Abstract Abstract
We explore the potential value of DNA barcode divergence for species delimitation in the genus Caryocolum Gregor & Povolný, 1954 (Lepidoptera, Gelechiidae), based on data from 44 European species (including 4 subspecies). Low intraspecific divergence of the DNA barcodes of the mtCOI (cytochrome c oxidase 1) gene and/or distinct barcode gaps to the nearest neighbor support species status for all examined nominal taxa. However, in 8 taxa we observed deep splits with a maximum intraspecific barcode divergence beyond a threshold of 3%, thus indicating possible cryptic diversity. The taxonomy of these taxa has to be re-assessed in the future. We investigated one such deep split in Caryocolum amaurella (Hering, 1924) and found it in congruence with yet unrecognized diagnostic morphological characters and specific host-plants. The integrative species delineation leads to the description of Caryocolum crypticum sp. n. from northern Italy, Switzerland and Greece. The new species and the hitherto intermixed closest relative C. amaurella are described in detail and adults and genitalia of both species are illustrated and a lectotype of C. amaurella is designated; a diagnostic comparison of the closely related C. iranicum Huemer, 1989, is added.
Keywords: Lepidoptera, Gelechiidae, Caryocolum, species delineation, integrative taxonomy, DNA barcode, morphology, Europe
Introduction
The genus Caryocolum Gregor & Povolný, 1954 is one of the most species-rich genera of European Gelechiidae (Huemer and Karsholt 2010). Having been revised in monographic papers (Klimesch 1953–1954, Huemer 1988), its taxonomy seemed well established. However, in the last decade new species were found in, e.g. Sicily, southern France and Greece (Bella 2008, Grange and Nel 2012, Huemer and Nel 2005, Huemer and Karsholt 2010) raising the number of described species to 51. Most of the species are considered indisputable based on their morphology and distinct biology – as far as known, these species are closely linked to Caryophyllaceae as their exclusive larval host-plant family. We investigate, for the first time in Caryocolum, the congruence of traditional morphological species delineation and molecular data from the COI barcode region for a vast majority of the European fauna, covering altogether 44 species, including four subspecies. Surprisingly, the potential for cryptic diversity proved extraordinarily high for a supposedly well-known genus and we newly describe one of the hitherto overlooked species.
Material and methods
Extensive generic descriptions and diagnoses of European species of Caryocolum have been published in several reviews, particularly Huemer and Karsholt (2010) and Huemer (1988), and are thus not repeated here.
Specimens. Our study is based on about 50 specimens of the Caryocolum amaurella (Hering, 1924) species-group and an uncounted number of European Caryocolum, exceeding 1000 specimens, but only partially used for genetic analysis (see below). Most of the material was traditionally set and dried or alternatively spread; a few specimens are only pinned. Genitalia preparations followed standard techniques (Robinson 1976) adapted for male genitalia of Gelechiidae and (some) female genitalia of Caryocolum by the so-called “unrolling technique” (Pitkin 1986, Huemer 1987).
DNA Barcodes. Full-length lepidopteran DNA barcode sequences are a 648 base-pair long segment of the 5’ terminus of the mitochondrial COI gene (cytochrome c oxidase 1). DNA samples (dried leg) were prepared according to the accepted standards. Legs from 250 specimens of Caryocolum were processed at the Canadian Centre for DNA Barcoding (CCDB, Biodiversity Institute of Ontario, University of Guelph) to obtain DNA barcodes using the standard high-throughput protocol described in deWaard et al. (2008). Sequences longer than 500 bp were included in the analysis. Successfully sequenced voucher specimens are listed in Suppl. material 1. Sequences were submitted to GenBank; further details including complete voucher data and images can be accessed in the public dataset “Lepidoptera of Europe Caryocolum” dx.doi.org/10.5883/DS-LECARY in the Barcode of Life Data Systems (BOLD; Ratnasingham and Hebert 2007). Degrees of intra- and interspecific variation in the DNA barcode fragment were calculated under Kimura 2 parameter (K2P) model of nucleotide substitution using analytical tools in BOLD systems v3.0. (http://www.boldsystems.org). A neighbour-joining tree of DNA barcode data of European taxa was constructed using Mega 5 (Tamura et al. 2011) under the K2P model for nucleotide substitutions.
Photographic documentation. Photographs of the adults were taken with an Olympus SZX 10 binocular microscope and an Olympus E 3 digital camera and processed using the software Helicon Focus 4.3 and Adobe Photoshop CS4 and Lightroom 2.3. Genitalia photographs were taken with an Olympus E1 Digital Camera from Olympus BH2 microscope.
Abbreviations of institutional collections
BMNH
TLMF
ZMUH
ZMUC
ZMUO
Results
Molecular analysis
Forty-four of 51 European species were successfully sequenced, resulting in a full-length barcode fragment for 191 specimens and more than 500 bp for further 26 specimens (Fig. 1, Table 1, Suppl. material 1). Nine shorter sequences were not included in the analysis and sequencing of 24 specimens failed. The maximum intraspecific K2P distance varies from 0% in several species to 6.27% in Caryocolum fibigerium. Ten species have a high maximum intraspecific divergence greater than 2%. In six species (newly described species excluded) with a medium divergence greater than 3% potential cryptic diversity should be investigated. Furthermore, the intraspecific divergence of more than 3% in Caryocolum schleichi, a species separated into 3 allopatric subspecies, is beyond variation typically found within species, supporting their status as valid species. The only other subspecies we have examined are nominotypical Caryocolum marmorea and the recently separated Caryocolum marmorea mediocorsa with a very low divergence of 0.3%.
Figure 1.
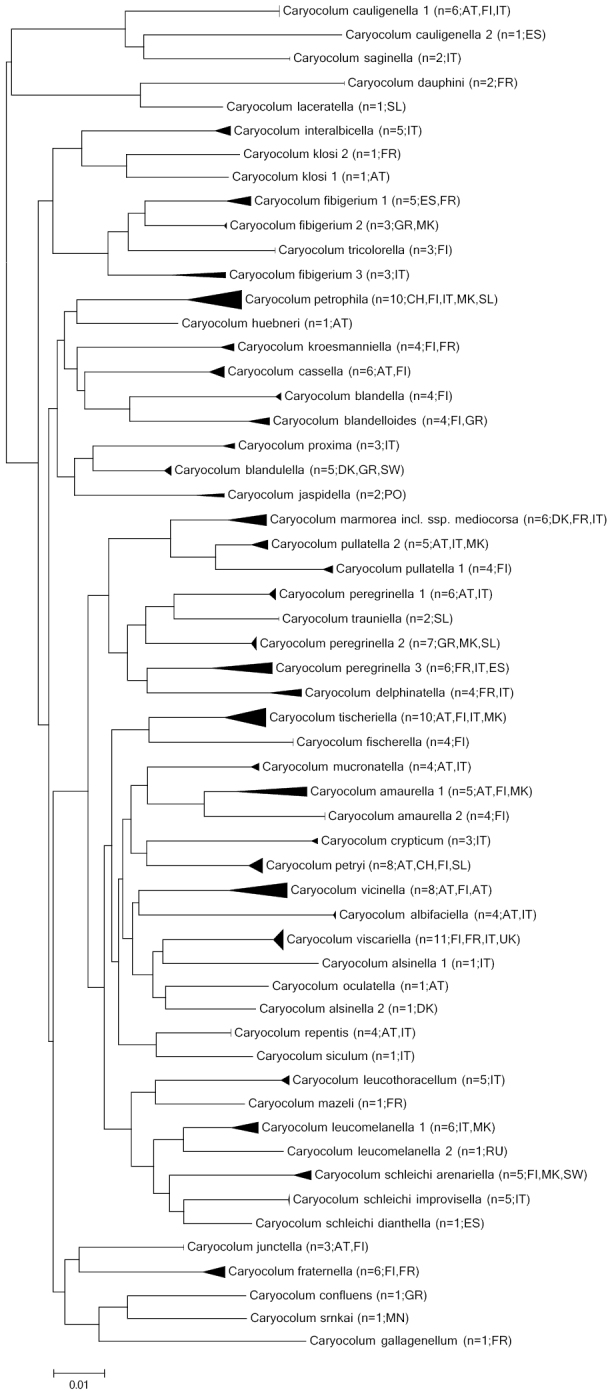
Neighbour-joining tree (Kimura 2 parameter, built with MEGA 5; cf. Tamura et al. 2011), with only sequences longer than 500 bp considered. The width of the triangles represents the sample size, and the depth the genetic variation within the cluster. Currently recognized conspecific taxa with maximum divergence greater than 3% are shown as separate clades. Source: DNA Barcode data from BOLD (Barcode of Life Database, cf. Ratnasingham and Hebert 2007).
