Abstract
Disrupted-in-schizophrenia-1 (DISC1) gene has been established as a risk factor for various neuropsychiatric phenotypes. Both coding and regulatory variants in DISC1 have been identified and associated with these phenotypes in genetic studies. MicroRNAs (miRNAs) are important regulators of protein coding genes. Since the miRNA-mRNA target recognition mechanism is vulnerable to disruption by DNA polymorphisms, we investigated whether polymorphisms in the DISC1 3′UTR affect binding of miRNAs and lead to allele-specific regulation of DISC1. We identified four predicted polymorphic miRNA target sites in the DISC1 3′UTR, and demonstrated that miR-135b-5p regulates the level of DISC1 mRNA. Moreover, DISC1 regulation by miR-135b-5p is allele specific: miR-135b-5p only binds to the major allele (A) of rs11122396, not to the minor allele (G). Thus, the G allele may be functionally related to the DISC1-associated phenotypes by abolishing regulation by miR-135b-5p, leading to elevated DISC1 levels.
Keywords: DISC1, microRNA, miR-135b-5p, neuropsychiatric disorder, allele-specific regulation
INTRODUCTION
Genetic variants of the Disrupted-in-schizophrenia-1 (DISC1) gene have been associated with various neuropsychiatric phenotypes.1 The initially identified translocation2 reduces DISC1 expression to half normal levels, suggesting haploinsufficiency as the mechanism of disease susceptibility in translocation carriers.3 In addition, expression quantitative trait loci affecting the expression of DISC1 have been identified4, 5 and shown to associate with age of onset in recurrent major depression.6 We hypothesized that differential regulation of DISC1 expression may contribute to the wide range of DISC1-associated disorders.
MicroRNAs (miRNAs) are small non-coding RNA molecules that repress gene expression by binding to their target mRNAs. There are no previous reports on miRNA regulation of DISC1, although in general miRNAs regulate about half of human genes. We were especially interested in putative polymorphic miRNA target sites in the DISC1 3′UTR that could lead to differential regulation of DISC1.
MATERIALS AND METHODS
RESULTS
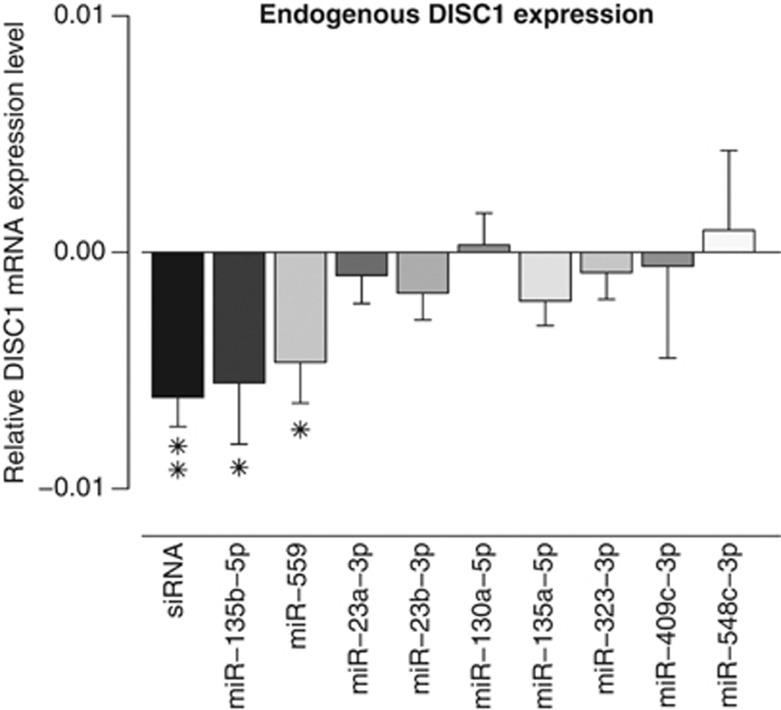
We identified four SNPs (rs11122396, rs980989, rs9308481, and rs11803088) within putative binding sites of nine human miRNAs (miR-23a-3p, miR-23b-3p, miR-130a-5p, miR-135a-5p, miR-135b-5p, miR-323-3p, miR-409-3p, miR-548c-3p, and miR-559) (Table 1) through bioinformatic target prediction (see Supplementary Methods). We first tested the effect of each of these miRNAs on endogenous DISC1 expression in vitro. We transfected a commercial miRNA precursor of each miRNA, a negative control miRNA, or a positive control siRNA, into HEK293FT cells and measured DISC1 mRNA levels by qRT-PCR. We found that two of the nine miRNAs, miR-135b-5p and miR-559, significantly reduced DISC1 mRNA expression: miR-559 by 23.7% (P=0.009) and miR-135b-5p by 16.2% (P=0.039), compared with the negative control miRNA (Figure 1). The endogenous expression of miR-135b-5p and miR-559 in HEK293FT cells was found to be low or undetectable, respectively (data not shown).
