Abstract
Background
Hepatic resection is the preferred treatment for huge hepatocellular carcinoma (>10 cm in diameter; H-HCC). However, the patients with H-HCC suffer from poor prognosis due to the early recurrence/metastasis. The underlying mechanism of H-HCC’s early recurrence/metastasis is currently not well understood.
Results
Here, we describe an Isobaric Tags for relative and absolute quantification (iTRAQ)-based quantitative proteomics approach to analyze the early recurrence/metastasis related proteins of H-HCC after radical resection through multidimensional chromatography coupled with tandem mass spectrometry (2DLC-MS/MS). The different protein expression profiles between the early recurrence/metastasis within 6 months(R/M≤6months) and late recurrence/metastasis within 6–12 months after surgery (R/M6-12months) were confirmed and might reveal different underlying molecular mechanisms. We identified 44 and 49 significantly differentially expressed proteins in the R/M≤6months group and the R/M6-12months group compared to the group who had no recurrence within 2 years post surgery (the NR/M group), respectively. Moreover, among those proteins, S100A12 and AMACR were down regulated in the R/M≤6months group but up-regulated in the R/M6-12months group; and this regulation was further confirmed in mRNA and protein level by Q-PCR, Western-Blot and Immunohistochemistry (IHC).
Conclusions
This current study presents the first proteomic profile of the early recurrence/metastasis of H-HCC. The results suggest that S100A12 and AMACR might be potential prognostic markers for predicting the early recurrence/metastasis of H-HCC after hepatectomy.
Keywords: Huge hepatocellular carcinoma (H-HCC), Early recurrence/metastasis, Quantitative proteomics, iTRAQ, Potential prognostic biomarker
Background
Hepatocellular carcinoma (HCC), the fifth most common cancer worldwide [1], is reported to be the second leading cause of cancer death in China [2]. Huge hepatocellular carcinoma (H-HCC) with the feature of diameter larger than 10 cm, is a special subtype of HCC. Despite the proven feasibility and safety of surgical resection for H-HCC [3,4], the prognosis of H-HCC patients remains poor, mainly due to the intra-hepatic recurrence and/or extra-hepatic metastasis. The incidence of recurrence/metastasis within 12 months in the residual liver of H-HCC patients who underwent curative resection, ranges from 50% to 70% [5-7]; and the median survival after recurrence/metastasis is only 13 months [8]. This has largely blocked the curative efficiency and long-term survival of hepatectomy.
In the last two decades, various molecular alterations have been found to correlate with early recurrence/metastasis of HCC [9-12]. However, the detailed underlying molecular mechanisms of the early recurrence/metastasis of H-HCC are still not well understood. The recurrence/metastasis of HCC is a complicated process, which is resulting from combined effects of multiple factors [13].
Studies focusing on individual gene or protein might be insufficient for elucidating the biological natures of the malignant behavior of tumor. Quantitative proteomics approaches, which are able to give an overview of the global protein profile alternation under pathological conditions, have been proposed to be extremely useful tools in studying the recurrence/metastasis behavior of cancer [14-18]. Several groups have been extensively applying the proteomics approach to elucidating the biological behaviors of HCC; Sun et al. have studied the differentially expressed proteins in tumor and adjacent non-tumor tissue samples, and found that Hcp70/Hsp90-organizing protein and heterogeneous nuclear ribonucleoproteins C1/C2 could be potential biomarkers in HCC [19]; Orimo et al. have studied the protein expression alternations in 45 surgically resected tissues with different degree of histological differentiation, and identified APC-binding protein EB1 (EB1) as a potential prognostic biomarker for HCC [20]. However, the traditional proteomics based approaches suffer from low throughput non-quantitative information coupled with difficulties in separating and/or detecting low abundant proteins [21], post translationally modified proteins [22,23], as well as those proteins with a pI value lower than 4 or higher than 9 [24].
Recently, high-throughput quantitative proteomic techniques have been developed [25-28]. In particular, isobaric tags for relative and absolute quantification (iTRAQ) labeling followed by nano liquid chromatography-mass spectrometry (NanoLC-MS/MS) is an extremely effective method for simultaneous quantitative comparison and analyzing the protein expression profile of multiple samples. Especially, this has been useful for studying disease associated and low abundance proteins [29-34] with good sensitivity. So far, several groups have reported the applying of iTRAQ based quantitative proteomics approach in the study of hepatic cancer, especially for the screening of diagnostic or prognostic protein biomarkers. He et al. have reported the serum biomarker screening of AFP negative HBV related HCC through the iTRAQ based approach [35]; Ko et al. have reported the iTRAQ based quantitative analysis of HCC cancer stem cell proteome [36]; Huang et al. have reported the iTRAQ based serum biomarker screening of the HCC micro-vascular invasion [37]; Qin et al., Yu et al. and Wang et al. have reported the screening of metastatic related proteins of HCC through iTRAQ based approach [33,38,39]; the iTRAQ based quantitative study of proteome change during HBV infection has also been reported [40,41]. However, the application of iTRAQ labeling in studying the molecular mechanisms and screening for biomarkers associated with the early recurrence/metastasis of H-HCC has not yet been reported to our knowledge.
In the present study, we applied the iTRAQ based quantitative proteomic approach (iTRAQ-2DLC-MS/MS) to quantitatively analyze the protein profiles and alternations of the early recurrence/metastasis in H-HCC after radical hepatic resection, and tried to identify the potential prognostic markers and reveal the underlying mechanism of the early recurrence/metastasis in H-HCC.
Results
The quantitative proteomics of the recurrence/metastasis of H-HCC
Here, the proteins were identified and quantified according to the following criteria: a protein is reported if a quantification ratio was obtained using at least 2 unique peptides with “unused” confidence cutoff (PortScore) >1.3 (95%); 1408 proteins (1818 proteins before grouping) were identified from 23767 distinct peptides; and 1279 unique proteins were quantified from samples analyzed on QSTAR Elite. A complete list of proteins identified in our study is shown in Additional file 1; the details of proteomic analysis including sequence coverage, score of unique peptides, quantification results with percentage variability are included in the list as well. The protein expression alternations in different groups were calculated from the intensity ratio of the iTRAQ reporter ions from the derived peptides. Full details of the dataset and supplementary can be accessed through ftp://epigenome.fudan.edu.cn/ by using FlashFXP.
Categorization and functional annotation of proteins quantitated in R/M≤6months and R/M6-12months
Functional annotations of all the identified proteins through IPA are shown in Additional file 2: Figure S1. Categories are based on primary localization, which include nucleus (17%), extracellular space (9%), cytoplasm (65%), plasma membrane (6%), while 3% of the identified proteins is currently not clearly classified (Additional file 2: Figure S1A). Moreover, molecular type of proteins was also analyzed, of which nearly half of the identified proteins are enzymes and transporters (Additional file 2: Figure S1B).
At the same time, functional annotation of all the identified proteins by Gene Ontology (GO) including biological process, sub-cellular localization and molecular function are shown in Additional file 2: Figure S2 and the top 10 involved functions were ranked in term of the enrichment.
