SUMMARY
Recent studies recognize a vast diversity of non-coding RNAs with largely unknown functions, but few have examined interspersed repeat sequences, which constitute almost half our genome. RNA hybridization in situ using CoT-1 (highly repeated) DNA probes detects surprisingly abundant euchromatin-associated RNA comprised predominantly of repeat sequences (“CoT-1 RNA”), including LINE-1. CoT-1-hybridizing RNA strictly localizes to the interphase chromosome territory in cis, and remains stably associated with the chromosome territory following prolonged transcriptional inhibition. The CoT-1 RNA territory resists mechanical disruption and fractionates with the non-chromatin scaffold, but can be experimentally released. Loss of repeat-rich, stable nuclear RNAs from euchromatin corresponds to aberrant chromatin distribution and condensation. CoT-1 RNA has several properties similar to XIST chromosomal RNA, but is excluded from chromatin condensed by XIST. These findings impact two “black boxes” of genome science: the poorly understood diversity of non-coding RNA and the unexplained abundance of repetitive elements.
INTRODUCTION
In recent years there has been a steady increase in studies demonstrating a diversity of RNA types, including small siRNAs, miRNAs, and piRNAs or many thousands of larger lncRNAs (reviewed in (Aalto and Pasquinelli, 2012; Rinn and Chang, 2012) that may regulate specific protein-coding genes. While most lncRNAs have no known function, specific lncRNAs have been shown to serve roles almost as diverse as that of proteins, ranging from forming a scaffold for a non-chromatin nuclear body to transcriptional regulation of specific loci (reviewed in (Geisler and Coller, 2013). While some non-coding RNAs (ncRNAs) are thought to interact with specific loci, RNA is not generally considered a broad component of chromatin or chromosomes.
Despite the expanding importance of ncRNAs and the “dark matter” transcriptome, the potential contribution of interspersed repeats has received little attention. Protein coding sequences make up only about two percent of the human genome (Lander et al., 2001), while interspersed repeats, including short and long interspersed nuclear elements (SINEs and LINEs) and simple sequence repeats (SSRs) comprise half or more (de Koning et al., 2011). LINEs and SINEs are among the most ancient sequences in the human genome, having survived evolution by vertical transfer, and are mostly intergenic, but they are also found in noncoding regions of most human genes (Goodier and Kazazian, 2008). There are examples of individual transposons regulating nearby genes (Goodier and Kazazian, 2008), however, the bulk of abundant repeats are widely believed to have no raison d’etre, except as evolutionary vestiges. Although the primary sequence of interspersed repeats is rarely constrained, their conserved presence, defined genomic organization (Chen and Manuelidis, 1989; Korenberg and Rykowski, 1988), and potential to form inter-molecular structures suggests potential functionality. In fact, organismal complexity is correlated with the proportion of repeats in the genome, rather than genes (Neguembor and Gabellini, 2010).
Our interest in exploring this understudied half of the genome is bolstered by considering how XIST RNA enacts X-chromosome silencing. The large XIST transcript (17kb) (Brown et al 1992) propagates across and “paints” the whole nuclear chromosome, yet is strictly localized within the boundary of the chromosome territory in cis (Clemson et al 1996). It has been suggested that when XIST RNA initiates a chromatin remodeling cascade, both canonical premRNA transcripts and repeat-rich hnRNAs (Hall et al., 2002) are silenced concomitant with chromosome condensation to form the heterochromatic Barr body (reviewed in (Hall and Lawrence, 2010; Wutz, 2011)). Interestingly, human and mouse XIST/Xist transcripts have little sequence conservation, yet function similarly. As detailed elsewhere, several aspects of XIST RNA biology implicate repetitive elements as important to chromosome regulation (Hall and Lawrence, 2010), including the competence of an autosome to be partially (Lyon, 1998) or comprehensively (e.g. (Jiang et al., 2013) silenced by XIST RNA.
Given the expectation that interspersed repeats are widely expected to be transcriptionally inert, and they do not uniquely map to the genome, they have been routinely removed or overlooked in most genomic analyses (Consortium, 2011). RNAs embedded in nuclear structure would likely be underrepresented by extraction protocols designed for cytoplasmic RNAs, and repetitive RNAs may form more complex and less soluble structures. Many studies have shown that even after extensive biochemical extraction, which removes most DNA and protein, much as yet undefined nuclear RNA remains (e.g. (Fey et al., 1986), thus some RNAs may resist extraction of even isolated nuclei. A means to circumvent the limitations of extraction-based and bioinformatic approaches is to examine the potential expression and distribution of repeat RNA in situ.
Here, in situ analyses show that RNA is broadly and stably associated with euchromatin and that the predominant component of this chromatin-associated RNA is surprisingly abundant CoT-1 RNA from interspersed repetitive elements, including L1. The unusual properties of CoT-1 RNAs are distinct from short-lived nascent transcripts and indicate CoT-1 repeat RNAs comprise a class of “chromosomal RNAs”, which persist long after transcriptional inhibition, and remain localized strictly with the interphase chromosome territory in cis. Further, we show the RNA is tightly associated with euchromatin and the non-chromatin scaffold, but it is excluded and silenced by XIST RNA on inactivated chromosomes. Finally, chromosomal RNA associated with euchromatin, comprised largely of CoT-1 repeat RNA, may help maintain open chromatin packaging.
RESULTS
CoT-1 repeat RNA is abundant in mammalian nuclei
CoT-1 DNA is routinely used to block non-specific hybridization of genomic probes to repeats, however here we use labeled CoT-1 DNA as a probe to detect RNAs containing high copy repeats (“CoT-1 RNA”). We previously showed that CoT-1 RNA hybridization provides a convenient assay to identify silent heterochromatic regions within nuclei by the absence of “hnRNA” hybridization signal (Clemson et al., 2006; Hall et al., 2002; Tam, 2004). CoT-1 “hnRNA” is broadly distributed throughout the nucleoplasm in all interphase cells, but is absent from nucleoli, cytoplasm and DAPI dense heterochromatic regions (Figure 1 & Figure S1A). This robust CoT-1 RNA signal is ubiquitous in all primary and cancer cell lines (mouse and human) and frozen tissue sections examined (Table S1). Several lines of evidence establish this signal is single-stranded RNA and not DNA: it is detected under non-denaturing conditions (which does not detect highly abundant DNA sequences) (Lawrence et al., 1989), it is eliminated by RNase A (Figure S1B–G), and the CoT-1 RNAs detach from chromatin and disperse to the cytoplasm at mitosis (Figure 1A).
Figure 1. CoT-1 RNA is expressed in all mammalian cells examined.
All blue signals are DAPI DNA. CoT-1 RNA is expressed in 100% of interphase cells. e.g. A) HeLa (M=metaphase), B) NIH3T3. Inserts: CoT-1 RNA exclusion from chromocenters (inset arrows) of indicated cell (arrow). C) Human ES cells and D) frozen human tissue sections. D–E) Linescan across nuclei shows CoT-1 RNA absence from peripheral heterochromatic compartment (yellow zones). F–G) XIST RNA (arrow) paints the inactive X chr in female cells, while CoT-1 RNA is silenced. (See Fig S1 & Table S1). Scale bar 5µm.