Table 1. Predicted polymorphic miRNA target sites of DISC1.
| SNP | Positiona | MAF CEU | Octamer (‘seed')b | Effect | SNP allele | Targeting miRNA (stem loop) | Stem loop location | Mature miRNA |
|---|---|---|---|---|---|---|---|---|
| rs11122396 | 230241891 | 0.042 | caAGCC[A]Taactc | C | A* | miR-135a-1 | 3p21 | hsa-miR-135a-5p |
| C | miR-135a-2 | 12q23.1 | ||||||
| C | miR-135b | 1q32 | hsa-miR-135b-5p | |||||
| G (ancestral) | None | |||||||
| rs980989 | 230242818 | 0.217 | tttACA[T]TCAtat | C | T | miR-409 | 13q32.3 | hsa-miR-409-3p |
| G* (ancestral) | None | |||||||
| rs9308481 | 230242929 | 0.225 | taaAAT[G]TGAagt | N | G* (ancestral) | miR-323 | 14q32 | hsa-miR-323-3p |
| aaaAT[G]TGAAgt | N | miR-23a | 19p13 | hsa-miR-23a-3p | ||||
| N | miR-23b | 9q22.3 | hsa-miR-23b-3p | |||||
| AAT[G]TGAA | N | miR-130a | 11q12 | hsa-miR-130a-5p | ||||
| A | None | |||||||
| rs11803088 | 230243392 | 0.069 | cttTTA[C]TTTtaa | N | C* (ancestral) | miR-559 | 2p21 | hsa-miR-559 |
| cttttA[T]TTTTAa | D | T | miR-548c | 12q14.2 | hsa-miR-548c-3p |
Predictions from Patrocles and PolymiRTS are shown, with miRNAs and SNPs predicted by both programs underlined. The SNP alleles of the wild-type DISC1 Lv construct are indicated with an asterisk (*). The effect of the SNP on the target site is denoted as follows: C=derived allele creates a novel miRNA binding site, N=derived allele disrupts a non-conservative miRNA binding site, D=disrupts a conservative miRNA binding site. MAF=minor allele frequency; CEU=HapMap CEPH population.
RefSeq build 36.3.
Genomic sequence 5′ to 3′.
Figure 1.
The effect of overexpression of nine miRNAs on endogenous DISC1 mRNA levels. MiR-135b-5p, miR-559, miR-23a-3p, miR-23b-3p, miR-130a-5p, miR-135a-5p, miR-323-3p, miR-409-3p, and miR-548c-3p were overexpressed in HEK293FT cells and relative levels of endogenous DISC1 and the housekeeping gene, GAPDH, measured using qRT-PCR. We compared the DISC1 expression levels after miRNA transfection with the DISC1 expression levels after transfection of a negative control miRNA. We performed three independent experiments and carried out linear regression analysis separately for each batch. We then used the obtained residuals in subsequent linear regression analysis including all the batches to obtain a P-value for all three experiments combined, taking the batch effect into account. Out of the nine miRNAs tested, miR-135b-5p and miR-559 reduced DISC1 mRNA levels significantly. An siRNA against DISC1 was used as a positive control. Results are expressed as the regression coefficient ±standard error. *P<0.05, **P<0.005.