Table 1.
Intraspecific mean K2P (Kimura 2 Parameter) divergences, maximum pairwise distances and distance to nearest neighbor.
| Species | Mean Intra-Sp | Max Intra-Sp | Nearest Neighbour | Nearest Species | Distance to NN |
|---|---|---|---|---|---|
| Caryocolum alsinella | 4.85 | 4.85 | PHLAE427-11 | Caryocolum oculatella | 3.81 |
| Caryocolum amaurella | 3.05 | 4.76 | LEATC402-13 | Caryocolum mucronatella | 5.21 |
| Caryocolum blandella | 0.16 | 0.3 | LEFIK150-10 | Caryocolum blandelloides | 5.78 |
| Caryocolum blandelloides | 0.4 | 0.81 | LEFIB755-10 | Caryocolum blandella | 5.78 |
| Caryocolum blandulella | 0.21 | 0.46 | LEATD656-13 | Caryocolum proxima | 3.94 |
| Caryocolum cassella | 0.42 | 0.61 | PHLAI019-12 | Caryocolum blandulella | 5.07 |
| Caryocolum cauligenella | 1.99 | 6.95 | PHLAA069-09 | Caryocolum saginella | 6.61 |
| Caryocolum confluens | N/A | N/A | PHLAF489-11 | Caryocolum srnkai | 4.54 |
| Caryocolum crypticum | 0.21 | 0.31 | LEATC-402-13 | Caryocolum mucronatella | 5.41 |
| Caryocolum dauphini | 0 | 0 | PHLAB900-10 | Caryocolum laceratella | 5.29 |
| Caryocolum delphinatella | 1.02 | 1.39 | PHLAI203-13 | Caryocolum marmorea mediocorsa | 4.57 |
| Caryocolum fibigerium | 3.4 | 6.27 | LEFIF467-10 | Caryocolum tricolorella | 4.67 |
| Caryocolum fischerella | 0 | 0 | LEFIC281-10 | Caryocolum tischeriella | 4.5 |
| Caryocolum fraternella | 0.47 | 1.7 | PHLAI156-12 | Caryocolum junctella | 4.55 |
| Caryocolum gallagenellum | N/A | N/A | PHLAI019-12 | Caryocolum blandulella | 6.54 |
| Caryocolum huebneri | N/A | N/A | LEFIJ1014-11 | Caryocolum petrophila | 4.88 |
| Caryocolum interalbicella | 0.4 | 0.77 | PHLAI156-12 | Caryocolum junctella | 5.55 |
| Caryocolum jaspidella | 1.08 | 1.08 | PHLAI019-12 | Caryocolum blandulella | 4.39 |
| Caryocolum junctella | 0 | 0 | LEFIF480-10 | Caryocolum fraternella | 4.55 |
| Caryocolum klosi | 4.25 | 4.25 | PHLAA055-09 | Caryocolum interalbicella | 5.56 |
| Caryocolum kroesmanniella | 0.31 | 0.61 | LEEUA184-11 | Caryocolum blandulella | 4.9 |
| Caryocolum laceratella | N/A | N/A | PHLAI447-13 | Caryocolum dauphini | 5.29 |
| Caryocolum leucomelanella | 1.47 | 3.79 | PHLAG331-12 | Caryocolum mazeli | 3.76 |
| Caryocolum leucothoracellum | 0.12 | 0.3 | PHLAG331-12 | Caryocolum mazeli | 4.24 |
| Caryocolum marmorea mediocorsa | 0 | 0 | LEEUA182-11 | Caryocolum marmorea | 0.3 |
| Caryocolum marmorea | 1 | 1.54 | PHLAI203-13 | Caryocolum marmorea mediocorsa | 0.3 |
| Caryocolum mazeli | N/A | N/A | LEATE421-13 | Caryocolum leucomelanella | 3.76 |
| Caryocolum mucronatella | 0.3 | 0.46 | PHLAE427-11 | Caryocolum oculatella | 4.87 |
| Caryocolum oculatella | N/A | N/A | LEEUA388-11 | Caryocolum alsinella | 3.81 |
| Caryocolum peregrinella | 3.58 | 5.69 | PHLAB899-10 | Caryocolum trauniella | 3.93 |
| Caryocolum petrophila | 0.97 | 2.26 | PHLAH147-12 | Caryocolum huebneri | 4.88 |
| Caryocolum petryi | 0.23 | 0.61 | PHLAD576-11 | Caryocolum repentis | 3.85 |
| Caryocolum proxima | 0.41 | 0.61 | PHLAI019-12 | Caryocolum blandulella | 3.94 |
| Caryocolum pullatella | 2.07 | 3.61 | LEATC292-13 | Caryocolum marmorea | 3.12 |
| Caryocolum repentis | 0 | 0 | PHLAE429-11 | Caryocolum siculum | 3.33 |
| Caryocolum saginella | 0 | 0 | LEFIJ778-10 | Caryocolum cauligenella | 6.61 |
| Caryocolum schleichi dianthella | N/A | N/A | PHLAD573-11 | Caryocolum schleichi improvisella | 3.42 |
| Caryocolum schleichi improvisella | 0.06 | 0.15 | PHLSA085-11 | Caryocolum schleichi dianthella | 3.42 |
| Caryocolum schleichi arenariella | 0.77 | 1.24 | PHLSA085-11 | Caryocolum schleichi dianthella | 3.74 |
| Caryocolum siculum | N/A | N/A | PHLAD576-11 | Caryocolum repentis | 3.33 |
| Caryocolum srnkai | N/A | N/A | PHLAG580-12 | Caryocolum confluens | 4.54 |
| Caryocolum tischeriella | 1.09 | 2.02 | PHLAD576-11 | Caryocolum repentis | 4.01 |
| Caryocolum trauniella | 0 | 0 | PHLAB622-10 | Caryocolum peregrinella | 3.93 |
| Caryocolum tricolorella | 0 | 0 | PHLAI014-12 | Caryocolum fibigerium | 4.67 |
| Caryocolum vicinella | 1.48 | 2.7 | PHLAF105-11 | Caryocolum leucomelanella | 5.36 |
| Caryocolum viscariella | 0.22 | 0.47 | LEEUA388-11 | Caryocolum alsinella | 4.16 |
Sequences of the COI barcode region of all analysed morphospecies reveal significant interspecific genetic distances with barcode gaps ranging from a minimum of 3.11% to the nearest neighbour (Caryocolum pullatella – Caryocolum marmorea) to a maximum of 6.61% (Caryocolum saginella – Caryocolum cauligenella).
Taxonomy
The Caryocolum amaurella species-group as defined by Huemer (1988) differs from other congeners mainly by the characteristic shape of the sacculus, which is unique in the genus. Until now it only included Caryocolum amaurella and Caryocolum iranicum (Huemer 1988, 1989b). Based on the DNA barcode divergence and diagnostic morphological characters combined with biological data we describe the new species Caryocolum crypticum. Due to the mix-up of Caryocolum crypticum with Caryocolum amaurella in recent identification guides the latter species is also re-described here in detail.
Caryocolum Gregor & Povolný, 1954
Caryocolum Gregor & Povolný, 1954: 87.
Type species. Gelechia leucomelanella Zeller, 1839: 138.
Caryocolum crypticum sp. n.
http://zoobank.org/5E1FB9E5-3A65-49C6-80BF-A5CA7C4FFF99
http://species-id.net/wiki/Caryocolum_crypticum
Figs 2–3 , 6–7 , 10–11 , 14–15
Figures 2–5.
Adults. 2 Caryocolum crypticum sp. n., holotype 3 Caryocolum crypticum sp. n., paratype, female, Greece 4 Caryocolum amaurella, male, Finland 5 Caryocolum amaurella, male, Austria.