The abundance of CoT-1 repeat RNA is indicated by a bright signal, broadly distributed through most of the nucleus, the intensity of which is close to or exceeds that of highly abundant nuclear RNAs such as rRNA or XIST RNA (Figure 2). For example, the linescan in Figure 2C–D illustrates that the intensity of CoT-1 RNA signal is similar to that of XIST RNA on the X-chromosome territory, but CoT-1 RNA is much more widely distributed (and thus abundant). Likewise, by measuring the average total RNA signal for rRNA and CoT-1 RNA per nucleus, we found CoT-1 RNA signal exceeds that of abundant nuclear rRNA (Figure 2E–H). The surprising abundance and distribution of this nuclear repeat RNA signal led us to investigate its stability and relationship to chromosome structure.
Figure 2. CoT-1 RNA appears highly abundant.
A) XIST RNA paints interphase X-chromosome territory. Scale bar 5µm. B) The DAPI dense Barr Body (arrow) defines the inactive X-chromosome painted by XIST RNA. C) Both CoT-1 and XIST RNAs are detected simultaneously with the same fluorochrome. D) Linescan (white line in C) through the Barr body (arrow) illustrates similar intensities for both RNAs (green). E–H) CoT-1 RNA is more abundant than rRNA. All images are the same exposure. E) CoT-1 RNA alone, F) nucleolar 18S rRNA alone (Insert is same cell with DAPI DNA included), G) CoT-1 RNA and 18S rRNA detected with same fluorochrome. H) Average total intensities were measured for each RNA signal (n=40).
CoT-1 RNA localization is tightly restricted to the parent chromosome territory
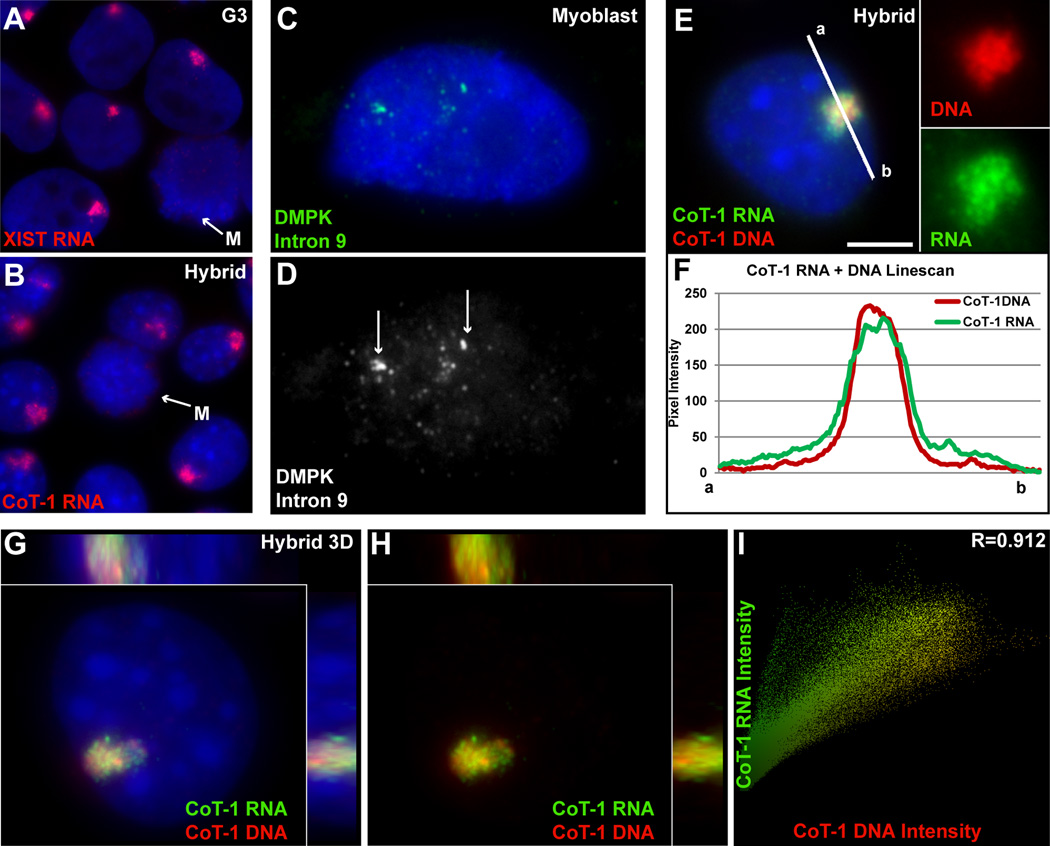
XIST RNA is unique in that it coats and is restricted to its parent chromosome territory in interphase nuclei and does not disperse to the surrounding nucleoplasm (Figure 3A), unlike mRNAs or many excised introns (Figure 3C–D)(Clemson et al., 1996; Johnson et al., 2000; Smith et al., 2007). This finding was a key to understanding that a non-coding RNA could have an unanticipated role in regulating chromatin structure and function. To investigate whether CoT-1 RNA showed a structural relationship to the chromosome of origin, we needed a strategy to examine ubiquitous repeats expressed from just an individual chromosome.
Figure 3. CoT-1 RNA localizes to the chromosome similar to XIST RNA.
A) XIST RNA is strictly localized to the inactive chromosome in interphase. B) Human CoT-1 RNA is also strictly localized to human Chr4 in all interphase hybrid cells. Both RNAs are released at Mitosis (M) (arrows). C–D) Excised introns do not localize to chromatin, and drift away from their transcription sites. Transcription foci denoted by arrows. E) CoT-1 DNA identifies human Chr4 in hybrid cells painted by CoT-1 RNA (channels separated at right). Scale bar 5µm. F) A linescan (white line in E) shows the RNA has a sharp border at the edge of the chromosome territory. G) 3D image of CoT-1 DNA and RNA on human Chr4 in a single hybrid nucleus, with side views. H) same image as G with DAPI DNA removed. I) CoT-1 DNA and RNA signals are colocalized. (See Fig S1 & S2 and Movies S1, S2 & S3).
To this end, we studied the localization and behavior of human CoT-1 RNA in mouse somatic hybrid cells (GM11687) containing a single human Chromosome 4 (Chr4). While mouse and human genomes have similar families and patterns of interspersed repeats, the primary sequences differ sufficiently that human CoT-1 does not hybridize to mouse DNA (or RNA) and vice versa. RNA FISH in the hybrid cells showed a striking result: that CoT-1 RNA is restricted to a tightly defined nuclear territory (Figure 3B). This is highly reminiscent of the XIST RNA territory over the inactive X-chromosome (Figure 3A). Simultaneous hybridization to CoT-1 DNA and RNA (in two different colors) demonstrates that both the RNA and DNA territories appear as sharply bordered structures with coincident boundaries (Figure 3E–F), and are highly colocalized by 3D imaging analysis (Figure 3G–H and Movies S1 & S2), similar to XIST RNA and its parent chromosome (Figure S1H, Figure S2A–B and Movie S3). Additionally, human CoT-1 RNA does not overlap with mouse (mCoT-1) RNA (Figure S2C), indicating they remain associated with their parental chromosomes.
