Abstract
Bacteria deploy a range of chemistries to regulate their behaviour and respond to their environment. Quorum sensing is one mean by which bacteria use chemical reactions to modulate pre-infection behaviour such as surface attachment. Polymers that can interfere with bacterial adhesion or the chemical reactions used for quorum sensing are thus a potential means to control bacterial population responses. Here we report how polymeric “bacteria sequestrants”, designed to bind to bacteria through electrostatic interactions and thus inhibit bacterial adhesion to surfaces, induce the expression of quorum sensing controlled phenotypes as a consequence of cell clustering. A combination of polymer and analytical chemistry, biological assays and computational modelling has been used to characterise the feedback between bacteria clustering and quorum sensing signaling. We have also derived design principles and chemical strategies for controlling bacterial behaviour at the population level.
Non-lethal means of targeting bacteria1,2, such as stimulation of host immune systems3,4, interference with cell adhesion5,6 or bacterial communication7,8, are emerging as attractive means to avoid resistance against antimicrobial therapies. Polymeric antimicrobials have been an increasing focus of attention in recent years owing to their ability to present multiple functionalities for detecting, binding and inactivating pathogens9-11. Examples now exist of polymers that can prevent cell growth in multi-drug resistant strains11, or which can sequester specific bacteria12-14, toxins15,16, and/or cell-signal molecules17-19.
Of special promise are materials that can prevent bacteria binding to hosts5,6, a prerequisite for most infections and particularly those related to invasive pathogens19. Two main strategies have been exploited, utilising either anti-fouling surfaces to inhibit bacterial adhesion directly20-22, or the display of multiple ligands that bind competitively to the surface of the bacteria thus inhibiting their attachment to host surface ligands12-14. Depending on the material design, one of the consequences of the latter approach is the aggregation of bacteria into clusters, a microenvironment where diffusion of nutrients and signals can be significantly affected.
A number of papers have now described significant effects of local concentration and spatial confinement, as well as molecule and bacteria diffusion, on bacterial cell-cell communication networks23-28. Bacterial communication, also known as Quorum Sensing (QS)29,30 is an important regulator of bacterial behaviour, including swarming, aggregation, production of exo-enzymes and toxins, as well as processes preceding infection such as surface colonisation and biofilm formation31-34. QS signaling in bacteria often involves complex feedback mechanisms, and is regulated by gene circuits and multiple interconnected control mechanisms29,35. This feedback between cell clustering and QS signaling has stimulated intense debate as to the nature of QS and whether it is always a population density response rather than a function of cell clustering and signal diffusion36,37.
We recently reported preliminary data that certain polymers can modulate the luminescence of Vibrio harveyi, a marine pathogen that responds to the QS signal AI-2 by producing light. These materials were designed to cluster bacteria while simultaneously reducing the concentration of AI-2, a component of the QS circuit of several bacteria38. Unlike conventional polymers able to only bind to the QS signals, and inhibit light production in a dose-dependent way, some of those polymers were able to induce luminescence in V. harveyi under specific experimental conditions, suggesting interdependence between bacteria clustering and QS response39.
We report here how a polymeric bacteria sequestrant, which induces bacterial aggregation through electrostatic interactions and with no functionalities to interfere with the QS signals, is able to induce QS-related responses in a range of bacteria. These include not only the model microorganism V. harveyi but also the human pathogens Escherichia coli and Pseudomonas aeruginosa. We employ synthetic and analytical chemistry, biological assays and computational modelling to demonstrate that QS-associated behaviour occurs as a direct consequence of bacteria clustering. Furthermore, the responses of V. harveyi as a model organism are simulated and compared against a representative “quorum quencher”, which should only bind to QS signals, and a “dual-action” polymer, with the ability to bind both the surface of bacteria and the signal molecules. The results give important insight into the unexpected consequences of feedback between bacteria clustering and QS signaling. Furthermore, the data suggest entirely new chemical design principles not only for novel anti-adhesive materials, but also for inducing consequences of QS responses that are beneficial, such as antibiotic production40,41.
Results
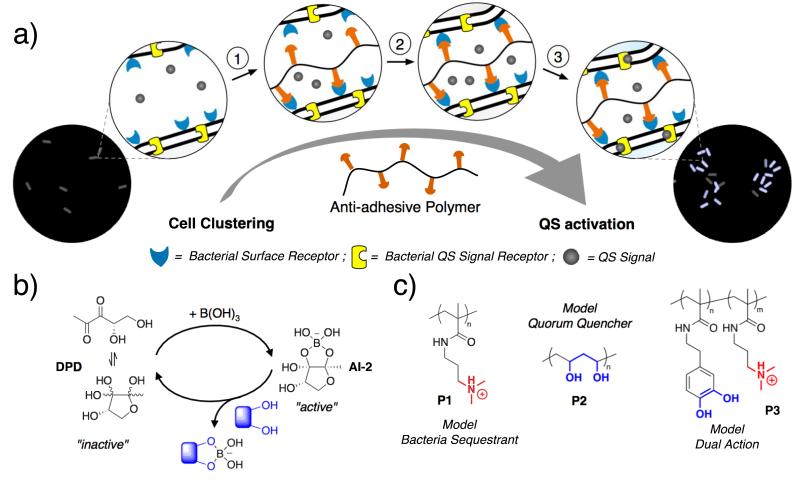
The starting hypothesis was that polymeric materials with the ability to aggregate bacteria into clusters would be able to induce the expression of QS controlled phenotypes (Figure 1a)39. We thus derived a model which predicted, from a phenomenological point of view, induction of a feedback loop into QS signaling by bacteria clustering, interrelating polymer (P) concentration, bacterial (B) aggregation and QS signals (S). Three classes of polymers were therefore defined, in order to predict all the potential interactions between polymers, bacteria and signals: a) “bacteria sequestrants”, that should only bind to bacteria, inducing cell clustering; b) “quorum quenchers”, that would only be able to bind the signals; and c) “dual-action” polymers, with the ability to bind both signals and bacteria. The predicted clustering and QS responses were validated against experimental data, using V. harvey and its AI-2 network (Figure 1b) as a model.
