Abstract
In recent years, multiple loci dispersed on the genome have been shown to be associated with coronary artery disease (CAD). We investigated whether these common genetic variants also hold value for CAD prediction in a large cohort of patients with familial hypercholesterolemia (FH). We genotyped a total of 41 single-nucleotide polymorphisms (SNPs) in 1701 FH patients, of whom 482 patients (28.3%) had at least one coronary event during an average follow up of 66 years. The association of each SNP with event-free survival time was calculated with a Cox proportional hazard model. In the cardiovascular disease risk factor adjusted analysis, the most significant SNP was rs1122608:G>T in the SMARCA4 gene near the LDL-receptor (LDLR) gene, with a hazard ratio for CAD risk of 0.74 (95% CI 0.49–0.99; P-value 0.021). However, none of the SNPs reached the Bonferroni threshold. Of all the known CAD loci analyzed, the SMARCA4 locus near the LDLR had the strongest negative association with CAD in this high-risk FH cohort. The effect is contrary to what was expected. None of the other loci showed association with CAD.
Keywords: CAD, FH, GWAS, association
Introduction
Familial hypercholesterolemia (FH) is an autosomal dominant disorder characterized by increased low-density lipoprotein cholesterol (LDL-C) levels and preponderance to coronary artery disease (CAD). The diagnosis is based on stringent clinical criteria or on the identification of mutations in the LDL-receptor (LDLR), apolipoprotein B or proprotein convertase subtilisin/kexin type 9 (PCSK9) gene. The frequency of heterozygosity is at least 1/500 in most European countries.1 By virtue of the elevated LDL-C levels, FH results in lipid accumulation in the arterial wall and as a consequence accelerated atherosclerosis.2, 3 If left untreated, 50% of male and 30% of female heterozygous FH patients will develop CAD before 60 years of age.4 The age of onset and severity of CAD varies considerably between FH patients, even among individuals who share an identical gene defect.5
Previously, we performed a retrospective multicentre cohort study of 2400 FH patients, of whom 782 patients (32.6%) had at least one cardiovascular event during an average follow up of 66 years.6 In this cohort, we demonstrated that LDL-C levels are more important than the LDLR mutation type in determining the age of onset of CAD.7 We also showed that classical risk factors including male gender, smoking, hypertension, type 2 diabetes, low levels of high-density lipoprotein cholesterol (HDL-C) and elevated lipoprotein(a) levels were independent risk factors for the development of CAD. However, these factors combined explained only 18.7% of the variation in CVD occurrence.8 Thus, a considerable part of the variability in CVD occurrence remains to be disentangled and common genetic variation might provide one of the explanations.
Recent large-scale genome-wide association (GWA) studies have revealed common genetic variations at 45 loci that moderately affect (hazard ratios (HR) varying between 1.1 and 2.0) the incidence of CAD in the general Caucasian population.9, 10, 11, 12
We set out to address whether common variations within the 45 previously identified loci by recent GWA studies are modifiers of CAD risk in a high-risk population of heterozygous FH cases.
Materials and methods
Ethics statement
Written informed consent was obtained from all patients. The Medical Ethics Review Board of each participating hospital approved the protocol, which complies with the Declaration of Helsinki.
Study design and study population
The Genetic Identification of Risk Factors in Familial Hypercholesterolemia (GIRaFH) is a retrospective multicenter cohort study. The study design and study population have been described elsewhere.6 Briefly, DNA samples from patient who, based on clinically oriented algorithms are anticipated to suffer from FH are being sent in to the central core molecular diagnostic laboratory by physicians working at one of the nationwide lipid clinics. LDLR gene variation was genotyped according to previously published methods.13 DNA of a total of 9300 hypercholesterolemic patients was stored in the DNA database at the time of initiation of the cohort. Only those cases from larger lipid clinics were selected (9188) for further analysis, as smaller clinics normally only send DNA samples of the rare, usually very serious FH cases. Of this set, 4000 cases were randomly selected. After review of medical records, a group of 2400 patients fulfilled the FH diagnostic criteria based on internationally established criteria and were included in the study.14 Phenotypic, CVD event and cause of death data were acquired from the medical charts. None of the study population received primary prevention in the form of beta-blockers or aspirin. CAD was defined as angina pectoris (AP), acute coronary syndrome (ACS), percutaneous coronary interventions (PCI) or coronary artery bypass grafting (CABG).