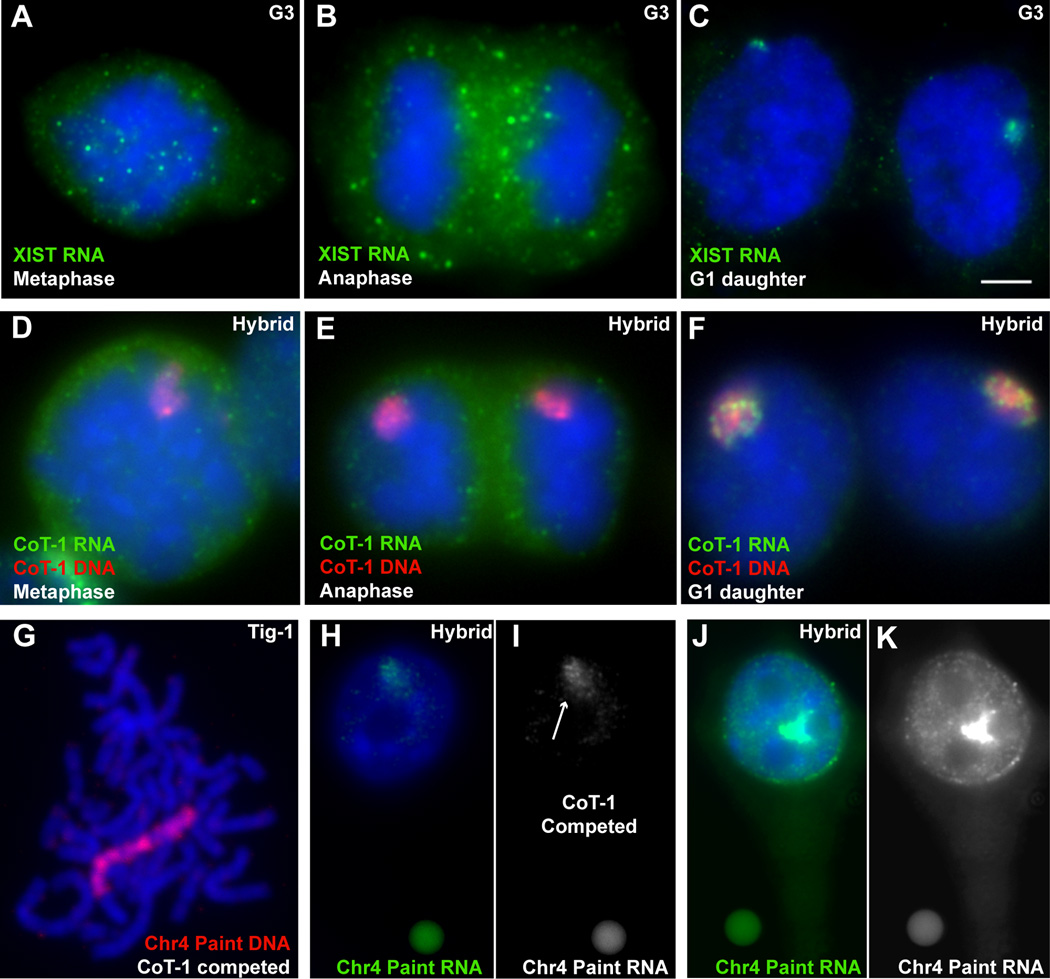
Although XIST RNA is a structural RNA, tightly bound to its parent chromosome in cis, it is released from chromatin in mitotic cells and re-synthesized in early G1 daughter (G1d) cells (Figure 4A–C). CoT-1 RNA behaves similarly, and disassociates from the chromosome at prophase, where it disperses in the cytoplasm, and is resynthesized in G1d cells (Figure 4D–F).
Figure 4. The majority of RNA associated with interphase chromosomes is repeat RNA, which is released at mitosis and resynthesized in G1.
A–C) XIST RNA is released at mitosis and re-synthesized in early G1d cells. Scale bar 5µm. D–F) CoT-1 RNA is also released at mitosis and re-synthesized in G1d. G) A Chr4 library probe effectively detects the unique single copy DNA sequences across the chromosome (with CoT-1 competition). H–I) The same Chr4 library probe detects only a weak RNA signal (arrow) over the Chr4 territory (with CoT-1 competition). J–K) More RNA is detected on the Chr4 territory when CoT-1 competition is removed. H–K are taken at the same exposure with standardized fluorescent beads (small round objects in images). (See Fig S2). Beads are 2.5µm.
Most RNA stably associated with interphase chromosomes is comprised of repeats
The abundance and localization of repeat RNA to its parent chromosome suggests that it may not be simply a byproduct of “genic” transcription. To further evaluate this, we compared the abundance of repeat RNAs associated with the interphase chromosome to the collective contribution of unique RNA from a whole Chr4 library.
Chromosome libraries are designed to detect specific sequences throughout a particular chromosome. Although libraries are depleted of repeats, they still contain some, requiring competition with cold CoT-1 DNA to enhance the library specificity (Figure 4G and Figure S2D–E). Using two commercially available Chr4-specific library probes to detect RNA in hybrid cell nuclei, we found the RNA territory over the human chromosome was only weakly defined (Figure 4H–I) in contrast to the well-defined, bright CoT-1 RNA territory over the same chromosome. This was not simply due to the complexity of the Chr4 library because it labels the unique sequences along the whole chromosome well by DNA FISH (Figure 4G). Instead, this suggests that the total amount of steady-state RNA that accumulates over the chromosome may be substantially less from unique sequences than from repetitive sequences. To further examine this, we removed the cold CoT-1 competitor from the hybridization reaction and this identical Chr4 library yielded a large increase in RNA signal over the territory (Figure 4J–K). This supports that the vast majority of RNA sequences associated with an interphase chromosome territory are repetitive in nature.
CoT-1 transcripts are stable following transcriptional inhibition
The relative prevalence of repeat-containing RNA with interphase chromosomes could be due to either a high transcription rate or accumulation of stable RNA. While transcriptional inhibitors can have complex effects on RNA metabolism (Bensaude, 2011), most nascent transcripts or introns are reported to be relatively short-lived with transcriptional inhibition. For example, the small nascent transcription focus of XIST intron is no longer detected 1 hour after inhibition, whereas the mature XIST RNA has a longer (~5–6 hour) half-life, which was part of the initial evidence for a novel type of chromosomal RNA (Clemson et al., 1996). To assess CoT-1 RNA stability when transcription is arrested (by multiple distinct mechanisms), we used three different transcription inhibitors: 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), Actinomycin-D (ActD) and α-amanatin (α-aman), in hybrid and human cells. Numerous experiments with the different inhibitors consistently demonstrated exceptional stability of CoT-1 RNA in interphase nuclei (see methods). In fact, because of this unusual stability, we relied on G1 daughter (G1d) cells to confirm that the inhibitors were working (i.e. preventing re-synthesis). As detailed in Figure 5 (and Figures S2 & S3), CoT-1 RNA was not re-synthesized in 90–100% of G1d cells in all three inhibitors, but remained robust 93–100% of nuclei that had not divided.
Figure 5. CoT-1 RNA localization is very stable under transcriptional inhibition.
A–B) CoT-1 RNA is re-synthesized early in G1d (arrows). C–H) Transcriptional inhibition using three inhibitors (C–H) prevents CoT-1 re-synthesis in 95–100% of G1d cells (arrows), but doesn’t significantly affect CoT-1 RNA levels in 93–95% of interphase cells. I–L) COL1A1 transcription foci are detected in 95% of interphase and G1d cells (arrows). (I–J) COL1A1 RNA foci are eliminated in 82% of cells after 5 hours in DRB, but only G1d cells (arrows) also lack CoT-1 RNA. (See Fig S2 & S3). All scale bars are 5µm.