Figure 1. QS induction in the AI-2 network.
a) Schematic representation of QS activation by “bacteria sequestrants” that promote bacteria clustering: ☉ Polymer binds to the surface of the bacteria via multivalent interactions. ◯ Bacteria are cross-linked as polymer interacts with different bacterium. ☽ Signal diffusion is limited maintaining a high concentration within the cell cluster b) Key components of the autoinducer-2 network, including 4,5-dihydroxy-2,3-pentanedione (DPD) and the active species formed in the presence of B(OH)-in the media. Mechanism of AI 2 quenching by competitive binding with diols. c) Structure of the polymers employed in this work: poly(N-[3-(dimethylamino)propyl] methacrylamide) (P1), poly(vinyl alcohol) (P2) and poly(N-dopaminemethacrylamide-co-N-[3-(dimethylamino)propyl] methacrylamide) (P3).
Poly(N-[3-(dimethylamino)propyl] methacrylamide) (P1), a cationic polymer that should bind to the surface of bacteria through electrostatic interactions, was synthesised as a representative “bacteria-sequestrant”. Controlled radical polymerisations (RAFT) were used to tune molar mass and the materials were characterised by NMR and GPC. The behaviour of bacteria in the presence of P1 was determined and compared to model polymers of the other classes. Because AI-2 in V. harveyi is a borate ester, and its concentration in solution can be reduced by competitive binding to the boric acid precursor with polymeric diols (Figure 1b), commercially available poly(vinyl alcohol) (P2) and poly(N-dopamine methacrylamide-co-N-[3-(dimethylamino)propyl] methacrylamide) (P3)39, were chosen as representative “quorum quenchers” and “dual-action” polymers respectively (Figure 1c).
The viability of V. harveyi in the presence of these polymers was assessed by monitoring cell growth during luminescence experiments. For the relevant duration of the experiment (0-8 h), before solvent evaporation in the well plates becomes significant, no differences in optical density of the cultures were observed in the absence and presence of increasing amounts of each polymer (see Supplementary Information, Figures S9b-S14b). In addition, viability of V. harveyi in the presence of P1, a polymer which might be expected to exhibit toxicity due to a higher content of tertiary amines42,43, was also investigated using nuclear staining and fluorescent microscopy. When compared against cultures in the absence of polymer (positive control) and cultures in the presence of methanol (negative control), the ratio between viable (green) and non-viable (red) bacteria in the presence of P1 was similar to that of the positive control (untreated bacteria) and significantly different from the negative control (see Supplementary Information, Figure S27). This indicated that P1 was not altering QS through a direct toxic response.
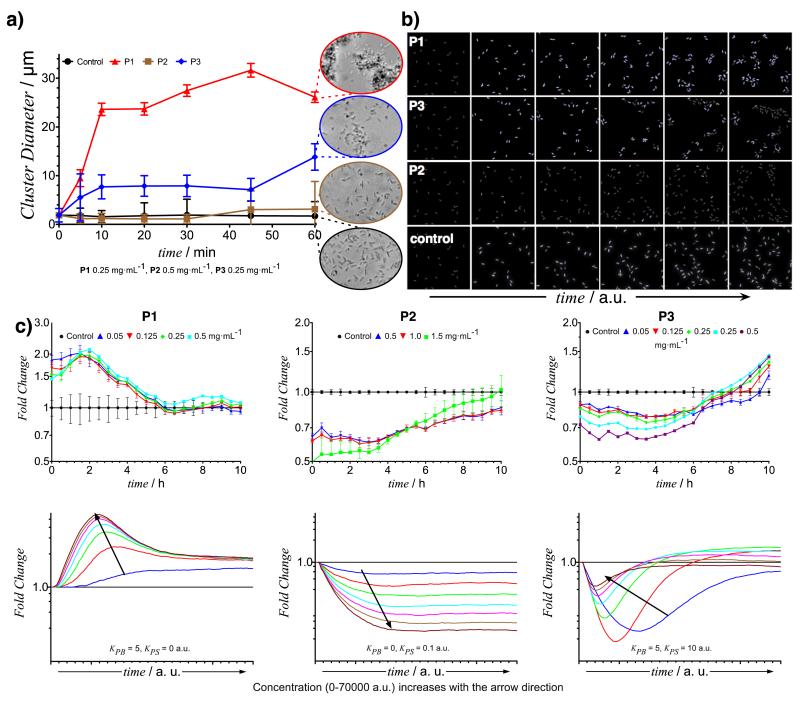
The ability of the polymers to cluster bacteria and their effect over QS networks was investigated against cultures of two strains of V. harveyi (MM32 and BB170). Initial experiments were carried out with V. harveyi MM32, which responds to exogenous AI-2 but does not produce the QS signal precursor, 4,5-dihydroxy-2,3-pentanedione (DPD). Concurrent with the experimental assays, polymer-bacteria interactions were simulated based on a simple affinity model. Cell aggregation experiments (Fig. 2a) showed a good match against the computationally predicted results (Fig 2b), with P1 inducing rapid bacterial clustering, P2 producing no apparent difference compared to bacterial suspensions alone, and P3 forming aggregates with bacteria at a similar rate to P1. Computationally predicted results were simulated 10 times using different randomisations, in order to obtain statistically reliable results. Effects were consistent within the 10 simulations. Initial conditions (cell positioning, random seeds, affinities and polymer concentrations) for the simulations under different polymers were identical (see Fig 2b and Supplementary Information, Figure S28-30).