SNP selection
On the basis of the latest published meta-analysis, a total of 45 single-nucleotide polymorphisms (SNPs) associated with CAD were identified.12 We did not include SNPs which were only associated with an intermediate trait such as lipid levels, type 2 diabetes or hypertension.
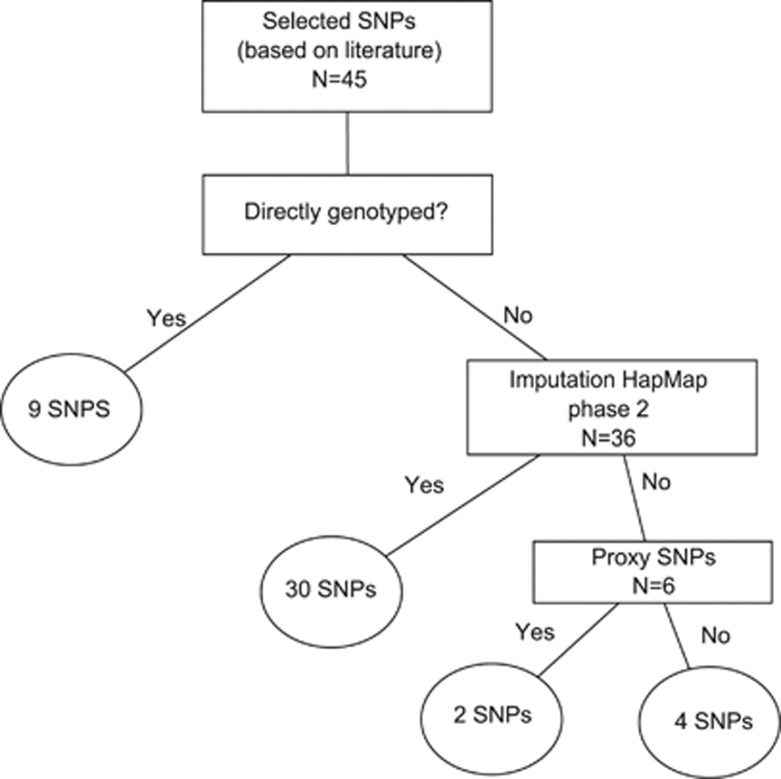
If these SNPs were not directly genotyped, imputed data, using MACH and the HapMap phase 2 data sets (build 36 release 22),15, 16, 17 were used. Finally if this failed proxies were looked up within a window of 500 kb (R2≥0.8).15, 16, 17 See Figure 1.
Figure 1.
SNP selection procedure.
Genotyping and imputation
For 1701 of the 2400 GIRaFH cases DNA was available for additional genotyping. Genotyping was performed using the 50 K gene-centric Human CVD BeadChip18 and genotypes were called using the BeadStudio software (Illumina, San Diego, CA, USA) and subjected to quality control filters at the sample and the SNP level. After genotyping, PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) was used to test the SNPs for population substructure which could introduce false-positive associations. This was done by means of multidimensional scaling implementation.19 In addition, the SNPs were subjected to additional quality control filters based on sample size and minor allele frequencies (MAF). Samples with a call rate of <95% were excluded from further analysis. Genetic markers with a MAF <1% were excluded from further analysis. An identity-by-state analysis was performed to ensure that only Caucasian individuals were included in the final association analyses.
Statistical analysis
With an effective sample size of 1701 cases and 483 events, the GIRaFH sample has 80% power to identify statistically significant associations for SNPs conferring a relative risk >2.2 and MAF >0.10. Differences between subgroups were tested with χ2 statistics or an independent sample t-test where appropriate. Triglycerides had a skewed distribution and therefore statistical analyses were performed on log-transformed data. The association of each SNP with event-free survival time was calculated with a Cox proportional hazard model in the R package ProbABEL20 The event-free survival time was defined as time from birth to date of CAD event, or when no event had occurred as time from birth to date of inclusion in the study. An additive genetic model was applied in the Cox model and classical cardiovascular risks were used as covariates.21 We corrected for factors that had previously been shown to be associated with CAD risk in this population: age, gender, smoking, type 2 diabetes, hypertension and body mass index (BMI). Analyses were performed for the loci previously reported to be associated with CAD. Of the 45 reported SNPs, data were available of 41 SNPs after imputation. Significance was defined as a P-value <0.05 divided by the number of SNPs tested, yielding a significance level of 1.22 × 10−3 (41 SNPs based on the literature). All analyses were also performed separately for males and females. Statistical analyses were performed using SPSS software (version 17; Chicago, IL, USA).
