Background: Regulation of microRNA activity independent of processing and biogenesis has not been demonstrated.
Results: The RNA-binding protein, TDP-43, interacts with mature miR-1/miR-206, limiting their RNA-induced silencing complex (RISC) association and activity.
Conclusion: RNA-binding proteins can selectively control microRNA activity by disrupting RISC incorporation.
Significance: This is the first known microRNA-protein interaction that controls microRNA activity independent of processing.
Keywords: Cardiac muscle, Gene Regulation, MicroRNA, RNA-binding Protein, RNA-Protein Interaction, Skeletal Muscle
Abstract
MicroRNA (miRNA) maturation is regulated by interaction of particular miRNA precursors with specific RNA-binding proteins. Following their biogenesis, mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) where they interact with mRNAs to negatively regulate protein production. However, little is known about how mature miRNAs are regulated at the level of their activity. To address this, we screened for proteins differentially bound to the mature form of the miR-1 or miR-133 miRNA families. These muscle-enriched, co-transcribed miRNA pairs cooperate to suppress smooth muscle gene expression in the heart. However, they also have opposing roles, with the miR-1 family, composed of miR-1 and miR-206, promoting myogenic differentiation, whereas miR-133 maintains the progenitor state. Here, we describe a physical interaction between TDP-43, an RNA-binding protein that forms aggregates in the neuromuscular disease, amyotrophic lateral sclerosis, and the miR-1, but not miR-133, family. Deficiency of the TDP-43 Drosophila ortholog enhanced dmiR-1 activity in vivo. In mammalian cells, TDP-43 limited the activity of both miR-1 and miR-206, but not the miR-133 family, by disrupting their RISC association. Consistent with TDP-43 dampening miR-1/206 activity, protein levels of the miR-1/206 targets, IGF-1 and HDAC4, were elevated in TDP-43 transgenic mouse muscle. This occurred without corresponding Igf-1 or Hdac4 mRNA increases and despite higher miR-1 and miR-206 expression. Our findings reveal that TDP-43 negatively regulates the activity of the miR-1 family of miRNAs by limiting their bioavailability for RISC loading and suggest a processing-independent mechanism for differential regulation of miRNA activity.
Introduction
MicroRNA (miRNA)2 biogenesis involves a series of processing steps, many of which are regulated by interaction of precursor forms of the miRNA with specific RNA-binding proteins. Of the over 1,000 miRNAs encoded in mammalian genomes, the majority are derived from primary microRNAs that encode two or more mature miRNAs within a common transcript. Although these polycistronic miRNAs share common transcriptional regulation, they often have varying, and sometimes even opposing, effects on cellular biology. How cells differentially regulate the activity of mature forms of co-transcribed miRNAs to achieve specific biological outcomes remains unknown. In particular, whether interaction of RNA-binding proteins with mature miRNAs regulates their activity has not been investigated.
Muscle development and homeostasis are regulated, in part, by polycistronic microRNAs. The miR-1 family, composed of miR-1 and miR-206, whose mature sequences are nearly identical, and the miR-133 family (1, 2) are highly conserved and are enriched in cardiac and skeletal muscle in species as distantly related as flies and humans (3–5) (see Fig. 1A). In mammals, up to three genomic loci encode bicistronic transcripts to produce miR-133 and either miR-1 or miR-206. In most higher vertebrates, the miR-1/miR-133a genomic locus was duplicated, with the expression of both loci being maintained in cardiac and skeletal muscle. A third locus encodes miR-206 and miR-133b and is uniquely expressed in skeletal, not cardiac, muscle.
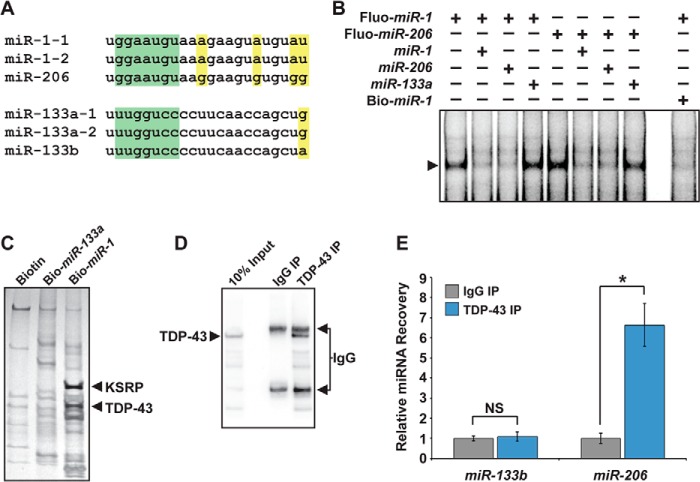
FIGURE 1.
TDP-43 interacts with the miR-1/miR-206 family, but not miR-133. A, sequence alignment of the miR-1/miR-206 family or the miR-133 family. miRNA seed sequence is highlighted in green. Residues that differ among family members are highlighted in yellow. B, EMSAs revealed an miRNA-protein complex (arrow) in undifferentiated C2C12 cell lysate that interacted with fluorescently labeled miR-1 or miR-206 probe (Fluo-miR-1, Fluo-miR-206). These interactions could be competed with unlabeled miR-1 or miR-206, but not with unlabeled miR-133a. 5′-biotinylated miR-1 (Bio-miR-1) also effectively competed for binding. C, eluates from negative control (Biotin), Bio-miR-133a, or Bio-miR-1 pulldowns were run on denaturing gels. Proteins enriched in the Bio-miR-1 pulldown lane were identified by mass spectrometry and included KSRP and TDP-43. D, Western analysis detecting TDP-43 protein following cross-linking and immunoprecipitation from C2C12 skeletal myoblasts using α-TDP-43 (TDP-43 IP), rabbit IgG (IgG IP), or 10% input showed efficient and specific recovery of TDP-43 protein. E, qRT-PCR to detect miR-206 and miR-133b after TDP-43 cross-linking and immunoprecipitation performed in D revealed preferential interaction of TDP-43 with miR-206 versus miR-133b. Results were normalized to pulldown with IgG (NS, not significant; *, p < 0.05).