Detailed analysis was performed with DRB in human fibroblasts to compare interphase CoT-1 RNA stability with mRNA transcription (COL1A1 and GAPDH) and with the relatively long-lived XIST RNA (Figure 5 and Figure S2 & S3). Five hours in DRB was sufficient to essentially eliminate COL1A1 RNA transcription foci in interphase Tig-1 nuclei (Figure 5I–L), with only 18% retaining a barely visible signal (Figure S2H–I), and this was also seen using intron probes. In contrast, the CoT-1 RNA, though somewhat reduced, remained in 100% of these same nuclei, and persisted longer than XIST RNA (Figure S2J–K). CoT-1, XIST, COL1A1 and GAPDH RNA all were absent and not re-synthesized in inhibited G1d cells (Figure 5K–L). Upon removal of the reversible DRB inhibitor, 100% of G1d cells re-expressed CoT-1 across the nucleus within an hour (Figure S2L–M). Taken together, the persistence of CoT-1 RNA in these transcriptionally-inhibited interphase cells is due to stability, not continued synthesis.
We used highly extended treatments with α-amanitin to further examine the stability of the RNA, and were surprised to see that a bright RNA signal remained after 16–32 hours. In fact, comparison of the RNA signal to a standard fluorescent bead showed the signal actually became brighter in most cells, at both concentrations (5 & 20µg/ml), seen in multiple experiments (Figure S3K). While this may relate to the extraordinary stability of the RNA, as considered in the Discussion, it is possible that this is due to increased synthesis of some repeat RNAs in response to stress. Since 18s rRNA (RNAPI) and 5s rRNA (RNAPIII) were seen in G1 daughter nuclei under conditions where CoT-1 RNA was not (Figure S3L–Q), this suggests that much of the CoT-1 RNA signal could be RNAPII regulated. However, the increased interphase expression with prolonged α-amanitin potentially implicates the involvement of RNAPIII. These results are consistent with other recent evidence that there is a complex interplay between RNAPII and RNAPIII transcription (Raha et al., 2010).
An important observation is that the repeat RNA consistently maintained its tight localization to the chromosome territory in cis, unlike poly-A and other nuclear RNAs, which typically redistribute if they persist following transcription arrest (discussed in (Hall et al., 2006). CoT-1 RNAs did not significantly disperse during short or long periods of DRB or α-amanitin (Figure 5A–H), suggesting a structural association with the chromosome territory. The only exception was that in ActD, some portion of the CoT-1 RNA drifted from the parent chromosome in hybrid cells (Figure S3A–D). This was also seen with XIST RNA in female fibroblasts (Figure S3E–H), suggesting the interesting possibility that this DNA intercalating drug displaces some chromosome-bound RNAs.
Collectively, data indicates the abundance of repeat RNA on chromosomes reflects a steady-state accumulation of stable RNAs on chromosome structure, rather than a high rate of their transcription.
5’ truncated L1 sequences are a prominent component of CoT-1 RNA
Since CoT-1 DNA will detect a mixture of repeat-containing transcripts, we sought to identify specific components of CoT-1 RNA and evaluate the relative expression of different repeat families. Using specific probes to several different repeat families (Table S2), we determined that satellite RNAs (alpha, SatII and SatIII) and several simple sequence repeats did not contribute appreciably to the broadly distributed nucleoplasmic CoT-1 RNA signal. In contrast, both L1 and Alu RNAs showed nucleoplasmic signal, although initial observations suggested their levels may differ. Comparison of overall expression of repeat families is complicated by differences in size, number and divergence of genomic repeats. To circumvent this and technical differences in probes, we developed a strategy based on the ratio of nuclear RNA to DNA signal as an indication of the amount of RNA detected per unit of DNA. Thus RNA:DNA ratios for L1s (5’ and 3”) and Alu (Table 2) were compared (see methods). We introduced fluorescent beads as an intensity standard (Figure S4) for more rigorous quantitative comparisons within and between slides.
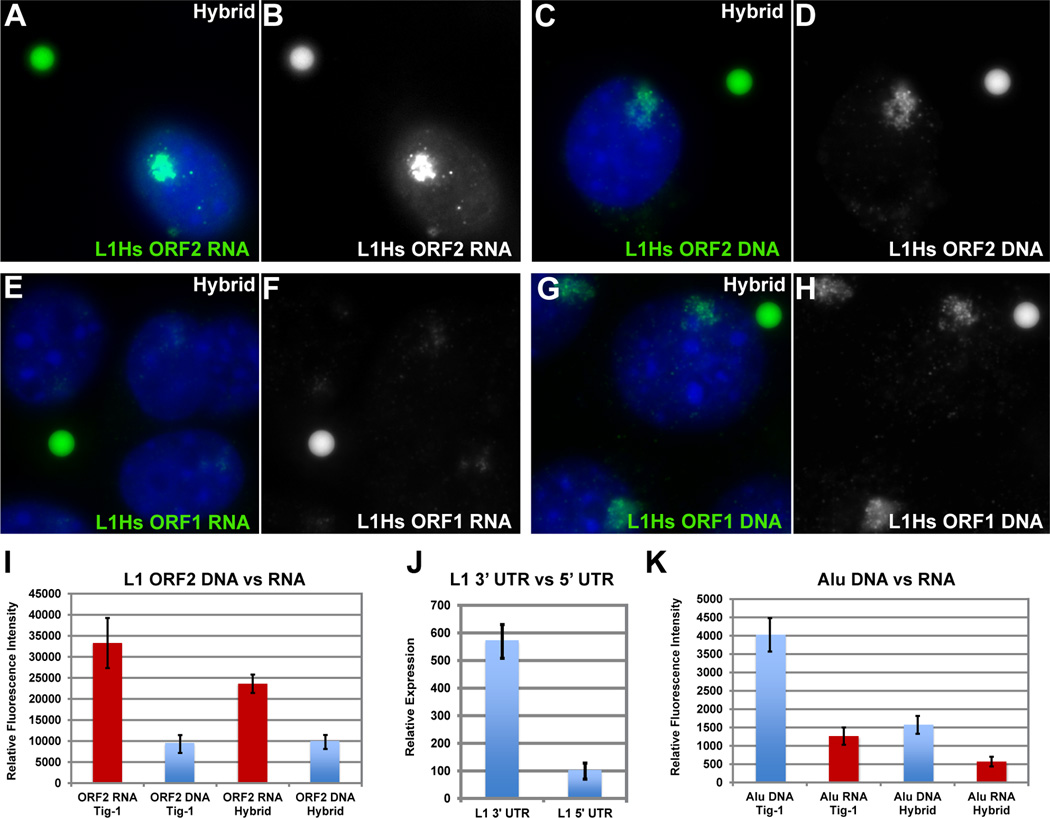
Full-length (transposable) LINEs (6 kb) have a canonical 5’ promoter transcribing two open reading frames (ORF1 and ORF2), encoding a reverse transcriptase and endonuclease that can “copy and paste” the retrotransposon elsewhere in the genome. Most of the ~500,000 human copies are truncated and lack the 5’ promoter, with the few full-length LINE1 (L1) elements suppressed by promoter methylation in somatic cells (Goodier and Kazazian, 2008; Lander et al., 2001). Thus, the vast majority of L1 are thought silent in normal cells. We first used a 4kb probe to ORF2 of human L1 (L1.3) to broadly assess for presence of L1 RNAs (Figure S5A).