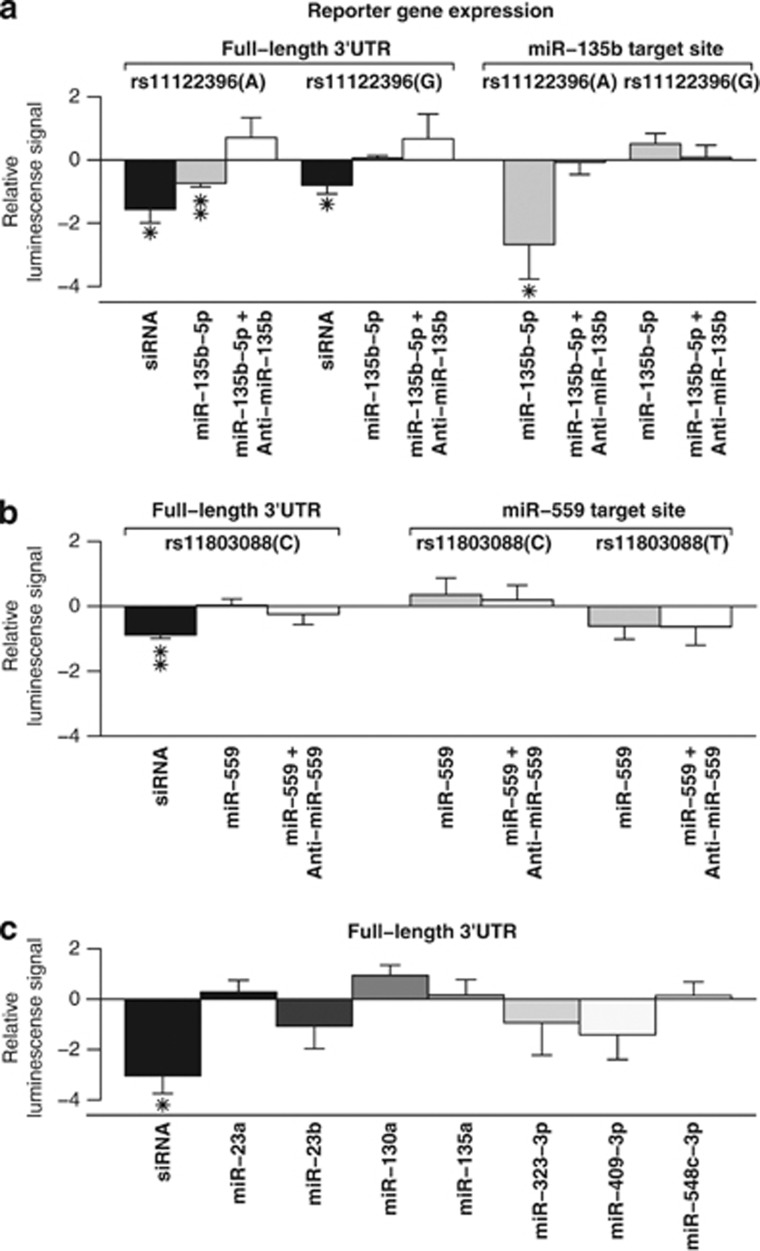
To determine whether these miRNAs regulate DISC1 levels by targeting the predicted sites, we used a reporter gene assay, which uses luciferase activity on a protein level as the output measure. We cloned either the full-length DISC1 Lv isoform 3′UTR (4.4 kb) or the miRNA binding sites with flanking sequences (∼60 nt) into a pmirGLO dual luciferase expression vector. These constructs were co-transfected individually into HEK293FT cells with either the miR-135b-5p or the miR-559 precursor, or with a positive control siRNA or a negative control miRNA. We found that expression from the DISC1 full-length 3′UTR construct was reduced 32.1% (P=0.003) by miR-135b-5p, and by 10.3%, (P=0.03) from the construct expressing the 60-nt miRNA binding site and flanking sequences (Figure 2a). The specificity of the repression of DISC1 expression by miR-135b-5p was further established by demonstrating that co-transfection of miR-135b-5p with anti-miR miRNA inhibitor against both endogenous miR-135b-5p and its miRNA precursor completely abolished the repression of DISC1 expression (Figure 2a). Altogether, these findings establish that miR-135b-5p regulates DISC1 mRNA expression by targeting the predicted site in the DISC1 3′UTR. In contrast, miR-599 overexpression did not affect the expression from the DISC1 full-length 3′UTR construct or the construct with its 60 nt binding site (Figure 2b). Likewise, none of the other seven miRNAs (miR-23a-3p, miR-23b-3p, miR-130a-5p, miR-135a-5p, miR-323-3p, miR-409-3p, and miR-548c-3p) with predicted binding sites in the DISC1 3′UTR were found to have any effect on expression from the full-length DISC1 3′UTR reporter construct (Figure 2c).