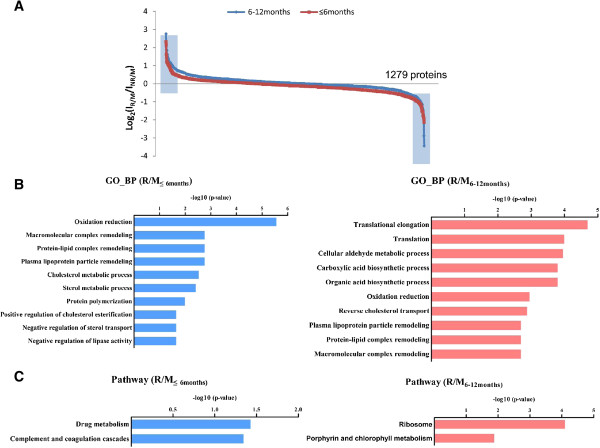
In this study, we designated the differentially expressed proteins with the fold change cutoff ratio < 0.5 or >2.0. Figure 1A shows iTRAQ fold changes of all identified proteins, and a small subset of differential expressed proteins in both R/M≤6months and R/M6-12months group compared to the NR/M group (indicated by the blue tag). 46 differentially expressed proteins (15 up-regulated and 31 down-regulated) were detected in the R/M≤6months group and 49 differentially expressed proteins (33 up-regulated and 16 down-regulated) were detected in the R/M6-12months group upon comparison with the NR/M group (Additional file 1: Table S1 and Table S2). Functional annotations of these differentially expressed proteins (compared to the NR/M group) were analyzed as follows: (A) Gene Ontology (GO) analysis to reveal the different involved biological functions in R/M≤6months and R/M6-12months (Figure 1B), (B) KEGG pathway enrichment analysis which reveals that the drug metabolism as well as complement and coagulation cascades were mainly involved in R/M≤6months; metabolism related signaling pathways (like ribosome metabolism, porphyrin and chlorophyll metabolism) were actively involved in the R/M6-12months group (Figure 1C) and (C) GO analysis of cell components and molecular function analysis to reveal the different characters of the early (R/M≤6months group) and late (R/M6-12months group) recurrence/metastasis patients (Additional file 2: Figure S3). All the differentially expressed proteins in R/M≤6months and R/M6-12months comparing to the NR/M group have been listed in Additional file 1: Table S1 and Table S2, respectively. Comparison of results from Additional file 1: Table S1 and Table S2, clearly reveals that over 80% of the protein expression alternations were different between R/M≤6months group and R/M6-12months group. It indicates that the early recurrence/metastasis might be induced by different factors compared to the late stage recurrence/metastasis; indicating that they are possibly undergoing different molecular mechanisms.
Figure 1.
The sub-cellular distribution and biological functional annotation of identified proteins. (A) The protein expression distributions of R/M≤6months and R/M6-12months; the differential expressed proteins in R/M≤6months and R/M6-12months comparing to the NR/M group was indicated by the blue tag. (B) Go analysis for biological functions of the differentially expressed proteins in R/M≤6months and R/M6-12months, the top 10 were shown in panel B. (C) KEGG pathway enrichment analysis of the differentially expressed proteins in R/M≤6months and R/M6-12months group (p < 0.1).
Differentially expressed proteins in R/M≤6months and R/M6-12months
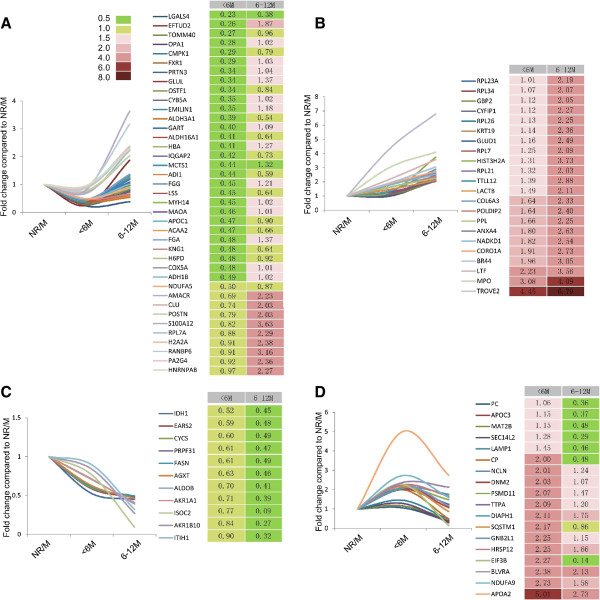
By carefully comparing the protein expression alternations, there are 4 types of trends according to the protein fold changes. The first type is the altered proteins decreased in the R/M≤6months group, but increased in the R/M6-12months group comparing to the NR/M control group; in these proteins, the enzymes related to the glycolysis (ADH1B, ALDH3A1, MAOA, ALDH16A1) and the proteins related to protein synthesis (RPL7A, PA2G4, MCTS1, EFTUD2) were involved in (Figure 2A). While ribosome proteins related to protein synthesis were found highly expressed in both R/M≤6months group and R/M6-12months group comparing to the NR/M group, such as RPL7, RPL26, RPL21, RPL34, RPL7, RPL23A etc. (Figure 2B). Figure 2C shows proteins decreased in both R/M≤6months group and R/M6-12months group compared to the NR/M control group, which are involved in the amino acid metabolic process (EARS2, IDH1, PASN, AGXT). Lastly, there is exists another type of differentially expressed proteins whose expression is increased in the R/M≤6months group, but decreased in the R/M6-12months group in comparison with the NR/M group and lying scattered in multiple signaling pathways (Figure 2D).
Figure 2.
Alternation tendency of proteins in R/M≤6months group and R/M6-12months group comparing to the NR/M group. (A) The proteins which are decreased in the R/M≤6months group, but increased in the R/M6-12months group comparing to the NR/M group; these proteins are mostly related with glycolysis and protein synthesis. (B) The proteins which are highly expressed in both R/M≤6months group and R/M6-12months group comparing to the NR/M group; these proteins are mostly involved in protein synthesis process. (C) The proteins which are lowly expressed in both R/M≤6months group and R/M6-12months group comparing to the NR/M group; these proteins are mostly related to the amino acid metabolic process. (D) The proteins which are increased in the R/M≤6months group, but decreased in the R/M6-12months group comparing to the NR/M group; these proteins are involved in multiple signaling pathways.
By carefully comparing the protein expression alternations, we filtered 8 proteins that altered their expression in both R/M≤6months group and R/M6-12months group relative to the NR/M group, which might closely relate to the metastatic propensity (Table 1). These proteins mostly belong to the secretory proteins category (Galectin-4 and S100A12), transport proteins (Lactotransferrin and Apo-lipoprotein A-II), transcription and translation factors (60-kDa-SS-A/Ro ribonucleoprotein and Eukaryotic translation initiation factor 3 subunit B, eIF3b), and antioxidant proteins (Biliverdin Reductase A and Alpha-methy-lacyl-CoA racemase, AMACR). Although these proteins have rarely been linked with cancer, the signaling pathways regulated by these proteins and their protein families play an important role in oncogenesis, and are extensively involved in the invasion and metastasis of tumor [42-49].
Table 1.
List of common proteins altered their expression in both R/M ≤6months group and R/M 6-12months group
| Accession number | %Cov | Protein name | iTRAQ ratio 117:121(R/M ≤6months ) | iTRAQ ratio 119:121(R/M 6-12months ) |
|---|---|---|---|---|
| LEG4_HUMAN |
33.1 |
Galectin-4 |
0.2291 |
0.3837 |
| S10AC_HUMAN |
27.2 |
Protein S100-A12 |
0.3052 |
3.6308 |
| AMACR_HUMAN |
42.9 |
Alpha-methylacyl-CoA racemase |
0.3585 |
2.2284 |
| TRFL_HUMAN |
18.6 |
Lactotransferrin |
2.2284 |
3.5645 |
| EIF3B_HUMAN |
13.1 |
Eukaryotic translation initiation factor 3 subunit B |
2.2699 |
2.1355 |
| BIEA_HUMAN |
17.6 |
Biliverdin reductase A |
2.3768 |
2.1281 |
| RO60_HUMAN |
13.9 |
60 kDa SS-A/Ro ribonucleoprotein |
4.4463 |
6.7920 |
| APOA2_HUMAN | 73 | Apolipoprotein A-II | 5.0119 | 2.729 |
In these proteins, we could clearly see that the protein S100A12 and AMACR show different alternation tendency between R/M≤6months group and R/M6-12months group; the protein S100A12, which has been reported to regulate the calcium metabolism and arachidonic acid metabolism in the neutrophil [50], is down regulated more than 3.3 folds in R/M≤6months group compared to the NR/M group, but up regulated more than 3.6 folds in R/M6-12months group relative to the NR/M group. AMACR, which has been reported to regulate the metabolism of fatty acid, cholalic acid [51] and closely associated with the differential diagnosis of prostatic cancer [52], was down regulated by over 2.8 folds in R/M≤6months group compared to the NR/M group, but up-regulated more than 2.2 folds in R/M6-12months group compared to the NR/M group. Since these two proteins have clearly different expression profiles in the R/M≤6months group and the R/M6-12months group, they might be interesting potential biomarkers for predicting early recurrence/metastasis after radical resection in H-HCC.