L1 ORF2 RNA signal was abundant through the nucleoplasm, but was not in the cytoplasm, nucleoli or heterochromatin, similar to CoT-1 RNA (Figure S5B–G). In hybrid cells, L1 RNA formed a bright, well-defined RNA territory, precisely overlapping the corresponding Chr4 DNA territory (Figure 6A–B), and also persisted following transcription arrest. Using the quantification strategy described above, L1 RNA signal was roughly four-fold more than its abundant DNA signal. The RNA:DNA ratio (range 2.3–4.5) was comparable in hybrid cells and normal fibroblasts (Figures 6A–D & I, and Table S3), hence L1 RNA level is not an aberration of hybrid cells (Figure S5H–M).
Figure 6. RNA from the 3’ end of L1 is a large component of the CoT-1 RNA signal.
L1 ORF2 RNA signal (A–B) is ~4X brighter than the double stranded L1 ORF2 DNA signal (C–D), while the L1 ORF1 RNA signal (E–F) is ~0.2X the DNA signal (G–H). I) L1 RNA/DNA signals were quantified using digital fluorimetry. J) qPCR confirms L1 ORF2 RNA is ~5–7X more expressed than ORF1 RNA. K) Alu RNA is ~3–4X dimmer than its DNA signal quantified by digital fluorimetry (See Fig S5 & S6).
Since full-length L1s are 5’ truncated, we measured the DNA:RNA ratios for probes to the 5’ or 3’ ends (Figure S5A and Table S3). L1 ORF2 RNA was consistently several-fold brighter than the corresponding DNA, whereas the opposite was true for the 5’ ORF1 (RNA:DNA ~1:4)(Figure 6E–H and Figure S6A). Preference for 3’ L1 RNA was confirmed using smaller probes of identical size (200nt) to the 5’ versus 3’ UTRs of L1Hs (Figure S6B–F), and was further supported by 5’ vs 3’ L1Hs RT-qPCR (Figure 6J). Consistent with other evidence that full-length L1Hs are silenced, results indicate 5’ truncated L1s are abundantly and stably associated with chromosome territories.
We also examined L1 expression in several human RNA deep sequencing datasets, and found more L1 reads mapping to the 3’ than to the 5’ end, and mostly in the sense direction (Figure S6G). However, L1 read frequency overall in these extraction-based RNA samples was lower than expected from our FISH analyses. For example, in normal pancreatic tissue, using a database of L1 consensus sequences from Repbase, we find only 100 reads per million mapping to L1. We also attempted to examine these repeat RNAs by dot blot or northern blot (Figure S6H) using standard Trizol RNA extraction and found only a low level of heterogeneously sized L1 transcripts. We suggest that the underrepresentation of these transcripts by most extraction-based methods may be due to the structural nature of this chromatin-associated RNA (discussed further below).
The copious nature of L1 nuclear RNA was reinforced by comparison to a substantially lower RNA:DNA ratio for the major human SINE, Alu, using oligo as well as larger probes (Table S2), measured in parallel samples with L1. While Alu RNA was across the nucleoplasm, Alu RNA signal was consistently several-fold less than its DNA signal in fibroblasts, and very weak in hybrid cells (Figure 6K, Figure S6I–P). Assuming roughly similar accessibility of sequences in situ, this marked difference in RNA:DNA ratios indicates representation of L1 sequences in nuclear RNA is substantially higher (Table S3). Bioinformatic analysis of L1 and Alu on human Chr4 shows similar copy number (151 full-length and 58,639 truncated L1s, and 54,993 Alu). Since 68% of Alus and 47% of L1s are in introns, genic transcription would not account for this difference. This further suggests the nuclear repeat RNA signal is not mere transcriptional noise, and it raises the possibility of differential regulation/roles of repeat families (see Discussion).
CoT-1 RNA structurally associates with the interphase territory but can be released by perturbation of the nuclear scaffold
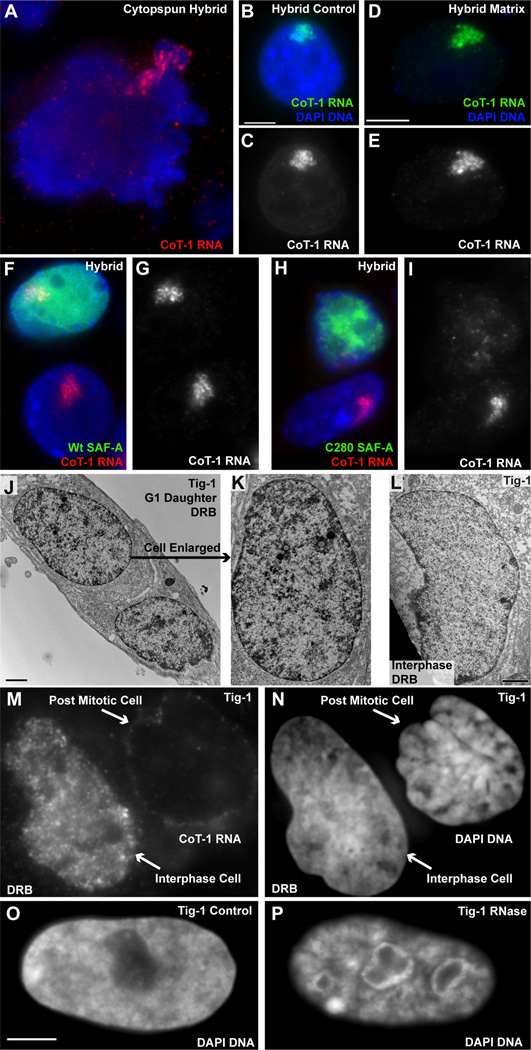
Strict localization of CoT-1 RNA to the territory in cis (even after transcription arrest) suggests it may be actively tethered to nuclear/chromosome structure, as opposed to passive accumulation of a non-transported RNA (or rapid degradation of released RNA). We found CoT-1 RNA localization resists mechanical disruption of nuclei by vigorous cytospinning (Figure 7A), and even after biochemical fractionation (removing >90% protein and DNA), the bright, precisely localized CoT-1 or L1 RNA territory remains undisturbed (Figure 7B–E & S7A–D). This suggests this RNA is bound to or embedded in a non-chromatin nuclear sub-structure, which was initially characterized as a complex network of multiple insoluble proteins and RNA (reviewed in (Nickerson, 2001). Similarly, XIST RNA, which is established to bind the X-chromosome, remains as an undisturbed nuclear RNA territory after removal of most chromatin (Figure S7E–H) (Clemson et al., 1996).
Figure 7. CoT-1 RNA associates with the nuclear scaffold and Cot-1 RNA loss is coincident with chromatin collapse.
A) CoT-1 RNA remains bound to human Chr4 in ruptured nuclei. B–C) CoT-1 RNA is localized in control cells, and D–E) remains localized after removal of ~95% of DNA and histones (in 100% of interphase cells). Exposure times are equal for both images. F–G) CoT-1 RNA remains localized in 88% of cells containing wt SAF-A-GFP, while 80% of cells transfected with C280-GFP mutant (H–I) release CoT-1 RNA. Images include neighboring untransfected cells. J–K) Chromatin collapse seen by EM in transcriptionally inhibited G1d cells compared to similarly inhibited interphase cells (L). M) Transcriptionally inhibited interphase cells retain stable CoT-1 RNA, while post-mitotic cells do not. N) Chromatin does not collapse in interphase cells, but more compacted post-mitotic cells were seen in treated (86%) versus control (12%) samples. O–P) Interphase chromatin collapses when unfixed cells are treated with RNase. Scale bars are 5µm except for EM images which are 2µm.