Targeted deletions of miR-1 or miR-133 in mice generally result in impaired cardiac development and function (6–11). Although miR-1 and miR-133 cooperate to repress smooth muscle gene expression in the heart (6, 7, 10, 11), miR-1 promotes differentiation of striated muscle progenitors, whereas miR-133 maintains the undifferentiated state in vitro (5, 12, 13). Deletion of one miR-1 locus or both miR-1 loci causes cardiac defects without a detectable skeletal muscle phenotype, likely due to the persistent expression of miR-206 in skeletal muscle (7, 8, 11). Targeted deletion of miR-206 alone does not disrupt muscle development or function in mice, but prevents efficient regeneration of neuromuscular junctions (NMJs) after acute nerve injury, leading to severe muscle loss (9). Deficiency of miR-206 in the SOD-1 mouse model of amyotrophic lateral sclerosis (ALS) also accelerates disease progression, which is characterized by skeletal muscle atrophy from motor neuron degeneration and NMJ disruption (9). This hastening occurs although miR-206 is not expressed in motor neurons, which are the primary cell type affected in ALS, and highlights the importance of bidirectional signaling between motor neurons and skeletal muscle for maintaining the NMJ. Similarly, in worms, muscle expression of miR-1 is important for retrograde signaling to the motor neuron resulting in NMJ maintenance (14). Although evidence of miR-1 or miR-206 dysregulation in mouse models or human cases of ALS has not been reported, skeletal muscle up-regulation of the miR-1/206 target, HDAC4, which inhibits muscle reinnervation, is positively correlated with ALS progression and severity (15).
The miR-1 and miR-133 family loci are under transcriptional control of key myogenic proteins including myogenin, MyoD, serum response factor (SRF), myocardin (MYOCD) (3, 4, 16), and myocyte-enhancing factor 2 (MEF-2) (17). Post-transcriptionally, several proteins regulate miR-1 family biogenesis including the KH-type splicing regulatory protein (KSRP) (18), Muscleblind-like splicing regulator (MBNL1) (19), and RNA-binding protein LIN28 (19). These factors presumably control miR-1 family levels in specific contexts. Given the extended half-life of miRNAs and the observations from deep-sequencing studies that the miR-1 family accounts for up to half of accumulated miRNAs in cardiac and skeletal muscles (20, 21), directly controlling the activity of these critical myogenic regulators and their differential activity as compared with miR-133 may be important to maintain muscle homeostasis. Although regulation of these bicistronic miRNAs has been studied at the level of post-transcriptional processing, proteins that differentially interact with and regulate the activity of these co-transcribed miRNAs have not been reported.
Here, we report that TDP-43, an RNA-binding protein that aggregates in individuals afflicted with ALS, physically associates with the mature form of the miR-1/miR-206 family of miRNAs in muscle cells, but not with the co-transcribed miR-133. Our results demonstrate that TDP-43 negatively regulates the activity of miR-1 and miR-206 in muscle through a physical interaction that limits their bioavailability for RNA-induced silencing complex (RISC) loading and offer a mechanism by which mature miRNAs can be differentially regulated at the level of their activity. These findings also establish, for the first time, a mechanistic link between TDP-43 and the miR-1/miR-206 family that may be an unappreciated component of ALS pathogenesis.
EXPERIMENTAL PROCEDURES
Electromobility Shift Assays
Cell extract (5 μg) was incubated with 1 pmol of fluor-conjugated miRNA (Integrated DNA Technologies) in 20 μl of 1× binding buffer (60 mm KCl, 10 mm HEPES, pH 7.6, 3 mm MgCl2, 5% glycerol, 1 mm DTT, 0.1 μg/μl tRNA, 5 μg/μl heparin) for 20 min at room temperature. Where indicated, 100 pmol of unlabeled or biotinylated miRNA competitor was added. Samples were separated on 6% polyacrylamide nondenaturing gel (without loading buffer in sample lanes) and imaged on a LI-COR Odyssey system.
RNA Pulldown
Undifferentiated C2C12 cells, grown in 10-cm dishes, were harvested in 1 ml of lysis buffer (10% glycerol, 20 mm Tris-HCl, pH 8.0, 0.2 mm EDTA, 0.1% Nonidet P-40, 0.5 m KCl) with fresh protease inhibitor (1 tablet/10 ml of lysis buffer, Complete protease inhibitor cocktail tablets, EDTA-free, Roche Applied Science). Lysate (30 mg) was precleared by incubating with streptavidin-conjugated Dynabeads (Life Technologies) on a rotator for 1 h at 4 °C. Lysate was incubated with 5′-end biotinylated miRNA (Integrated DNA Technologies) on a rotator overnight at 4 °C. Complexes were pulled down by incubation with streptavidin-conjugated Dynabeads on a rotator for 1 h at 4 °C. Beads were then washed extensively with lysis buffer, boiled in Laemmli buffer, separated on 4–12% gradient polyacrylamide gels (Bio-Rad), and stained with SilverQuest kit (Invitrogen).
