Abstract
Because activated oncogenes like Ras have traditionally been thought as promoting unrestrained proliferation, the concept of oncogene-induced senescence has been, and still is, controversial. The counter-intuitive notion that activation of oncogenes leads to the prevention of cellular proliferation has initially been fueled by in vitro studies using ectopic expression of activated Ras in primary fibroblasts. While these initial studies demonstrated unambiguously the existence of a new type of cellular senescence, induced by oncogenes in an ex-vivo system, questions were raised about the physiological relevance of this process. Indeed, recent technical advances in mouse modeling for cancer have suggested that the occurrence of Ras-induced senescence is highly dependent on the cellular context, as well as the level of expression of activated Ras, and may not be pertinent to the study of human cancer initiation and/or progression. However, our increased knowledge of the molecular basis for cellular senescence has led to a better understanding of the molecular events modulating cancer progression in vivo. Recent studies have not only clearly established the incidence of cellular senescence in pre-neoplasic lesions, but also its role as a potential tumor-suppressor mechanism in vivo. Here, we review the recent and exciting new findings regarding the physiological relevance of Ras-induced senescence, and discuss their implications in terms of cancer therapy.
Keywords: Senescence, cell cycle, cancer, chromatin, Rb, SAHF, SASP
Cellular Senescence: Phenotypes and Markers
Senescence was first a functional characterization of a specific cellular state. A senescent cell is a cell that has exited the cell cycle permanently, and is incapable of resuming proliferation, even upon mitogenic cues [1,2]. This functional definition of cellular senescence is in contrast to cellular quiescence, which also represents a G0 cell cycle exit, but is reversible upon pro-proliferative stimuli [3]. While this definition of cellular senescence has been helpful in delineating a functional classification within cell cycle exit, markers that would allow the visualization of senescent cells, regardless of the trigger driving senescence, needed to be identified.
The most obvious feature of a cell that has undergone senescence in culture is a typical flat and enlarged morphology. This phenotype is consistent with cell cycle withdrawal along with a sustained active metabolism. Obviously, this phenotype cannot be used as a means to discriminate which cells are senescent in vivo. As an alternative approach to identify senescent cells, the use of protein markers that would mark senescent cells specifically was investigated. Of note, until the molecular basis for cellular senescence became better understood, the correlation between the presence of these marks and the senescent process was largely circumstantial (see below). These approaches led however to the identification of several ways to visualize senescent cells, both in vivo and in vitro.
Senescence–Associated β-Galactosidase Positivity
A breakthrough in the research surrounding cellular senescence and its physiological relevance was the identification of an enzymatic activity that could allow the detection of senescent cells: indeed, senescent cells exhibit what seems to be a unique and senescence-specific β-galactosidase activity in acidic conditions (pH 6.0) [4]. This serendipitous finding has helped the field tremendously. Indeed, this activity, called senescence associated β-galactosidase activity (SA-β-Gal) has been shown to consistently allow the identification of senescent cells in vitro, upon replicative senescence or stress induced senescence, as well as in vivo, within tissues that are believed to contain a high proportion of senescent cells [5]. For example, primary fibroblasts subjected to serial passaging or infected with activated Ras or skin samples isolated from old donors, stain positive for SA-β-Gal [4,6]. Consistent with this observation, GLB1, a lysosomal β-D-galactosidase, active under acidic conditions, was recently shown to accumulate upon serial passaging in primary fibroblasts, and to be required for the increased β-galactosidase associated with the onset of senescence [7]. Importantly, knock-down of GLB1 does not prevent cell cycle exit upon serial passaging, suggesting that the increase in SA-β-Gal activity is a consequence rather than a driver of senescence. Of note, GLB1 is a lysosomal protein, hence any physiological situation, other than senescence, that results in the increase of lysosomes/ lysosomal activity within the cell, can also lead to increased SA-β-Gal activity. For these reasons, it is conceivable that, despite its undeniable usefulness so far in the senescence field, SA–β-Gal positivity may not be a marker absolutely specific for cellular senescence.
Senescence Associated Heterochromatic Foci
One insight in both the genesis of senescence as well as the identification of senescent cells came from a recent study which demonstrated that nuclei of senescent cells are subjected to a dramatic reorganization, as evidenced by the emergence of dense foci called SAHF (for Senescence Associated Heterochromatic Foci) [8] that are brightly stained with 4’-6-Diamidino-2-phenylindole (DAPI). Indeed, at the onset of replicative or oncogene induced senescence, specific loci are embedded into heterochromatin, which is the dense subset of chromatin, believed be in a closed conformation, thus preventing access to the transcriptional machinery. These loci are then assembled to form large sub-nuclear structures that can easily be visualized using antibodies directed against markers of constitutive heterochromatin, such as histone H3 Lysine 9 tri-methylation (H3K9me3), hypoacetylated histones and HP1, as well as other chromatin associated marks such as macroH2A and HMGA proteins and depletion of linker histone H1 [9–11]. Consistent with the heterochromatic nature of these modifications, SAHF are resistant to digestion by nucleases. While such foci are easily visualized in human cells (as well as in human tissues, see below), their detection in mouse cells is hindered by the fact that rodent pericentric loci are readily decorated by the aforementioned chromatin marks, regardless of the senescence status. SAHF are believed to comprise genomic loci encoding E2F repressor targets, pro-proliferative proteins, thus preventing their accessibility by the transcription machinery at the onset of senescence [8]. In addition, it has been suggested that each chromosome condenses into one single SAHF, where the genes to be repressed are found in the interior or immediate periphery of the corresponding focus [11].