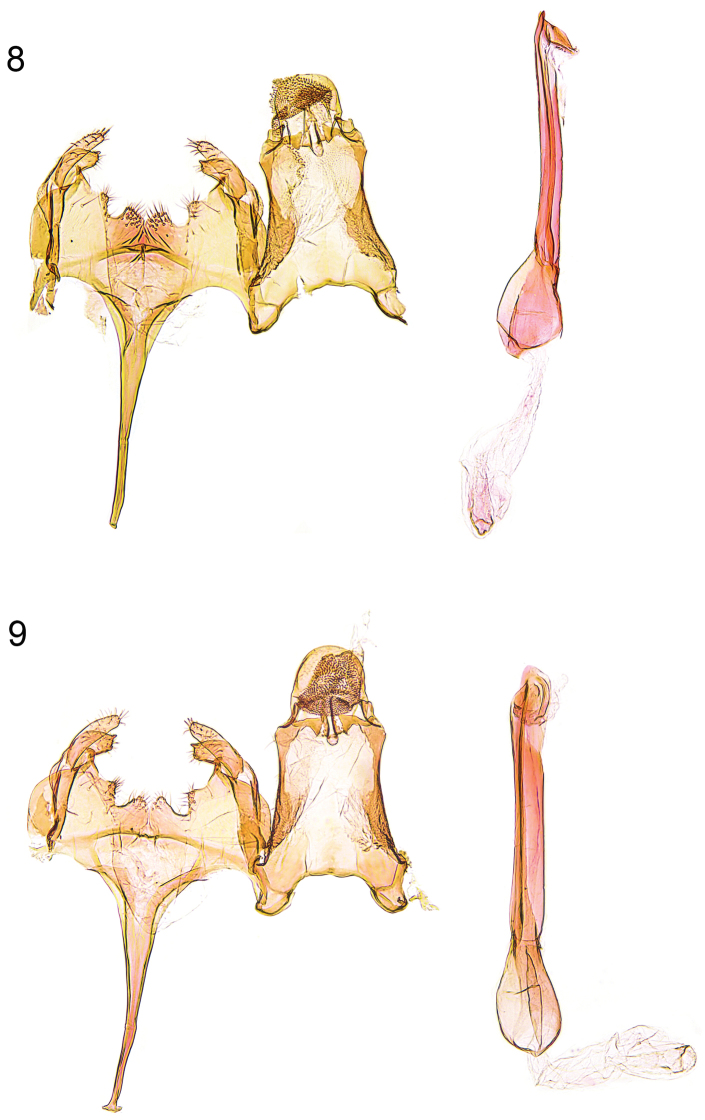
Figures 6–7.
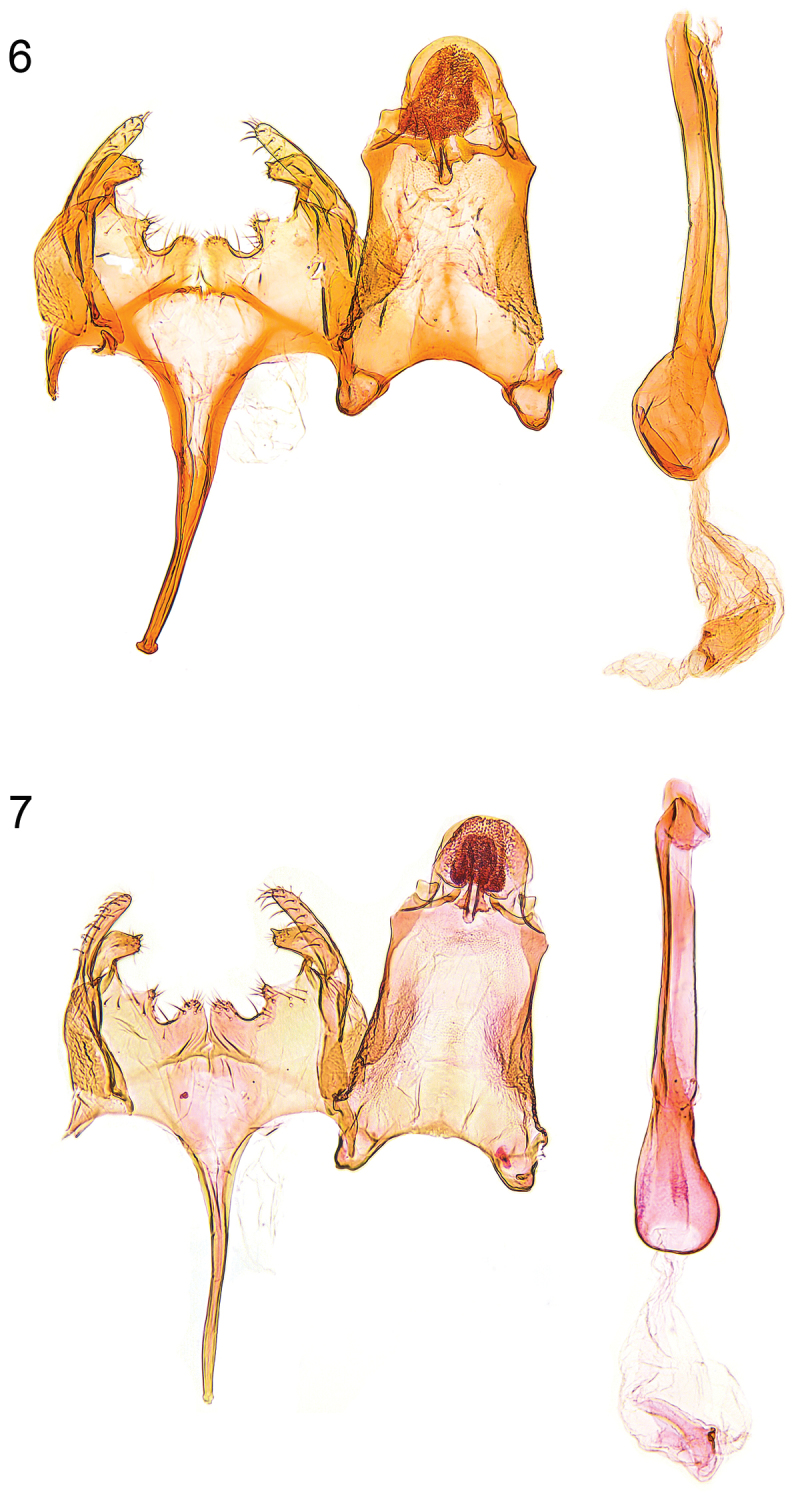
Male genitalia. 6 Caryocolum crypticum sp. n., paratype, Italy, slide GU 86/041 P.Huemer 7 Caryocolum crypticum sp. n., paratype, Italy, slide GEL 1215 P.Huemer.
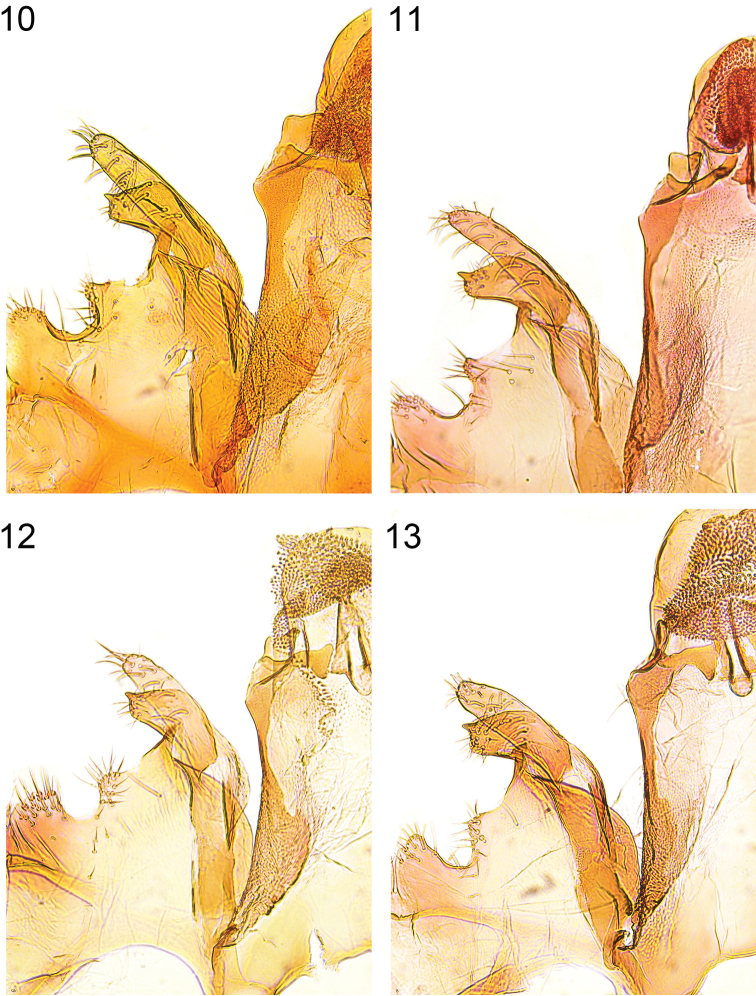
Figures 10–13.