Mass Spectrometry
Gel pieces at the appropriate molecular weight were isolated with a fresh razor blade and dehydrated by two 10-min washes with 25 mm ammonium bicarbonate, 70% acetonitrile. The disulfide bonds were reduced with 10 mm DTT in 25 mm ammonium bicarbonate for 45 min at 50 °C; reduced cysteines were alkylated by 50 mm iodoacetamide in 25 mm ammonium bicarbonate for 60 min at room temperature in the dark. Pieces were washed twice for 5 min with 25 mm ammonium bicarbonate, 70% acetonitrile. Proteins were trypsin-digested overnight at 37 °C. Supernatants were collected, each piece was incubated in 50% acetonitrile, 5% formic acid for 10 min, and supernatants were collected again. The total supernatant was dried on a SpeedVac and reconstituted in 0.1% formic acid. The samples were desalted using Zip-Tip C18 cartridge columns (Millipore) and run at the University of California San Francisco (UCSF) Mass Spectrometry Facility.
TDP-43 Cross-linking and Immunoprecipitation (CLIP)
TDP-43 CLIP was as described (22). Briefly, C2C12 cells were irradiated in a UV cross-linker once for 400 ml/cm2 and again for 200 mJ/cm2, harvested in 1× PXL (0.1% SDS, 0.5% Nonidet P-40, 0.5% deoxycholate, in 1× PBS) with protease inhibitors (1 tablet/10 ml of lysis buffer, Complete protease inhibitor cocktail tablets, EDTA-free, Roche Applied Science) and RNasin (250 units/ml; Promega), and treated with DNase for 5 min at 37 °C. Lysates were centrifuged at 10,000 × g at 4 °C for 10 min to remove cell debris and then precleared with protein A Dynabeads (Life Technologies) at 4 °C for 60 min. An input sample of each lysate was set aside for subsequent analysis. Each lysate was then divided and incubated with 5 μg/ml of either α-TDP-43 (Proteintech) or rabbit IgG (Millipore) on a rotator overnight at 4 °C. Complexes were pulled down by incubation with protein A Dynabeads on a rotator for 2 h at 4 °C. Beads were washed twice with 1× PXL, washed twice with 5× PXL (0.1% SDS, 0.5% Nonidet P-40, 0.5% deoxycholate, in 5× PBS), and boiled to reverse UV cross-linking. Co-precipitated RNAs were extracted and analyzed by quantitative RT-PCR (qRT-PCR).
Drosophila Studies
All flies were maintained on standard fly medium. The TDP-43Q367X Drosophila line was generously provided by Fen-Biao Gao (University of Massachusetts Medical School) (23). All other lines were from the Bloomington Stock Center. Wild-type control flies were W1118. Transgene expression was achieved using the UAS-GAL4 system (dpp-GAL4, Bloomington Stock Center). Crosses were performed at 18 °C unless otherwise specified. Drosophila of the appropriate genotype were anesthetized with CO2 and then placed in isopropyl alcohol for 1 min and euthanized. Flies were allowed to air dry, and wings were removed and embedded into Canada Balsam (Sigma).
Luciferase Reporter Assays
C2C12 cells were maintained in DMEM, 10% FBS and transfected in triplicate with the indicated vectors, miRNA mimics (Life Technologies, miR-1: PM10660; miR-206: PM10409 or Pre-miR miRNA Precursor Negative Control #1), and siRNAs (Sigma, Mission siRNA Universal Negative Control #1: SIC001; siTDP-43: SAS1-Mm01-00198818) using Lipofectamine 2000 (Life Technologies). Total DNA/RNA was equivalent for each transfection condition. After 20 h, cells were harvested, and luciferase activity was measured in duplicate using the Dual-Luciferase reporter system (Promega) on a Victor 1420 multilabel counter (PerkinElmer). Luciferase activity was normalized to the activity of a co-transfected Renilla reporter.
AGO2 RNA Immunoprecipitation
C2C12 cells were maintained in DMEM, 10% FBS and transfected with the indicated siRNAs (Sigma, Mission siRNA Universal Negative Control #1: SIC001; siTDP-43: SAS1-Mm01-00198818) using Lipofectamine 2000 (Life Technologies). Argonaute2 (AGO2) RNA immunoprecipitation (IP) was performed as described (29). Cells were harvested in AGO2 lysis buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm MgCl2, 0.5% Nonidet P-40, and 5 mm DTT) containing protease inhibitors (1 tablet/10 ml of lysis buffer, Complete protease inhibitor cocktail tablets, EDTA-free, Roche Applied Science) and RNasin (250 units/ml; Promega) after 24 h. Lysates were centrifuged at 10,000 × g at 4 °C for 10 min to remove cell debris and then precleared with protein G Dynabeads (Life Technologies) at 4 °C for 60 min. An input sample of each lysate was set aside for subsequent analysis. Each lysate was divided and incubated with 5 μg/ml of α-AGO2 (Abnova) or mouse IgG (Millipore) on a rotator overnight at 4 °C. Complexes were pulled down by incubating with protein G Dynabeads on a rotator for 1 h at 4 °C. Beads were washed as follows: two times with lysis buffer; three times with lysis buffer with 900 mm NaCl and 1% Nonidet P-40; and three times with lysis buffer. Washed beads and input samples were treated with DNA digestion solution (40 mm Tris-HCl, pH 8.0, 10 mm MgSO4, 1 mm CaCl2, 200 units/ml RNasin, and 0.04 units/ml DNase I (Promega)) at 37 °C for 20 min.
qRT-PCR
Total RNA was extracted from cells or protein A or G beads with TRIzol (Life Technologies). Reverse transcription was performed using the Superscript III First-Strand Synthesis SuperMix for qRT-PCR Kit (catalog number 11752-050, Life Technologies) for mRNA quantitation or the TaqMan microRNA RT Kit (catalog number 4366596, Life Technologies) for miRNA quantitation. PCR was performed in triplicate on an ABI 7900HT (Applied Biosystems) using the following TaqMan expression assays (Life Technologies): miR-1, 000385; miR-206, 000510; miR-133b, 002247; Igf-1, Mm00439560_m1; and HDAC4, Mm01299558-g1, and was analyzed with SDS software (Life Technologies).