Since CoT-1 RNA fractionates with the non-chromatin scaffold, we used a dominant negative mutant of Scaffold Attachment Factor A, SAF-A/hnRNPU (C280)(Figure S7I) (Fackelmayer et al., 1994), to generally disrupt the scaffold. Strikingly, transfection with C280 readily released CoT-1 RNA (in ~80% of hybrid cells), which is then clearly seen dispersed through the nucleoplasm (Figure 7H–I and Figure S7J–N). Full-length SAF-A did not have this effect (88% remain localized) (Figure 7F–G & Figure S7L) and there was no indication of apoptosis (data not shown). Since this mutant will impact the complex nuclear scaffold, we do not yet know how direct the relationship of SAF-A and repeat RNAs is (see Discussion). However, the fact that CoT-1 RNA can be released from the territory shows that its normal localization is not via passive accumulation but involves a reversible mechanism for tethering to the parent nuclear chromosome.
Early studies suggested that chromatin associated RNAs may be more resistant to extraction (Bynum and Volkin, 1980), and our results indicate repeat RNAs are nuclear embedded and likely under-represented by traditional methods. Supporting this, we find the RNA:DNA ratio for L1 when hybridized in situ is 4:1, but less than 1:1500 following extraction and qRT-PCR (Figure S7O & Table S3). This may also explain why L1 ORF2 RNA is poorly represented in datasets compared to RNA FISH (Figure S6G and S7P–Q).
Loss of CoT-1 RNA is associated with chromatin condensation
In contrast to XIST RNA’s association with Xi heterochromatin, CoT-1 RNA distributes across euchromatic regions but is excluded from heterochromatin, as apparent for mouse peri-centromeric heterochromatin (Figure 1B), and the peripheral heterochromatic compartment (Tam et al., 2002) (Figure 1D–E). We previously showed that exclusion of CoT-1 RNA provides a convenient assay for chromosome silencing (Hall et al., 2002) and that this “CoT-1 RNA hole” overlapped the condensed Barr Body, which we showed was a repeat-rich silent core of the chromosome (Clemson et al., 2006; Hall et al., 2002)(Figure 1F–G). While CoT-1 RNA has been assumed to reflect nascent pre-mRNAs, results here indicate repeat RNAs are more widespread and associated with active chromosomes. Thus, chromosome inactivation by XIST involves widespread silencing of CoT-1 RNA and overall chromosome condensation.
This suggests the possibility that CoT-1 RNA loss may be associated with chromatin condensation in heterochromatin. To further investigate this, we examined post-mitotic G1 daughter (G1d) cells which lack CoT-1 RNA following transcriptional inhibition, compared to two types of controls: inhibited interphase cells (which retain the stable CoT-1 RNAs but not short-lived or pre-mRNAs, Figure 5K–L) and untreated normal G1d cells (which re-synthesize CoT-1 RNA). Using mitotic shake-off to enrich for G1d cells, we noted more condensed DNA regions within treated nuclei, and examined them by both fluorescence and electron microscopy. Electron microscopy confirmed a consistent increase in dense chromatin clumps in most DRB treated G1d cells (Figure 7J–K & S8B,D), not seen in similarly treated interphase nuclei (Figure 7L) nor in untreated G1d controls (Figure S8A,C); additionally we noted two types of unusual condensed structures not seen in controls (Figure S8E–F). We quantified these same cultures by light microscopy, where 77% of DRB treated G1d cells exhibited larger, more numerous condensed clumps of chromatin, compared to only 16% of controls. Similar results were seen in inhibited asynchronous cultures, which showed two-fold more paired G1 daughter cells with condensed clumps of chromatin (Figure 7M–N & S8G–H). The chromatin of interphase cells remained more uniform in topography, with less regions of high-density DNA compared to inhibited G1d nuclei (Figure S8M–O), and this was also evidenced by less homogeneous distribution of chromatin proteins (Figure S8I–L). Examination of the Chr4 DNA territory in inhibited G1d hybrid cells showed the territory partially decondensed but retained a large mass of condensed chromatin within it (Figure S8P–Q).
The lack of chromatin collapse in interphase cells (which retain CoT-1 RNA) (Figure 7L–N) indicates that condensation is not due to transcriptional inhibition per se nor the absence of nascent or short lived nuclear RNAs. Alternatively, it could be due to lack of re-synthesis of stable nuclear RNAs or proteins necessary to open chromatin. To address this, we used RNase in unfixed cells to determine if removal of CoT-1 and other nuclear RNA could cause interphase chromatin to condense (Figure S8R–S). Chromatin collapse was immediately visible (within 20 minutes); ~60% of cells were markedly affected as seen in Figure 7O–P. This is consistent with earlier observations of RNase (Nickerson et al., 1989), but collective findings here support that abundant, stable, repeat-rich RNAs are widely associated with euchromatic chromosome territories and likely promote more open chromatin packaging.
DISCUSSION
These findings impact two “black boxes” of genome biology: the unexplained prevalence of repetitive elements interspersed throughout higher-order genomes and the poorly understood diversity of non-coding RNAs. In situ visualization of RNA using CoT-1 DNA as a probe reveals that abundant RNA is broadly distributed with chromatin and remains stable and localized to the parent chromosome long after transcriptional arrest. Recent evidence indicates that specific ncRNAs may bind a subset of chromosomal loci. Findings here support a distinct but potentially related concept: that RNA generally and broadly associates with nuclear chromosome territories, where it remains stable and localized independent of ongoing transcription. This suggests RNA is a fundamental component of chromosome biology, rather than only a product of it. Like XIST RNA, CoT-1 RNA detaches from mitotic chromosomes, yet it tightly adheres to the interphase chromosome structure in cis, even after nuclear fractionation. Hence, CoT-1 RNAs and XIST RNA can be considered “chromosomal RNAs” that likely bridge chromatin with insoluble non-chromatin structural elements (see Graphical Abstract). While speculative, repetitive sequences would be well suited to form an intermolecular lattice or structural element, since long-non-coding RNAs can have an “architectural” role and intermolecular RNA duplexes were long ago noted in nuclear RNA (Fedoroff et al., 1977). Such RNAs may also play a role in higher-order chromatin packaging linked to regulation. In contrast to XIST RNA which triggers chromosome condensation, CoT-1 transcripts specifically distribute across euchromatin, where they may promote an open chromatin state.