Functional analysis of the protein expression alternations in the recurrence/metastasis of H-HCC
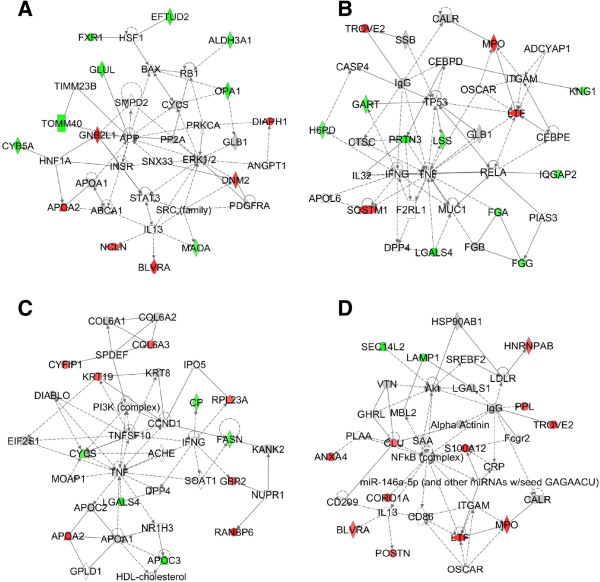
To analysis the roles of the protein expression alternations in the recurrence/metastasis of H-HCC, we used the IPA software to study the key signaling pathways and networks of those differentially expressed proteins. The analyzed results suggest that different signaling pathways are indeed involved in the early (R/M≤6months group) and late (R/M6-12months group) recurrence/metastasis patients, although there are common signaling pathways involved in as well. The IPA software identified that the altered protein expressions in the R/M≤6months group patients are mostly involved in ERK1/2 signaling pathway and TNF signaling pathway, but the altered protein expressions in the R/M6-12months group patients are mostly involved in TNF signaling pathway and NFκB signaling pathway.
In the R/M≤6months group patients, we could see that 14 proteins (including 6 up-regulated proteins and 8 down-regulated proteins) were involved in the ERK1/2 signaling pathway (Figure 3A); and 13 proteins (including 4 up-regulated proteins and 9 down-regulated proteins) involved in TNF signaling pathway (Figure 3B). In the R/M6-12months group patients, 12 proteins (including 7 up-regulated proteins and 5 down-regulated proteins) involved in the TNF signaling pathway (Figure 3C); and 13 proteins (including 11 up-regulated proteins and 2 down-regulated proteins) involved in the NFκB signaling pathway (Figure 3D). Although, all of these 3 signaling pathways are actively associated with cancers [53-55], as describing above, only the TNF signaling pathway is extensively involved in both the early and late recurrence/metastasis of H-HCC. The reported functions of TNF signaling pathway in carcinogenesis include tumor initiation, tumor cell proliferation, tumor angiogenesis, and enhancing the invasive property of tumor cells [55,56]; therefore, it is not surprising that the TNF signaling pathway is involved in both groups. Interestingly, although both the ERK1/2 signaling pathway and the NFκB signaling pathway are reported to associated with cancer development, during malignant transformation and metastasis [53,54], only the ERK1/2 signaling pathway is enriched in the early (R/M≤6months) recurrence/metastasis patients. ERK1/2 is known an extremely important regulator of cell growth and proliferation [54], therefore, the early recurrence/metastasis H-HCC cells probably have stronger growth and proliferation abilities than late recurrence/metastasis H-HCC cells; but the detailed underlying mechanisms should be further studied.
Figure 3.
The key signaling pathways involved in the early recurrence/metastasis (R/M≤6months) and late recurrence/metastasis (R/M6-12months) of H-HCC. (A) The alternations of the ERK1/2 signaling pathway in the R/M≤6months group, including 6 up-regulated proteins (red labeling) and 7 down-regulated (green labeling) proteins. (B) The alternations of the TNF signaling pathway in the R/M≤6months group, including 4 up-regulated proteins (red labeling) and 9 down-regulated proteins (green labeling). (C) The alternations of the TNF signaling pathway in the R/M6-12months group, including 7 up-regulated proteins (red labeling) and 5 down-regulated proteins (green labeling). (D) the alternations of the NFκB signaling pathway in the R/M6-12months group, including 9 up-regulated proteins (red labeling) and 2 down-regulated (green labeling) proteins.
The verification of S100A12 and AMACR expression alternation
As mentioned above, the protein S100A12 and AMACR were reversely expressed in the R/M≤6months group (early recurrence/metastasis) and the R/M6-12months group (late recurrence/metastasis). Therefore, they might be potential interesting biomarkers to predict the early and late recurrence/metastasis in H-HCC. Here, we took liver sections from H-HCC patients to verify their expression profiles and predicting potentials in a larger scale. Combing with the patient case history, the expression profiles of both S100A12 and AMACR in different groups were confirmed by Q-PCR at the mRNA level and by Western-blot and immunohistochemistry in the protein level.
As shown in Figure 4A, the gene expression of S100A12 and AMACR were down-regulated 2.5 fold and 14.2 fold in the R/M≤6months group (n = 20 patients), respectively; but up-regulated 5.1 fold and 5.38 fold in R/M6-12months group (n = 20 patients), respectively. As shown in Figure 4B, the protein expression of S100A12 and AMACR are down regulated 1.75 fold and 16.6 fold in the R/M≤6months group (n = 20 patients), respectively; but up-regulated 2.14 fold and 1.38 fold in the R/M6–12 months group (n = 20 patients), respectively. These results thus fitted well with the quantitative proteomics study results.
Figure 4.
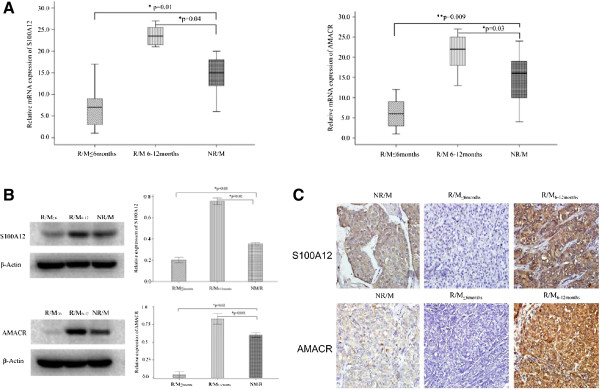
Verification of differentially expressed proteins in different stage of recurrence/metastasis in huge hepatocellular carcinoma patients. (A) The mRNA expression level of S100A12 and AMACR (p < 0.05, paired T-test); (B) The protein expression level of S100A12 and AMACR when validated by Western-Blot (p < 0.05, paired T-test); (C) The immunohistochemistry staining results of S100A12 and AMACR (with 400x magnification). Both S100A12 and AMACR are strongly over-expressed on the patient’s samples from R/M6-12months group, but down regulated on the patient’s samples from R/M≤6months group when it is comparing with the control samples.
To further confirm the protein expression and localization profiles of S100A12 and AMACR, the immunohistochemistry was performed on patient’s liver sections. As shown in Figure 4C, both the expression intensity and the positive rate (Tables 2 and 3) of the S100A12 and AMACR were down regulated in the R/M≤6months group but up regulated in the R/M6-12months group, compared to the NR/M group respectively.
Table 2.
Immunohistochemistry assessment of the S100A12 expression level on patient’s samples with different recurrence/metastasis stage
| |
|
S100A12 expression |
|
|
|
|---|---|---|---|---|---|
| Groups | Numbers | Negative | Positive | Positive expression rate (%) | P values a) |
| R/M≤6months group |
20 |
16 |
4 |
20.0 |
0.048 |
| R/M6-12months group |
20 |
3 |
17 |
85.0 |
0.008 |
| NR/M group | 20 | 9 | 11 | 55.0 | |
a)P value listed above stands for the significance of the S100A12 positive expression rate when R/M≤6months group and R/M6-12months group compared with NR/M group respectively. P value less than 0.05 is taking as significant difference with Chi-square test.