Details of male genitalia (vinculum-valva-complex). 10 Caryocolum crypticum sp. n., paratype, Italy, slide GU 86/041 P.Huemer 11 Caryocolum crypticum sp. n., paratype, Italy, slide GEL 1215 P.Huemer 12 Caryocolum amaurella, Finland, slide GU 14/1373 P.Huemer 13 Caryocolum amaurella, Finland, slide GU 14/1374 P.Huemer.
Figures 14–15.
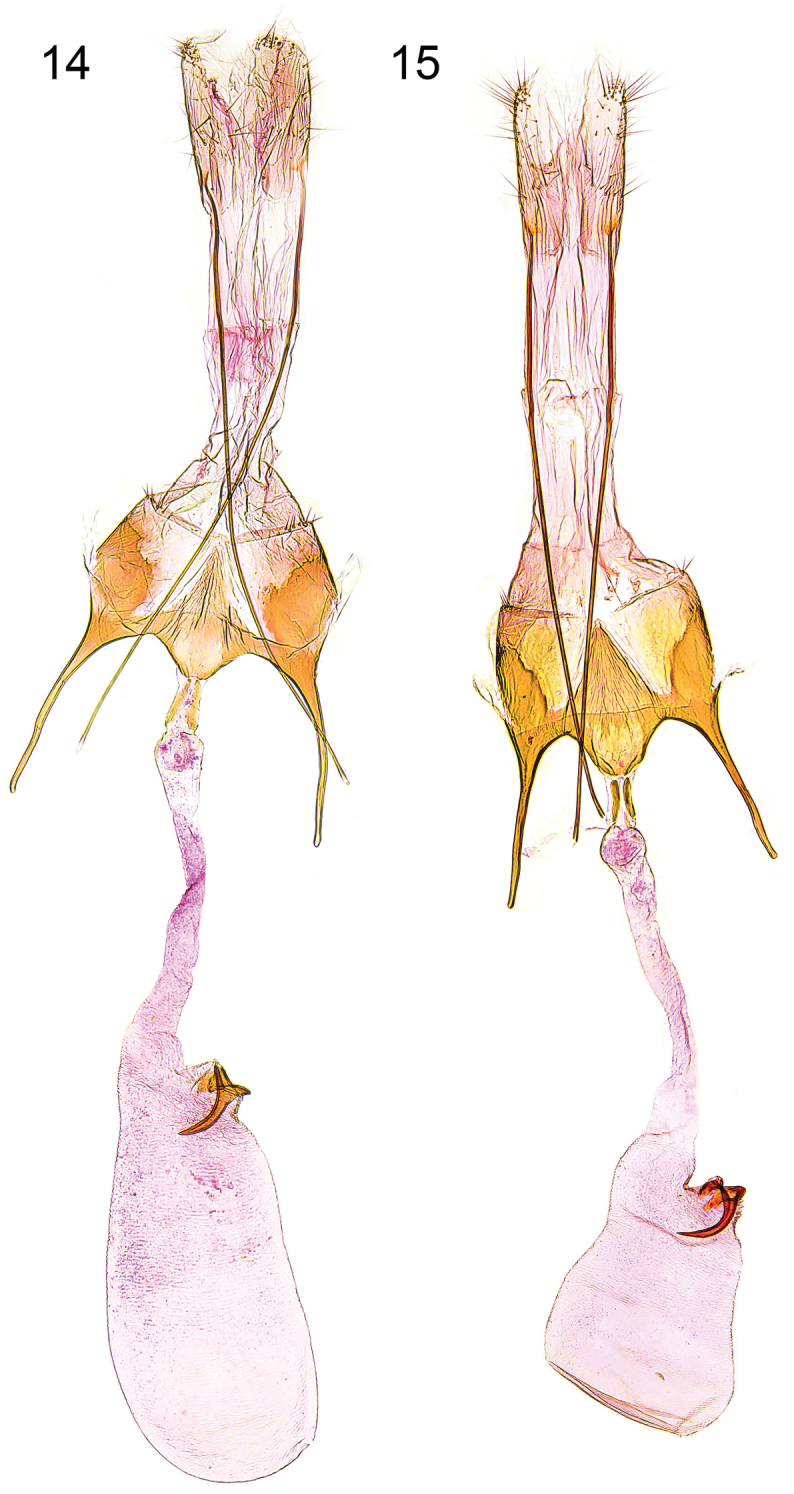
Female genitalia. 14 Caryocolum crypticum sp. n., holotype, slide GEL 1234 P.Huemer 15 Caryocolum crypticum sp. n., paratype, Italy, slide GEL 1232 P.Huemer.
Type material.
Holotype: ♀ (Fig. 2), Italia sept., Teriolis merid., Laatsch, 1000 m, 29.6.1987 e.l. (Silene otites 10.5.), leg. Huemer, slide GEL 1234 ♀ (TLMF).
Paratypes. Italy: 1 ♂, South Tyrol, Vinschgau, Schleiser Leiten, 1350 m, 6.7.2013, leg. Huemer, slide GEL 1215, dna barcode id TLMF Lep 12313 (TLMF); 1 ♂ [without abdomen], same data (TLMF); 1 ♀, same data, but 18.8.2013, slide GEL 1232, dna barcode id TLMF Lep 11883 (TLMF); 1 ♀, same data, but dna barcode id TLMF Lep 11882 (TLMF); 1 male [without abdomen], 8 ♀, same data, but 7.9.2013 (TLMF); 1 ♂, South Tyrol, Taufers, 1300 m, 22.8.1978, leg. Burmann, slide GU 86/041 P. Huemer (TLMF). Switzerland: 2 ♀, Wallis, Martigny-Rosel, 460 m, 28.6.–14.7.1983 e.l. (Silene otites), leg. Whitebread (Naturhistorisches Museum Basel, Switzerland). Greece: 1 ♀, Larisa, Ossa Oros, 1.5 km N Spilia, 940 m, 13.6.1988 e.l. (Silene nutans), leg. Huemer (TLMF).
Diagnosis.