All described variants will be submitted to the following public variant database Leiden Open Variation Database (LOVD)3 (http://databases.lovd.nl/shared/genes/).
Results
Genotyping and imputation
Of the 1701 DNA samples, 7 individuals did not cluster appropriately in the IBS, reflecting non-Caucasoid origin, and were consecutively excluded, leaving a total of 1694 DNA samples for analysis. A total of 38 978 SNPs met our quality control steps. No subjects were excluded because of low call rate. The genomic inflation factor was close to 1 (λ=1.07), indicating that the influence of population substructure and genotyping errors was negligible. Using HapMap, we were able to impute up to 2.5 million SNPs for all individuals. Out of the 45 SNPs, 9 were directly genotyped and 30 were imputed. A proxy of 2 of the remaining 6 SNPs (rs12205331:C>T→ rs12197124:C>T (R2=1.0) and rs9369640:C>A→rs7751826:C>T (R2=1.0) could be found in LD with the lead SNP. So a total of 9 SNPs was genotyped directly, 30 SNPs were imputed, for two SNPs, proxy SNPs were found and 4 SNPs could not be found within the imputed data, because they were not available in the reference panel or they were poorly imputed. We only analyzed the 41 SNPs that were available after imputation.
Demographic data
Demographic data of the 1694 study cases are listed in Table 1. The average age at inclusion was higher in the CAD group. The mean age of onset of CAD was 49.1 (standard deviation; SD 10.7) years and the mean event-free survival in individuals without CAD was 47.3 (SD 12.6) years. During follow-up, 28% of our cohort developed CAD. Cardiovascular risk factors were significantly more prevalent in CAD cases than in controls. Treatment-naive LDL-C levels at the time of inclusion in the cohort did not differ between patients with and without CAD. All first visits to the lipid clinic took place between March 1969 and November 2002.
Table 1. Baseline characteristics.
|
CAD+ |
CAD− |
||
|---|---|---|---|
| n=482 | n=1212 | P-value | |
| Age first visit lipid clinic (years) | 50.5 (11.1) | 42.2 (12.3) | <0.0001 |
| Age last visit lipid clinic (years) | 57.1 (11.2) | 46.6 (12.5) | <0.0001 |
| Male gender | 301 (62) | 516 (42) | <0.0001 |
| Diabetes mellitus | 54 (11) | 45 (4) | <0.0001 |
| Hypertension | 79 (16) | 77 (6) | <0.0001 |
| History of smoking | 365 (75) | 766 (63) | <0.0001 |
| Body mass index, kg/m2 | 25.7±3.2 | 24.9±3.6 | <0.0001 |
| Family history CVD | 204 (42) | 607 (50) | 0.005 |
| Treatment-naive lipids levels: | |||
| Total cholesterol, mmol/l | 9.6 (2.1) | 9.4 (1.9) | 0.146 |
| LDL-cholesterol, mmol/l | 7.0 (1.9) | 7.2 (1.8) | 0.152 |
| HDL-cholesterol, mmol/l | 1.1 (0.3) | 1.2 (0.4) | <0.0001 |
| Triglyceridesa, mmol/l | 1.8 (1.3–2.5) | 1.5 (1.0–2.0) | <0.0001 |
| LDL-receptor mutation provenb | 207 (43) | 655 (54) | 0.017 |
| Age of CAD onset, years | 49.1±10.7 | Na | Na |
| Age of start statin use | 48.8±11.5 | 40.9±12.6 | <0.0001 |
Abbreviations: CAD, coronary artery disease; CVD, cardiovascular disease; LDL, low-density lipoprotein; HDL, high-density lipoprotein.
Values are given as number (percentage) or mean±SD unless indicated otherwise.
Mean±interquartile range.