Table 3.
Immunohistochemistry assessment of the AMACR expression level on patient’s samples with different recurrence/metastasis stage
| |
|
AMACR expression |
|
|
|
|---|---|---|---|---|---|
| Groups | Numbers | Negative | Positive | Positive expression rate (%) | P values a) |
| R/M≤6months group |
20 |
17 |
3 |
15.0 |
0.041 |
| R/M6-12months group |
20 |
2 |
18 |
90.0 |
0.014 |
| NR/M group | 20 | 10 | 10 | 40.0 | |
a)P value listed above stands for the significance of the AMACR positive expression rate when R/M≤6months group and R/M6-12months group compared with NR/M group respectively. P value less than 0.05 is taking as significant difference with Chi-square test.
Combining with the patient’s case history and the expression profiles of S100A12 and AMACR, these two proteins could well distinguish the early (within 6 months) and late (from 6 to 12 months) recurrence/metastasis of H-HCC. They might be potential interesting biomarkers for the predicting of early recurrence/metastasis of H-HCC, but the underlying molecular mechanisms needs to be further dissected.
Discussions
As reported in our previous clinical studies [57], the early recurrence/metastasis of H-HCC is affected by multiple factors. The independent risk factors for the early recurrence/metastasis occurred within 6 months, and those factors for the late recurrence/metastasis are different and occurred between or after 6 to 12 months. Therefore, the underlying molecular mechanism of the early (occurred within 6 months) and late (occurred from 6 months to 12 months) recurrence/metastasis might be different.
The recent rapid development of proteomic techniques allow a full-scale illustration of the protein expression profile alteration under a particular disease circumstance, and thus in identifying important disease-related protein biomarkers or therapeutic targets [33,35-41]. The HCC associated proteomics is believed to further facilitate the studies on systematic screening of the molecular mechanisms involved in HCC recurrence/metastasis, and in the development of prognosis biomarkers for HCC recurrence/metastasis.
In this paper, we applied the iTRAQ based quantitative proteomics approach to study the overall protein profile alternations in the early and late recurrence/metastasis of H-HCC. Here, we successfully identified 46 and 49 differentially expression proteins in the R/M≤6months group and R/M6-12months group compared to the NR/M group, respectively; however, we fished out only 8 proteins that altered its expression in both R/M≤6months group and R/M6-12months group. In our study, we pooled the samples to standardize the reading since the individual sample is likely to give highly divergent and incoherent results; while with pooled samples one could lose information on individual patient variability and also might mess up the readings if there are sub-groups. However, many of the studies utilizing clinical samples face similar problems, which to certain extent limit the development of clinical proteomics.
Among those identified proteins, over 80% of the expression altered proteins were different between the R/M≤6months group and R/M6-12months group, which suggested different molecular mechanisms operate during the early (within 6 months) and late (from 6 to 12 months) recurrence/metastasis of H-HCC. This finding fitted well with our initial hypothesis. Therefore, those H-HCC patients who had recurrence/metastasis at different time points as per our classification (early or late) might need completely different medical treatment. In such a case, it is necessary to identify novel biomarkers and therapeutic targets for distinguishing, predicting and treating the early and late recurrence/metastasis, which will certainly provide important guidelines for identifying H-HCC patients with high risk of early recurrence/metastasis.
The indentified protein expression alterations involve several biological processes, including carbohydrate metabolism, lipid metabolism, cell-cell communication, apoptosis, proliferation, anti-oxidation, transcription and translation, which are confirming that the malignant behavior of HCC cells is controlled by complicated signaling process. Some of the identified proteins have also been described to be associated with HCC recurrence/metastasis. For example, CK19 is up-regulated 2.35 folds in R/M6–12 months group compared with NR/M group; it has been reported to be highly expressed in HCC tissues, and the patients with positive CK19 results had a higher incidence of pulmonary metastasis within 1 month after receiving surgical resection [58]. Periostin (PN) is a secreted matrix glycoprotein, which has been reported to be closely related with the metastatic potential and prognosis of HCC after surgery [59]; Clusterin has been reported to promote the HCC metastasis via regulating TGF-β1-smad3 signaling and then induction of the epithelial-mesenchymal transition (EMT) process [60]. In our study, the expressions of those proteins have also proved to be altered in the R/M6-12months group patients. Therefore, it is clearly establishes that our quantitative proteomics approach is suitable for studying the recurrence/metastasis of H-HCC.
In our clinical, more than 50% of H-HCC patients have recurrence/metastasis within 6 months after radical surgery [58]. However, we did not have any techniques or biomarkers available to early predict the recurrence/metastasis risky of H-HCC patients so far. Predicting early the recurrence/metastasis potential of the H-HCC patients will improve the prognosis. In our study, we identified that protein S100A12 and AMACR were reversely expressed in the early (within 6 months, down regulated) and late (from 6 to 12 months, up regulated) recurrence/metastasis H-HCC patients; and further validated these findings from clinical samples at the level of both mRNA and protein expression. The recurrence/metastasis of HCC is closely related with clinical parameters such as the size of tumor, tumor capsule, tumor boundaries, portal vein tumor thrombosis (PVTT), intraoperative ascites, cirrhotic nodule, heteroploidy, PCNA (Proliferating Cell Nuclear Antigen) expression and others. Our study shows that the low expression of AMACR is related with the absence of tumor capsule, indistinct tumor boundary as well as presence of portal vein tumor thrombosis (PVTT) and intraoperative ascites, which fits with the earlier recurrence/metastasis of AMACR lower expression group.
S100A12 is a new member of S100 protein family (calcium binding protein family); the gene encoding S100A12 was localized on the human chromosome 1q21 [61], and is normally expressed in neutrophil granulocytes, with low expression in both lymphocytes and monocytes [50]. Its reported function involves calcium regulation and arachidonic acid metabolism in the neutrophile granulocytes [50]; it has also been reported that the expression of S100A12 was up-regulated both at mRNA and protein levels in the patients with colorectal carcinoma relative to healthy volunteers [62]. The AMACR encoding gene is localized on chromosome 5p13 [51]; it is probably involved in the metabolism of fatty acid and cholalic acid [51]. Previous studies have reported the application of AMACR in the diagnosis of prostatic cancer [62,63] and there are also a few studies that reported AMACR could be used as biomarker to distinguish the hepatocellular carcinoma and the benign tissue [64,65]. However, these two proteins were rarely reported to be associated with the prognosis of HCC. In this study, we not only prove that the S100A12 and AMACR were associated with the recurrence/metastasis of H-HCC, but also clearly demonstrate that these two proteins have completely inverse expression profiles between the early (within 6 months, down regulated) and late (from 6 to 12 months, up regulated) recurrence/metastasis of H-HCC, even considering a larger “n” number of clinical samples as employed in this study. Therefore, they might be potentially interesting biomarkers for distinguishing and predicting the early and late recurrence/metastasis of H-HCC. However, the exact molecular mechanisms of how these two proteins involved in the pathogenesis and progression of H-HCC should be further verified and studied. It has been reported that the abnormal expression of S100A12 is closely related with inflammation and vascular invasion of tumor cell [66], over inflammation and vascular invasion are closely related with tumor recurrence/metastasis. Meanwhile, the low expression of AMACR was independently associated with the development of metastasis and lethal in prostate cancer through regulating lipid metabolism and nuclear receptor activity [67]. Whether or not these reported functions are involved in the here reported HCC recurrence/metastasis needs further studies.
Overall, we have used iTRAQ based quantitative proteomics approach to study the protein expression profile alternations of the recurrence/metastasis of Huge HCC in different stage, and identified potentially interesting biomarkers for early distinguishing and predicting the recurrence/metastasis behaviors of H-HCC.