These findings point to the import of interspersed repeat sequences in chromatinassociated RNA, raising many next questions about this poorly studied fraction of the genome. Britten and Davidson (1971) long ago hypothesized that repeats function in genome regulation, stating “A concept that is repugnant to us is that about half of the DNA of higher organisms is trivial or permanently inert…” (Britten and Kohne, 1968). Our motivation to study repeats stemmed from their poorly explained abundance and wide distribution in conserved patterns, suggesting to us a potential role in coordinate regulation of the genome. We show repeat sequences are not only prevalent in chromosome-associated RNA, but they are the predominant component of hnRNA stably associated with chromosomes. Retention on the parent chromosome and longer-nuclear half-life, likely contribute to the greater level of repeat RNAs detected on the chromosome versus the collective non-repetitive sequences in the whole chromosome library. Hence, the level of RNA on the chromosome will be disproportionate to the transcription rate and cytoplasmic abundance. The in situ analyses here revealed both unexpected nuclear abundance and unusual cellular properties of CoT-1 RNAs, distinct from those expected for nascent pre-mRNAs or “genic” transcripts, including localization to the chromosome territory which persisted for over a day after transcription arrest. To minimize any chance that an inhibitor induced stabilization, we tested inhibitors with three distinct mechanisms, with similar results. Interestingly, it has been reported that the many thousands of uncharacterized “lncRNAs” are rich in (or related to) interspersed repeat elements (Hadjiargyrou and Delihas, 2013; Kapusta et al., 2013), and many show prolonged stability (Clark et al., 2012).
Full characterization of the heterogeneous class of CoT-1 repeat RNAs will require improved nuclear RNA extraction methods and RNA sequencing, however we were able to identify L1 RNA as a major component. The prevalence of 5’ truncated L1 is consistent with evidence that full-length transposable L1s are silenced in normal cells (Goodier and Kazazian, 2008), and is distinct from the reported transient expression of a few full-length L1s during mouse X-chromosome silencing (Chow et al., 2010). A recent study found that highly abundant, truncated L1 elements contain internal and 3’ promoters, are expressed at low-levels in a cell-type specific manner, and may be resistant to extraction (Faulkner et al., 2009). Our in situ analysis with large probes, and preliminary Northern analyses (also limited by extraction) showed most L1 transcripts are large (>kb), thus these could include lncRNAs, or even intron-derived sequences. Importantly, the vast majority of introns contain interspersed repeats, and most introns still have no known function. While excised introns are generally thought to rapidly degrade, a very recent study in xenopus oocytes found that highly stable RNAs representing excised introns accumulate in the germinal vesicle (Gardner et al., 2012). While the xenopus oocyte is a distinct biological system, it is an interesting possibility that repeat-rich sequences in excised introns could accumulate on chromosomes.
To minimize the technical obstacles to studying repeats, we needed to develop new approaches, such as the use of RNA:DNA ratios (and fluorescent beads as intensity standards) to assess the relative expression of repeat families and control for differences between probes used. Results indicated L1 RNA is abundant on the chromosome, more so than Alu RNA (which shows some cytoplasmic component). Goodrich and colleagues have reported that SINE RNAs (mouse B2 and human Alu) are upregulated by RNAP III upon heat shock, and then bind RNAP II to broadly repress transcription (Espinoza et al., 2004; Mariner et al., 2008). Studies here used normal unstressed cells, however under prolonged transcriptional inhibition (with α-amanitin), CoT-1 RNA signal intensity increased consistently, thus we are investigating whether some component(s) of CoT-1 RNA are up-regulated upon inhibition of RNAPII. Interestingly, HERV-H retroviruses, a component of CoT-1 DNA, has been reported to be highly expressed specifically in pluripotent stem cells (Santoni et al., 2012). Thus, we believe the collective evidence supports that studies of genome biology should include the “repeat genome” in which distinct repeat families likely will show differential regulation and different functions in a cell-type specific manner.
Our demonstration that the CoT-1 (and L1) RNA “territories” resist mechanical (or transcriptional) disruption, but can be released to disperse indicates their accumulation is not passive, but involves a reversible mechanism(s). That the release was triggered by perturbation of a nuclear scaffold factor further supports the biochemical fractionation results which indicate repeat RNAs are tethered with a non-chromatin scaffold. Since the latter is likely a complex structure (not simple polymers), we do not yet have a full understanding of how the dominant negative C280 mutant disrupts CoT-1 RNA localization. Preliminary evidence indicates the relationship may not be direct and multiple factors may anchor the RNA, thus the role of SAF-A with RNA and chromatin is the subject of a separate study. Nonetheless, our findings identify cis-localized CoT-1 RNA as a major component of the undefined RNAs previously suggested to be part of an insoluble non-chromatin structure (e.g. (Fey et al., 1986). The structural nature of repetitive RNAs may well make them more difficult to extract by methods designed for the cytoplasm. We are working to identify improved extraction methods for further analyses, however molecular hybridization in situ avoided this issue, and was essential to study the relationship of these RNAs to the nuclear chromosomes.
A key point is that the CoT-1 RNA distributes specifically over euchromatin, and several of our results suggest euchromatin-associated RNA helps promote open chromatin packaging, as recently suggested for repetitive RNAs in Drosophila (Schubert et al., 2012). CoT-1 and XIST chromosomal RNAs show inverse relationships to active and inactive chromosomes, respectively. We previously reported that the condensed Barr Body is rich in CoT-1 DNA (Clemson et al., 2006), although it is devoid of CoT-1 RNA (Hall et al., 2002). A priori, this “CoT-1 RNA hole” could reflect clustering of already silent DNA repeats. However, evidence here supports that CoT-1 RNA is abundant on active chromosomes, and thus we suggest that XIST RNA silences or displaces repeat RNAs as well as genic transcription. This is consistent with prior evidence that the CoT-1 RNA “hole” is spatially and temporally separable from gene silencing (Chaumeil et al., 2006; Clemson et al., 2006). DNA condensation occurs as CoT-1 RNA is lost from the inactivating chromosome. This is one of several observations that lead us to propose that euchromatin associated RNAs (ecRNAs) may promote open chromatin, whereas heterochromatin associated RNAs (hcRNAs) trigger increased condensation. Our results with RNase indicate that removal of ecRNAs leads rapidly to chromatin condensation or “collapse”, however our results do not address if this is specific to repeat RNAs, the most prominent component of ecRNA. However, the involvement of interspersed repeats (sequences common on all chromosomes) in chromosome condensation and silencing is further suggested by XIST RNA’s ability to comprehensively silence an autosome (Jiang et al., 2013).
In sum, the repetitive “junk” genome is worthy of study, not only as DNA, but as a major part of the “dark matter” transcriptome. As only 5% of promoters are with canonical genes (Venters and Pugh, 2013), we are far from understanding genome expression and regulation. This work contributes to what may emerge as a next revolution in genome science: the relationship of chromosomal RNAs and repetitive elements to each other and to genome packaging, regulation, and evolution.
EXPERIMENTAL PROCEDURES
Cell culture and treatments
DM1 Myoblasts, HT1080 G3 (Hall et al., 2002), Wi38, Tig-1, NIH3T3, GM11687 were grown under conditions recommended by supplier (ATCC, DMI, Myoblasts from Charles Thorton, Wellstone Muscular Dystrophy Cooperative Research Center, Rochester, NY). Inhibitors: Actinomycin D (5–20µg/ml), 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) (40–80µg/ml) or α-amanitin (5–50µg/ml) (dissolved in DMSO) were added to culture media (4–6 hours, also 16–32 hrs for α-amanitin). Nocodazole (100ng/mL) arrested cells were released into inhibitors for 4–6 hrs to score inhibited G1 daughter cells. Cytospin: Interphase cells were trypsinized and cytospun at 8000–10000 RPM onto glass coverslips, prior to fixation. Matrix-prep: Digestion of DNA and histones were performed as described previously (Clemson et al., 1996). RNase: Un-fixed cells on coverslips were permeabilized with 0.1% triton-X in CSK buffer (4°C for 3min) then treated with 5µL/mL DNase-free RNase (Roche #11119915001) in CSK (20min) or in PBS (3 hours, 37°C) for fixed cells. SAF-A: 2µg/mL GFP-tagged full length and C280 SAF-A were used to transfect cells with Lipofectamine 2000 (Invitrogen). Cells fixed after 16, 24, 48, or 72 hours. Highly over-expressed cells were not scored.