Western Analyses and ELISAs
Flash-frozen mouse skeletal muscle samples (quadriceps femoris) were homogenized using a Bullet Blender (Next Advance) in radioimmunoprecipitation assay buffer containing protease inhibitors, and protein was quantitated using a Micro BCA protein assay kit (Thermo Scientific, 23235). For Western analyses, 20 μg of each sample was diluted 1:1 in Laemmli buffer and run on 4–20% gradient SDS-PAGE gels (Bio-Rad). After transfer to Immobilon-FL (Millipore) membrane and blocking in Odyssey blocking buffer (LI-COR), blots were probed with the following antibodies: rabbit monoclonal α-HDAC4 (Cell Signaling, catalog number 2072; 1:1000), rabbit polyclonal α-TDP-43 (Proteintech, catalog number 10782-2-AP; 1:500), or mouse monoclonal α-GAPDH (Abcam, ab8245; 1:5000) diluted in Odyssey blocking buffer and 0.01% Tween 20 overnight at 4 °C. Blots were washed in PBST (PBS, 0.05% Tween 20) and incubated in the appropriate IRDye-conjugated secondary antibody (LI-COR) for 1 h at room temperature, washed in PBST, and imaged and quantitated on a LI-COR Odyssey system. For ELISAs, samples were diluted to 1 μg/μl and assayed in triplicate using the IGF-1 mouse ELISA kit (Abcam, ab100695) according to the manufacturer's instructions. Colorimetric signal was measured using a Victor 1420 multilabel counter (PerkinElmer).
RESULTS
The miR-1 Family Physically Interacts with TDP-43
To identify proteins that physically interact with and might regulate activity of the miR-1/miR-206 family, but not the miR-133 family (Fig. 1A), we performed RNA electrophoretic mobility shift assays (EMSAs) seeking proteins that uniquely bind and alter the migration of these miRNAs. We used fluorescently labeled mature miR-1 and protein lysates from undifferentiated C2C12 cells, a mouse skeletal myoblast cell line. We found a prominent band representing an miRNA-protein complex in C2C12 lysates incubated with labeled miR-1 that was effectively lost with the addition of excess unlabeled miR-1 or miR-206, but not with unlabeled miR-133 (Fig. 1B). The same band was observed when fluorescently labeled miR-206 was incubated with C2C12 lysates and could be competed with either miR-1 family member, but not with miR-133 (Fig. 1B). We concluded that a protein or complex of proteins in C2C12 cells preferentially interacts with the mature form of the miR-1/miR-206 family, but not miR-133, in vitro.
To determine the identity of the proteins that interacted with the miR-1 family in EMSAs, we used biotinylated mature miR-1 to affinity-purify the interacting proteins. To ensure that biotinylated miR-1 could be used as a bait, we confirmed that biotinylating miR-1 did not affect its ability to compete with labeled miR-1 for interaction with the protein complex (Fig. 1B). Proteins that co-precipitated with biotinylated miR-1 were separated on denaturing polyacrylamide gels (Fig. 1C), and mass spectrometry was used to identify the bands that emerged in miR-1, but not control or miR-133a, pulldowns. The major proteins identified were KSRP and TDP-43. Both of these RNA-binding proteins physically interact with components of the microprocessor or RISC (18, 24). Although KSRP is required for biogenesis of the miR-1 family in specific contexts (18), the effects of TDP-43 on the miR-1 family have not been reported.
To test whether the interaction of miR-1/miR-206 with TDP-43 occurs in a cellular context, we performed CLIP of TDP-43 from undifferentiated C2C12 cells, which express miR-206 and miR-133b, but not abundant miR-1 or miR-133a (Fig. 1D). Despite cross-linking, qRT-PCR of the co-precipitated RNA showed that miR-133b was not recovered from the TDP-43 CLIP (Fig. 1E). In contrast, miR-206 was immunoprecipitated with TDP-43 in myoblasts (Fig. 1E). These results confirm that the TDP-43-miR-1 family interaction occurs endogenously in muscle cells.