Conclusions
We have applied the iTRAQ based quantitative proteomics approach to study the overall protein profile alternations in the early (within 6 months) and late (from 6–12 months) recurrence/metastasis of huge hepatocellular carcinoma after radical resection. The results proved different protein alternation profiles and different signaling pathways involving in the early and late recurrence/metastasis of H-HCC. Meanwhile, we identified and further verified S100A12 and AMACR might be considered as potentially interesting prognostic markers for predicting the early recurrence/metastasis of H-HCC after hepatectomy.
Methods
Sample collection
H-HCC patients who had undergone surgery at the liver center of the First Affiliated Hospital of Fujian Medical University of China from April 2008 to December 2010 were evaluated for inclusion in the current study. The absence of intrahepatic recurrence/metastasis in the residual liver was monitored by ultrasonography (US), computer tomography (CT) scan and angiography. CT scan was also carried out to rule out distant metastasis, including lung, brain and bone. The H-HCC patients were monitored for recurrence/metastasis every one month in the first year after surgery and every 3 months after 1 year post operation, including serum AFP, US and CT scan. Enrollment Eligibility Criteria: (1) The patient was diagnosed with HCC by post-operative pathological examination; (2) Pre-operative serum HBs Ag positive, but HBc Ab negative; (3) Subject to the standard radical resection [68]: no distal metastasis was revealed in both pre-and intra-operative examination; no lesion was found in the rest of the liver during intra-operative ultrasonic scan; no visible cancer embolus in the hepatic portal vein or primary venous branch; no cancer cell was found in the incisal margin at the post-operative pathological examination; no recurrent/metastatic lesion was found at the ultrasonic and CT scan during the return visit after 2 months of surgery; (4) the elevated pre-operative serum AFP should decline to the normal level after 2 months post operation; (5) The patient did not undergo any other intervention or therapies before surgery.
The tissues of patients were divided into 3 groups according to the time of recurrence/metastasis after operation: the patients who had recurrence/metastasis within 6 months after operation (R/M≤6Months group, n = 20); the patients whose recurrence/metastasis occurred between 6 and 12 months after operation (R/M6-12months group, n = 20); the patients who had no recurrence/metastasis within 2 years of operation (NR/M group, n = 20). Fresh tissues were collected at the time of surgery from patients with HBV associated H-HCC for liquid nitrogen preservation after washing, and part of the tissues were formalin embedded and stored for immunohistochemistry. The project was approved for the using of human biopsy by the Institution Review Board of the First Affiliated Hospital of Fujian Medical University. The written consent was received from all participants in this study.
Protein preparation and iTRAQ labeling
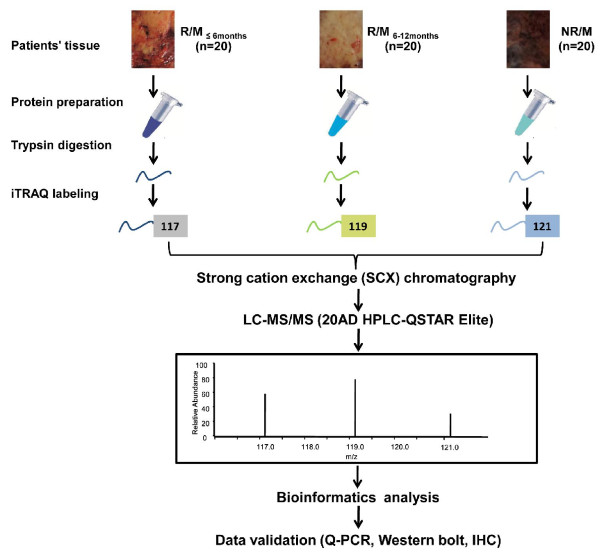
Ten milligrams of each liver tissue were lysed in the protein extraction buffer (150 mM NaCl, 10 mM Tris, 5 mM EDTA, 1% Triton X-100, 5% glycerol, and 0.1% SDS, pH 7.2) after smash in liquid nitrogen, and then incubated at 4°C for 30 min. After centrifugation at 12,000 rpm at 4°C for 30 min, the supernatant was collected, and the protein concentration was determined according to the BCA protocol (Beyotime, China). Samples were further normalized based on protein concentration. The iTRAQ labeling was performed according to the manufacturer’s protocol (AB SCIEX, USA). Briefly, 100 μg proteins of each group were precipitated with cold acetone for 1 hour at −20°C, and then re-suspended in 20 μl dissolution buffer. After protein reduction and alkylation followed by overnight digestion with trypsin, the peptides were labeled with the iTRAQ regents for 1 hour at room temperature. The iTRAQ regents 117, 119 and 121 were used to label the peptides from R/M≤6Months group, R/M6-12months group and NR/M group respectively. Then the samples were mixed with equal amounts, and desalted with the Sep-Pak Vac C18 cartridges and dried in a vacuum centrifuge. The workflow of our study was presented in Figure 5.
Figure 5.
Outline of the iTRAQ based quantitative proteomic strategy. iTRAQ labeling was carried out by using tumor tissues from the patients who had recurrence/metastasis within 6 months after operation (R/M≤6months group, n = 20); the patients whose recurrence/metastasis occurred between 6 and 12 months after operation (R/M6-12months group, n = 20); and the patients who had no recurrence/metastasis within 2 years of operation (NR/M group, n = 20). Samples were digested by trypsin, and the peptides from R/M≤6months, R/M6-12months, and NR/M were labeled by iTRAQ reagents 117, 119 and 121, respectively. After labeling, peptides from all three samples were combined and fractionated by SCX chromatography. Each fraction was then analyzed by LC-MS/MS on a QSTAR Elite mass spectrometers. The identified potential interesting targets were further validated by Q-PCR, Western bolt and immunohistochemistry (IHC).
2DLC-MS/MS analysis
The mixed peptides were separated by strong cation exchange (SCX) chromatography using a polysulfoethanyl column (2.1 mm × 100 mm, 5 μm, 200 Å, The Nest Group, USA) at a flow rate of 200 μL/min for 60 min with a gradient of 0–80% Buffer B (10 mM KH2PO4 in 25% acetonitrile, 350 mM KCl, pH 2.6) in Buffer A (10 mM KH2PO4 in 25% acetonitrile, pH 2.6) on a 20 AD HPLC system (Shimadzu, Japan), and a total of 20 SCX fractions were collected.
The mixed peptides (desalted with a PepMap C18 cartridge) were further separated by nano-HPLC (20 AD, Shimadzu, Japan) on the secondary RP analytical column (ZORBAX 300SB-C18 column, 150 mm × 100 μm, 5 μm, 300 Å, USA). Peptides were subsequently eluted using the following gradient conditions with phase B (95% ACN with 0.1% formic acid): 5-35% B (0–90 min), 35–80% B (90–95 min), 80–5% B (100–105 min) and 5–0% B (105-120 min); the flow rate was maintained at 300 μL/min. Electrospray voltage of 2.3 kV versus the inlet of the mass spectrometer was used.
A hybrid quadrupole time-of-flight mass spectrometer (QStar hybrid LC/MS/MS Q-TOF, AB SCIEX, USA) was operated in data-dependent mode to switch automatically between MS and MS/MS acquisition. MS spectra were acquired across the mass range of 400–1800 m/z in high resolution mode using 250 ms accumulation time per spectrum. Tandem mass spectral scanned from 100–2000 m/z in high sensitivity mode with collision induced dissociation (CID). The four most intense precursors were selected for fragmentation per cycle with dynamic exclusion time of 9 s.
Data analysis
The MS/MS data was searched against UniProtKB/ Swiss-Prot FASTA (it was released October 15, 2011 and consists of 20238 human sequences). Protein identification and iTRAQ quantitation were performed with ProteinPilot software (Version 4.0, AB SCIEX, USA). The user-defined search parameters included iTRAQ labeling at N-terminus and lysine residues, cysteine modification by methyl methanethiosulfonate (MMTS) and digestion by trypsin. For iTRAQ quantitation, the peptide was automatically selected with the Pro Group algorithm to calculate the reporter peak area, error factor (EF) and p value. A decoy database search strategy was adopted to estimate the FDR for peptide identification. In our study, a strict unused confidence cutoff >1.3 was used for protein identification; proteins with at least two peptides (confidence >95%) were used for quantification. The results were then exported into Microsoft Excel for manual data interpretation. The proteins were considered to be differentially expressed if their iTRAQ ratios were >2 or <0.5 in the R/M≤6Months group and R/M6-12months group relative to the NR/M group (P < 0.05) [69].