Figure 2. Effect of polymers on V. harveyi MM32 behaviour.
Aggregation of bacteria in the presence of polymers, measured in a Coulter Counter® and by optical microscopy (a) were in good agreement with those predicted by the computational stochastic simulation, observed from simultaneous screenshots of simulated bacteria cultures in the absence and presence of polymers (b). Mean value and polydispersity index are reported. Similarly, QS signalling in the presence of polymers, measured by luminescence (c, top) was in good agreement with that predicted by the computational model (c, bottom). “Bacteria sequestrants” (P1) enhanced luminescence (Fold Change ≥ 1) throughout the duration of the experiment (c, left). “Quorum quenchers” (P2) reduced luminescence (Fold Change ≤ 1) during the same timeframe (c, middle). For “dual-action” polymers (P3) both induction and quenching were observed (c, right). Mean value and standard deviation are reported. Two-way Anova analysis of experimental results indicates that significant differences in fold change are observed, as polymer concentration increases, for the relevant duration of the experiment (0-8 h). See Supplementary Information for further details.
We then considered the effects of clustering on QS response as reported by luminescence. Taking into consideration feasible diffusion rates and affinities for the interactions between bacteria, signals and polymers, we predicted changes in luminescence with addition of “bacteria sequestrants” (P1), “quorum quenchers” (P2), and “dual-action” polymers (P3). As apparent from Fig. 2c, when compared to a control in the absence of polymers, P1 induced an increase in light production in V. harveyi MM32 cultures throughout the duration of the experiment (Fold change in luminescence≥ 1), despite not being targeted to QS and lacking the functionalities to interfere with the signals. On the other hand, P2 was able to reduce luminescence during the same time (Fold change ≤ 1). As expected, P3 howed a dual mode of behaviour, with the ability either to enhance or reduce light production dependent on specific polymer concentration and time (bacteria density). For P1, the absolute change in luminescence was higher at later stages of the experiments, when cell numbers were higher (Figure S8), but the relative difference (Fold change) in luminescence was higher at earlier stages. Relative variations in luminescence at early stages of bacterial growth can appear exaggerated in cases of low initial values of luminescence, since the timescale for aggregation is considerably smaller than that for light production. Therefore, the effects of cell clustering were most apparent at an early time in the experiment, and became less pronounced as bacterial growth matches density and viscosity within clusters. Thus, at early time periods, it was expected that slow diffusion of signals from the cell clusters enabled bacteria to sense a higher concentration of QS signals more rapidly. Indeed, during the key timescales of the experiment, (i.e. 0-8 h, after which cell numbers increase markedly), the effect of polymer on QS controlled luminescence matched well with that predicted by the theoretical model.
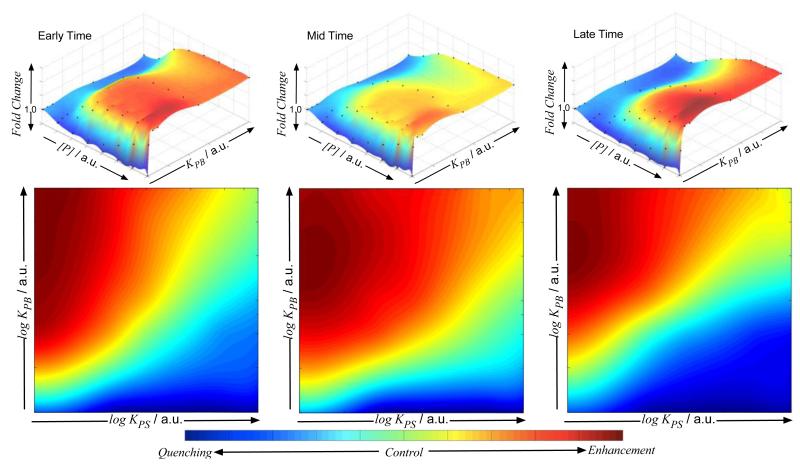
To evaluate further the feedback between the ability of polymers to induce aggregation and the QS controlled light production, several simulations were performed, where the affinities towards bacteria (KPB) and signals (KPS) of P3 were systematically varied. Affinities were investigated over a three order of magnitude range, and different combinations of KPB and KPS were simulated (see Supplementary Information for further details). As can be seen in Figure 3, the potential of a polymer to inhibit or enhance light production was highly dependent on polymer concentration, experiment time and the polymer affinities for signals and bacteria. Variations in any of these parameters were predicted to lead to, and indeed showed, marked changes in QS signaling as manifest in light production. For instance, polymers with high affinity for bacteria enhanced light production regardless of their concentration and the time of the analysis, even if they showed a high affinity towards the signals.