The molecular events leading to the formation of SAHF are still under investigation, but several proteins have been demonstrated to contribute to the formation of SAHF: The proteins shown to be required for Ras-induced formation of SAHF include the histone chaperones HIRA and Asf1, HP1γ, HMGA proteins, and the H3K9me3 methyl-transferases Suv39h1/h2 [10,12,13]. Importantly, these reports have also suggested that the establishment of SAHF is required for the cell cycle withdrawal observed upon pro-senescence signals (see below). In addition, since all chromatin modifications identified in SAHF have now been shown to be reversible, the irreversible nature of senescence is likely to rely on additional molecular mechanisms yet to be identified.
Other markers
The search for additional markers with perhaps greater specificity for senescence has recently been aided by the genetic elucidation of key signaling pathways activated upon induction of senescence. For instance, many proteins that promote cell cycle exit are up-regulated at the onset of senescence, including p19ARF in mouse cells, and p16INK4A and p15INK4B in mouse and human cells [14–16]. These markers have been used to identify cells subjected to cellular senescence in human preneoplasic lesions or aging tissues [14,17]. One caveat regarding the use of these markers, however, is that many of them do not discriminate between senescence and any other form of growth arrest.
Finally, additional markers significantly up-regulated at the onset of replicative or oncogene-induced senescence have recently been identified. Despite our incomplete understanding of the functional significance of their up-regulation at the onset of senescence, the increase of such markers has so far proven to be specific to senescent cells in vitro as well as in senescent tissues, as opposed to other types of cell cycle arrest. These include Dcr2, Dec1, Mcl1, among others [14,18–20]. More studies need however to be conducted in order to definitely establish whether these markers are found in all senescent cells regardless of the cellular context.
Triggers for Cellular Senescence
Replicative Senescence in human and mouse
As mentioned above, cellular senescence can be triggered by several stimuli besides oncogene activation. Historically, cellular senescence was first evidenced by the exhaustion of the proliferation potential of primary cells in vitro, leading to the notion of replicative senescence [21]. Later, ectopic expression of activated Ras [6], as well as administration of agents that cause DNA double strand breaks [22] were shown to induce a cell cycle withdrawal in primary cells that is phenotypically indistinguishable from replicative senescence. The fact that cells subjected to distinct pro-senescence stimuli acquire phenotypes that are not noticeably discernible from each other strongly suggests that common molecular mechanisms are engaged under these distinct situations. Among the types of senescence inducers, serial passaging and oncogene activation are the most studied.
Telomere erosion upon serial passaging is believed to account for replicative senescence of primary human cells, a hypothesis supported by the delayed senescence and extended replicative life span of fibroblasts expressing ectopic telomerase [23,24]. The molecular mechanism connecting telomere erosion to cellular senescence was recently elucidated. Upon serial passaging, telomeres of primary somatic cells shorten. Once telomere erosion reaches a critical point where the number of telomere repeats is too low to support capping by the protein complex Shelterin [25], chromosome ends are then recognized as DNA breaks thus activating a DNA damage checkpoint [26]. Such DNA damage recognition is believed to trigger the onset of replicative senescence through the activation of the ATM/p53 pathway [26].
In mouse cells, however, serial passaging leads to senescence through a different process. Indeed, mouse telomeres are much longer than human telomeres. In addition, telomerase is constitutively expressed in mouse somatic cells, thus allowing the stabilization of telomere length throughout serial passaging. Despite these differences, mouse embryonic fibroblasts still senesce upon serial passaging, suggesting the existence of a telomere-independent “culture stress” that would account for senescence [27]. Importantly, it was recently demonstrated that culturing mouse embryonic fibroblasts in low oxygen conditions (3% O2) obliterates replicative senescence [28]. Consistent with the specific sensitivity of murine cells to high oxygen levels, MEFs accumulate DNA damage in 20% O2, but not in 3% O2. This effect of high oxygen is believed to be mediated by the accumulation of Reactive Oxygen Species (ROS), which are detrimental to the integrity of nucleic acid molecule. Altogether, these observations suggest that DNA damage may be the main factor underlying replicative senescence, in human as well as in murine primary cells, even though this DNA damage occurs via different pathways. Consistent with a functional link between DNA damage checkpoint and the onset of cellular senescence is the observation that exposure to DNA damaging agents also directly promotes the accumulation of senescent cells [22]. In conclusion, one potential function of cellular senescence is to prevent the proliferation of cells that have accumulated deleterious mutations.
