UNC-97 proteins. Note that a complex of the four proteins can be detected with HA-UNC-112 WT, but not with HA-UNC-112 D382V. Molar ratios of the amount of endogenous PAT-4 co-immunoprecipitated with HA-UNC-112, using either WT or D382V UNC-112, are indicated. B, HA-UNC-112 D382V co-precipitating relatively more myc-PAT-4 P257L than myc-PAT-4 wild type. Lysates were prepared from transgenic worms co-expressing HA-UNC-112 D382V plus myc-PAT-4 WT, or HA-UNC-112 D382V plus myc-PAT-4 P257L, and the HA-tagged proteins were immunoprecipitated using anti-HA-coupled beads. After washing, eluted proteins were tested by immunoblotting for the presence of myc-PAT-4. Molar ratios of the amount of myc-PAT-4 WT or myc-PAT-4 P257L co-immunoprecipitated with HA-UNC-112 D382V are indicated.
We next examined the ability of PAT-4 P257L to restore the ability of UNC-112 D382V to bind to PAT-4 in a similar co-immunoprecipitation experiment. Protein lysates were prepared from transgenic animals co-expressing either HA-UNC-112 D382V and myc-PAT-4 WT, or HA-UNC-112 D382V and myc-PAT-4 P257L, and immunoprecipitated with anti-HA. The UNC-112 D382V was detected with anti-HA and the co-precipitated PAT-4 detected with anti-myc. As shown in Fig. 4B, HA-UNC-112 D382V could precipitate four times more myc-PAT-4 P257L than myc-PAT-4 WT, when molar ratios were compared. Therefore, PAT-4 P257L that restores binding to UNC-112 D382V in a yeast two-hybrid assay and restores ability of UNC-112 D382V to localize to integrin adhesion sites also restores the ability to bind to UNC-112 D382V in an in vivo protein complex.
DISCUSSION
Beginning with a mutant version (D382V) of UNC-112 that cannot bind to wild-type PAT-4, we isolated missense mutants of PAT-4 that can bind to this mutant UNC-112. Of 37 mutant clones isolated, 5 of them had single amino acid changes. The others had multiple changes (2–5 mutations each). Site-directed mutagenesis permitted us to identify the single missense mutations responsible for the phenotype in 4 additional mutant clones. All together, we identified 9 single missense mutations that permit PAT-4 to bind to UNC-112 D382V (Fig. 1). All 9 of these single amino acid changes reside in the protein kinase domain of PAT-4, the region of PAT-4 shown previously to be required for binding to UNC-112. All 9 residues affected by the suppressor mutations cluster in two regions on the surface of a homology model structure of PAT-4, based on the crystal structure of its human ortholog, ILK. The reported ILK structure is actually a complex of ILK bound to α-parvin, the ortholog of nematode PAT-6. Significantly, when PAT-4 was substituted for ILK in the ILK·α-parvin complex, α-parvin does not overlap or cover either cluster of suppressor residues on PAT-4. Thus, we suggest that the surfaces of PAT-4 on which the suppressor mutations reside constitute an independent binding surface for UNC-112 (Fig. 2).
Although it has been reported that a UNC-112·PAT-4·PAT-6 ternary complex can form, our structural insight suggests that a single molecule of PAT-4 can interact simultaneously with one molecule each of PAT-6 and UNC-112. Verification of this model awaits co-crystallization of PAT-4 with UNC-112.
Our previous data support a model in which one of the functions of the interaction of PAT-4 with UNC-112 is to promote opening up of UNC-112 so that it can bind to the cytoplasmic tail of PAT-3 (β-integrin) (1). In agreement with this model, we have shown here that in vivo, expression of one of the PAT-4 missense mutants permits UNC-112 D382V to localize to integrin adhesion sites (Fig. 3B). Previous reports also suggest that another function of the interaction of PAT-4 with UNC-112 is to recruit PAT-6 and UNC-97. Here, we show that the immunoprecipitation of HA-tagged UNC-112 from nematode lysates co-precipitates PAT-4, PAT-6, and UNC-97 (Fig. 4A). This is the first evidence of formation of this four-protein complex in vivo. In contrast, immunoprecipitation of HA-tagged UNC-112 D382V co-precipitates 10-fold less PAT-4 and no detectable PAT-6 or UNC-97. However, co-expression of UNC-112 D382V together with one of the PAT-4 missense mutants (P257L) restores UNC-112·PAT-4 complex formation. The recruitment of UNC-97 and PAT-6 by PAT-4 is essential for formation of huge protein complexes at the base of the integrin adhesion sites. We have already described that downstream of UNC-97 there are proteins that bind directly or indirectly to myosin at the M-line (20, 30). For example, UNC-97 interacts with UNC-98, and UNC-98 interacts with myosin (20); UNC-97 interacts with LIM-9 (FHL), LIM-9 interacts with UNC-96, and UNC-96 interacts with myosin (30). Recently, we have described that downstream of PAT-6 is CPNA-1, which links to UNC-89 and several other M-line proteins (32). At this time, we have not yet identified proteins that reside downstream of either UNC-97 or PAT-6 exclusively at the dense bodies.