Mutations of the LDL receptor that have been examined according to Umans-Eckenhausen MA et al.13
SNPs and risk of CAD
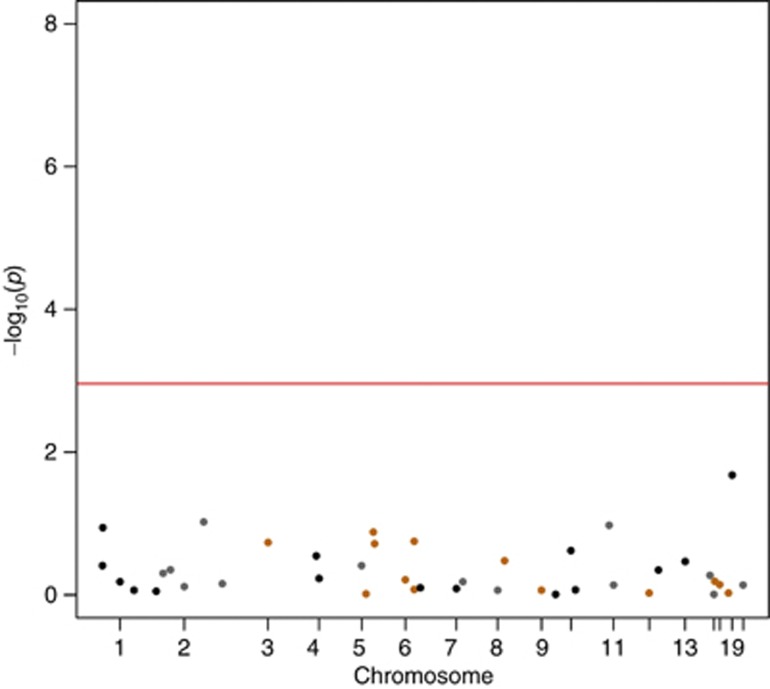
None of the 41 SNPs reached a significant P-value after the Bonferroni correction (P<1.11 × 10−3 ) (see Table 2). The best performing SNP in the cardiovascular risk factors adjusted analysis was rs1122608:G>T, in the SMARCA4 gene near the LDLR gene, with HR 0.74 (CI 0.49−0.99) and P-value 0.021 (Figure 2). No differences were observed in gender specific analysis (data not shown).
Table 2. The effect of the 41 SNPs analyzed in our study on CAD in FH.
| SNP | proxy | Chr | Pos | RA | RAF | HR | 95% CI lower | 95% CI upper | P-value | Nearest gene | Genotyped/imputed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17464857 | 1 | 220829332 | G | 0.11 | 1.0339 | 0.57 | 1.50 | 0.89 | MIA3 | Imputed | |
| rs17114036 | 1 | 56735409 | G | 0.11 | 1.4222 | 0.98 | 1.86 | 0.11 | PPAP2B | Imputed | |
| rs11206510 | 1 | 55268627 | C | 0.17 | 1.0886 | 0.89 | 1.28 | 0.39 | PCSK9 | Genotyped | |
| rs602633 | 1 | 109623034 | T | 0.19 | 1.0458 | 0.85 | 1.24 | 0.66 | SORT1 | Genotyped | |
| rs4845625 | 1 | 152688691 | T | 0.44 | 1.0139 | 0.86 | 1.17 | 0.86 | IL6R | Genotyped | |
| rs6725887 | 2 | 203454130 | T | 0.14 | 1.2882 | 0.02 | 2.56 | 0.70 | WDR12 | Imputed | |
| rs515135 | 2 | 21139562 | T | 0.15 | 1.0793 | 0.86 | 1.30 | 0.50 | APOB | Imputed | |
| rs6544713 | 2 | 43927385 | T | 0.33 | 1.0643 | 0.90 | 1.22 | 0.45 | ABCG5-ABCG8 | Imputed | |
| rs1561198 | 2 | 85663500 | C | 0.46 | 0.9765 | 0.82 | 1.13 | 0.77 | VAMP5-VAMP8-GGCX | Imputed | |
| rs2252641 | 2 | 145517931 | T | 0.48 | 1.5015 | 1.03 | 1.97 | 0.10 | ZEB2-AC074093.1 | Imputed | |
| rs9818870 | 3 | 139604812 | T | 0.16 | 0.2871 | −1.55 | 2.13 | 0.18 | MRAS | Imputed | |
| rs1878406 | 4 | 148613114 | T | 0.15 | 0.8840 | 0.66 | 1.11 | 0.29 | EDNRA | Imputed | |
| rs7692387 | 4 | 156854759 | A | 0.2 | 0.0230 | −13.48 | 13.53 | 0.59 | GUCY1A3 | Imputed | |
| rs273909 | 5 | 131695252 | G | 0.15 | 0.8902 | 0.63 | 1.15 | 0.39 | SLC22A4-SLC22A5 | Imputed | |
| rs12205331 | rs12197124 | 6 | 35029419 | T | 0.21 | 0.6550 | 0.10 | 1.21 | 0.13 | ANKS1A | Imputed |
| rs10947789 | 6 | 39282900 | C | 0.22 | 0.0001 | −13.46 | 13.46 | 0.19 | KCNK5 | Imputed | |
| rs4252120 | 6 | 161063598 | C | 0.3 | 0.9838 | 0.83 | 1.14 | 0.84 | PLG | Imputed | |
