Abstract
Exposure to lead (Pb) during childhood can result in learning disabilities and behavioral problems. Although described in animal models, whether Pb exposure also alters neuronal differentiation in the developing brains of exposed children is unknown. Here, we investigated the effects of physiologically relevant concentrations of Pb (from 0.4 to 1.9μM) on the capacity of human embryonic stem cells (hESCs) to progress to a neuronal fate. We found that neither acute nor chronic exposure to Pb prevented hESCs from generating neural progenitor cells (NPCs). NPCs derived from hESCs chronically exposed to 1.9μM Pb throughout the neural differentiation process generated 2.5 times more TUJ1-positive neurons than those derived from control hESCs. Pb exposure of hESCs during the stage of neural rosette formation resulted in a significant decrease in the expression levels of the neural marker genes PAX6 and MSI1. Furthermore, the resulting NPCs differentiated into neurons with shorter neurites and less branching than control neurons, as assessed by Sholl analysis. DNA methylation studies of control, acutely treated hESCs and NPCs derived from chronically exposed hESCs using the Illumina HumanMethylation450 BeadChip demonstrated that Pb exposure induced changes in the methylation status of genes involved in neurogenetic signaling pathways. In summary, our study shows that exposure to Pb subtly alters the neuronal differentiation of exposed hESCs and that these changes could be partly mediated by modifications in the DNA methylation status of genes crucial to brain development.
Keywords: Heavy metal, lead, human embryonic stem cell, neuronal differentiation, DNA methylation
Lead (Pb) is a pervasive environmental neurotoxin that does not degrade or lose potency over time, contaminates population centers worldwide, and has no safe level of exposure for children (Kuehn, 2012). Although Pb poisoning is a preventable disease, each year there continue to be thousands of new cases in the United States, where about 500,000 children under 5 years old have blood Pb levels (BLLs) greater than the Centers for Disease Control and Prevention (CDC) threshold level of 5 μg/dL (0.25μM), at which counseling and/or treatment are initiated (www.cdc.gov/nceh/lead/). Early-life environmental exposure to Pb is especially toxic to the developing brain and can severely alter neurological structure and function (Bellinger, 2013; Kuehn, 2012; Toscano and Guilarte, 2005). Thus, chronic exposure to Pb during gestation or as a young child, even at BLLs below the CDC threshold level, can result in disrupted cognitive abilities, attention deficit hyperactivity-like disorders, lowered Intelligence Quotient (IQ), increased delinquency, changes in activity level, and altered sensory function (references in Senut et al., 2012). A growing body of evidence is showing that gestational exposure to Pb not only may have damaging neurological effects that last well into adulthood (Brubaker et al., 2009; Cecil et al., 2008; Stokes et al., 1998), but may also increase the susceptibility to a variety of neurological diseases with onset in later life, such as Alzheimer's disease and schizophrenia (Basha et al., 2005; Guilarte et al., 2012; Mazumdar et al., 2012).
Milestone animal studies have identified the synapse as a target of Pb neurotoxicity by demonstrating that chronic Pb exposure can reduce the density of synapses, lessen the levels of synaptic/neural proteins, and alter presynaptic function and vesicular release (reviewed in Neal and Guilarte, 2010; White et al., 2007). Pb can also have detrimental effects on the proliferation, differentiation, and survival of newborn neurons (Davidovics and DiCicco-Bloom, 2005; Giddabasappa et al., 2011; Gilbert et al., 2005; Huang and Schneider, 2004; Jaako-Movits et al., 2005; Kermani et al., 2008; Verina et al., 2007; Zurich et al., 2002), thereby altering neurogenesis, a crucial step in brain development. Whether similar changes occur in the developing brains of Pb-exposed children is yet unknown, but in vitro studies reported altered activity of protein phosphatases known to regulate synaptic plasticity and inhibition of Ca2+ channels neurotransmission following Pb exposure in primary human fetal neurons (Rahman et al., 2011) and human embryonic kidney cells (Peng et al., 2002), respectively.
Although the cellular and molecular underpinnings of Pb neurotoxicity are not well understood, in vitro and in vivo animal models have helped identifying several pathways potentially involved in Pb neurotoxicity, such as disruption of calcium signaling, oxidative stress, and altered expression of brain-specific genes (Sanders et al., 2009). Because alterations in epigenetic determinants help define the transcriptional pathways that contribute to both synapse function (Sultan and Day, 2011) and neurogenesis (Jobe et al., 2012), they have recently emerged as potential intermediaries in Pb neurotoxicity. DNA methylation, which occurs when DNA methyltransferases add a methyl group to the 5 carbon position on the cytosine pyrimidine ring (5-methylcytosine), is an epigenetic modification that is essential to the regulation of gene expression and is most commonly associated with gene silencing (Elango and Yi, 2011). An earlier report showed that in humans maternal bone Pb levels were inversely associated with cord blood methylation levels in long interspersed nuclear element 1 (LINE-1) and Alu1, two elements frequently analyzed in genomic DNA methylation studies (Pilsner et al., 2009). Since then, a scant number of epidemiological studies addressing Pb-induced changes in DNA methylation have provided further evidence that exposure to Pb can induce hypomethylation or hypermethylation of various gene regulatory sequences (Hanna et al., 2012; Kovatsi et al., 2010; Pilsner et al., 2009; Wright et al., 2010). Animal studies have also advanced the DNA methylome as a potential target of Pb exposure by showing alterations in the expression levels of genes involved in DNA methylation, including DNA methyltransferases, methyl-cytosine-phosphate-guanine (Me-CpG) binding protein-2 (MeCP2), methionine synthase, as well as global hypomethylation of cytosines (Bihaqi et al., 2011; Dosunmu et al., 2012; Schneider et al., 2013; Wu et al., 2008b). Taken together, these data provide compelling evidence that changes in the DNA methylation status of genes that are crucial to brain development may mediate some of the neurotoxic effects observed after early-life exposure to Pb.