Functional analysis
The biological function of all the identified proteins was analyzed in the online Gene Ontology (GO) Term mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper). The function and pathway annotations of identified proteins were analyzed by Ingenuity Pathways Analysis (IPA) software (version 7.5), which is based on the Ingenuity Pathways database. The involved signaling pathways and networks were ranked in term of the enrichment of the differentially expressed proteins.
Immunoblotting
The tissue samples from patients were further taking for immunoblotting. The tissue samples were lysed in extraction buffer (150 mM NaCl, 10 mM Tris, 5 mM EDTA, 1% Triton X-100, 5% glycerol, and 0.1% SDS, pH 7.2), then were centrifuged at 22,000 rpm, 4°C for 30 min. The supernatants were collected, and the protein concentration was determined according to BCA protocol. Equal amount of proteins from each patient were applied for the immunoblotting experiments. 40 μg proteins per patients were analyzed on 12.5% SDS-PAGE, and transferred on PVDF membrane (transfer buffer: 25 mM Tris, 192 mM glycine, 20% methanol, pH8.3) by the Bio-Rad Semidry apparatus with a constant current of 300 mA for 1 hour or 30 min according to the size of the protein. Afterwards, the membrane was blocked in blocking buffer (1 × PBS, 0.5% Tween-20 with 5% nonfat milk) for 2 hours, and then further blotted with the primary antibody (monoclonal anti-S100A12 antibody, 1:500 dilution, Abcam; monoclonal anti-AMACR antibody, 1:1000 dilution, Abcam; monoclonal anti-β-actin antibody, 1:1000 dilution, Santa Cruz) in 4°C overnight and HRP conjugated secondary antibody in room temperature for 2 hrs. Afterwards, the membrane was incubated with Chemi-luminescent Detection Reagent (Pierce) for 5 min, and then was exposed to X-ray film. All of the above mentioned experiments were independently repeated 3 times.
Immunohistochemistry of Liver sections
The expression profile of the protein S100A12 and AMACR in liver section of the patients was investigated by the two-step Envision™ plus staining technique. The paraffin-embedded liver sections were de-waxed and re-hydrated through incubating in EDTA (1 mM) solution with 3 minutes for high-pressure antigen retrieval. Afterwards, the liver section was incubated with the primary antibody (mouse monoclonal anti-human S100A12 antibody, 1:500 dilution, Abcam; mouse monoclonal anti-human AMACR antibody, 1:1000 dilution, Abcam) at 4°C overnight, and then washed by PBS buffer for 3 times (5 min of each) at room temperature. It further followed by incubating with HRP-conjugated secondary antibodies (Fuzhou Maxim Biotech Inc, China) at room temperature for another 1 hour. Finally, the liver sections were subjected to DAB coloration and hematoxylin re-staining. The results were independently assessed by two pathologists double-blindly.
Q-PCR analysis
Trizol homogenization buffer (Takara Inc.) was added into the tissue samples to extract the total RNAs, which were then reversely transcribed into cDNAs (Reverse transcription was conducted according to the manufacture’s protocol from Takara). The β-actin was taken as the internal reference gene. The obtained cDNAs were used as the template for the Q-PCR analysis of the S100A12 and AMACR genes in a total reaction system of 50 μL (including 1 μL of the cDNA template, 1 μL of the upstream primer, 1 μL of the down stream primer, 25 μL of the SYBR green super mix and 22 μL of the RNase free dH2O). The reaction conditions were as follows: 94°C, 3 min, 1 cycle; followed by 40 cycles of 95°C 15 s, 62°C 15 s (S100A12) or 60°C 15 s (AMACR), 60°C 40 s. The relative fluorescence intensity from 55°C to 95°C (0.5°C increments every 10 sec) was collected to plot the melting curve, so as to survey the formation of primer dimmers and the non-specific amplification. The relative gene expression was calculated according to the Livak method (2-ΔΔCt). The experiments were repeated 3 times independently. The sequence information of the primers was as follows:
S100A12, forward primer 5′-ATTAGGCTGGGAAGATGACAAA-3′, and the reverse primer 5′-GCTTCAGCTCACCCTTAGAGAG-3′;
AMACR, forward primer 5′-ATTTGGCTTTGTCAGGTGTTCT-3′, and the reverse primer 5′- GCGGTCAAAAAGAGCCATTAT-3′;
β-actin, forward primer 5′-CCACTGGCATCGTGATGGAC-3′, and the reverse primer 5′- GCGGATGTCCACGTCACACT-3′.
Abbreviations
2DLC-MS/MS: Multidimensional chromatography tandem mass spectrometry; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Competing interests
The authors declare no competing interests.
Authors′ contributions
XH and YZ performed the iTRAQ experiments; XX and XL performed the data analysis and paper writing; YZ, JZ and BX performed the clinical sample collection; XH, YZ, YG and ZC performed the validation experiments; XL, AH and JL designed the experiment. All authors read and approved the final manuscript.
Supplementary Material
Tables of Identified Proteins.
Supporting Information.
Contributor Information
Xinhui Huang, Email: hxhui24@163.com.
Yongyi Zeng, Email: lamp1973@medmail.com.cn.
Xiaohua Xing, Email: xingxiaohua2006@163.com.
Jinhua Zeng, Email: jinhuaz2013@163.com.
Yunzhen Gao, Email: gaoyunzhen2003@yahoo.com.cn.
Zhixiong Cai, Email: caizhixiong1985@163.com.
Bo Xu, Email: xubo-1128@163.com.
Xiaolong Liu, Email: xiaoloong.liu@gmail.com.
Aimin Huang, Email: aimin@fjmu.edu.cn.
Jingfeng Liu, Email: drjingfeng@126.com.
Acknowledgements
This work is supported by the key project of National Scinece and technology of China (Grant No. 2012ZX10002010-001-006, and Grant No. 2012ZX10002016-013), the National Natural Science Foundation of China (Grant No. 31201008), the key project of Science and Technology Department of Fujian Province (Grant No. 2011Y0022), the Natural Science Foundation of Fujian Province (Grant No. 2011 J01162), the University Foundation of Fujian Province (Grant No. JK2011021), and the University Natural Science Foundation of Jiangsu Province (Grant No. 12KJB180013).