Chiswell et al. (33) have reported the crystal structure of the ankyrin repeats of human ILK (PAT-4) bound to the first LIM domain (of five total LIM domains) of PINCH (UNC-97). We have noticed that the ILK ankyrin-PINCH LIM1 complex is similar in size and shape to the cell cycle inhibitor p19INK4d (34) (Fig. 6). Actually, the reported structure is of p19INK4d bound to cyclin D-dependent kinase Cdk6. Although there is no evidence that the ankyrin repeats of ILK (PAT-4) are bound to the kinase domain of ILK (PAT-4), we thought it would be reasonable to substitute our homology model of PAT-4 kinase for CDK6 and the ankyrin-PINCH LIM1 complex for p19INK4d (Fig. 7A). Note that the ankyrin-PINCH LIM1 complex (and thus orthologous structures from PAT-4 and UNC-97) do not overlap or cover the sites of the PAT-4 suppressor mutations. However, the LIM1 domain of PINCH (UNC-97) is “near” the suppressor mutation residue Phe-262. When α-parvin (PAT-6) is docked back onto this model of PAT-4, we note that 7 of 9 of the residues mutated in the suppressor mutants (Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, Lys-356, and Asn-453) are on the surface between α-parvin and PINCH, on the face opposite from the binding site of the ankyrin repeats (Fig. 7, B and C). Therefore, even with these additional proteins bound to PAT-4, the binding surface defined by the PAT-4 suppressor mutations will be unobstructed to bind to UNC-112. Again, all of this speculation awaits confirmation from PAT-4·UNC-112 and larger complex crystal structures.
FIGURE 7.
Residues involved in PAT-4 suppressor mutations on the surface of a homology model of PAT-4 are unobstructed after PAT-4 has bound to α-parvin (PAT-6) and to an ankyrin-LIM domain complex. A, two views of a model of PAT-4 (ILK) (magenta) with ankyrin repeats of ILK (light green) and PINCH LIM1 (light blue). These models were generated by substituting the ankyrin-PINCH LIM1 structure from 3F6Q (33) for the inhibitor in the human CDK6/p19INK4D structure (34). Note that ankyrin repeats-PINCH LIM1 complex does not overlap or cover the sites of the PAT-4 suppressor mutations. However, the LIM1 domain of PINCH is near the suppressor mutation Phe-262. B, two views of the composite model: PAT-4 (magenta), parvin (3KMW, dark blue), ankyrin repeats (3F6Q, light green) and PINCH LIM1 (3F6Q, light blue). Note that ankyrin-PINCH LIM1 binds on the opposite surface of PAT-4 as parvin. C, two views of the same complex shown in B, with PAT-4 in space fill mode. The suppressor mutations are shown in green. Note that a majority of the suppressor mutations (Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, Lys-356, and Asn-453) are on the surface between the parvin and PINCH LIM1, on the face opposite the ankyrin repeats.
Acknowledgments
We thank Andy Fire (Stanford University) for C. elegans expression vectors and Kozo Kaibuchi (Nagoya University) for bacterial and yeast expression vectors.
This study was supported by American Heart Association Grant 11GRNT7820000.

This article contains supplemental Fig. 1.
P. Barrett, personal communication.
- ILK
- integrin-linked kinase.