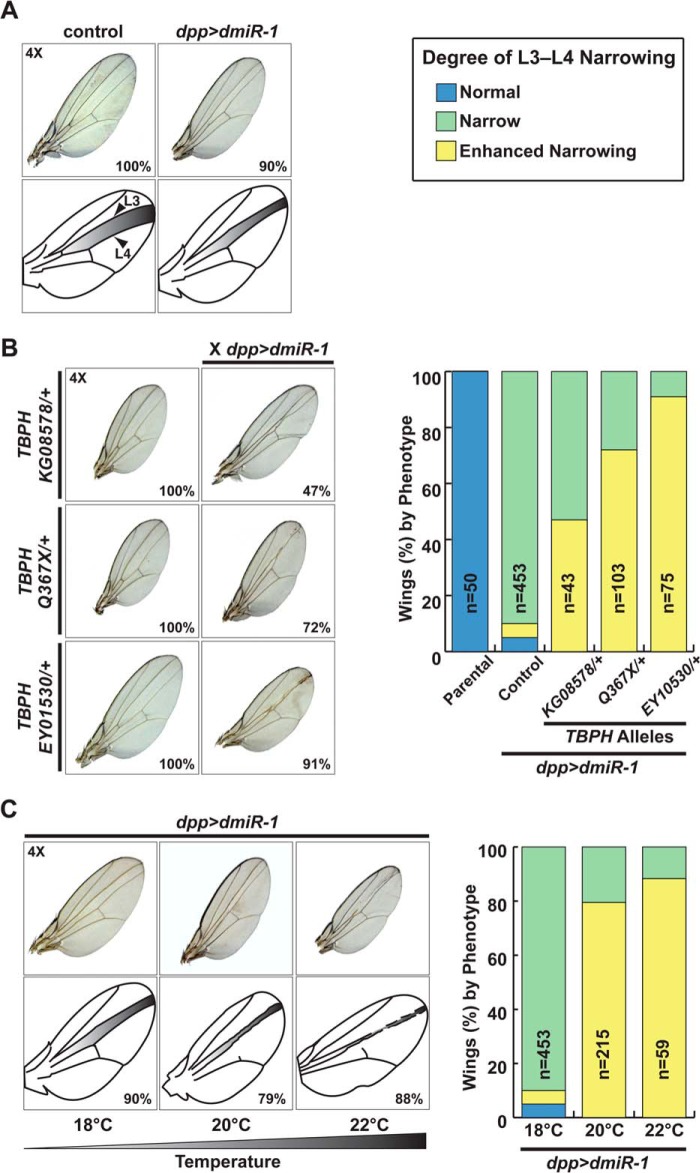
Loss of TDP-43 Enhances miR-1 Effects in Drosophila
To validate the interaction between TDP-43 and miR-1 in a more in vivo context, and to determine the consequences on miR-1 function, we turned to the Drosophila system. In Drosophila wings, misexpression of Drosophila miR-1 (dmiR-1) under the control of a decapentaplegic (dpp) promoter leads to decreased long vein 3-4 (L3-L4) intervein distance (Fig. 2A). This system has been used to identify proteins that genetically interact with dmiR-1 by scoring for the loss or gain of intervein distance in wings of offspring generated from crosses to mutant lines (25). We obtained three fly lines harboring hypomorphic or null alleles of TBPH, the Drosophila ortholog of Tdp-43, which each had normal wing morphology in the heterozygous state. When crossed to the dpp-GAL4::UAS-dmiR-1 (dpp>dmiR-1) fly line, resulting offspring also heterozygous for TBPH mutants had an enhanced narrowing of L3-L4 intervein distance (Fig. 2B). Although penetrance varied among the three TBPH alleles, the consistently enhanced wing phenotype indicated that the effect was due to loss of Tdp-43 and not to background or allele-specific effects. Furthermore, the combination of reduced Tdp-43 levels and misexpression of dmiR-1 in the fly wing phenocopied the effects of even higher levels of dmiR-1 transgene expression induced by a temperature-responsive dpp>dmiR-1 allele at 22 °C (Fig. 2C). These experiments reveal that a conserved interaction between miR-1 and TDP-43 occurs in vivo. Furthermore, the fact that reduced Tdp-43 dosage enhanced the effects of dmiR-1 expression in the fly wing suggests that Tdp-43 may dampen miR-1 activity.
FIGURE 2.
Loss of TBPH, the Drosophila Tdp-43 ortholog, increases miR-1 activity in fly wings. A, wings from control or dppGAL4::UAS-dmiR-1-expressing flies (dpp>dmiR-1) show that miR-1 expression causes decreased L3-L4 intervein distance (25). B, fly wings from intercrosses of three different TBPH hypomorphic lines to dpp>dmiR-1 flies showed enhancement of the dpp>dmiR-1 phenotype. Quantitation of wing phenotypes from parental, dpp>dmiR-1 (Control), or dpp>dmiR-1 X TBPH hypomorph crosses is shown. C, wing morphology in flies carrying the temperature-sensitive dpp>dmiR-1 allele housed at 18, 20, or 22 °C. Quantitation of wing phenotypes is shown. Narrow is defined as L3-L4 intervein distance of dpp>dmiR-1 at 18 °C; Enhanced Narrowing is defined as ≥50% further reduction in L3-L4 intervein distance as compared with dpp>dmiR-1 at 18 °C.
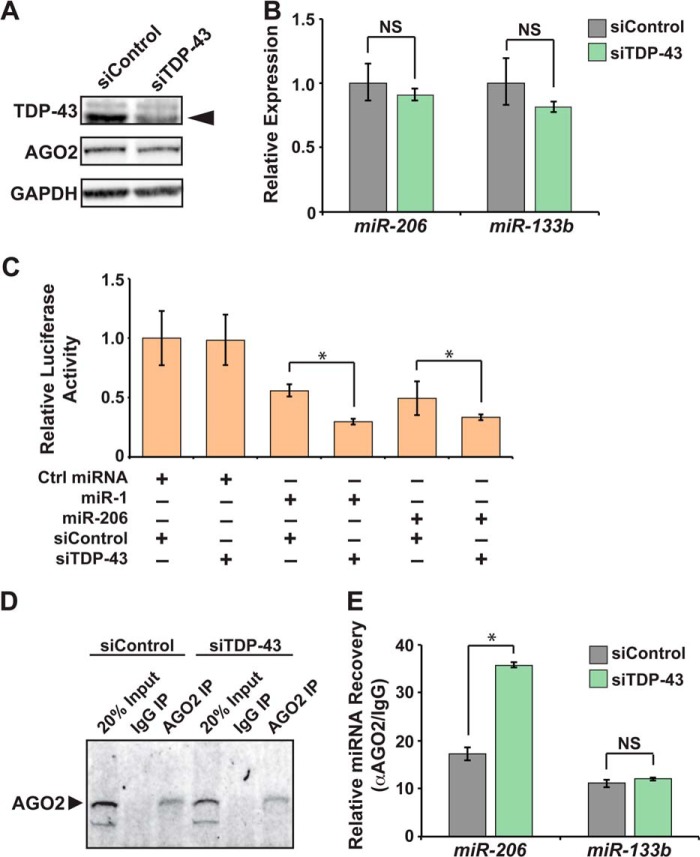
Loss of TDP-43 Increases miR-1/miR-206 Family Activity in Skeletal Myoblasts
To directly test whether miR-1/miR-206 activity was suppressed by TDP-43, we returned to C2C12 skeletal myoblasts where we depleted TDP-43 using a targeted siRNA. Efficient TDP-43 knockdown was confirmed by Western analysis (Fig. 3A). Levels of mature miR-206 and miR-133b, which are the most abundant family members expressed in skeletal myoblasts, were comparable in control and TDP-43-depleted cells (Fig. 3B), indicating that loss of TDP-43 did not affect expression or biogenesis of these miRNAs. We used these conditions to measure activity of the miR-1/miR-206 family in luciferase assays. In the presence of TDP-43, miR-1 was able to repress expression of a luciferase reporter with a validated miR-1 family binding site in the 3′-UTR (4). Depleting TDP-43 increased miR-1 repression of this reporter (Fig. 3C) without affecting baseline luciferase expression in the presence of a nontargeting control miRNA. TDP-43 similarly enhanced miR-206-mediated reporter repression (Fig. 3C), demonstrating that loss of TDP-43 enhances activity of both miR-1 family members.