Figure 2.
The effect of overexpression of miRNAs on reporter gene activity. (a) We measured relative levels of luciferase activity (Firefly/Renilla ratio) after transfecting HEK293FT cells with either the full-length 3′UTR of DISC1 (full-length 3′UTR) or the predicted miR-135b-5p target site sequence (miR-135b-5p target site) with the two alternative alleles of rs11122396 together with miR-135b-5p precursor. We observed a significant reduction in the reporter gene activity with the A allele constructs, but not the G allele constructs. When anti-miR-135b was co-transfected with miR-135b-5p, reporter gene activity was restored to the control miRNA levels. (b) Similar experiment with the two alleles of rs11803088 within the predicted miR-559 target site. MiR-559 did not have a significant effect on the reporter gene expression with either of the two alleles. (c) Similar experiment with miR-23a-3p, miR-23b-3p, miR-130a-5p, miR-135a-5p, miR-323-3p, miR-409-3p, or miR-548c-3p and the full-length 3′UTR construct harboring the wild type-alleles at miRNA binding sites. None of these miRNAs reduced the reporter gene activity significantly. For all the analyses, the miRNA effect was compared with the negative control miRNA effect. We performed a minimum of two independent experiments for each miRNA and carried out linear regression analysis separately for each batch. We then used the obtained residuals in subsequent linear regression analysis including all the batches to obtain a P-value for all experiments combined taking the batch effect into account. An siRNA against DISC1 was used as a positive control. Results are expressed as the regression coefficient ±standard error. *P<0.05, **P<0.005.
As polymorphisms may disrupt miRNA binding sites, leading to predisposition to disease phenotypes,7, 8 we next investigated the putative allele specificity of miR-135b-5p binding. The HEK293FT cell line is homozygous for the derived allele A of rs11122396, predicted to create a novel binding site for miR-135b-5p (Table 1). We created constructs with the DISC1 G allele at rs11122396, to determine whether the repression of DISC1 expression by miR-135b-5p is allele specific. Strikingly, miR-135b-5p had no effect either on the luciferase activity of the full-length 3′UTR with G at rs11122396 (P=0.49), or on the construct with the ∼60-nt miRNA binding site with flanking sequences insert (P=0.18) (Figure 2a), indicating that miR-135b-5p binding is specific to the derived A allele at rs11122396 of DISC1. Thus, the G allele may be functionally related to the DISC1-associated phenotypes by abolishing regulation by miR-135b-5p, leading to elevated DISC1 levels.