REFERENCES
- 1. Qadota H., Moerman D. G., Benian G. M. (2012) A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (kindlin) to integrin adhesion sites. J. Biol. Chem. 287, 28537–28551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zamir E., Geiger B. (2001) Components of cell-matrix adhesions. J. Cell Sci. 114, 3577–3579 [DOI] [PubMed] [Google Scholar]
- 3. Zamir E., Geiger B. (2001) Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114, 3583–3590 [DOI] [PubMed] [Google Scholar]
- 4. Lo S. H. (2006) Focal adhesions: what's new inside. Dev. Biol. 294, 280–291 [DOI] [PubMed] [Google Scholar]
- 5. Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. (2002) Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 18, 637–706 [DOI] [PubMed] [Google Scholar]
- 6. Ervasti J. M. (2003) Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 278, 13591–13594 [DOI] [PubMed] [Google Scholar]
- 7. Francis G. R., Waterston R. H. (1985) Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101, 1532–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waterston R. H. (1988) in The Nematode Caenorhabditis elegans (Wood W. B., ed) pp. 281–335, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 9. Moerman D. G., Williams B. D. (2006) in WormBook (the C. elegans Research Community, ed) / 10.1895/wormbook, 1.81.118050483 [DOI] [Google Scholar]
- 10. Qadota H., Benian G. M. (2010) Molecular structure of sarcomere-to-membrane attachment at M-lines in C. elegans muscle. J. Biomed. Biotechnol. 2010, 864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Warner A., Qadota H., Benian G. M., Vogl A. W., Moerman D. G. (2011) The Caenorhabditis elegans paxillin ortholog, PXL-1, is required for pharyngeal muscle contraction and for viability. Mol. Biol. Cell 22, 2551–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rogalski T. M., Mullen G. P., Gilbert M. M., Williams B. D., Moerman D. G. (2000) The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J. Cell Biol. 150, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G., Williams B. D. (2002) C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 12, 787–797 [DOI] [PubMed] [Google Scholar]
- 14. Lin X., Qadota H., Moerman D. G., Williams B. D. (2003) C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr. Biol. 13, 922–932 [DOI] [PubMed] [Google Scholar]
- 15. Norman K. R., Cordes S., Qadota H., Rahmani P., Moerman D. G. (2007) UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev. Biol. 309, 45–55 [DOI] [PubMed] [Google Scholar]
- 16. Williams B. D., Waterston R. H. (1994) Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124, 475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hobert O., Moerman D. G., Clark K. A., Beckerle M. C., Ruvkun G. (1999) A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J. Cell Biol. 144, 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meves A., Stremmel C., Gottschalk K., Fässler R. (2009) The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol. 19, 504–513 [DOI] [PubMed] [Google Scholar]
- 19. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller R. K., Qadota H., Landsverk M. L., Mercer K. B., Epstein H. F., Benian G. M. (2006) UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J. Cell Biol. 175, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) Curr. Proto. Bioinform. 15, 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Fukuda K., Gupta S., Chen K., Wu C., Qin J. (2009) The pseudoactive site of ILK is essential for its binding to α-parvin and localization to focal adhesions. Mol. Cell 36, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meng E. C., Pettersen E. F., Couch G. S., Huang C. C., Ferrin T. E. (2006) Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yochem J., Gu T., Han M. (1998) A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149, 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nonet M. L., Grundahl K., Meyer B. J., Rand J. B. (1993) Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73, 1291–1305 [DOI] [PubMed] [Google Scholar]
- 29. Wilson K. J., Qadota H., Benian G. M. (2012) Immunofluorescent localization of proteins in Caenorhabditis elegans muscle. Methods Mol. Biol. 798, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qadota H., Mercer K. B., Miller R. K., Kaibuchi K., Benian G. M. (2007) Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell 18, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitani S. (1995) Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev. Growth Differ. 37, 551–557 [DOI] [PubMed] [Google Scholar]
- 32. Warner A., Xiong G., Qadota H., Rogalski T., Vogl A. W., Moerman D. G., Benian G. M. (2013) CPNA-1, a copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans. Mol. Biol. Cell 24, 601–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiswell B. P., Zhang R., Murphy J. W., Boggon T. J., Calderwood D. A. (2008) The structural basis of integrin-linked kinase-PINCH interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 20677–20682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brotherton D. H., Dhanaraj V., Wick S., Brizuela L., Domaille P. J., Volyanik E., Xu X., Parisini E., Smith B. O., Archer S. J., Serrano M., Brenner S. L., Blundell T. L., Laue E. D. (1998) Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature 395, 244–250 [DOI] [PubMed] [Google Scholar]