| rs9369640 | rs7751826 | 6 | 12900977 | C | 0.37 | 0.9236 | −3.59 | 5.44 | 0.97 | PHACTR1 | Imputed |
| rs2048327 | 6 | 160783522 | T | 0.37 | 0.8940 | 0.73 | 1.06 | 0.18 | SLC22A3-LPAL2-LPA | Imputed | |
| rs12190287 | 6 | 134256218 | C | 0.38 | 1.0986 | 0.73 | 1.46 | 0.61 | TCF21 | Imputed | |
| rs2023938 | 7 | 19003300 | C | 0.12 | 1.1024 | 0.37 | 1.83 | 0.79 | HDAC9 | Imputed | |
| rs11556924 | 7 | 129450732 | C | 0.4 | 0.9822 | 0.83 | 1.14 | 0.82 | ZC3HC1 | Genotyped | |
| rs264 | 8 | 19857460 | G | 0.14 | 0.9500 | 0.72 | 1.18 | 0.66 | LPL | Genotyped | |
| rs2954029 | 8 | 126560154 | T | 0.45 | 0.9869 | 0.84 | 1.14 | 0.86 | TRIB1 | Genotyped | |
| rs579459 | 9 | 135143989 | C | 0.25 | 0.9838 | 0.81 | 1.16 | 0.85 | ABO | Imputed | |
| rs3217992 | 9 | 21993223 | C | 0.37 | 1.0781 | 0.93 | 1.23 | 0.33 | CDKN2BAS1 | Imputed | |
| rs12413409 | 10 | 104709086 | A | 0.07 | 1.0449 | 0.60 | 1.49 | 0.85 | CYP17A1-CNNM2-NT5C2 | Imputed | |
| rs2047009 | 10 | 43859919 | G | 0.44 | 0.9702 | −1.63 | 3.57 | 0.98 | CXCL12 | Imputed | |
| rs11203042 | 10 | 90979089 | T | 0.44 | 0.9130 | 0.76 | 1.06 | 0.24 | LIPA | Genotyped | |
| rs9326246 | 11 | 116116943 | C | 0.08 | 0.9456 | 0.64 | 1.25 | 0.72 | ZNF259-APOA5-APOA1 | Imputed | |
| rs974819 | 11 | 103165777 | T | 0.18 | 0.4280 | −0.58 | 1.44 | 0.11 | PDGFD | Imputed | |
| rs3184504 | 12 | 110368991 | C | 0.44 | 0.9943 | 0.84 | 1.15 | 0.94 | SH2B3 | Genotyped | |
| rs9515203 | 13 | 109847624 | C | 0.31 | 2.8507 | 0.67 | 5.03 | 0.34 | COL4A1-COL4A2 | Imputed | |
| rs9319428 | 13 | 27871621 | G | 0.33 | 1.0639 | 0.90 | 1.22 | 0.45 | FLT1 | Imputed | |
| rs7173743 | 15 | 76928839 | T | 0.45 | 0.9068 | 0.60 | 1.21 | 0.53 | ADAMTS7 | Imputed | |
| rs17514846 | 15 | 89217554 | C | 0.46 | 0.9977 | 0.85 | 1.15 | 0.98 | FURIN-FES | Genotyped | |
| rs2281727 | 17 | 2064695 | G | 0.32 | 1.0620 | 0.80 | 1.32 | 0.65 | SMG6 | Imputed | |
| rs12936587 | 17 | 17484447 | A | 0.47 | 1.0324 | 0.86 | 1.20 | 0.72 | RAI1-PEMT-RASD1 | Imputed | |
| rs15563 | 17 | 44360192 | G | 0.48 | 0.9946 | 0.85 | 1.14 | 0.94 | UBE2Z | Imputed | |
| rs1122608 | 19 | 11024601 | T | 0.21 | 0.7415 | 0.49 | 0.99 | 0.02 | LDLR | Imputed | |
| rs9982601 | 21 | 34520998 | C | 0.21 | 0.6858 | −1.43 | 2.80 | 0.73 | Gene desert (KCNE2) | Imputed |
Abbreviations: CAD, coronary artery disease; FH, familial hypercholesterolemia; SNPs, single-nucleotide polymorphisms.