FIGURE 3.
TDP-43 suppresses activity of miR-1 and miR-206 by inhibiting their association with AGO2. A, Western blots to detect TDP-43 and AGO2 in C2C12 cells transfected with nontargeting control siRNA (siControl) or siRNA directed against Tdp-43 (siTDP-43). Arrow indicates 43-kDa band corresponding to TDP-43. GAPDH levels show equal protein loading. B, miR-206 and miR-133b levels in C2C12 cells transfected with nontargeting control siRNA (siControl) or siRNA directed against Tdp-43 (siTDP-43) as detected by qRT-PCR and normalized to U6 levels. C, luciferase reporter assays measuring activity of miR-1 or miR-206 in control (siControl) or TDP-43-depleted (siTDP-43) cells. Ctrl miRNA, control miRNA. D, Western analysis detecting AGO2 protein following immunoprecipitation from TDP-43-depleted (siTDP-43) or control (siControl) C2C12 cells using α-AGO2 (AGO2 IP), mouse IgG (IgG IP), or 20% input showed efficient and specific recovery of AGO2 protein. E, miRNA levels recovered by AGO2 RNA IP from C2C12 cells transfected with an siRNA targeting Tdp-43 (siTDP-43) were compared with those transfected with a nontargeting control siRNA (siControl) (NS, not significant; *, p < 0.05).
TDP-43 Disrupts Association of miR-1/miR-206 with AGO2
Because miRNA activity requires interaction with the RISC, one potential mechanism by which TDP-43 negatively regulates miR-1 family activity may be through inhibiting their RISC association, thus decreasing their availability to repress target mRNA sequences. To test this hypothesis, we immunoprecipitated the major RISC component, AGO2, from control or TDP-43-depleted C2C12 cells (Fig. 3D). Western analysis confirmed that TDP-43 depletion did not affect AGO2 expression (Fig. 3A). qRT-PCR of the co-precipitated RNA showed that TDP-43 depletion increased the amount of miR-206 associated with AGO2 without affecting the levels of co-precipitated miR-133b (Fig. 3E). Thus, loss of TDP-43 enhanced miR-206 incorporation into the RISC.
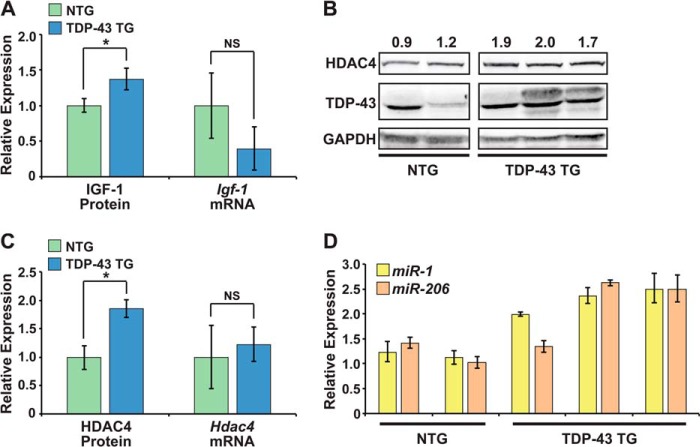
TDP-43 Overexpression Decreases miR-1/miR-206 Family Activity in Transgenic Mice
Multiple TDP-43 transgenic mouse lines have been generated to model ALS. We took advantage of an existing transgenic mouse line with human TDP-43 overexpressed primarily in skeletal muscle to analyze the effects on endogenous miR-1 family activity in vivo (26). Overexpression of the human TDP-43 transgene in muscle results in variable levels of both high and low molecular weight TDP-43 species (Fig. 4B) (26). Igf-1 and Hdac4 are reported targets of the miR-1 family in skeletal muscle, and miR-1 modulation affects protein levels of these two targets, without altering their mRNA expression (5, 27). Using ELISA, we examined protein levels of IGF-1 in skeletal muscle from 3.5-week-old transgenic (TDP-43 TG) and nontransgenic (NTG) littermates, before the onset of muscle weakness (Fig. 4A). We found that transgenic mice had higher protein levels of both TDP-43 and the miR-1 family target, IGF-1 (Fig. 4, A and B). However, IGF-1 protein tended to be elevated in the muscle of transgenic mice without a corresponding increase in Igf-1 mRNA (Fig. 4A). This type of discordant mRNA and protein expression can be a feature of altered miRNA function and is consistent with a model where TDP-43 limits the activity of the miR-1/miR-206 family in skeletal muscle, leading to increased IGF-1 translation.
FIGURE 4.