References
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Zeng HM, Zheng RS, Zhang SW, He J. Cancer incidence and mortality in china, 2007. Chin J Cancer Res. 2012;24:1–8. doi: 10.1007/s11670-012-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Qiu FZ, Wu ZD, Zhang BX. Hepatectomy for huge hepatocellular carcinoma in 634 cases. World J Gastroenterol. 2006;12:4652–4655. doi: 10.3748/wjg.v12.i29.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau KH, Ruo L, Shia J, Padela A, Gonen M, Jarnagin WR, Fong Y, D’Angelica MI, Blumgart LH, DeMatteo RP. Outcome of partial hepatectomy for large (>10 cm) hepatocellular carcinoma. Cancer. 2005;104:1948–1955. doi: 10.1002/cncr.21415. [DOI] [PubMed] [Google Scholar]
- Choi GH, Han DH, Kim DH, Choi SB, Kang CM, Kim KS, Choi JS, Park YN, Park JY, Kim do Y, Han KH, Chon CY, Lee WJ. Outcome after curative resection for a huge (>or = 10 cm) hepatocellular carcinoma and prognostic significance of gross tumor classification. Am J Surg. 2009;198:693–701. doi: 10.1016/j.amjsurg.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Ng KM, Yan TD, Black D, Chu FC, Morris DL. Prognostic determinants for survival after resection/ablation of a large hepatocellular carcinoma. HPB (Oxford) 2009;11:311–320. doi: 10.1111/j.1477-2574.2009.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Wei AC, Cleary SP, Yang I, McGilvray ID, Gallinger S, Grant DR, Greig PD. Prognosis and results after resection of very large (>or = 10 cm) hepatocellular carcinoma. J Gastrointest Surg. 2007;11:589–595. doi: 10.1007/s11605-007-0154-7. [DOI] [PubMed] [Google Scholar]
- Lee SG, Hwang S, Jung JP, Lee YJ, Kim KH, Ahn CS. Outcome of patients with huge hepatocellular carcinoma after primary resection and treatment of recurrent lesions. Br J Surg. 2007;94:320–326. doi: 10.1002/bjs.5622. [DOI] [PubMed] [Google Scholar]
- Guo K, Kang NX, Li Y, Sun L, Gan L, Cui FJ, Gao MD, Liu KY. Regulation of HSP27 on NF-kappaB pathway activation may be involved in metastatic hepatocellular carcinoma cells apoptosis. BMC Cancer. 2009;9:100. doi: 10.1186/1471-2407-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama T, Takahashi M, Nakanishi K, Yokoo H, Kamachi H, Kobayashi N, Ozaki M, Todo S. alpha-fetoprotein, vascular endothelial growth factor receptor-1 and early recurrence of hepatoma. World J Gastroenterol. 2012;18:340–348. doi: 10.3748/wjg.v18.i4.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103:307–312. doi: 10.1002/cncr.20774. [DOI] [PubMed] [Google Scholar]
- Wang SM, Ooi LL, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2011;17:6040–6051. doi: 10.1158/1078-0432.CCR-11-0557. [DOI] [PubMed] [Google Scholar]
- Bozzuto G, Ruggieri P, Molinari A. Molecular aspects of tumor cell migration and invasion. Ann Ist Super Sanita. 2010;46:66–80. doi: 10.4415/ANN_10_01_09. [DOI] [PubMed] [Google Scholar]
- Liang S, Xu Z, Xu X, Zhao X, Huang C, Wei Y. Quantitative proteomics for cancer biomarker discovery. Comb Chem High Throughput Screen. 2012;15:221–231. doi: 10.2174/138620712799218635. [DOI] [PubMed] [Google Scholar]
- Nikolov M, Schmidt C, Urlaub H. Quantitative mass spectrometry-based proteomics: an overview. Methods Mol Biol. 2012;893:85–100. doi: 10.1007/978-1-61779-885-6_7. [DOI] [PubMed] [Google Scholar]
- Uto H, Kanmura S, Takami Y, Tsubouchi H. Clinical proteomics for liver disease: a promising approach for discovery of novel biomarkers. Proteome Sci. 2010;8:70. doi: 10.1186/1477-5956-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Hudler P, Gorsic M, Komel R. Proteomic strategies and challenges in tumor metastasis research. Clin Exp Metastasis. 2010;27:441–451. doi: 10.1007/s10585-010-9339-7. [DOI] [PubMed] [Google Scholar]
- Sun W, Xing B, Sun Y, Du X, Lu M, Hao C, Lu Z, Mi W, Wu S, Wei H, Gao X, Zhu Y, Jiang Y, Qian X, He F. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol Cell Prot. 2007;6:1798–1808. doi: 10.1074/mcp.M600449-MCP200. [DOI] [PubMed] [Google Scholar]
- Orimo T, Ojima H, Hiraoka N, Saito S, Kosuge T, Kakisaka T, Yokoo H, Nakanishi K, Kamiyama T, Todo S, Hirohashi S, Kondo T. Proteomic profiling reveals the prognostic value of adenomatous polyposis coli-end-binding protein 1 in hepatocellular carcinoma. Hepatology. 2008;48:1851–1863. doi: 10.1002/hep.22552. [DOI] [PubMed] [Google Scholar]
- Qin S, Ferdinand AS, Richie JP, O’Leary MP, Mok SC, Liu BC. Chromatofocusing fractionation and two-dimensional difference gel electrophoresis for low abundance serum proteins. Proteomics. 2005;5:3183–3192. doi: 10.1002/pmic.200401137. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche S, Santoni V, Rabilloud T. Evaluation of nonionic and zwitterionic detergents as membrane protein solubilizers in two-dimensional electrophoresis. Proteomics. 2003;3:249–253. doi: 10.1002/pmic.200390037. [DOI] [PubMed] [Google Scholar]
- Kameshita I, Ishida A, Fujisawa H. Analysis of protein-protein interaction by two-dimensional affinity electrophoresis. Anal Biochem. 1998;262:90–92. doi: 10.1006/abio.1998.2728. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Li Z, Tan XF, Lim TK, Mao Y, Lin Q. iTRAQ based quantitative proteomics approach validated the role of calcyclin binding protein (CacyBP) in promoting colorectal cancer metastasis. Mol Cell Prot. 2013;12:1865–1880. doi: 10.1074/mcp.M112.023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, Chen X, Zeng D, Tong A, Chen L, Wei Y, Huang C. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenot E, Urbach S, Vigy O, Seveno M, Galeotti N, Nguyen G, Bockaert J, Marin P. Quantitative proteomic analysis reveals protein expression changes in the murine neuronal secretome during apoptosis. J Proteomics. 2012;77:394–405. doi: 10.1016/j.jprot.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Tonack S, Aspinall-O’Dea M, Jenkins RE, Elliot V, Murray S, Lane CS, Kitteringham NR, Neoptolemos JP, Costello E. A technically detailed and pragmatic protocol for quantitative serum proteomics using iTRAQ. J Proteomics. 2009;73:352–356. doi: 10.1016/j.jprot.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Chang KP, Yu JS, Chien KY, Lee CW, Liang Y, Liao CT, Yen TC, Lee LY, Huang LL, Liu SC, Chang YS, Chi LM. Identification of PRDX4 and P4HA2 as metastasis-associated proteins in oral cavity squamous cell carcinoma by comparative tissue proteomics of microdissected specimens using iTRAQ technology. J Proteome Res. 2011;10:4935–4947. doi: 10.1021/pr200311p. [DOI] [PubMed] [Google Scholar]
- Haura EB, Muller A, Breitwieser FP, Li J, Grebien F, Colinge J, Bennett KL. Using iTRAQ combined with tandem affinity purification to enhance low-abundance proteins associated with somatically mutated EGFR core complexes in lung cancer. J Proteome Res. 2011;10:182–190. doi: 10.1021/pr100863f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Tan HT, Lim KH, Lim TK, Khoo A, Tan IB, Yeoh KG, Chung MC. Identification and functional validation of caldesmon as a potential gastric cancer metastasis-associated protein. J Proteome Res. 2013;12:980–990. doi: 10.1021/pr3010259. [DOI] [PubMed] [Google Scholar]
- Masui O, White NM, DeSouza LV, Krakovska O, Matta A, Metias S, Khalil B, Romaschin AD, Honey RJ, Stewart R, Pace K, Bjarnason GA, Siu KW, Yousef GM. Quantitative proteomic analysis in metastatic renal cell carcinoma reveals a unique set of proteins with potential prognostic significance. Mol Cell Prot. 2013;12:132–144. doi: 10.1074/mcp.M112.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Chen Q, Sun C, Wang C, Peng Q, Xie L, Liu Y, Li S. High-throughput screening of tumor metastatic-related differential glycoprotein in hepatocellular carcinoma by iTRAQ combines lectin-related techniques. Med Oncol. 2013;30:420. doi: 10.1007/s12032-012-0420-8. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Prot. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- He X, Wang Y, Zhang W, Li H, Luo R, Zhou Y, Li C, Liao M, Huang H, Lv X, Xie Z, He M. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. Neoplasma. 2014;61:10. [PubMed] [Google Scholar]