Figure 2.
Manhattan plot containing the entire set of analyzed SNPs (41) associated with CAD. The Manhattan plot presents the –log10 (P-value) of all the SNPs analyzed in the current analysis adjusted for cardiovascular risk factors (age, gender, smoking, type 2 diabetes, hypertension and BMI). Four of the 45 SNPs associated with CAD were not available for analysis in our data after imputation. The red line represents the P-value indicating statistical significance, taking into account the Bonferroni correction for multiple testing.
Discussion
We tested the hypothesis that common genetic variants that were previously shown in GWA studies to be associated with CAD risk in the general population, might affect the risk of CAD in a high-risk cohort of FH patients. As previously reported, established risk factors do associate with the risk of CAD in FH patients.8, 22 However, none of the tested CAD-associated SNPs significantly modified the risk of CAD in our FH cohort in analyses unadjusted or adjusted for established cardiovascular risk factors. The lowest observed P-value of association was for a SNP in the SMARCA44 gene, near the LDLR gene (in adjusted analysis; P=0.021 ); however, it showed a paradoxically protective effect.
FH patients are known to be at high CAD risk. Other patient cohorts with high risk are those with established cardiovascular disease and those with type 2 diabetes. Of these three patient categories, the effect of 9p21 variants on survival had been tested only in those with established CAD. In a prospective observational study including 846 Caucasoid cases who underwent CABG, the 9p21 SNP rs10116277:G>T was independently associated with all-cause mortality during 5 years follow-up after surgery.23 Homozygotes for the minor allele of this SNP had an increased risk of all-cause mortality (HR 1.7; CI 1.1−2.7). The SNP even remained associated with outcome after adjustment for the Euroscore, a score commonly used to predict CVD outcome after CABG. In contrast, in a larger cohort of patients with established CAD (>8000 patients), a haplotype block with eight of the strongest 9p21 SNPs was associated with better prognosis in whites but not in blacks or Hispanics.24 Moreover, in line with our findings, the HRs for prognosis among the risk alleles were in opposite direction compared with the published HRs for CAD/MI risk in the Caucasoid population for both the two most widely reported 9p21 SNPs; rs2383207:A>G and rs10757278:A>G, G alleles were 0.75 (0.60−0.93) and P=0.0083 and 0.81 (0.66−1.0) P=0.05. However, a less commonly reported linkage disequilibrium consisting of six 9p21 SNPs was associated with worse prognosis.24 Compared with these two studies in high-risk populations, our study adds a considerable number of events in a high-risk populations; a total of 482 CAD events occurred during follow-up in our cohort, whereas the studies by Muehlschlegel et al23 and Gong et al24 reported analyses on 38 and 134 CVD events, respectively. In summary, the only two other studies that addressed the effect of 9p21 SNPs showed conflicting results in high-risk populations.
Previous work conducted by our group to determine the genetic modifiers of CVD risk among FH patients showed that the G20210A polymorphism in the protrombin gene was strongly associated with significantly increased CVD risk.14 However, in that publication, the threshold to reach statistical significance was rather lenient. In this paper, a P-value <0.001 was considered statistically significant; however, applying the Bonferroni correction, which is common practice nowadays would suggest a P-value <0.00076. None of the SNPs reached the a priori determined value for statistical significance. Because the reported protrombin variant is not considered to be a CAD risk SNP its effect on survival was not calculated in our current analysis. In current analysis, only 1701 samples of the original 1940 were available. A selection bias has not taken place. This is merely a reflection of the usage of the DNA. Baseline characteristic are similar in both studies.