Figure 3. Effect of relative binding affinities over light production.
The effect that polymer affinity for signals (KPS) and bacteria (KPB) has over light production was predicted by the model. In the presence of weak polymer-bacteria interactions, “dual-action” polymers (P3) quench light production regardless of the polymer concentration [P] and time at which light production was evaluated (top). As polymer bacteria affinity (KPB) increases, the overall outcome of the polymer interference changes, and enhancement of light production is expected at higher polymer concentrations. In addition, the overall light production depends on the relative intensities (KPB and KPS) of both affinities as well as the time at which light production is evaluated (bottom). In order to obtain “dual-action” polymers that consistently quench light production, polymers with low affinity towards the bacteria (KPB) are required. Initial conditions (cell positioning, random seeds) for the simulations are identical. Time = 5000, 15000 and 30000 a.u. were selected as representative early, mid and late time respectively for the simulations. Top: Polymer affinity for signals, KPS 0.1 a.u., was selected as a representative value. See Supplementary Information for further details.
The effects bacterial density and growth rate have on the activity of “dual-action” polymers were also investigated. Polymer affinities for bacteria and signals were fixed and simulations with different initial densities of bacteria (B0) or different growth rates, as expressed by bacteria doubling time (T), were performed (see Supplementary Information for further details). When the initial density of bacteria (B0) was reduced by an order of magnitude (Figure S41), P3 was able to induce QS signaling throughout the duration of the simulation and regardless of the concentration of polymers, as opposed to the “dual-action” exhibited when the starting number of bacteria was higher (Figure S40). Similarly, the ability of P3 to induce or inhibit light production was significantly affected in the presence of bacteria growing at different rates (Figure S40 vs Figure S42).
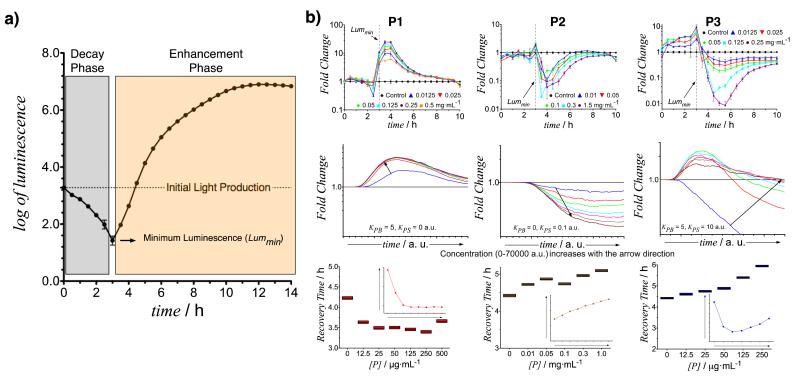
The interaction of polymers and bacteria was also investigated using the V. harveyi BB170 strain which is capable of producing DPD. In this case, light production in the absence of polymers shows two phases (Fig. 4a). In the first phase, luminescence decreased as the bacteria responded to the lower concentrations of DPD in the sample media as opposed to the pre-culture medium, prior to their production of endogenous DPD. When a threshold of DPD concentration was achieved, a new phase was attained wherein light production increased as a function of DPD concentration.
Figure 4. Effect of polymers on V. harveyi BB170 luminescence.
Luminescence in V. harveyi BB170 shows two phases. A decay phase where luminescence is reduced and an enhancement phase, after the minimum concentration of AI-2 for luminescence induction is reached (a). Polymer interference with QS signalling, as measured by luminescence (b, top) was in good agreement with that modelled (b, medium). “Bacteria sequestrants” (P1) enhanced luminescence (Fold Change ≥ 1) (b, left); “quorum quenchers” (P2) reduced light production (Fold Change ≤ 1) (b, middle); and for “dual-action” polymers (P3) both induction and quenching of luminescence were observed (b, right). Additionally, differences in the time necessary to recover the initial intensity of luminescence could be noted (b, bottom). P1 induced earlier light production and P2 delayed the recovery time. In this case, P3 behaved as a quorum quencher and delayed the onset of luminescence. This behaviour was well predicted by the model (b, bottom, inset). Mean value and standard deviation are reported. Two-way Anova analysis of experimental results indicates that significant differences in fold change are observed, as polymer concentration increases, for the relevant duration of the experiment (0-8 h). See Supplementary Information for further details.
Despite the differences in light production profile for both strains, the effect that all 3 classes of polymers had on BB170 QS signalling (Figure 4b, top and middle), as measured by luminescence, was very similar to that described for MM32 (Figure 2c, top). Throughout the duration of the experiments, “bacteria sequestrants” (P1) were able to induce light production, “quorum quenchers” (P2) reduced the overall production of light and “dual-action” polymers (P3) showed both induction and quenching of luminescence. The effect was weaker during the decay phase (time lower than time required for Lummin), as the concentration of AI-2 will be well below the detection threshold for most of this period.