Cell Fixation, FISH and IF
Fixation: Our standard fixation protocols have been detailed previously (Johnson et al., 1991; Tam et al., 2002). Human tissue blocks were cryosectioned, then fixed. The absence of cytoplasmic CoT-1/L1 RNA was confirmed using fixations that preserve cytoplasmic RNAs (Clemson et al., 1996; Hall et al. 2009). See Figure S4 legends for standardized beads. FISH: All DNA probes (1µg/reaction) nick-translated using biotin-11-dUTP or digoxigenin-16-dUTP (Roche). Oligos are end labeled with Biotin, Fluoroscein, or Alexa 594 (Invitrogen). RNA FISH and simultaneous RNA/DNA detection have been described previously (Johnson et al., 1991; Tam et al., 2002). Oligos are hybridized at 15% formamide. FISH probes: 4kb human L1.3 ORF2, and L1.3 ORF1 (from J. Moran, UMich), PCR generated 3’ and 5’ L1Hs UTR, and 2kb DMPK intron 9 (Table S2), introns (non-repetitive) for XIST, COL1A1 and GAPDH were PCR generated and cloned into pSC-A (Stratagene) (Table S2), 10kb human XIST pG1A,10kb 18S rDNA (from G. Stein, UMMS), 330bp Human Alu pPD39 clone (ATTC), human and mouse CoT-1 DNA (Roche and Invitrogen respectively), and two Chr4 paints (Qbiogene & Cytocell). Oligos used were 54-mer Poly-dT, 55-mer 5s rRNA, and 33-mer human Alu (Table S2). Immunofluorescence: Slides incubated with appropriate dilution of primary antibody in 1%BSA, 1×PBS and 1U/ul RNasin, for 1 hour at 37C, then washed, and immunodetected using 1:500 dilution of conjugated (Alexa 488 or Alexa 594, Invitrogen) secondary (anti-goat, mouse or rabbit) antibody, in 1×PBS with 1% BSA. Antibodies: HP-1 Gamma (Chemicon), Histone H3K9me3 (Upstate) Rad 21 (Abcam).
Microscopy and Image Analysis
An Axiovert 200 or an Axiophot Zeiss microscope was used, equipped with a 100X PlanApo objective (NA 1.4) and Chroma 83000 multi-bandpass dichroic and emission filter sets (Brattleboro, VT), set up in a wheel to prevent optical shift, with an Orca-ER camera (Hamamatsu, NJ) or a cooled charge-coupled device (CCD) camera (200 series, Photometrics). Where required, a narrow band-pass fluorescein filter was inserted to correct for any bleed-thru of Texas-red fluorescence into the fluorescein channel. Most experiments were carried out a minimum of 3 times, with typically 50–100 cells scored in each experiment. Key results were confirmed by at least two independent investigators. Images were minimally enhanced for brightness and contrast to resemble what was seen by eye through the microscope (unless otherwise noted in the text/legends). Digital imaging software (Volocity from Perkin Elmer & Metamorph) was used to quantify signals (see Supplement for details).
Supplementary Material
HIGHLIGHTS.
Abundant euchromatin-associated chromosomal RNA is predominantly CoT-1 repeat RNA
Stable CoT-1 RNA remains localized to the chromosome long after transcription arrest
CoT-1 RNA fractionates with non-chromatin scaffold and disperses upon its disruption
Repeat-rich chromosomal RNA is silenced by XIST RNA and may promote open euchromatin
ACKNOWLEDGEMENTS
This work was supported by NIH grant #GM053234 to J.B.L. D.M.C. is supported by NIH/NCI NRSA 1F32CA154086. We wish to thank John Moran for the gift of L1 ORF1 and ORF2 plasmids and Laurie Lizotte for general technical assistance. A special thanks to Lara Strittmatter from the UMMS Electron Microscopy core for the EM fixation and analysis. The EM Core facility is supported by NIH Award # S10RR027897.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: J.B.L and L.L.H conceived the project. J.B.L., L.L.H. and D.M.C designed and analyzed experiments and wrote the manuscript. L.L.H., D.M.C, M.B., A.V.G, and N.M. performed all CoT-1 (repeat) in situ and quantification experiments. L.L.H. and M.B. performed XIST in situ experiments. D.M.C. performed RNA extraction experiments. J.B.L., H.J.K., and F.O.F. designed, performed and analyzed SAF-A experiments.
REFERENCES
- Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Current opinion in cell biology. 2012;24:333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. Repeated sequences in DNA: Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Bynum JW, Volkin E. Chromatin-associated RNA: differential extraction and characterization. Biochimica et biophysica acta. 1980;607:304–318. doi: 10.1016/0005-2787(80)90083-0. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TL, Manuelidis L. SINEs and LINEs cluster in distinct DNA fragments of Giemsa band size. Chromosoma. 1989;98:309–316. doi: 10.1007/BF00292382. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TEP. A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:1–21. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Fedoroff N, Wellauer PK, Wall R. Intermolecular duplexes in heterogeneous nuclear RNA from HeLa cells. Cell. 1977;10:597–610. doi: 10.1016/0092-8674(77)90092-7. [DOI] [PubMed] [Google Scholar]
- Fey EG, Ornelles DA, Penman S. Association of RNA with the cytoskeleton and the nuclear matrix. JCell Sci. 1986;(Suppl.5):99–119. doi: 10.1242/jcs.1986.supplement_5.6. [DOI] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes & development. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Hadjiargyrou M, Delihas N. The Intertwining of Transposable Elements and Non-Coding RNAs. Int J Mol Sci. 2013;14:13307–13328. doi: 10.3390/ijms140713307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. XIST RNA and architecture of the inactive X chromosome: implications for the repeat genome. Cold Spring Harb Symp Quant Biol. 2010;75:345–356. doi: 10.1101/sqb.2010.75.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CV, Primorac D, McKinstry M, McNeil JA, Rowe D, Lawrence JB. Tracking COL1A1 RNA in Osteogenesis Imperfecta: Splice-defective transcripts initiate transport from the gene but are retained within the SC-35 domain. J Cell Biol. 2000;150:417–431. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: Implications for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS genetics. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Neguembor MV, Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics. 2010;2:271–287. doi: 10.2217/epi.10.8. [DOI] [PubMed] [Google Scholar]
- Nickerson J. Experimental observations of a nuclear matrix. J Cell Sci. 2001;114:463–474. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, Imhof A, Langst G. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Molecular cell. 2012;48:434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- Smith K, Byron M, Johnson C, Xing Y, Lawrence JB. Defining early steps in mRNA transport: Mutant mRNA in Myotonic Dystrophy Type I is blocked at entry into SC-35 domains. Journal of Cell Biology. 2007 doi: 10.1083/jcb.200706048. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R, Shopland LS, Johnson CV, McNeil J, Lawrence JB. Applications of RNA FISH for visualizing gene expression and nuclear architecture". Vol 260. New York: Oxford University Press; 2002. [Google Scholar]
- Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatic unlike most human telomeres. Journal of Cell Biology. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53–58. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.