Oncogene-induced senescence
Similar to what has been reported upon serial passaging, cellular senescence has been proposed to halt proliferation of primary cells subjected to oncogene activation, for example upon Ras mutation [6]. In this case, cells withdraw from the cell cycle rapidly following expression of the activated oncogene, and acquire all phenotypes initially observed in cells driven to senescence by serial passaging or DNA damage. This process has since been dubbed “oncogene-induced senescence” (OIS) [29,30]. While OIS was first evidenced upon ectopic expression of activated Ras, ectopic expression of other oncogenes, including E2F1, Raf, mos, cdc6 and cyclin E, were later shown to result in the same cellular effects [31–33]. Similarly, overexpression of MEK induces senescence in primary mouse fibroblasts [34]. In addition, inactivation of the tumor suppressors PTEN or Nf1, leads to the accumulation of cells expressing markers of senescence [35,36]. Together, these observations strongly suggested that aberrant mitogenic signaling in primary cells could lead to permanent cell cycle arrest and cellular senescence.
However, why do some oncogenes trigger senescence and others do not remains unexplained, and the molecular bases for this difference are likely to lie within the molecular basis for senescence by contrast to quiescence. One possibility is that activation of Ras or its downstream targets engages specific biological pathways when hyperactivated, which are not engaged by other oncogenes. Activation of these specific pathways is then essential for the occurrence of senescence. Importantly, the downstream effectors of Ras that are required to trigger senescence are beginning to be unraveled (see below).
From Ras activation to oncogene-induced senescence
Cellular senescence in mouse cells is primarily driven by p19ARF, which serves as a sensor for activation of an oncogenic signal and triggers the p53 response [37]. Consistent with the notion that p19ARF, by contrast to p16INK4a, mediates Ras-induced senescence in mouse cells, murine fibroblasts derived from p19ARF null animals failed to arrest upon forced expression of activated K-Ras, while p16INK4a null primary mouse fibroblasts remain susceptible to oncogene-induced senescence [15,38,39]. By contrast, in human cells, Ras-induced senescence apparently involves a p53 response that is independent of p14ARF (the human homolog of p19ARF) [40–42].
The molecular pathway leading from activation of Ras to cellular senescence was believed to be a simple activation of the p53/p19ARF and/or p16INK4A pathways until recently. Although these proteins are likely to be major player in permanent cell cycle withdrawal elicited upon Ras activation, recent and exciting studies have demonstrated that other unsuspected actors contribute to the induction of the senescent phenotype downstream of Ras. Basically, the pathways engaged by activated Ras to induce permanent cell cycle withdrawal can be divided into three major groups, which surprisingly seem to intersect. The first pathway is transcriptional repression of pro-proliferative genes. The second pathway involves the DNA damage response elicited upon oncogene activation in primary cells, and the third pathway engaged upon Ras-activation senescence is the specific secretion program that has recently been identified to play a specific and essential role in cellular senescence. These are reviewed in turn below.
Transcriptional repression of E2F target genes
Consistent with the notion that senescent cells permanently suppress transcription of pro-proliferative genes become heterochromatinized in SAHF upon Ras-activation in primary cells [8]. As presented earlier, these structures are believed to embed E2F target pro-proliferative loci, and harbor most features of constitutive heterochromatin. Repression of some E2F targets relies on Rb and specific E2F factors, while repression of other E2F targets depends on the Rb-related p107 or p130 proteins. It is important to note that not all E2F targets are similarly repressed upon cell cycle exit, as revealed in quiescent cells [43]. Importantly, the range of E2F target genes that are silenced upon Ras activation has not been fully investigated. While Rb is clearly required in human cells for Ras to induce senescence [41,44], its contribution to senescence in mouse cells may be masked by the related p107 and p130 pocket proteins. Indeed, in mouse primary fibroblasts, inactivation of both Rb and one of the two other pocket proteins is required for cells to bypass replicative or Ras-induced senescence [45–47]. Importantly, acute somatic inactivation of Rb was sufficient in mouse primary fibroblasts to escape senescence, suggesting that the partial redundancy between Rb and its related proteins may be established early during development in the absence of a functional Rb allele [48].