The effect of polymers on BB170 QS signaling was also reflected in the duration of the decay phase and the time taken for bacteria to sense a concentration of AI-2 above the threshold. During this decay phase, light production was reduced because luciferase production was switched off while bacteria re-adapted to the low concentration of AI-2 after dilution. As the population of bacteria increased, the amount of AI-2 in solution increased accordingly, so that QS signaling could recover. This effect was easily monitored from the light production plots, by measuring the time taken by the bacterial suspension to recover a significant level of light intensity, for instance, the initial value of light production (Figure 4a).
Interestingly, by confining cells into clusters, “bacteria sequestrants” (P1) were able to induce an earlier activation of QS signaling, as a consequence of the local higher concentration of AI-2. Conversely, “quorum quenchers” (P2) were able to delay the time needed to do so, as the concentration of AI-2 in solution was reduced. For “dual-action” polymers (P3) a combination of both effects was to be expected. In the reported example, P3 showed an overall quenching effect increasing the time needed for the recovery of the initial luminescence (Figure 4 b, bottom).
To investigate the generality of this effect and the potential for directing QS controlled phenotypes in relevant human pathogens, further experiments with E. coli and P. aeruginosa, were performed. E. coli lux-based acylhomoserine lactone (AHL) biosensors, JM109::pSB107544, and JM109::pSB53645, that produce light in response to N-(3-oxododecanoyl)-L-homoserine lactone (OdDHL)46 and N-butyryl-homoserine lactone (BHL)47 respectively, were selected as representative E. coli strains. P. aeruginosa PA01 pqsA CTX-lux::pqsA48, that produces light in response to 2-heptyl-3-hydroxy-4(1H)-quinolone, usually termed as Pseudomonas quinolone signal (PQS)49, was selected as a representative P. aeruginosa strain.
Aggregation of these strains in the presence of P1 was very fast, and a dose dependent increase in optical density of the cultures could be observed as soon as P1 was added (Figures S19-S26, time 0). In addition, the growth of both E. coli strains in the presence of P1 was significantly compromised, notably in the case of pS536 reporter. This lack of growth had an impact in luminescence production for these strains. As P1 concentration increased, the production of light decreased, in agreement with the decreased viability of E. coli in the presence of P1. With higher polymer concentrations, recovery of light production was observed as a consequence of light induction being triggered by clustering (Figures S20 and S22). P. aeruginosa showed better viability in the presence of P1, and induction of light production in the presence of “bacteria-sequestrant” P1 was clearly observed, particularly at higher polymer concentrations (Figures S23-S26).
Discussion
The initial finding that polymers intended to suppress QS could in fact enhance cell signaling (as reported by light production)39, was unexpected and suggested that bacteria confinement into clusters could be responsible for QS induction, thus producing an effect opposite to that desired.
Spatial confinement is inherent to QS as cell density helps regulate bacteria behaviour. Significantly, recent papers23,24,28 have suggested that confined individual bacterium can show QS-type behaviour. This behaviour has been computationally anticipated50 in studies suggesting that programmable compartmentalisation is a Turing-complete mechanism, and can thus be a potentially useful tool for the control of population responses in Synthetic Biology. In addition, as noted in the case of Pseudomonas species, QS signal gradients have a ‘context-dependent’ action, with limited effects on biofilm growth in liquid culture but pronounced and significant effects on confined cell communities attached to surfaces i.e. when bacteria and signals are in close proximity27.
Therefore, to understand how QS could be activated by “bacteria sequestrants”, we derived a phenomenological synthetic biology model in order to simulate and predict QS controlled luminescence, as a function of binding affinities towards bacteria and signals, in two different mutants of V. harveyi. We utilised the MM32 strain, which responds to, but cannot produce, its QS signal (DPD, the AI-2 precursor) and the BB170 strain, which is capable of synthesising DPD and which thus introduces natural variability and non-linearity into the system.
We thus synthesised a model “bacteria-sequestrant”, P1, intended to bind to the surface of bacteria through electrostatic interactions. The ability of P1 to aggregate bacteria into clusters was confirmed by measuring cluster size and by optical microscopy. In addition, P1 induced light production throughout the incubation assay, despite not being targeted to QS and having no specific functionalities to interfere with the signals. Taking into account the ability of the polymers to cluster bacteria, the predicted relative affinity of the monomer units within the polymers for the bacteria39, the behaviour of V. harveyi in the presence of P1 was well predicted by the computational model.
In contrast, the behaviour of V. harveyi in the presence of P2, a model “quorum-quencher” was markedly different to that in the presence of P1. In addition, a combination of both responses could be observed using P3. We termed P3 a “dual-action” polymer as it incorporated both cationic groups to bind to the surface of bacteria, inducing cluster formation, and diols capable of binding the boronic acid needed to activate AI-2. The numbers of monomer units i.e. the components in each repeating section of the polymers able to ‘bind’ signals or cells, were broadly similar across P1-P3 (degrees of polymerisation 100-400) and no significant differences in response were observed when P1 of different molecular weight were employed (Figures S6 and S18). Nevertheless, from a phenomenological point of view, there was good correlation between simulated and experimental QS responses in the presence of these polymers.