DISCUSSION
The rs11122396 G allele of DISC1, which we have shown here to disrupt miR-135b-5p binding, has a frequency of 4.4% in the European population,9 raising the possibility that differential miRNA binding caused by DNA sequence polymorphisms may be a relatively common source of variation of gene expression in humans. The major allele A of rs11122396 is the derived allele that introduces a miRNA-135b-5p binding site, implying that the ancestral G allele may be a risk allele that abolishes miRNA regulation. Unlike in Mendelian diseases where the risk allele is usually the derived allele, in common diseases the ancestral allele sometimes functions as a risk allele under modern environmental conditions (see Di Rienzo and Hudson10), for example in the case of the ɛ4 allele of APOE in coronary artery disease11 and Alzheimer's disease.12
Based on the publicly available BrainSpan Atlas mRNA and miRNA sequencing data (www.brainspan.org), miR-135b-5p and DISC1 are co-expressed in the human brain with the highest levels in the cerebellar cortex and mediodorsal nucleus of thalamus. These brain regions are involved in the regulation of cognitive and affective processes disturbed in neuropsychiatric diseases.13, 14 Thus far, up to 44 DISC1 isoforms have been reported in adult and fetal human brain and in lymphoblastoid cell lines.15 According to current annotations, four isoforms carry the four SNPs of this study. Previously, rs11122396 was associated with schizophrenia16 through a rare haplotype. Moreover, the rs11122396 SNP is in complete linkage disequilibrium (LD) in the European population with rs3737597 (G2879A),9 which has been associated with schizophrenia and recurrent major depression by several independent studies.6, 16, 17 In contrast, no association was detected between the rs11122396 SNP and autism spectrum disorders in a family-based study sample.18 These reports imply that variants in the 3′UTR of DISC1 may have an important regulatory role in neuropsychiatric phenotypes. We conjecture that variable regulation of DISC1 by miR-135b-5p in the brain may predispose to neuropsychiatric phenotypes, as has been proposed with, for example, miR-185 in the case of the 22q11.2 microdeletions and schizophrenia.19
In conclusion, we show that miR-135b-5p regulates the level of DISC1 expression in vitro in an allele-specific manner, and suggest that the disturbed miRNA-mRNA target recognition mechanism may contribute to the wide range of phenotypic associations of DISC1, in accord with comparable natural variations in miRNA target sequences that have been shown to have major phenotypic consequences.7, 8, 20
Acknowledgments
We thank Brendan Battersby and Joni Nikkanen for experimental and Jeffrey Barrett for statistical advise. This study was supported by the Helsinki Graduate Program in Biotechnology and Molecular Biology, Synaptic Systems dissecting brain function in health and disease (EU FP7- 242167) (MR), Orion-Farmos Research Foundation, Biomedicum Helsinki Foundation, Research Foundation of the University of Helsinki, Maud Kuistila Memorial Foundation (HK), Helsinki Biomedical Graduate Program (MM and IS), Academy of Finland Research Fellowship (IH and WH), the Academy of Finland Center of Excellence in Complex Disease Genetics (213506 and 129680), Academy of Finland (251217), and the Sigrid Juselius Foundation (IH and SR).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry. 2011;16:1096–1104, 1063. doi: 10.1038/mp.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Macintyre DJ, Hamilton G, et al. Association of DISC1 variants with age of onset in a population-based sample of recurrent major depression. Mol Psychiatry. 2012;18:745–747. doi: 10.1038/mp.2012.117. [DOI] [PubMed] [Google Scholar]
- Abelson JF, Kwan KY, O'Roak BJ, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- De Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM. Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels. Hum Mut. 1994;4:178–194. doi: 10.1002/humu.1380040303. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Baillieux H, De Smet HJ, Paquier PF, De Deyn PP, Marien P. Cerebellar neurocognition: insights into the bottom of the brain. Clin Neurol Neurosurg. 2008;110:763–773. doi: 10.1016/j.clineuro.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Van der Werf YD, Jolles J, Witter MP, Uylings HB. Contributions of thalamic nuclei to declarative memory functioning. Cortex. 2003;39:1047–1062. doi: 10.1016/s0010-9452(08)70877-3. [DOI] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Varilo T, Kestila M, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- Saetre P, Agartz I, De Franciscis A, et al. Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr Res. 2008;106:237–241. doi: 10.1016/j.schres.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Ylisaukko-Oja T, Hennah W, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.