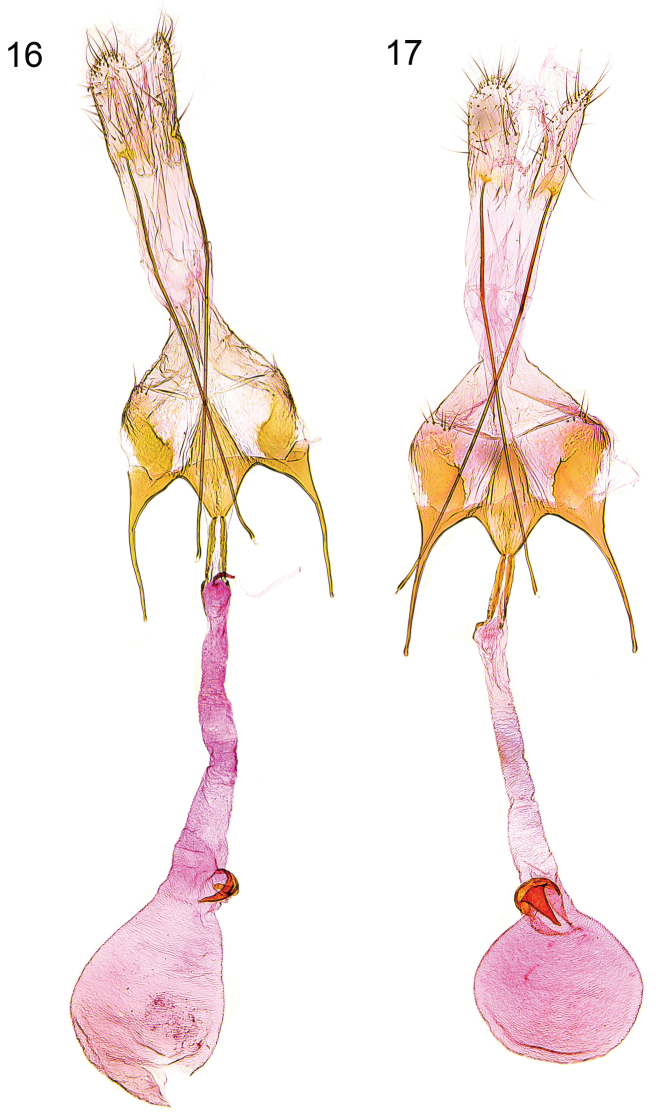
Caryocolum crypticum sp. n. is externally similar to several other species of the genus and can be best recognized by the largely unmarked forewings with cream costal and tornal spots. From its closest relatives Caryocolum amaurella and Caryocolum iranicum it differs by the rusty brown distal half of the thorax and the concolorous tegulae, the dark brown forewings with rusty brown scales, and the cream colours of the costal and tornal spots. The male genitalia of Caryocolum crypticum are very similar to those of Caryocolum amaurella but the valva is more slender and slightly longer (see Figs 6–7, 10–11 versus 8–9, 12–13). The similar Caryocolum iranicum differs by the shape of the sacculus with almost straight dorsal margin (see Huemer 1989b: Figs 14–16). However, the most striking diagnostic characters of the new species are found in the female genitalia which differ from Caryocolum amaurella particularly by the short lateral sclerites of the ductus bursae and the much longer and more slender signum hook (see Figs 14–15 versus 16–17). The female genitalia furthermore differ from Caryocolum iranicum by the weakly cup-shaped rather than funnel-shaped antrum, shorter lateral sclerites of the ductus bursae, and the shorter apophysis anterior which is almost twice the length of segment VIII in Caryocolum iranicum.
Figures 16–17.
Female genitalia. 16 Caryocolum amaurella, Finland, slide GU 14/1372 P.Huemer 17 Caryocolum amaurella, Finland, slide GU 14/1371 P.Huemer.
Description.
Adult (Figs 2–3). Wingspan 10.5-14 mm. Segment 2 of labial palpus with a few cream-coloured scales on inner and upper surface, blackish brown on outer and lower surface; segment 3 almost black with light tip. Antenna black, indistinctly lighter ringed. Head with light yellow frons and black neck; thorax blackish brown with rusty brown posterior part; tegulae rusty brown except for blackish brown base. Forewing blackish brown, mottled with some rusty brown, particularly in proximal half; supplementary black spots in fold and in cell obscure; costal and tornal spot small, cream, separated. Hindwing light grey.
Variation. No variation observed except for size, which differs considerably in two reared specimens from Italy and Greece.
Male genitalia (Figs 6–7, 10–11). Uncus subovate; tegumen stout; transtilla membranous; valva moderately short and slender, digitate, apex rounded; sacculus short, with angular ventral and weakly convex dorsal margin, apically pointed; posterior margin of vinculum with deep medial emargination and slight medial incision, two pairs of short processes developed; saccus long, comparatively broad at base, distal part gradually tapered; phallus long and slender, weakly curved, with some minute cornuti apically.
Female genitalia (Figs 14–15). Segment VIII without processes, subgenital plate sub-triangular, with numerous narrow folds, separated from sclerotized lateral plates by membranous zone; apophysis anterior about length of segment VIII; antrum short, about one quarter length of apophysis anterior, nearly cup-shaped; posterior part of ductus bursae with pair of short sclerites, extending to middle of apophysis anterior, and with two tiny sclerites anteriorly; signum with crescent-shaped base, long and slender, strongly bent hook.
Molecular data. The intraspecific divergence of the barcode region is low with mean intraspecific divergence of 0.21% and maximum intraspecific divergence of 0.31% (n=3). The distance to the nearest neighbour Caryocolum mucronatella is 5.41%, the divergence to the morphologically closest Caryocolum amaurella is 6.82%.
Etymology.
The name “crypticum” refers to the cryptic morphology of the species and is derived from the latinized adjective crypticus.
Distribution.
The species is known from widely separated localities in northern Italy, Switzerland and Greece, indicating a more widespread distribution in Sub-Mediterranean and Mediterranean Europe. However, the host-plants are much more widespread, ranging to northern Europe in the north and to Central Asia in the east. No sympatric occurrence with Caryocolum amaurella is reported though the two taxa can occur close to one another in the Alps.
Bionomics.
The larva has been found in early spring, feeding in the stem of Silene otites (L.) Wibel (Caryophyllaceae) (Burmann 1990) and Silene nutans L. (Huemer 1989) but detailed descriptions of feeding habits and larval morphology are missing. The adult occurs from early July (reared material dates from mid-June to mid-July) to September and it is attracted to light. Caryocolum crypticum prefers xerophilous steppes and rocky habitats with sparse vegetation. Vertical distribution: from about 500 to 1300 m, restricted to mountainous areas.
Remarks.
Huemer (1988) already examined females reared from Silene otites in Switzerland by Whitebread but in the absence of males considered them as deviating Caryocolum amaurella.
The majority of collected material belongs to females whereas Caryocolum amaurella is mainly known from the male sex. This may indicate differences in attraction to artificial lights or a female-biased sex ratio in Caryocolum crypticum.
Caryocolum amaurella
(Hering, 1924)
http://species-id.net/wiki/Caryocolum_amaurella
Figs 4–5 , 8–9 , 12–13 , 16–17
Figures 8–9.
Male genitalia. 8 Caryocolum amaurella (Hering), Finland, slide GU 14/1373 P.Huemer; 9 Caryocolum amaurella, Finland, slide GU 14/1374 P.Huemer.
Lita amaurella Hering 1924: 82, Figs 11–12.
Lita viscariae Schütze 1926: 171.
Material examined.
Lectotype ♂[with nine labels]: ‘Fennia Ab Bromarf’ ‘R. F:tius’ ‘21.7.21’ [piece of celluloid where genitalia was mounted] ‘Type ♂’ [red] ‘Lita amaurella m. det. Mart. Hering ♂’ ‘Mus. Zool. H:fors spec. typ. No 7016 Lita amaurella Hering’ ‘Lita amaurella m. ♂ Sch.-Armatur Bromarf 21.7.21 Fabritius’ ‘LECTOTYPE O. Karsholt design.’.