The goal of the model was not only to understand the feedback between aggregation and light production, but also to derive design principles for QS control. Therefore, we performed a series of simulations where the relative affinities of P3 towards bacteria and signal were systematically varied. As can be seen in Figure 3, in order to design efficient polymeric materials that can cluster bacteria, “dual-action” polymers have to be considered where the balance between the affinity towards the bacteria (KPB) and the affinity towards the signal (KPB) prevents induction of QS controlled phenotypes (Green to blue color in the graphs). In a similar way, the model predicted, and experiments showed, that “quorum quenchers” designed to reduce the expression of QS controlled behaviour should also exhibit a very low affinity for bacterial surfaces in order to retain their intended effects on bacterial populations. Most notably, the models and experiments showed that small changes in initial bacterial density and growth rates could ‘tip the balance’ to strongly opposing effects, such that either luminescence enhancement or quenching could be seen for the same polymer under very similar conditions. This variability in conditions is likely to be most apparent in therapeutic applications of polymers, where the numbers and growth for pathogens will differ significantly across patients, or the degree of infection.
The model developed herein was designed to be ‘agnostic’ to the nature of the bacteria, as well as the type of response triggered by QS. In principle, therefore, any bacteria behaviour under QS control such as the production of exoenzymes and toxins, or biofilm formation31,32,34, could be triggered if cell clustering is induced by “bacteria sequestrants”.
We therefore investigated if the reported enhancement of light production by a “bacteria-sequestrant” could also be detected using different bacteria and different signaling molecules. Experiments under the same conditions optimised for V. harveyi were performed using E. coli and P. aeruginosa luminescence reporters for HSLs and PQS respectively. These signals are significantly different to AI-2 in terms of their chemical functionality, and the QS response of the microorganisms are not synchronised in the way that V. harveyi responds to AI-2. Despite these differences, and the lower viability of E. coli strains in the presence of P1, the ability of this polymer to induce light production as a consequence of aggregation was also observed, establishing the generality both of the QS/polymer/bacteria feedback model and the mechanism of activity of the “dual-action” polymers.
Conclusions
In conclusion, we have shown how polymeric “bacteria sequestrants”, with high affinity for bacterial surfaces, have the ability to interfere with non-targeted signaling pathways such as QS in a range of prokaryotes. We have defined a theoretical and practical framework for understanding bacterial responses to QS-interference in the presence of polymers with the ability to bind bacteria and/or signaling molecules, which should aid the development of novel non-antibiotic anti-infectives.
Given that many bacteria attach to host surfaces prior to colonisation and invasion, our data suggests that materials designed to interfere with infection pathways should be designed so that they do not promote unwanted effects in cell signaling and QS. Significantly, the results show that materials which promote bacteria clustering induce unexpected responses in QS controlled phenotypes, and that these responses can be better modulated through control of the affinity towards both bacteria and signals. As a corollary the combined model/experiment approach enables experimental data to be obtained regarding spatial effects on QS, which can be interrogated through computational models, which in turn can feedback into materials design. This combined chemistry/computation approach should enhance our understanding of QS in complex environments. In turn, the ability to utilise specific chemical design principles to control cell behaviour should facilitate the development of antimicrobials that avoid selection pressure and inform synthetic biology strategies wherein QS is used to induce production of valuable metabolites.
Methods
Aggregation Assay
A single colony of V. harveyi grown on Luria Bertani (LB) agar plates was used to inoculate 2 mL LB medium containing chloramphenicol (10 μg/ml), and kanamycin (50 μg/ml) in the case of BB170. The bacteria were grown with aeration at 30 °C overnight. Boron depleted Assay Broth (AB) medium was then inoculated with this preculture to give a bacterial suspension with an OD600 of 1.0. Aliquots of this culture were then mixed with known volumes of stock solutions of polymers in Dulbecco’s Phosphate Buffer Saline (DPBS). The values of polymer concentration reported for the aggregation experiments correspond to the polymer concentrations in these suspensions. To measure cluster size, these bacterial suspensions were added to a Coulter Counter flow cell filled with H2O (<14 mL) to obtain an obscuration of 8-12%. Cluster size was then measured at different time intervals. For optical microscopy analysis, aliquots (10 μL) of the bacterial suspensions, in the absence and presence of polymers, were collected after 60 min, mounted on a glass slide with a cover slip on top and examined with an optical microscope. See Supplementary Information for further details.
Microbiological assays
A single colony of V. harveyi grown on LB agar plates was used to inoculate 2 mL LB medium containing chloramphenicol (10 μg/ml), and kanamycin (50 μg/ml) in the case of BB170. The bacteria were grown with aeration at 30 °C overnight. Boron depleted AB medium was then inoculated with this preculture (5000:1). For MM32 boric acid was added to a final concentration of 400 μM, and DPD was added to a final concentration of 22 μM. For BB170 boric acid was added to a final concentration of 22 μM. 180 μL of the inoculated medium were placed in each of the wells of a 96 well plate and combined with 20 μL of the samples to be analysed. Each compound was tested over at least 3 different concentrations. Light production and optical density (600 nm) were recorded at 30 °C every 30 minutes for at least 10 hours in a 96-well plate, after which time solvent evaporation became a significant issue. The experiments were carried out in triplicate and the plotted curves are derived from the mean value. The normalized luminescence was calculated by dividing the light output by the optical density at each time point.