How does Ras activation signal towards Rb and p53, both of which are essential regulators of cell cycle progression? In addition to Ras-mediated accumulation of p16INK4A, Ras activation also leads, in mouse cells, to the accumulation of the p19ARF protein, which then prevents Mdm2-degradation of p53, and leads to cell cycle arrest. The molecular links driving the accumulation of p16INK4A and p19ARF, both products of the INK4A locus, at the onset of senescence, have been at least part elucidated with the discovery that the polycomb protein Bmi1 represses transcription of the INK4A locus in normal culture conditions but is released upon oncogenic stress or serial passaging in primary cells [49,50]. p38 activation, which believe to be a downstream event of Ras activation, was shown to lead to a cascade of phosporylation/activation events results in the activation of 3pK (also known as MAPKAP kinase 3), which phosphorylates Bmi1, promoting its release from chromatin [51] (see Figure 1). Whether other target genes in addition to those present in the INK4A locus contribute to Bmi1 biological effects as they relate to senescence is unknown. More importantly, recent results have indicated that, at the onset of senescence, histones present within the INK4A locus become de-methylated on H3K27 (histone H3, lysine 27). Trimethylation of H3K27 (H3K27me3) serves as a recruitment mark for PRC1, the polycomb repressor complex that contains Bmi1. The molecular basis for the observed H3K27 de-methylation of the INK4A locus at the onset of senescence relies on Ras-mediated down-regulation of Ezh2, the H3K27 histone methyltransferase, as well as Ras-mediated up-regulation of JMJD3, an H3K27me3 demethylase that is recruited to the INK4A locus at the onset of senescence [48,50,51].
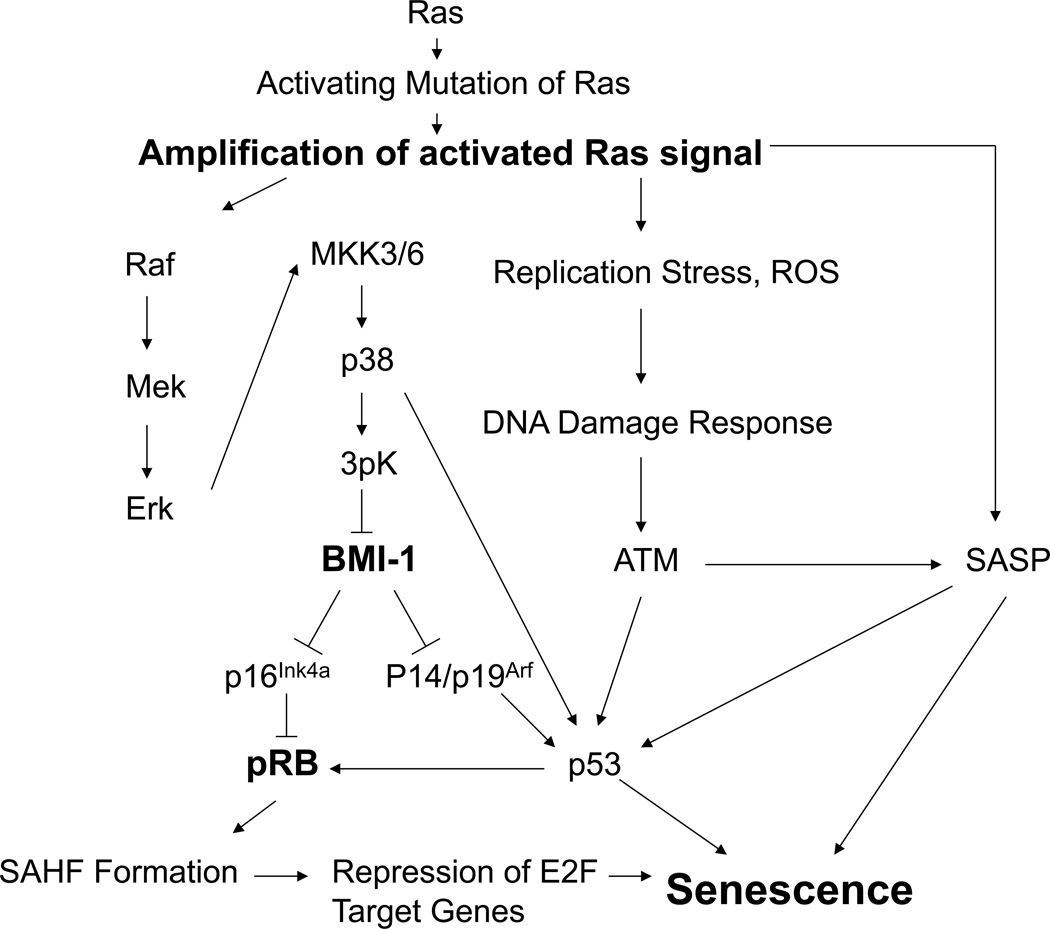
Figure 1. Schematic representation of the pathways involved in Ras-induced senescence and their potential crosstalks in the mouse.
Oncogene Induced Senescence (OIS) can initially be driven an activating mutation in Ras oncogene and eventual amplification of activated Ras signal. Mutated Ras then induces OIS through the cooperation of the MapK signaling cascade, the DNA damage response pathway and the SASP response. Activation of the MapK cascade leads to p38 activation through a series of phosphorylation events, which then activates MapKap Kinase 3 (3pK), which in turn phosphorylates Bmi1. This phosphorylation, along with chromatin modifications within the Ink4a locus, releases Bmi11 from the Ink4a locus allowing transcription of p16Ink4a and p19Arf. Upregulation of p16Ink4a proteins leads to SAHF formation and therefore repression of E2F target genes. Additionally, amplification of the activated Ras signal leads to the generation of Reactive Oxygen Species (ROS) and replication stress thereby engaging the DNA Damage Response. This response may promote senescence directly though up-regulation p53 in a direct manner, or through the activation of the SASP response. In addition, the DNA Damage Response also contributes to senescence through p53. For details and references, see corresponding section in the text.