- Ko CH, Cheng CF, Lai CP, Tzu TH, Chiu CW, Lin MW, Wu SY, Sun CY, Tseng HW, Wang CC, Kuo ZK, Wang LM, Chen SF. Differential proteomic analysis of cancer stem cell properties in hepatocellular carcinomas by isobaric tag labeling and mass spectrometry. J Proteome Res. 2013;12:3573–3585. doi: 10.1021/pr4004294. [DOI] [PubMed] [Google Scholar]
- Huang C, Wang Y, Liu S, Ding G, Liu W, Zhou J, Kuang M, Ji Y, Kondo T, Fan J. Quantitative proteomic analysis identified paraoxonase 1 as a novel serum biomarker for microvascular invasion in hepatocellular carcinoma. J Proteome Res. 2013;12:9. doi: 10.1021/pr3011815. [DOI] [PubMed] [Google Scholar]
- Wang C, Guo K, Gao D, Kang X, Jiang K, Li Y, Sun L, Zhang S, Sun C, Liu X, Wu W, Yang P, Liu Y. Identification of transaldolase as a novel serum biomarker for hepatocellular carcinoma metastasis using xenografted mouse model and clinic samples. Cancer Lett. 2011;313:154–166. doi: 10.1016/j.canlet.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Yu Y, Pan X, Ding Y, Liu X, Tang H, Shen C, Shen H, Yang P. An iTRAQ based quantitative proteomic strategy to explore novel secreted proteins in metastatic hepatocellular carcinoma cell lines. Analyst. 2013;138:4505–4511. doi: 10.1039/c3an00517h. [DOI] [PubMed] [Google Scholar]
- Feng H, Li X, Niu D, Chen WN. Protein profile in HBx transfected cells: a comparative iTRAQ-coupled 2D LC-MS/MS analysis. J Proteomics. 2010;73:1421–1432. doi: 10.1016/j.jprot.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Feng H, Wang M, Chen WN. iTRAQ-coupled 2D LC-MS/MS analysis of secreted proteome of HBV-replicating HepG2 cells: potential in biomarkers for prognosis of HCC. Curr Microbiol. 2010;61:280–284. doi: 10.1007/s00284-010-9608-3. [DOI] [PubMed] [Google Scholar]
- Bee A, Ke Y, Forootan S, Lin K, Beesley C, Forrest SE, Foster CS. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12:2061–2065. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Nagy N, Bronckart Y, Camby I, Legendre H, Lahm H, Kaltner H, Hadari Y, Van Ham P, Yeaton P, Pector JC, Zick Y, Salmon I, Danguy A, Kiss R, Gabius HJ. Galectin-8 expression decreases in cancer compared with normal and dysplastic human colon tissue and acts significantly on human colon cancer cell migration as a suppressor. Gut. 2002;50:392–401. doi: 10.1136/gut.50.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology. 2003;38:1242–1249. doi: 10.1053/jhep.2003.50457. [DOI] [PubMed] [Google Scholar]
- Pogue-Geile K, Geiser JR, Shu M, Miller C, Wool IG, Meisler AI, Pipas JM. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich GA, Ilarregui JM. Conveying glycan information into T-cell homeostatic programs: a challenging role for galectin-1 in inflammatory and tumor microenvironments. Immunol Rev. 2009;230:144–159. doi: 10.1111/j.1600-065X.2009.00787.x. [DOI] [PubMed] [Google Scholar]
- Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34:1430–1448. doi: 10.1016/S1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. J Leukoc Biol. 2001;69:986–994. [PubMed] [Google Scholar]
- Schmitz W, Albers C, Fingerhut R, Conzelmann E. Purification and characterization of an alpha-methylacyl-CoA racemase from human liver. Eur J Biochem. 1995;231:815–822. doi: 10.1111/j.1432-1033.1995.tb20766.x. [DOI] [PubMed] [Google Scholar]
- Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102:639–644. doi: 10.1038/sj.bjc.6605530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Huang X, Wei W, Ya N, Zeng J, Zeng Y, Ma C, Chi M, Wu Y, Li Y, Huang Y, Zhang X, Huang A, Liu J. A mathematical model to predict short-term recurrence and metastasis of primary hepatocellular carcinoma larger than 10 cm in diameter. Hepatogastroenterology. 2013;60:225–230. doi: 10.5754/hge. [DOI] [PubMed] [Google Scholar]
- Wang ZS, Wu LQ, Yi X, Geng C, Li YJ, Yao RY, Hu WY, Han B. CK19 can be used to predict the early recurrence and prognosis of HBV-related hepatocellular carcinoma patients with low AFP serum concentration after R0 radical hepatectomy. Zhonghua Zhong Liu Za Zhi. 2012;34:753–758. doi: 10.3760/cma.j.issn.0253-3766.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Lv Y, Wang W, Jia WD, Sun QK, Li JS, Ma JL, Liu WB, Zhou HC, Ge YS, Yu JH, Xia HH, Xu GL. High-level expression of periostin is closely related to metastatic potential and poor prognosis of hepatocellular carcinoma. Med Oncol. 2013;30:385. doi: 10.1007/s12032-012-0385-7. [DOI] [PubMed] [Google Scholar]
- Lau SH, Sham JS, Xie D, Tzang CH, Tang D, Ma N, Hu L, Wang Y, Wen JM, Xiao G, Zhang WM, Lau GK, Yang M, Guan XY. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- Garcia AF, Garcia W, Nonato MC, Araujo AP. Structural stability and reversible unfolding of recombinant porcine S100A12. Biophys Chem. 2008;134:246–253. doi: 10.1016/j.bpc.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Thierolf M, Hagmann ML, Pfeffer M, Berntenis N, Wild N, Roessler M, Palme S, Karl J, Bodenmuller H, Ruschoff J, Rossol S, Rohr G, Rosch W, Friess H, Eickhoff A, Jauch KW, Langen H, Zolg W, Tacke M. Towards a comprehensive proteome of normal and malignant human colon tissue by 2-D-LC-ESI-MS and 2-DE proteomics and identification of S100A12 as potential cancer biomarker. Proteomics Clin Appl. 2008;2:11–22. doi: 10.1002/prca.200780046. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Fanger GR, Woda BA, Banner BF, Algate P, Dresser K, Xu J, Chu PG. Expression of alpha-methylacyl-CoA racemase (P504s) in various malignant neoplasms and normal tissues: astudy of 761 cases. Hum Pathol. 2003;34:792–796. doi: 10.1016/S0046-8177(03)00268-5. [DOI] [PubMed] [Google Scholar]
- Helal Tel A, Radwan NA, Abdel Kader Z, Helmy NA, Hammad SY. Role of alpha methylacyl-coenzyme A racemase in differentiating hepatocellular carcinoma from dysplastic and nondysplastic liver cell lesions. Ann Diagn Pathol. 2012;16:330–334. doi: 10.1016/j.anndiagpath.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Mozos A, Sole M, Miquel R, Petit A, Nadal A, Fernandez PL. Usefulness and limitations of alpha-methylacyl-CoA racemase expression in the hepatobiliary system and pancreas: a wide tissue microarray analysis of normal and neoplastic epithelia. Anal Quant Cytol Histol. 2010;32:261–268. [PubMed] [Google Scholar]
- Dabritz J, Langhorst J, Lugering A, Heidemann J, Mohr M, Wittkowski H, Krummenerl T, Foell D. Improving relapse prediction in inflammatory bowel disease by neutrophil-derived S100A12. Inflamm Bowel Dis. 2013;19:1130–1138. doi: 10.1097/MIB.0b013e318280b1cd. [DOI] [PubMed] [Google Scholar]
- Barry M, Dhillon PK, Stampfer MJ, Perner S, Ma J, Giovannucci E, Kurth T, Mucci LA, Rubin MA. alpha-Methylacyl-CoA racemase expression and lethal prostate cancer in the Physicians’ Health Study and Health Professionals Follow-up Study. Prostate. 2012;72:301–306. doi: 10.1002/pros.21432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, Hamada K, Nakayama H, Ishitsuka H, Miyamoto T, Hirabayashi A, Uchinmura S, Hamamoto Y. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- Garbis SD, Tyritzis SI, Roumeliotis T, Zerefos P, Giannopoulou EG, Vlahou A, Kossida S, Diaz J, Vourekas S, Tamvakopoulos C, Pavlakis K, Sanoudou D, Constantinides CA. Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (iTRAQ) with two-dimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2008;7:3146–3158. doi: 10.1021/pr800060r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables of Identified Proteins.
Supporting Information.