TDP-43 overexpression decreases miR-1 family activity in mouse muscle. A, average levels of IGF-1 protein, determined by ELISA, and Igf-1 mRNA, detected by qRT-PCR, in hindlimb skeletal muscle of 3.5 week old, male, nontransgenic (NTG, n = 2) or TDP-43 transgenic (TDP-43 TG, n = 3) littermates. B, Western blots to detect HDAC4 and TDP-43 protein in hindlimb skeletal muscle of 3.5 week old, male, nontransgenic (NTG) or TDP-43 transgenic (TDP-43 TG) littermates. Relative HDAC4 quantity, as determined by densitometry, is shown above. GAPDH levels show equal protein loading. C, average levels of HDAC4 protein and Hdac4 mRNA in samples analyzed in A and B. D, relative levels of miR-1 and miR-206 detected by qRT-PCR using the same samples analyzed in A–C (NS, not significant; *, p < 0.05).
We performed Western analyses to compare levels of a second miR-1 family target, HDAC4, in the same skeletal muscle preparations in which we measured TDP-43 and IGF-1 (Fig. 4B). Similarly, HDAC4 protein was elevated in TDP-43 TG skeletal muscle without an increase in Hdac4 mRNA (Fig. 4C). Thus, two well validated miR-1 family targets were up-regulated at the protein, but not mRNA level in TDP-43 transgenic muscle, in agreement with our model of reduced miR-1/206 activity. Elevated HDAC4 may be particularly relevant to the progressive muscle weakness observed in these TDP-43 transgenic mice as HDAC4 is an established negative regulator of muscle innervation (9). Indeed, the reinnervation defect observed in muscle of miR-206-null mice was attributed to up-regulation of this critical miR-1 family target (9).
Interestingly, this apparent decrease in miR-1 family activity occurred despite elevated levels of both miR-1 and miR-206 in TDP-43 TG muscle (Fig. 4D), further highlighting the dampening effect that TDP-43 has on miR-1 family activity in muscle. Together, these results support the idea that miR-1 family activity is decreased in TDP-43 transgenic muscle, resulting in increased translation of miR-1 family targets.
DISCUSSION
Here we show a unique physical and genetic interaction between TDP-43, an ALS disease protein, and the miR-1 family of muscle miRNAs that negatively regulates miR-1 family activity. TDP-43 decreased activity of mature miR-1 and miR-206, but not the co-transcribed miR-133 family, by preventing the bound miRNAs from associating with the RISC. Consequently, TDP-43 overexpression in skeletal muscle led to increased protein levels of the miR-1 family targets, IGF-1 and HDAC4. This increase was observed despite unchanged Igf-1 and Hdac4 mRNA expression and elevated miR-1 family levels. To our knowledge, a selective mature miRNA-protein interaction that limits miRNA activity, independent of miRNA biogenesis, has not been reported and suggests that the differential activity of mature miRNAs, including bicistronically encoded miRNAs, such as miR-1 and miR-133, can be regulated by selective interaction with RNA-binding proteins.
The predilection of TDP-43 for miR-1/miR-206 was observed in both in vitro miRNA pulldowns as well as in vivo CLIP experiments. Furthermore, TDP-43 depletion in myoblasts led to increased interaction of AGO2 with miR-206, but not miR-133b. TDP-43 overexpression in vivo up-regulated IGF-1 and HDAC4, proteins whose synthesis is normally repressed by the miR-1 family (5, 27). The extent to which TDP-43 affects activity of other miRNAs must be determined, but our results reveal a unique mechanism that may modulate the activity of mature miRNAs, independent of miRNA transcription or biogenesis. Because the miR-1 family promotes differentiation and the miR-133 family keeps muscle in a less mature, more proliferative state (5, 12, 13), the TDP-43-miR-1 family interaction may be important to control the balance of these co-transcribed miRNA families to promote development and maintain adult muscle homeostasis.
miR-206 expression increases in response to muscle denervation (9). However, the elevated miR-206 that we observed in transgenic muscle is unlikely to be a response to motor neuron death because these young animals had not yet become symptomatic. Furthermore, miR-1 levels were elevated in TDP-43 transgenic muscle, although miR-1 transcription is not affected by denervation (9). Instead, it is possible that interaction with TDP-43 stabilizes miR-1 and miR-206, preventing their degradation or clearance, and leading to their accumulation, while also limiting their RISC-associated activity. This is consistent with the observation that miR-1 and miR-206 levels greatly exceed those of miR-133 in mature muscle (20, 21). Further studies are required to assess the effect of TDP-43 on miRNA stability and turnover.
Given that TDP-43 is an ALS disease protein (28) and deleting miR-206 in mice exacerbates the phenotype of an ALS mouse model (9), we aimed to elucidate the consequence of the TDP-43-miR-1 family interaction in skeletal muscle. Dysregulation and aggregation of TDP-43 are commonly observed in motor neurons of ALS subjects, regardless of genetic etiology, but have not been described in skeletal muscle. Considering the ubiquitous expression of TDP-43 and other known genetic ALS determinants, some occurrences of ALS might be accompanied by skeletal muscle TDP-43 abnormalities. This could decrease the activity of the miR-1 family in affected muscle, thus altering retrograde signaling at the NMJ through dysregulation of both HDAC4 (9) and MEF-2 (14), and ultimately contributing to motor neuron demise. With our finding that TDP-43 overexpression increases protein levels of the miR-1/miR-206 target, HDAC4, whose dysregulation correlates with ALS disease progression (15), further interrogation of TDP-43 levels, localization, and activity in skeletal muscle of individuals with ALS at various stages of the disease seems warranted.
Acknowledgments
We thank Gary Howard for editorial comments and Bethany Taylor for help with manuscript and figure preparation. Mass spectrometry was performed in the UCSF Mass Spectrometry Facility.