Our study did not address the underlying explanation for the rather counterintuitive result. The top SNP in our analysis, rs1122608:G>T, had a protective effect, contrary to the latest papers. However, smaller reports have reported a protective effect of this variant for CAD and PAD.25, 26 Martinelli et al25 suggest that the effect they observed is due to the lipid effects of the variants.25, 26
The effect of common CAD-associated genetic variants on general population samples has also yielded conflicting results. In a study among ∼3000 cases and 3000 controls, a gene score comprising nine SNPs was associated with CAD risk.27 People in the top quintile for this gene score had a twofold increased risk of MI compared with those in the bottom quintile corrected for age, sex and ancestry. In line, SNP-based risk score designed in a Finnish cohort of 30 725 participants free of CVD was associated with the risk of a first CAD event, with a relative risk of 1.7 between the highest and lowest quintiles of the score adjusted for traditional risk factors; sex, LDL-C, HDL-C, current smoking, BMI, systolic and diastolic blood pressure, blood pressure treatment an prevalent type 2 diabetes.28 In contrast, Paynter et al29 did not observe an independent association between genetic risk factors and CAD risk in a cohort of 19 313 initially healthy women during 12.3 years follow-up. A risk score based on 12 SNPs was clearly associated with CAD risk after adjustment for age. However, after additional adjustment for other traditional risk factors this association disappeared.
We have previously shown that the type of LDLR mutations underlying FH, variation in LDL-C levels, and established classical risk factors explain 21.3% of the variation in CAD risk. Our current analysis suggests that environmental and/or unknown genetic factors may have a role. As there was no standardized information available on lifestyle factors such as dietary habits and physical activity, the effect of environmental factors and their potential interaction with genetic variants could not be studied, but it is unlikely that this could explain significant proportions of the remaining 80% of CAD risk prediction. At maximum, the common genetic variants we tested explained only 10% of the heritability of the trait, if we consider the heritability estimates of 40% for CAD to be correct.10 Much of the ‘missing heritability' is expected to be explained by common variants not yet identified, rare variants, structural variants and copy number variations.
Several aspects of the design of our study need to be considered. The major strength of this study is the unparalleled cohort size, the detailed information on CAD events during follow-up and the high CAD event rate. However, our study is hampered by several limitations. First, power calculations suggest that our study was at the limit of the power needed to detect statistically significant associations with CAD, as the studied SNPs were previously shown to have a moderate-effect (RR 1.1−1.7). In addition, the MAF of some of the SNPs were lower then used in the power calculation. Second, the majority of the CAD SNPs analyzed were not directly genotyped but imputed using HapMap and we cannot rule out misclassifications. However, Southam et al17 have shown that imputation of common variants is generally very accurate. Finally, the patients who were included in this study were referred to a Lipid Clinic. In theory, patients with the most detrimental genetic profiles might have died before referral. Therefore, the effect of genetic variants associated with a more severe CAD phenotype or early death could have been underestimated or missed.
Conclusion
In this high-risk cohort of patients with FH, common SNPs shown to be associated with CAD risk in the general population could not be associated with the disease.
Acknowledgments
This work was supported by Bloodomics consortium, European Union 6th Framework Programme (LSHM-CT-2004-503485), Ipse Movet, the Lifetime Achievement Award of the Dutch Heart Foundation (2010T082) to JJPK and grants from the British Heart Foundation and the National Institute for Health Research, England to WHO. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The authors declare no conflict of interest.