Simulation methods
Due to the spatial and time scales of the system, a mesoscopic lattice based model and an agent based approach was employed. Analysis of the results was carried out at the phenomenological level, i.e. capturing the characteristic effect of the three types of polymers. Modelled parameters were refined subsequently against measurable overall effects (i.e. bacterial binding and luminescence production). The starting boundary conditions for the model were set so that any deviations from control experiments were caused by the polymers manipulating the immediate extracellular environment of the bacteria. Three types of objects were considered in the model, bacteria (B), polymers (P) and signal molecules (S). The size of each B was fixed to occupy a square of 2×2 arbitrary lattice spaces. The sizes of S and P were considered to be negligibly small. One unit lattice space could thus contain a quarter of B and unlimited numbers of S and P. Two types of changes were considered to take place in the system, chemical binding and diffusion of the objects. The reactions and diffusion of the different species were modelled using a Gillespie algorithm.
Three types of binding reactions were delineated in the model: binding between S and B, binding between B and P, and binding between P and S. Each of these interactions was considered to be reversible. In addition, delimiting conditions for the model were: i) B have separate binding sites for S and P; ii) each quadrant of B has a number of S binding sites, denoted as BS and a number of P binding sites, BP. Similarly, each P has PS binding sites for S and 2 B binding sites, PB. Each S could thus only bind to one binding site (either B or P) and would not be available for other reactions once bound, until the reverse reaction occurs and the molecules and binding sites became free again. The same constraint was applied for P-B binding. We considered that S could only bind with P or B that are in the same lattice, and that bound S would move with P or B without further activation of the QS network. Individual P were considered to bind with B inside the same lattice space. For S, once a single P was bound to a single B they were considered to be fixed in a binding interaction over the timescale of the experiment. P bound to B were able to bind with other B in the neighbouring lattice; in such a case the B-P-B complex moved as a single unit until associative interactions were lost. This mechanism represented key multiple binding interactions that could lead to bacterial clustering and aggregation. The values of binding affinities and diffusion rates are dependent on the concentrations of the different objects in the local environment (See Supplementary Information for further details).
Supplementary Material
Acknowledgements
We thank the UK EPSRC and BBSRC (Grants EP/G042462/1, EP/D022347/1, D021847/1, EP/I031642/1, EP/J004111/1 and BB/F01855X/1) and the University of Nottingham, U.K., for funding. We thank Bonnie Bassler (Department of Molecular Biology, Princeton University) for the gift of V. harveyi strains. We would like to thank our SynBioNT colleagues (www.synbiont.net) for helpful discussions and in particular Benjamin Davis and Lee Cronin for their advice and suggestions in the wider SynBioNT collaboration.
Footnotes
Additional information
Supplementary information and chemical characterisation accompany this paper at. Reprints and permission information is available online at /.
References
- 1.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. Fresh Approaches to Anti-Infective Therapies. Science Transl. Med. 2012;4(140sr2) doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock REW, Nijnik A, Philpott DJ. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012;10:243–254. doi: 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- 4.Wei P, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm P, Vejborg RM, Hancock V. Prevention of bacterial adhesion. Appli. Microb. Biotechnol. 2010;88:451–459. doi: 10.1007/s00253-010-2805-y. [DOI] [PubMed] [Google Scholar]
- 6.Bricarello DA, Patel MA, Parikh AN. Inhibiting host-pathogen interactions using membrane-based nanostructures. Trends Biotechnol. 2012;30:323–330. doi: 10.1016/j.tibtech.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann GF, Park J, Janda KD. Bacterial quorum sensing: a new target for anti-infective immunotherapy. Expert Opin. Biol. Ther. 2008;8:719–724. doi: 10.1517/14712598.8.6.719. [DOI] [PubMed] [Google Scholar]
- 8.Park J, et al. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 2007;14:1119–1127. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock REW, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotech. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 10.Nederberg F, et al. Biodegradable nanostructures with selective lysis of microbial membranes. Nat. Chem. 2011;3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, et al. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nano. 2009;4:457–463. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- 12.Alexander C, Pasparakis G, Cockayne A. Control of bacterial aggregation by thermoresponsive glycopolymers. J. Am. Chem.Soc. 2007;129:11014–11015. doi: 10.1021/ja074349z. [DOI] [PubMed] [Google Scholar]
- 13.Shepherd J, et al. Hyperbranched poly(NIPAM) polymers modified with antibiotics for the reduction of bacterial burden in infected human tissue engineered skin. Biomaterials. 2011;32:258–267. doi: 10.1016/j.biomaterials.2010.08.084. [DOI] [PubMed] [Google Scholar]
- 14.Lee D-W, Kim T, Park I-S, Huang Z, Lee M. Multivalent Nanofibers of a Controlled Length: Regulation of Bacterial Cell Agglutination. J. Am. Chem. Soc. 2012;134:14722–14725. doi: 10.1021/ja306802m. [DOI] [PubMed] [Google Scholar]