Ras-induced accumulation of p16INK4A, mediated by the MEK/MAPK pathway, leads to the inhibition of Cdk4/6, and the hypophosphorylation of Rb and its related proteins [32,54]. This leads in turn to the recruitment of a pocket-protein/E2F repressors/chromatin modifiers complex at E2F target genes, and ultimately the permanent silencing of these loci [18,55–57]. Recent studies using loss of function experiments have suggested that the heterochromatinization of E2F target loci is indeed essential for Ras-induced senescence in vivo. First, Braig and colleagues have generated compound mice genetically inactivated for the H3K9 histone methyl-transferases Suv3-9h1 and h2, and expressing activated N-Ras in lymphocytes (Eμ-N-RasG12D). Surprisingly, concomitant inactivation of Suv3-9h1/h2 and Ras activation leads to a dramatic acceleration of cancer progression in vivo, which is consistent with the inability of Suv3-9h1/h2 deleted splenocytes to undergo senescence upon activated Ras expression ex vivo [12]. This function of Suv3-9h1/2 may be attributed to its requirement for the generation of H3K9me3-rich environment within SAHF, although a direct role of Suv3-9 enzymes on the heterochromatinization of E2F target loci remains to be formally demonstrated [58]. Although this mouse model may not recapitulate faithfully any naturally occurring human cancer, the results of this study represent the first in vivo evidence that cellular senescence can serve as a tumor suppressor mechanism. It is intriguing to note that primary mouse fibroblasts inactivated for Suv39h1 and h2 remain susceptible to replicative senescence, suggesting that replicative and Ras-induced senescence may not use a completely overlapping molecular machinery [8,59]. Alternatively, this discrepancy could reflect cell context specificity for the establishment of senescence (see below).
Activation of a DNA damage response
In addition to the transcriptional repression of E2F target genes, a second pathway that is believed to mediate Ras-induced senescence is the DNA damage pathway. Indeed, recent studies demonstrated that acute activation of the Ras pathway, through ectopic expression of activated Ras or overexpression of mos, triggers a DNA damage response pathway in human primary cells [33,60,61]. Specifically, oncogene activation in these cells leads to the generation of DNA damage foci and the accumulation of proteins involved in DNA damage response. In these studies, oncogene activation induces aberrant DNA replication events, including asymmetric replication fork progression and fork collapse, both events leading to replication stress and subsequent DNA damage. Importantly, these studies also report that activation of the DNA damage response is required for oncogene-induced senescence, as supported by the lack of senescence upon genetic or chemical inactivation of CHK2, ATM or p53 [33,60]. Of note, it has also been shown that Ras activation leads to the accumulation of ROS, further linking replicative and oncogene-induced senescence to DNA damage at the molecular level [62]. Supporting a physiological relevance for DNA damage as associated with senescence, precancerous lesions seems to accumulate more DNA damage markers (along senescence markers) than the corresponding full-blown tumors. However, if DNA damage is the results of Ras activation inducing hyper-replication, it remains unclear why would full-blown tumors display less DNA damage than their corresponding pre-cancerous lesions.
The potential crosstalk between the two above-mentioned pathways (permanent silencing of pro-proliferative genes through heterochromatinization and induction of a DNA damage response) in the establishment of Ras-induced senescence remains unclear. The demonstration that genetic inactivation of either of these pathways prevents senescence suggests that they both are essential. However, this notion is difficult to reconcile with the fact that activation of either one of these pathways is sufficient to promote senescence. One possibility would be that DNA damage can signal towards heterochromatinization of E2F target loci, and conversely, permanent transcriptional repression of E2F target could trigger activation of the DNA damage response pathway. Alternatively, it is possible that oncogenic stress signals through different pathways in a cell context and oncogene specific manner. For example, in human cells, p16INK4A was shown to be dispensable for cdc6-induced senescence, by contrast to what had been shown for Ras-induced senescence [33].