References
- Goldstein JL, Sobhani MK, Faust JR, et al. Heterozygous familial hypercholesterolemia: failure of normal allele to compensate for mutant allele at a regulated genetic locus. Cell. 1976;9:195–203. doi: 10.1016/0092-8674(76)90110-0. [DOI] [PubMed] [Google Scholar]
- Huijgen R, Vissers MN, Defesche JC, et al. Familial hypercholesterolemia: current treatment and advances in management. Expert Rev Cardiovasc Ther. 2008;6:567–581. doi: 10.1586/14779072.6.4.567. [DOI] [PubMed] [Google Scholar]
- Van Aalst-Cohen ES, Jansen AC, Tanck MW, et al. Diagnosing familial hypercholesterolaemia: the relevance of genetic testing. Eur Heart J. 2006;27:2240–2246. doi: 10.1093/eurheartj/ehl113. [DOI] [PubMed] [Google Scholar]
- Stone NJ, Levy RI, Fredrickson DS, et al. Coronary artery disease in 116 kindred with familial type II hyperlipoproteinemia. Circulation. 1974;49:476–488. doi: 10.1161/01.cir.49.3.476. [DOI] [PubMed] [Google Scholar]
- Sijbrands EJ, Westendorp RG, Defesche JC, et al. Mortality over two centuries in large pedigree with familial hypercholesterolaemia: family tree mortality study. BMJ. 2001;322:1019–1023. doi: 10.1136/bmj.322.7293.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AC, van Aalst-Cohen ES, Tanck MW, et al. The contribution of classical risk factors to cardiovascular disease in familial hypercholesterolaemia: data in 2400 patients. J Intern Med. 2004;256:482–490. doi: 10.1111/j.1365-2796.2004.01405.x. [DOI] [PubMed] [Google Scholar]
- Souverein OW, Defesche JC, Zwinderman AH, et al. Influence of LDL-receptor mutation type on age at first cardiovascular event in patients with familial hypercholesterolaemia. Eur Heart J. 2007;28:299–304. doi: 10.1093/eurheartj/ehl366. [DOI] [PubMed] [Google Scholar]
- Jansen AC, van Aalst-Cohen ES, Tanck MW, et al. The contribution of classical risk factors to cardiovascular disease in familial hypercholesterolaemia: data in 2400 patients. J Intern Med. 2004;256:482–490. doi: 10.1111/j.1365-2796.2004.01405.x. [DOI] [PubMed] [Google Scholar]
- Peden JF, Hopewell JC, Saleheen D, et al. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivapalaratnam S, Motazacker MM, Maiwald S, et al. Genome-wide association studies in atherosclerosis. Curr Atheroscler Rep. 2011;13:225–232. doi: 10.1007/s11883-011-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2012;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umans-Eckenhausen MA, Defesche JC, Sijbrands EJ, et al. Review of first 5 years of screening for familial hypercholesterolaemia in the Netherlands. Lancet. 2001;357:165–168. doi: 10.1016/S0140-6736(00)03587-X. [DOI] [PubMed] [Google Scholar]
- Jansen AC, van Aalst-Cohen ES, Tanck MW, et al. Genetic determinants of cardiovascular disease risk in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2005;25:1475–1481. doi: 10.1161/01.ATV.0000168909.44877.a7. [DOI] [PubMed] [Google Scholar]
- Hao K, Chudin E, McElwee J, et al. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 2009;10:27. doi: 10.1186/1471-2156-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam L, Panoutsopoulou K, Rayner NW, et al. The effect of genome-wide association scan quality control on imputation outcome for common variants. Eur J Hum Genet. 2011;19:610–614. doi: 10.1038/ejhg.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AC, van Aalst-Cohen ES, Tanck MW, et al. Genetic determinants of cardiovascular disease risk in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2005;25:1475–1481. doi: 10.1161/01.ATV.0000168909.44877.a7. [DOI] [PubMed] [Google Scholar]
- Khot UN, Khot MB, Bajzer CT, et al. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003;290:898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- Muehlschlegel JD, Liu KY, Perry TE, et al. Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation. 2010;122:S60–S65. doi: 10.1161/CIRCULATIONAHA.109.924233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Beitelshees AL, Cooper-Dehoff RM, et al. Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:169–178. doi: 10.1161/CIRCGENETICS.110.959296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Girelli D, Lunghi B, et al. Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood. 2010;116:5688–5697. doi: 10.1182/blood-2010-03-277079. [DOI] [PubMed] [Google Scholar]
- Murabito JM, White CC, Kavousi M, et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet. 2012;5:100–112. doi: 10.1161/CIRCGENETICS.111.961292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter NP, Chasman DI, Buring JE, et al. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]