- 15.Rai P, et al. Statistical pattern matching facilitates the design of polyvalent inhibitors of anthrax and cholera toxins. Nat. Biotechnol. 2006;24:582–586. doi: 10.1038/nbt1204. [DOI] [PubMed] [Google Scholar]
- 16.Piletska EV, et al. Attenuation of Vibrio fischeri Quorum Sensing Using Rationally Designed Polymers. Biomacromolecules. 2010;11:975–980. doi: 10.1021/bm901451j. [DOI] [PubMed] [Google Scholar]
- 17.Bush K, et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner AL, et al. A Multivalent Probe for AI-2 Quorum-Sensing Receptors. J. Am. Chem. Soc. 2011;133:15934–15937. doi: 10.1021/ja207556d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossart P, Sansonetti PJ. Bacterial invasion: The paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 20.Hook AL, et al. Combinatorial discovery of polymers resistant to bacterial attachment. Nat. Biotechnol. 2012;30:868–875. doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith RS, et al. Vascular Catheters with a Nonleaching Poly-Sulfobetaine Surface Modification Reduce Thrombus Formation and Microbial Attachment. Science Transl. Med. 2012;4:153ra132. doi: 10.1126/scitranslmed.3004120. [DOI] [PubMed] [Google Scholar]
- 22.Epstein AK, Wong T-S, Belisle RA, Boggs EM, Aizenberg J. Liquid-infused structured surfaces with exceptional anti-biofouling performance. Proc. Natl. Acad. Sci. USA. 2012;109:13182–13187. doi: 10.1073/pnas.1201973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastrup CJ, et al. Spatial localization of bacteria controls coagulation of human blood by ‘quorum acting’. Nat. Chem. Biol. 2008;4:742–750. doi: 10.1038/nchembio.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carnes EC, et al. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 2010;6:41–45. doi: 10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberghini S, et al. Consequences of relative cellular positioning on quorum sensing and bacterial cell-to-cell communication. FEMS Microbiol. Lett. 2009;292:149–161. doi: 10.1111/j.1574-6968.2008.01478.x. [DOI] [PubMed] [Google Scholar]
- 26.Goryachev AB. Understanding Bacterial Cell-Cell Communication with Computational Modeling. Chem. Rev. 2011;111:238–250. doi: 10.1021/cr100286z. [DOI] [PubMed] [Google Scholar]
- 27.Flickinger ST, et al. Quorum Sensing between Pseudomonas aeruginosa Biofilms Accelerates Cell Growth. J. Am. Chem. Soc. 2011;133:5966–5975. doi: 10.1021/ja111131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boedicker JQ, Vincent ME, Ismagilov RF. Microfluidic Confinement of Single Cells of Bacteria in Small Volumes Initiates High-Density Behavior of Quorum Sensing and Growth and Reveals Its Variability. Angew. Chem. Int. Ed. 2009;48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowery CA, Dickerson TJ, Janda KD. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 2008;37:1337–1346. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 30.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J. Roy. Soc. Interface. 2009;6:959–978. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 32.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams P. Quorum sensing, communication and cross-kingdom signaling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 34.Williams P, Winzer K, Chan WC, Camara M. Look who’s talking: communication and quorum sensing in the bacterial world. Phil. Trans. Roy. Soc. B-Biol. Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antunes LCM, Ferreira RBR. Intercellular communication in bacteria. Crit. Rev. Microbiol. 2009;35:69–80. doi: 10.1080/10408410902733946. [DOI] [PubMed] [Google Scholar]
- 36.Fuqua C, Greenberg EP. Listening in on bacteria: Acyl-homoserine lactone signaling. Nat. Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 37.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 38.Federle MJ, Bassler BL. Interspecies Communication in Bacteria. J. Clin. Investigation. 2003;112:1291–1299. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue X, et al. Synthetic Polymers for Simultaneous Bacterial Sequestration and Quorum Sense Interference. Angew. Chem. Int. Ed. 2011;50:9852–9856. doi: 10.1002/anie.201103130. [DOI] [PubMed] [Google Scholar]
- 40.Barnard AML, et al. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Phil. Trans. Roy. Soc. B-Biol. Sci. 2007;362:1165–1183. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fineran PC, Slater H, Everson L, Hughes K, Salmond GPC. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 2005;56:1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x. [DOI] [PubMed] [Google Scholar]
- 42.Kenawy E-R, Worley SD, Broughton R. The Chemistry and Applications of Antimicrobial Polymers: A State-of-the-Art Review. Biomacromolecules. 2007;8:1359–1384. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- 43.Engler AC, et al. Emerging trends in macromolecular antimicrobials to fight multi-drug-resistant infections. Nano Today. 2012;7:201–222. [Google Scholar]
- 44.Winson MK, et al. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 45.Swift S, et al. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas-aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winson MK, et al. Multiple N-Acyl-L-Homoserine Lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas-aeruginosa. Proc. Natl. Acad. Sci. USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diggle SP, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krasnogor N, et al. An Appealing Computational Mechanism Drawn from Bacterial Quorum Sensing. Bull. Eur. Assoc. Theor. Computer Sci. 2005;85:135–148. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.