Senescence –Associated Secretory Phenotype
Finally, a third pathway, essential for cellular senescence and engaged upon Ras activation is the Senescence–Associated Secretory Phenotype (SASP, also known as SMS). It has long been recognized that senescent cells secrete specific proteins, that function in promoting transformation of the surrounding cells, and at the same time in reinforcing their own senescent phenotype [63]. Recently, several studies have used genetic screen to identify cytokines or other secreted proteins that are essential for the establishment or maintenance of the senescent phenotype upon oncogene activation. Among those secreted proteins, interleukin-8 (IL-8), IL-6, IGFBP-7, and PAI-1 are secreted at the onset of senescence and can inhibit proliferation of cells by overriding mitogenic signals [64–66]. In addition, the up-regulation of specific receptors (including CXCR2 and PLA2R) or was shown to contribute to oncogene-induced senescence [20,67]. By releasing such proteins in the stroma, senescent cells can modulate tumor progression in a non-cell autonomous manner, an observation that may be of great importance in future potential therapeutic approaches. By contrast to their ability to reinforce the senescent status of the cells that produce them, some of these secreted proteins are also known to promote tumor progression. For example, IL-6 and IL-8 can induce invasion while MMP-3 or prevent differentiation [68].
The relationship between the secretory phenotype of senescent cells and the other molecular aspects of Ras-induced senescence, including DNA damage and the repression of pro-proliferative genes, is just starting to be uncovered. For example, a recent study demonstrated that SASP is being detected upon persistent activation of the DNA damage response [69]. In this study, the authors demonstrate that the senescent associated secretory phenotype is being activated downstream of the NBS/ATM/CHK2 components of the DNA damage response, but upstream of p53. Loss-of-function experiments have suggested that SASP is essential for Ras to induce senescence, through the C/EBPβ transcription factor [65], further reinforcing the molecular crosstalk between secretion of these cytokines and Ras induced senescence.
Altogether, these results indicated that Ras activation is able to engage multiple pathways, which converge to the permanent withdrawal from the cell cycle, likely through common pathways and numerous crosstalk mechanisms. The observation that genetic disruption of any of these pathways allows the bypass of Ras-induced senescence coupled with the demonstration that activation of any of these pathways is sufficient to induce cellular senescence, at least in vitro, strongly suggests that these three different arms of the response to Ras activation are intimately linked at the molecular level. Exploring the molecular links between these different pathways may reveal new and unsuspected triggers for the restoration of cellular senescence in transformed cells as a therapeutic approach.
Uncoupling immortalization and sensitivity to Ras-induced transformation
Since oncogene-induced senescence is thought to be a barrier to cellular transformation, cells that cannot undergo senescence are expected to be more susceptible to transformation. Based on the results obtained using mouse fibroblasts genetically inactivated for p53 or the INK4A locus, it has been suggested that preventing senescence invariably sensitizes cells to Ras-induced transformation [6,15]. Several subsequent studies involving different mouse models of cancer have reinforced this notion. However, these studies invariably used p53 or INK4A genetic inactivation as a mechanism allowing the bypass of senescence. Despite a clear requirement for either p53 or products of the INK4A locus in oncogene-induced senescence, it is important to bear in mind that these proteins all carry additional functions, which may account for their ability to prevent cancer initiation/progression.
The identification of additional proteins that are absolutely required for Ras-induced transformation has allowed to test experimentally if immortalization can be uncoupled from sensitivity to transformation. While the absence of several proteins that are required for Ras-induced senescence, including the downstream kinase PRAK [70], leads to sensitivity to Ras-induced transformation, this is not the case for all proteins that are essential for Ras-induced senescence. Indeed, Peeper et al. have demonstrated that although fibroblasts deleted for Rb and the related pocket proteins p107 and/or p130 are refractory to cellular senescence, they are not readily transformed upon ectopic expression of activated Ras [46]. Recent results suggest that downregulation of the p53/p21 pathway, for example through overexpression of Tbx2, is a necessary cooperative event for Ras-induced transformation of pocket-protein deleted cells [71]. Similarly, deletion of Sin3B, an essential cofactor for E2F factors mediated repression, allows the bypass of Ras-induced senescence, but is not sufficient to confer sensitivity to Ras-mediated transformation [56]. These observations therefore demonstrate that the relief of the senescence block does not automatically confer susceptibility to oncogenic transformation and suggest that senescence per se is not the only barrier to sensitivity to oncogene-induced transformation.
Ectopic expression of activated Ras versus activation of endogenous Ras: a question of dose?
Many of the findings reported above have been based on ectopic expression of activated Ras as an experimental system to induce senescence in primary cells, rather than mutation of the endogenous Ras gene. Because Ras (or its downstream target Raf) is mutated in more than a third of all human cancers, it seemed logical to develop mouse models of cancer investigating the consequences of activation of the endogenous Ras protein, in order to mimic what happens in human cancer. To do so, Tuveson, Jacks and colleagues, and McMahon and colleagues as well as Lodgson and colleagues, generated conditional knock-in alleles for activated K-Ras (K-RasG12D) or activated BRaf (BRafV600E) [72–75]. Briefly, the activated allele is inserted in the endogenous locus, but its transcription is prevented by the insertion of a stop cassette flanked by two loxP sites (LSL), so that only upon expression of the Cre recombinase is the activated allele expressed from the corresponding promoter. These models are then likely to closely reflect the phenotypic consequences of acute somatic mutation of the K-Ras or BRaf allele, which are believed to represent early events in tumorigenesis.