Finland: 1 ♂, Ab, Naantali, 25.8.1965, leg. Karvonen, slide Karsholt 2719; 2 ♂, N, Ekenäs, 17.–20.7.1980, leg. Fibiger; 1 ♂, N, Helsinki, 25.7.1982, leg. Schnack; 1 ♂, N, Borgå lk., Tirmo, 19–20.7.1980, leg. Fibiger; 5 ♂, same data, but 1.–2.8.1982, leg. Schnack; 2 ♂, U, Sluntle, 18.–31.7.1982, leg. Karsholt (all ZMUC); 6 ♂, 5 ♀, U, Porvoo, 6698:3426 Ånäs, e.l. 2012 (Lychnis viscaria), leg. Hirvonen (ZMUO); 4 ♂, V, Dragsfjärd, 664:3249, 2008, leg. Mutanen & Välimäki (ZMUO); 1 ♂, U, Hanko, 6642:3289, 2007, leg. Mutanen & Välimäki (ZMUO). Sweden: 2 ♂, Sk, Maglehen, 10.7.1965, leg. Svensson (TLMF, ZMUC); 1 ♂, Sm, Högsby, 13.7.1968, leg. Johansson; 1 ♂, Öl, Ödeshög, 17.7.1972, leg. Karsholt, slide Karsholt 1806; 1 ♂, St. Alvar, Tornrör, 25.7.1997, leg. Hendriksen, slide Hendriksen 1953; 2 ♂, Öl, Gårdby, 2.8.1999, leg. Hendriksen, slide Hendriksen 2411, 2415; 1 ♂, same data, but 22.7.2000; 1 ♂, Gtl., Hejnum Häller, 30.7.1977, leg. Hendriksen, slide Hendriksen 1944; 1 ♂, Ög, Ödeshög, 17.7.1972, leg. Karsholt; Upl., Film, 12.7.1995, leg. Hendriksen (all ZMUC). Norway: 2 ♂, On, Vinstra, 19.–29.7.1983, leg. Karsholt & Michelsen, slide Karsholt 4294, 4295; 2 ♀, same data, but 4.–5.7.1987, leg. Karsholt, slide Hendriksen 2099; 2 ♂, same data, but 9.8.1996, leg. Hendriksen (all ZMUC). Denmark: 1 ♂, Bornholm, Rø, 7.1892, leg. Gudmann, slide Wolff 2593; 5 ♂, 1 ♀, same data, but 28.7.1978, leg. Schnack, slide Schnack 1118; 1 ♂, Bornholm, Gudhjem, 1 ♂, 2 ♀, 29.6–3.7.1920, leg. Gudmann, slide Wolff 2625, 3682; 1 ♂, 1 ♀, same data, but e.l. 5.1921 (Lychnis viscaria), bred 21. & 28.6.1921, leg. Gudmann, slide Wolff 3681(all ZMUC); 6 ♂, 8 ♀, Bornholm, Hammeren, 18.7.1977, leg. Karsholt & Schnack, slide Hendriksen 1767, Karsholt 2948 TLMF, ZMUC); 2 ♂, same data, but 25.7.1977, leg. Schnack; 4 ♂, same data, but 16. –25.7.1978, leg. Schnack; 4 ♂, same data, but 19.–22.7.1979, leg. Hendriksen; 6 ♂, same data, but 29 –30.7.1981, leg. Hendriksen, slide Hendriksen 385, 561, 722; Bornholm, Randkløve, 1 ♂, 22.7.1977, leg. Schnack; Bornholm, 1 ♀, Ringe Bakker, 16.7.1978, leg. Schnack (all ZMUC). Germany: 1 ♂, 1 ♀, Lausitz, Umg. Bautzen, e.l. 1935 (Lychnis viscaria), leg. Starke (BMNH); 2 ♂, Thüringen, Bad Blankenburg, 14.7.1964, leg. Steuer (TLMF); 1 ♂, Thüringen, Bad Blankenburg, 8.7.1972, leg. Steuer (TLMF). France: 2 ♂, Alpes Maritimes, Col de la Cayolle, 2200–2300 m, 29.–30.7.2005, leg. Skou, slide Hendriksen 5364 (ZMUC). Austria: 1 ♂, Niederösterreich, Jauerling, 24.7.1935 (TLMF); 2 ♂, Oberösterreich, Windischgarsten, Veichltal, 23.7.1976, leg. Wimmer (TLMF); 1 ♂, Oberösterreich, Waldhausen, Schwarzenberg, 6.8.1997, leg. Wimmer (TLMF); 9 ♂, Kärnten, St. Jakob im Lesachtal, Mussen E, 1680–1800 m, 4.8.1999, leg. Huemer & Erlebach (TLMF). Slovakia: 1 ♀, Pol’ana, 28.7.1989, leg. Patocka (ZMUC). Macedonia: 4 ♂, NP Mavrovo, Korab, Korabska jezero, Kobilino pole, 2080–2180 m, 28.7.–1.8.2011, leg. Huemer & Tarmann (TLMF). Turkey: 2 ♂, 1 ♀, prov. Sivas, 10 km W Görün, 1650 m, 27.7.1989, leg. Esser & Fibiger, slide Huemer GU 90/130, GU 91/215; 4 ♂, prov. Erzerum, Kop Pass, 1750 m, 15.–16.9.1993, leg. Fibiger, slide Hendriksen 2889, 2894; 1 ♂, prov. Erzincan, Kizildaĝ, Geçidi, 2100 m, 19.8.1993, leg. Schepler, slide Hendriksen 2384 (all ZMUC).
Diagnosis.
See above.
Description.
Adult (Figs 4–5). Wingspan 10–14 mm. Segment 2 of labial palpus bone-white on inner and upper surface, blackish grey on outer and lower surface; segment 3 almost black with light tip. Antenna black, indistinctly lighter ringed. Head with light yellow frons and black neck; thorax and tegula black mottled with brown. Forewing blackish grey mottled with some light brown; base black; two indistinct black spots in fold; one oblique spot above it and one in cell; some white scales before and after these spots; costal and tornal spot small, white, rarely fused. Hindwing light grey.
Variation. The colour of the forewings varies from greyish to blackish. Worn specimens look lighter than fresh ones. Sometimes there are no white scales in the middle of the wing.
Male genitalia (Figs 8–9, 12–13). Uncus subovate; tegumen stout; transtilla membranous; valva short, moderately stout, apex rounded; sacculus short, with angular ventral and convex dorsal margin, apically pointed; posterior margin of vinculum with deep medial emargination and slight medial incision, two pairs of short processes developed; saccus long, comparatively broad at base, distal part gradually tapered; phallus long and slender, weakly curved, with some minute cornuti apically.
Female genitalia (Figs 16–17). Segment VIII without processes, subgenital plate sub-triangular, with numerous narrow folds, separated from sclerotized lateral plates by membranous zone; apophysis anterior slightly longer than segment VIII; antrum moderately short, about one-third to one-quarter length of apophysis anterior, broadly funnel-shaped; posterior part of ductus bursae with pair of lateral sclerites, extending to anterior third of apophysis anterior, and with two tiny sclerites anteriorly; signum with crescent-shaped base, short and stout, strongly bent hook.
Molecular data. The intraspecific divergence of the barcode region is high with mean intraspecific divergence of 3.01% and maximum intraspecific divergence of 4.62% (n=9). The distance to the nearest neighbour Caryocolum mucronatella is 5.21%, the divergence to the morphologically closest Caryocolum crypticum is 6.82%. The extraordinary high intraspecific divergence with 4 haplotypes is partially related to geographical pattern. However, we also found two haplotypes within one population in Finland and morphology does not support cryptic diversity.
Distribution.
With certainty known from scattered records from northern and Central Europe and Turkey. All the specimens from north of the Alps that we have been able to cross-check are correctly attributed to Caryocolum amaurella. However, recent records from Ukraine (Bidzilya and Budashkin 2009) and Russia (southern Ural Mountains) (Junnilainen et al. 2010) have to be re-examined due to a possible mix-up with Caryocolum crypticum. Records from Switzerland are dubious, and at least in one instance refer to the new species, whereas those from France (Nel 2003) are confirmed (see Huemer and Karsholt 2010, Fig. 154c).
Bionomics. The larva has been recorded feeding on Silene viscaria (L.) Jess (= Lychnis viscaria L. (Caryophyllaceae) (Huemer and Karsholt 2010), while the other stated host-plants, namely Silene otites (L.) Wibel (Burmann 1990) and Silene nutans L. (Huemer 1989a), refer to Caryocolum crypticum. Schütze (1926, 1931) gives a detailed account of the life-history. The larva feeds in April and May in the young terminal leaves which are – without spinning – attached to a tube where the larva is hidden. Dark frass is frequently extruded at the tip of the larval dwelling. Later it bores into the stem and the shoots often become swollen and stunted. Pupation takes place on the ground in a cocoon among debris. The adult occurs from late June to early September and it is attracted to light. Caryocolum amaurella is restricted to warm and sunny habitats such as dry meadows and pastures. Vertical distribution: from lowland localities to about 2200 m in the Alps.
Remarks.