This work was supported by the National Institutes of Health/NIGMS/Biomedical Technology Research Centers Grant P41GM103481 to the UCSF Mass Spectrometry Facility; The March of Dimes (to I. N. K.); the National Science Foundation (to A. H.); the National Institutes of Health/NHLBI Grant R01HL057181 (to D. S.); the William Younger Family Foundation (to D. S.); the L. K. Whittier Foundation (to D. S.); the Eugene Roddenberry Foundation (to D. S.); the American Heart Association Grant 11SDG5190024 (to K. N. I.); the Amyotrophic Lateral Sclerosis Association Grant 679Y22 (to K. N. I.); and the National Institutes of Health/National Center for Research Resources Grant RR18928 to The Gladstone Institutes.
This article was selected as a Paper of the Week.
- miRNA
- microRNA
- miR
- microRNA
- RISC
- RNA-induced silencing complex
- NMJ
- neuromuscular junction
- ALS
- amyotrophic lateral sclerosis
- KSRP
- KH-type splicing regulatory protein
- CLIP
- cross-linking and immunoprecipitation
- IP
- immunoprecipitation
- qRT-PCR
- quantitative RT-PCR
- L
- long vein
- TG
- transgenic
- NTG
- nontransgenic.
REFERENCES
- 1. Cordes K. R., Srivastava D., Ivey K. N. (2010) MicroRNAs in cardiac development. Pediatr. Cardiol. 31, 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gagan J., Dey B. K., Dutta A. (2012) MicroRNAs regulate and provide robustness to the myogenic transcriptional network. Curr. Opin. Pharmacol. 12, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwon C., Han Z., Olson E. N., Srivastava D. (2005) MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 18986–18991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao Y., Samal E., Srivastava D. (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 [DOI] [PubMed] [Google Scholar]
- 5. Chen J.-F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D.-Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu N., Bezprozvannaya S., Williams A. H., Qi X., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heidersbach A., Saxby C., Carver-Moore K., Huang Y., Ang Y.-S., de Jong P. J., Ivey K. N., Srivastava D. (2013) microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife 2, e01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y., Ransom J. F., Li A., Vedantham V., von Drehle M., Muth A. N., Tsuchihashi T., McManus M. T., Schwartz R. J., Srivastava D. (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 129, 303–317 [DOI] [PubMed] [Google Scholar]
- 9. Williams A. H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J. L., Bassel-Duby R., Sanes J. R., Olson E. N. (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wystub K., Besser J., Bachmann A., Boettger T., Braun T. (2013) miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet 9, e1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei Y., Peng S., Wu M., Sachidanandam R., Tu Z., Zhang S., Falce C., Sobie E. A., Lebeche D., Zhao Y. (2014) Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 24, 278–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivey K. N., Muth A., Arnold J., King F. W., Yeh R.-F., Fish J. E., Hsiao E. C., Schwartz R. J., Conklin B. R., Bernstein H. S., Srivastava D. (2008) MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao P. K., Missiaglia E., Shields L., Hyde G., Yuan B., Shepherd C. J., Shipley J., Lodish H. F. (2010) Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 24, 3427–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simon D. J., Madison J. M., Conery A. L., Thompson-Peer K. L., Soskis M., Ruvkun G. B., Kaplan J. M., Kim J. K. (2008) The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruneteau G., Simonet T., Bauché S., Mandjee N., Malfatti E., Girard E., Tanguy M. L., Behin A., Khiami F., Sariali E., Hell-Remy C., Salachas F., Pradat P. F., Fournier E., Lacomblez L., Koenig J., Romero N. B., Fontaine B., Meininger V., Schaeffer L., Hantaï D. (2013) Muscle histone deacetylase 4 upregulation in amyotrophic lateral sclerosis: potential role in reinnervation ability and disease progression. Brain 136, 2359–2368 [DOI] [PubMed] [Google Scholar]
- 16. Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S., Lodish H. F. (2006) Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 103, 8721–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu N., Williams A. H., Kim Y., McAnally J., Bezprozvannaya S., Sutherland L. B., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. U.S.A. 104, 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2009) The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rau F., Freyermuth F., Fugier C., Villemin J.-P., Fischer M.-C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D., Wahbi K., Day J. W., Fujimura H., Takahashi M. P., Auboeuf D., Dreumont N., Furling D., Charlet-Berguerand N. (2011) Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 18, 840–845 [DOI] [PubMed] [Google Scholar]
- 20. Rao P. K., Toyama Y., Chiang H. R., Gupta S., Bauer M., Medvid R., Reinhardt F., Liao R., Krieger M., Jaenisch R., Lodish H. F., Blelloch R. (2009) Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 105, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen M., Hansen J. H., Hedegaard J., Nielsen R. O., Panitz F., Bendixen C., Thomsen B. (2010) MicroRNA identity and abundance in porcine skeletal muscles determined by deep sequencing. Anim. Genet. 41, 159–168 [DOI] [PubMed] [Google Scholar]
- 22. Darnell R. (2012) CLIP (cross-linking and immunoprecipitation) identification of RNAs bound by a specific protein. Cold Spring Harb. Protoc. 2012, 1146–1160 [DOI] [PubMed] [Google Scholar]
- 23. Lu Y., Ferris J., Gao F.-B. (2009) Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol. Brain 2, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ling S.-C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. King I. N., Qian L., Liang J., Huang Y., Shieh J. T. C., Kwon C., Srivastava D. (2011) A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Dev. Cell 20, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stallings N. R., Puttaparthi K., Luther C. M., Burns D. K., Elliott J. L. (2010) Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol. Dis. 40, 404–414 [DOI] [PubMed] [Google Scholar]
- 27. Elia L., Contu R., Quintavalle M., Varrone F., Chimenti C., Russo M. A., Cimino V., De Marinis L., Frustaci A., Catalucci D., Condorelli G. (2009) Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 120, 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 29. Wang W. X., Wilfred B. R., Hu Y., Stromberg A. J., Nelson P. T. (2010) Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 16, 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]