When primary MEFs were isolated from LSL-K-RasV12 mice and infected with Cre recombinase encoding viruses, a very surprising result was observed. In contrast to the cellular senescence phenotype elicited upon ectopic activated Ras expression [6], activation of Ras expressed at endogenous levels caused enhanced proliferation of primary MEFs [73]. In addition, cells expressing activated Ras at endogenous levels were partially transformed, in that they were not contact inhibited. Even more surprising was the observation that the canonical pathways downstream of Ras were actually less activated in cells that express activated Ras driven by its endogenous promoter compared to wild type cells [73]. This observation has led to the hypothesis that activated Ras, when expressed at endogenous levels, leads to attenuation of its own effector pathways, although the molecular bases underlying this effect remain largely unknown at this point. The observation that sustained Ras activation, when expressed from its own promoter, leads to attenuation of its effector pathways has been confirmed in a different model system, namely the knock-down of NF1 in human cells [36]. In this case, NF1, a negative regulator of Ras activity, is knocked down in human cells, which leads to endogenous Ras activation. However, a few passages after NF1 downregulation, the downstream effectors of Ras become suppressed, through a negative feedback involving different modulators of Ras activity [36]. Intriguingly, these cells become senescent, by contrast to the primary MEFs expressing activated Ras presented above. However, in other models and cellular contexts, including the mice in which endogenous BRaf is activated in the lung epithelium, or the activated K-Ras allele driven by its endogenous promoter in acinar cells, downstream targets of Ras or Raf, including MEK or Erk phosphorylation, are highly activated in early preneoplasic lesions compared to the adjacent normal tissues [74,75]. In addition, these early lesions are positive for different markers of senescence, making the notion that senescence is caused by a shut-down of the Ras signaling pathway unlikely in these cases.
The discrepancy in the levels of activation of Ras-downstream targets reported in these different model systems could be reconciled by the hypothesis that the levels of Ras expression may be modulated early during the oncogenic transformation process. Indeed, one may hypothesize that, while expression of activated Ras at endogenous levels does not lead to senescence and hyperactivation of its downstream targets in primary fibroblasts, in other cellular contexts, including in cells susceptible to Ras-mediated transformation, the Ras signal may be amplified, reaching levels comparable to those observed upon ectopic expression of Ras. In this case, Ras activation would first result in proliferation, as observed in K-RasV12 knock-in fibroblasts, for several passages until Ras signals become amplified to a point where it is then able to trigger a senescence response, as observed in metaplasia or PanIN lesions from K-RasV12 knock-in mice, or in early lung adenoma from K-RasV12 mice or BRaf knock-in mice [14,74,75]. This would also be consistent with the report that benign lesions harboring BRaf mutations have proliferated for at least 10–15 passages following the acquisition of the mutation, before these cells withdraw from the cell cycle and become senescent.
This hypothesis implies that (i) different levels of expression of activated Ras drive different responses in vivo, ranging from proliferation to cellular senescence and (ii) levels of Ras activation are modulated throughout cancer initiation and progression (see Figure 1). Indeed, the levels of activation/expression of Ras or its downstream effectors have long been known to modulate its effects [76]. In vivo also, the notion that different levels of Ras activation/expression lead to different outcomes has been well-established. For example, Chodosh and colleagues have generated an inducible transgene driving the expression of activated Ras in mammary epithelial cells [77]. This transgene allowed the precise regulation of Ras levels, so that activated Ras can be expressed from levels comparable to those of endogenous Ras or levels comparable to gross overexpression, simply by changing the concentration of doxycycline in the drinking water. When the mammary glands expressed very high levels of activated Ras, cellular senescence was observed. By contrast, low levels of activated Ras, comparable to endogenous levels, ultimately gave rise to mammary tumors. Importantly, the levels of Ras expression in these tumors was extremely high, strongly suggesting that tumorigenesis required i) inactivation of the senescence pathway and ii) spontaneous upregulation of activated Ras expression. Although the physiological contribution of Ras to mammary tumor remains to be formally proven in humans, this set of experiments further strengthens the existence of cell context and dose-specific effects of activated Ras on senescence and tumorigenesis. In addition, a recent study using mouse pancreas models for cancer demonstrated that lesions including PanIN and PDAC found in activated K-Ras knock-in mice display a Ras activity much higher than in the corresponding normal tissue, suggesting that Ras upregulation or hyperactivation is an early event during tumorigenesis in this model [74]. The fact that Ras levels are increased during tumorigenesis was previously reported in an independent mouse model of pancreatic cancer, although in this case, amplification of the activated Ras allele was only documented in full-blown tumors [78]. It would be interesting to document at what point during tumorigenesis the activated Ras allele is amplified. Based on our hypothesis, we would anticipate that Ras amplification is a very early event in cancer initiation.