Lita amaurella was described from an unspecified number of specimens of both sexes (‘♂, ♀’) from Finland (Bromarf) (Hering 1924). In order to stabilize nomenclature, a male, labelled as type, in ZMUH is here designated as lectotype (see data above). Lita viscariae was described from 67 specimens reared from Silene viscaria from Eastern Germany (near Rachlau) (Schütze 1926). No type material was traced during this and earlier studies (Huemer 1988), but the original descriptions and topotypical material leave no doubt about the identity.
Turkish specimens of Caryocolum amaurella examined by us differ from European specimens of this species by the thorax with rusty brown posterior part and the rusty brown tegulae with blackish brown base, similar to Caryocolum crypticum, and they are thus hardly separable from the latter on external characters. The genitalia of both sexes of Caryocolum amaurella from Turkey agree in all details with those of European Caryocolum amaurella and, because no contradicting genetic data is currently available, we consider them as belonging to that species.
One of the examined specimens of Caryocolum amaurella from Turkey was collected in the same locality (Kizildaĝ Geçidi, prov. Erzincan) as a specimen Caryocolum iranicum in ZMUC. The latter species, which is only known from a few specimens, differs, as stated above, in characters of the male genitalia.
Discussion
The genus Caryocolum is a rare example of European Microlepidoptera which has gained significant attention from specialists during the last decades. Several monographic papers, from Klimesch (1953–54) to Huemer and Karsholt (2010), are a sound base for a stable taxonomy and a pre-requisite to test congruence of classical morphologically-driven species delineation with that of molecular data. DNA barcoding has evolved as a widely accepted method for preliminary species delimitation (Monaghan et al. 2009, Hendrich et al. 2010, Kekkonen and Hebert 2014) and therefore the animal DNA barcode region seemed an appropriate genetic marker to be used for this purpose. Indeed, barcoding resulted in an excellent support for all of the 44 studied species with a distinct barcode gap to the nearest neighbour ranging from about 3% to nearly 7% interspecific divergence.
Intraspecific variation shows a different pattern. The majority of species has a low (<2%) maximum intraspecific divergence and thus seems taxonomically well defined. However, a remarkable number of species (8 species, nearly one quarter of all, 9 species with only one sample not considered) is characterized by maximum divergence exceeding 3% (Fig. 1). Such deep intraspecific splits often suggest the possibility of cryptic diversity (for examples in Lepidoptera, see Dinca et al. 2011, Hausmann et al. 2009, Huemer and Hebert 2011, Huemer et al. 2012, Huemer et al 2013, Kaila and Mutanen 2012, Landry and Hebert 2013, Mutanen et al. 2012a, b, 2013, Segerer et al. 2011, Wilson et al. 2010). A morphological cross-check in one of these taxa, Caryocolum amaurella, proved the existence of a hitherto overlooked species with validity independently supported by morphology, biological data, and the DNA barcode. The potential of DNA barcoding for screening of cryptic diversity is obvious in this case, where morphological characters, particularly the normally well-separated male genitalia, are weak and thus have been neglected so far. Although deep intraspecific splits may alternatively refer to mitochondrial introgression, historical polymorphism or Wolbachia infection (Hurst and Jiggins 2005, Funk and Omland 2003), there is a considerable possibility of further cryptic diversity in the genus. In Caryocolum schleichi it seems most appropriate that the three sequenced subspecies should be considered as different species since host-plants and genitalia morphology differ as well (see i.e. Huemer and Karsholt 2010). The subspecies of Caryocolum schleichi are geographically isolated making their delimitation both rather artificial and very sensitive to the species concept applied (Mutanen et al. 2012c). An integrative revision of this group is in preparation by the authors. In contrast, the expected low divergence in subspecies is reflected by a very low divergence in Caryocolum marmorea and its subspecies Caryocolum marmorea mediocorsa. Diagnostic morphological characters seem present in further taxa from first examined samples, namely Caryocolum fibigerium and Caryocolum peregrinella with a maximum intrapecific divergence of 6.27% and 5.69% related to three deep phylogeographic splits in both species. Similar deep splits are observed in Caryocolum alsinella and in Caryocolum cauligenella. For all these taxa with subtle character differences a careful re-examination of morphology has to be undertaken in the future.
Supplementary Material
Acknowledgments
We are particularly grateful to Paul Hebert and his team at the Canadian Centre for DNA Barcoding (Guelph, Canada), whose sequencing work was enabled by funding from the Government of Canada to Genome Canada through the Ontario Genomics Institute. We are also grateful to the Ontario Ministry of Research and Innovation and to NSERC for their support of the BOLD informatics platform.
Stefan Heim (TLMF) is acknowledged for his kind assistance with photographic work. We thank Robert J. Heckford (Plymouth, GB), Petri Hirvonen (Porvoo, Finland), Thierry Varenne (Nice, France), Christian Wieser (Klagenfurt, Austria) and Josef Wimmer (Steyr, Austria) for providing material for our examination, and Lauri Kaila (ZMUH) for access to the type of Lita amaurella.
We are particularly indebted to the Promotion of Educational Policies, University and Research Department of the Autonomous Province of Bolzano - South Tyrol for helping to fund the project “Genetic biodiversity archive - DNA barcoding of Lepidoptera of the central Alpine region (South, East and North Tyrol)”. Furthermore fundings from inatura Erlebnis Naturschau (Dornbirn, Austria) are acknowledged.
Last, but not least, we thank Martin Corley (Faringdon, UK) for linguistic improvement of the manuscript.
Citation
Huemer P, Karsholt O, Mutanen M (2014) DNA barcoding as a screening tool for cryptic diversity: an example from Caryocolum, with description of a new species (Lepidoptera, Gelechiidae). ZooKeys 404: 91–111. doi: 10.3897/zookeys.404.7234
Supplementary material
Sample information for specimens included in this study.
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Peter Huemer, Ole Karsholt, Marko Mutanen
Data type: species data
Explanation note: Process IDs are sequence identifiers in BOLD; Sample IDs are specimen identifiers; BINs are Barcode Identification Numbers in BOLD. Details of collecting data, images, sequences, and trace files for the barcoded specimens are available in the public BOLD dataset “DS-LECARY”, accessed at dx.doi.org/10.5883/DS-LECARY.