In conclusion, Ras activation in vivo can lead to preneoplastic lesions displaying marker of cellular senescence, only when a threshold of Ras expression is attained, likely through spontaneous amplification of the locus or hyperactivation. Such lesions can then evolve into frank carcinoma if the molecular pathways for senescence are disrupted. Thus, cellular senescence induced by Ras is likely to be a physiological process that contributes to preventing tumorigenesis in vivo. A better understanding of the molecular basis for Ras-mediated senescence as well as the events that permit cells to escape senescence, should allow the delineation of new therapeutic approaches aimed at restoring the senescence pathway in human tumors.
Acknowledgements
We thank Drs. Lawrence Gardner, Nabeel Bardeesy, Dafna Bar-Sagi, Martin McMahon and the members of the David laboratory for helpful discussion. We apologize to any colleague whose work could not be cited due to space limitations. TD is supported by a predoctoral NIH training grant CA009161. Work in the David lab is supported by the American Federation for Aging Research (AFAR) and The American Cancer Society (ACS).
References
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J. Replicative senescence: an old lives' tale? Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 3.Blagosklonny MV. Cell senescence: hypertrophic arrest beyond the restriction point. J Cell Physiol. 2006;209:592–597. doi: 10.1002/jcp.20750. [DOI] [PubMed] [Google Scholar]
- 4.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 6.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 7.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 8.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 9.Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J Cell Biol. 2006;175:869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Chen W, Adams PD. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 13.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 15.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 16.Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandinetti KB, David G. Sin3B: an essential regulator of chromatin modifications at E2F target promoters during cell cycle withdrawal. Cell Cycle. 2008;7:1550–1554. doi: 10.4161/cc.7.11.6052. [DOI] [PubMed] [Google Scholar]
- 19.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 22.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 23.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 24.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 25.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 26.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 27.Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 28.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 30.Braig M, Schmitt CA. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2006;66:2881–2884. doi: 10.1158/0008-5472.CAN-05-4006. [DOI] [PubMed] [Google Scholar]
- 31.Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20:273–285. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 34.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 38.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 39.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 40.Brookes S, Rowe J, Ruas M, Llanos S, Clark PA, Lomax M, James MC, Vatcheva R, Bates S, Vousden KH, et al. INK4a-deficient human diploid fibroblasts are resistant to RAS-induced senescence. EMBO J. 2002;21:2936–2945. doi: 10.1093/emboj/cdf289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei W, Hemmer RM, Sedivy JM. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol Cell Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 43.Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, Scime A, Rudnicki MA, Young R, Dynlacht BD. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei W, Herbig U, Wei S, Dutriaux A, Sedivy JM. Loss of retinoblastoma but not p16 function allows bypass of replicative senescence in human fibroblasts. EMBO Rep. 2003;4:1061–1066. doi: 10.1038/sj.embor.7400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peeper DS, Dannenberg JH, Douma S, te Riele H, Bernards R. Escape from premature senescence is not sufficient for oncogenic transformation by Ras. Nat Cell Biol. 2001;3:198–203. doi: 10.1038/35055110. [DOI] [PubMed] [Google Scholar]
- 47.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 50.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, Kubben N, Holzer B, Ludwig S, Rapp UR. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem. 2005;280:5178–5187. doi: 10.1074/jbc.M407155200. [DOI] [PubMed] [Google Scholar]
- 52.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, Sun P. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–3403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grandinetti KB, Jelinic P, DiMauro T, Pellegrino J, Fernandez Rodriguez R, Finnerty PM, Ruoff R, Bardeesy N, Logan SK, David G. Sin3B expression is required for cellular senescence and is up-regulated upon oncogenic stress. Cancer Res. 2009;69:6430–6437. doi: 10.1158/0008-5472.CAN-09-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 59.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 60.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 61.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heyworth PG, Knaus UG, Settleman J, Curnutte JT, Bokoch GM. Regulation of NADPH oxidase activity by Rac GTPase activating protein(s) Mol Biol Cell. 1993;4:1217–1223. doi: 10.1091/mbc.4.11.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krtolica A, Campisi J. Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int J Biochem Cell Biol. 2002;34:1401–1414. doi: 10.1016/s1357-2725(02)00053-5. [DOI] [PubMed] [Google Scholar]
- 64.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 66.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Augert A, Payre C, de Launoit Y, Gil J, Lambeau G, Bernard D. The M-type receptor PLA2R regulates senescence through the p53 pathway. EMBO Rep. 2009;10:271–277. doi: 10.1038/embor.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 71.Vormer TL, Foijer F, Wielders CL, te Riele H. Anchorage-independent growth of pocket protein-deficient murine fibroblasts requires bypass of G2 arrest and can be accomplished by expression of TBX2. Mol Cell Biol. 2008;28:7263–7273. doi: 10.1128/MCB.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 73.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 74.Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, Bi Y, Grote T, Longnecker DS, Logsdon CD. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–1082. 1082, e1071–e1076. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pritchard CA, Samuels ML, Bosch E, McMahon M. Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol Cell Biol. 1995;15:6430–6442. doi: 10.1128/mcb.15.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 78.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]