References
- Bella S. (2008) Caryocolum siculum sp. n. (Gelechiidae), feeding on Gypsophila (Caryophyllaceae) in Sicily. Nota lepidopterologica 31: 69-75 [Google Scholar]
- Bidzilya AV, Budashkin YI. (2009) New records of Microlepidoptera from Ukraine. Proceedings Zoological Museum Kiev Taras Shevchenko National University 5: 14-28 [In Russian] [Google Scholar]
- Burmann K. (1990) Beiträge zur Microlepidopteren-Fauna Tirols. XIV. Caryocolum Gregor & Povolný, 1954 (Insecta: Lepidoptera, Gelechiidae). Berichte des naturwissenschaftlich-medizinischen Vereins Innsbruck 77: 171-184 [Google Scholar]
- deWaard JR, Ivanova NV, Hajibabaei M, Hebert PDN. (2008) Assembling DNA Barcodes: Analytical Protocols. In: Cristofre M. (Ed) Methods in Molecular Biology: Environmental Genetics. Humana Press Inc., Totowa, USA, 275-293 [DOI] [PubMed] [Google Scholar]
- Dinca V, Lukhtanov VA, Talavera G, Vila R. (2011) Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nature Communications 2: 324. doi: 10.1038/ncomms1329 [DOI] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. (2003) Species-level paraphyly and polyphyly: Frequency, causes and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics 34: 397-423. doi: 10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Grange JC, Nel J. (2012) Caryocolum dauphini n. sp., un endémique du Sud-Ouest alpin découvert dans le Parc national du Mercantour (Gelechiidae, Gnorimoschemini). Oreina 17: 22-23 [Google Scholar]
- Gregor F, Povolný D. (1954) Systematische und zoogeographische Studie über die Gruppe der Arten Gnorimoschema Busck mit Rücksicht auf die richtige Diagnostik des Schädlings Gnorimoschema ocellatellum Boyd. Zoologické a Entomologické Listy 3: 83–97, pl. 7, map [Google Scholar]
- Hausmann A, Hebert PDN, Mitchell A, Rougerie R, Sommerer M, Edwards T, Young CJ. (2009) Revision of the Australian Oenochroma vinaria Guenée, 1858 species-complex (Lepidoptera: Geometridae, Oenochrominae): DNA barcoding reveals cryptic diversity and assesses status of type specimen without dissection. Zootaxa 2239: 1-21 [Google Scholar]
- Hendrich L, Pons J, Ribera I, Balke M. (2010) Mitochondrial Cox1 sequence data reliably uncover patterns of insect diversity but suffer from high lineage-idiosyncratic error rates. PLoS ONE 5: . doi: 10.1371/journal.pone.0014448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering EM. (1924) Beitrag zur Kenntnis der Microlepidopteren-Fauna Finlands. Notulae Entomologicae 4: 75-84 [Google Scholar]
- Huemer P. (1987) Eine modifizierte Genitalpräparationstechnik für die Gattung Caryocolum. Mitteilungen der Schweizerischen Entomologischen Gesellschaft 60: 207-211 [Google Scholar]
- Huemer P. (1988) A taxonomic revision ofCaryocolum (Lepidoptera: Gelechiidae). Bulletin of the British Museum of Natural History (Entomology) 57: 439-571 [Google Scholar]
- Huemer P. (1989a) Bemerkenswerte Funde von Caryocolum-Arten aus den Südalpen und dem Mediterraneum (Lepidoptera, Gelechiidae). Nachrichtenblatt der Bayerischen Entomologen 38: 37-40 [Google Scholar]
- Huemer P. (1989b) Neue und wenig bekannte Arten der Gattung Caryocolum Gregor & Povolný, 1954, aus Südwestasien. Mitteilungen der Münchner Entomologischen Gesellschaft 79: 127-142 [Google Scholar]
- Huemer P. (2013) Die Schmetterlinge Österreichs (Lepidoptera). Systematische und faunistische Checkliste. Studiohefte 12, 304 pp [Google Scholar]
- Huemer P, Karsholt O. (2010) Gelechiidae II (Gelechiinae: Gnorimoschemini. In: Huemer P, Karsholt O, Nuss M. Microlepidoptera of Europe. Vol. 6 Apollo Books, Stenstrup, 586 pp. [Google Scholar]
- Huemer P, Nel J. (2005) Caryocolum mazeli sp. n., a new species from southern France (Lepidoptera, Gelechiidae). Bulletin de la Société Entomologique de France 110: 125-127 [Google Scholar]
- Huemer P, Elsner G, Karsholt O. (2013) Review of the Eulamprotes wilkella species-group based on morphology and DNA barcodes, with descriptions of new taxa (Lepidoptera, Gelechiidae). Zootaxa 3746: 069-100 [DOI] [PubMed] [Google Scholar]
- Huemer P, Hebert PDN. (2011) Cryptic diversity and phylogeography of high alpine Sattleria— a case study combining DNA barcodes and morphology (Lepidoptera: Gelechiidae). Zootaxa 2981: 1-22 [Google Scholar]
- Huemer P, Zlatkov B, Baixeras J. (2012) Dichrorampha dinarica, new species, a century of confusion in European lepidopterology (Lepidoptera: Tortricidae) resolved by combining morphology and DNA barcoding. Zootaxa 3389: 41-50 [Google Scholar]
- Hurst GDD, Jiggins FM. (2005) Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proceedings of the Royal Society Biological Sciences Series B 272: 1525-1534. doi: 10.1098/rspb.2005.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junnilainen J, Karsholt O, Nupponen K, Kaitila JP, Nupponen T, Olschwang V. (2010) The gelechiid fauna of the Southern Ural Mountains, part II: list of recorded species with taxonomical notes (Lepidoptera: Gelechiidae). Zootaxa 2367: 1-68 [Google Scholar]
- Kaila L, Mutanen M. (2012) DNA barcoding and morphology support the division of Elachista nuraghella sensu auct. (Lepidoptera: Elachistidae: Elachistinae) into two vicariant species. Zootaxa 3343: 57-68 [Google Scholar]
- Kekkonen M, Hebert PDN. (2014) DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Molecular Ecology Resources. doi: 10.1111/1755-0998.12233 [DOI] [PMC free article] [PubMed]
- Klimesch J. (1953–1954) Die an Caryophyllaceen lebenden europäischen Gnorimoschema Busck (= Phthorimaea Meyr.)-Arten. Zeitschrift der Wiener Entomologischen Gesellschaft 38 (1953): 225–239, 272,–282, 311,–319; 39 (1954): 273,–288, 335,–341, 357–362 [Google Scholar]
- Landry JF, Hebert PDN. (2013) Plutella australiana (Lepidoptera: Plutellidae), an overlooked diamondback moth revealed by DNA barcodes. Zookeys 327: 43-63. doi: 10.3897/zookeys.327.5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan MT, Wild R, Elliot M, Fujisawa T, Balke M, Inward DJG, Lees DC, Ranaivosolo R, Eggleton P, Barraclough TG, Vogler AP. (2009) Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Systematic Biology 58: 298-311. doi: 10.1093/sysbio/syp027 [DOI] [PubMed] [Google Scholar]
- Mutanen M, Aarvik L, Landry J-F, Segerer A, Karsholt O. (2012a) Epinotia cinereana (Haworth, 1811) bona sp., a Holarctic tortricid distinct from E. nisella (Clerck, 1759) (Lepidoptera: Tortricidae: Eucosmini) as evidenced by DNA barcodes, morphology and life history. Zootaxa 3318: 1-25 [Google Scholar]
- Mutanen M, Aarvik L, Huemer P, Kaila L, Karsholt O, Tuck K. (2012b) DNA barcodes reveal that the widespread European tortricid moth Phalonidia manniana (Lepidoptera: Tortricidae) is a mixture of two species. Zootaxa 3262: 1-21 [Google Scholar]
- Mutanen M, Hausmann A, Hebert PDN, Landry J-F, deWaard J, Huemer P. (2012c) Allopatry as a Gordian knot for taxonomists: patterns of barcode divergences in arctic-alpine Lepidoptera. PLoS ONE 7: . doi: 10.1371/journal.pone.0047214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutanen M, Kaila L, Tabell J. (2013) Wide-ranging barcoding aids discovery of one-third increase of species richness in presumably well-investigated moths. Scientific Reports 3: . doi: 10.1038/srep02901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel J. (2003) Microlépidoptères nouveaux ou rarement signalés de la fauna de France (Lepidoptera). Bulletin de la Société Entomologique de France 108: 81-86 [Google Scholar]
- Ratnasingham S, Hebert PDN. (2007) BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular Ecology Notes 7: 355-364. doi: 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze KT. (1926) Lita viscariae n. sp. Deutsche entomologische Zeitschrift Iris 40: 171-175 [Google Scholar]
- Schütze KT. (1931) Die Biologie der Kleinschmetterlinge unter besonderer Berücksichtigung ihrer Nährpflanzen und Erscheinungszeiten. Frankfurt/Main, 235 pp [Google Scholar]
- Segerer AH, Haslberger A, Grünewald T. (2011. [“2010”]) Olethreutes subtilana (Falkovich, 1959): Unexpected occurrence of an ‘eastern’ leaf roller in Central Europe, uncovered by DNA barcoding (Tortricidae: Olethreutinae). Nota lepidopterologica 33: 197–206 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731-2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Landry J-F, Janzen DH, Hallwachs W, Nazari V, Hajibabaei M, Hebert PDN. (2010) Identity of the ailanthus webworm moth (Lepidoptera, Yponomeutidae), a complex of two species: evidence from DNA barcoding, morphology and ecology. ZooKeys 46: 41-60. doi: 10.3897/zookeys.46.406 [Google Scholar]
- Zeller PC. (1839) Versuch einer naturgemässen Eintheilung der Schaben. Isis, Leipzig: 1839: 167-220 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample information for specimens included in this study.
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Peter Huemer, Ole Karsholt, Marko Mutanen
Data type: species data
Explanation note: Process IDs are sequence identifiers in BOLD; Sample IDs are specimen identifiers; BINs are Barcode Identification Numbers in BOLD. Details of collecting data, images, sequences, and trace files for the barcoded specimens are available in the public BOLD dataset “DS-LECARY”, accessed at dx.doi.org/10.5883/DS-LECARY.