Very few studies have analyzed the mechanisms of Pb-induced human brain dysfunction at the cellular levels mainly due to the insufficient number of cell lines or tissues available for analysis. Embryonic stem cells (ESCs), and their subsequent differentiation into neural progenitor cells (NPCs) and neurons, have enabled researchers to establish pluripotent cell-based model systems to study developmental neurotoxicity of chemical agents and heavy metals (Wobus and Loser, 2011). Although ESCs have been shown to reproduce a wide range of requisite neural and neuronal characteristics, their use in the study of pharmacological and toxicological agents is just starting to be explored. To this date, studies examining the effects of Pb exposure on the neural differentiation of stem cells have mostly been aimed at mouse ESCs and shown that Pb can alter the potential of such cells to differentiate into neurons (Baek et al., 2011; Visan et al., 2012; Zimmer et al., 2011). Two recent studies using human embryonic stem cells (hESCs) or their neural derivatives found that Pb can hinder the proliferation of human amniotic fluid stem cells (Gundacker et al., 2012) and impair the migration of hESC-derived neural crest cells (Zimmer et al., 2012). However, further investigation is needed to better understand the adverse effects that Pb may have on human brain development and their underlying mechanisms.
In this report, we used the in vitro differentiation of hESCs into NPCs and subsequently neurons to study the early developmental neurotoxic effects of Pb levels similar to BLLs measured in Pb-exposed children. We show that exposure of hESCs to physiologically relevant Pb levels not only affects their subsequent differentiation into neurons but also induces rapid methylation changes in the CpG sites of specific genes key to neuronal growth. Understanding whether and how the DNA methylome and other epigenetic regulators cooperate to generate the neurodevelopmental effects of Pb exposure will reveal novel molecular pathways of Pb neurotoxicity and will have implications for prevention and therapeutic intervention.
MATERIALS AND METHODS
Maintenance and Culture of hESCs
In these experiments, we used the WA09 hESC line (passages 26–53; WiCell Research Institute, Madison, WI) (Thomson et al., 1998) that is NIH-approved and included in the National Stem Cell Registry. Human ESCs were maintained on a feeder layer of irradiated mouse embryonic fibroblasts (GlobalStem, Rockville, MD) in hESC medium. The hESC culture medium consisted of 20% knockout serum replacement, 2mM L-Glutamine, 1% nonessential amino acids, 1% penicillin/streptomycin (all products from Life Technologies Corporation, Carlsbad, CA), 4 ng/ml human fibroblast growth factor basic (FGF-2; Peprotech, Rocky Hill, NJ), and 0.1μM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO) in DMEM/F12 (Life Technologies Corporation). Cultures were maintained at 37°C with 5%CO2 in a humidified incubator, and medium was changed daily. Colonies were passaged every 4–6 days by mechanical dissection. The pluripotent state of hESCs was regularly assessed by immunofluorescence detection for pluripotency markers, such as Oct4, Nanog, and Lin28.
Differentiation of hESCs Into NPCs
Neural induction of hESCs was performed according to previously published protocols (Chang et al., 2010; Zhang et al., 2001; Fig. 1A). Briefly, hESC colonies were mechanically dissociated, transferred in 100 mm bacterial plates and maintained as aggregates (embryoid bodies, EBs) for 4 days in a medium composed of 50% hESC medium and 50% N2 medium. N2 medium was made of 1% N2 supplement (Life Technologies Corporation) and 1% penicillin/streptomycin in DMEM/F12. On culture day 4, EBs were transferred to a new bacterial plate and cultured for an additional 4 days in N2 medium supplemented with 3μM retinoic acid (Sigma-Aldrich). On day 8, and over the next 3 days, EBs were cultured in N2 medium, containing 20 ng/ml FGF-2 (N2/FGF-2; Peprotech). EBs were subsequently transferred to 6-well plates coated with 20μg/ml fibronectin (Life Technologies Corporation) and maintained in N2/FGF-2 medium for an additional 8 days. Cells differentiating as neuroectodermal cells organized into neural-tube-like rosettes, which were then dissociated and plated in N2/FGF-2 medium on 10 μg/ml polyornithine/5 μg/ml laminin-coated dishes.
FIG. 1.
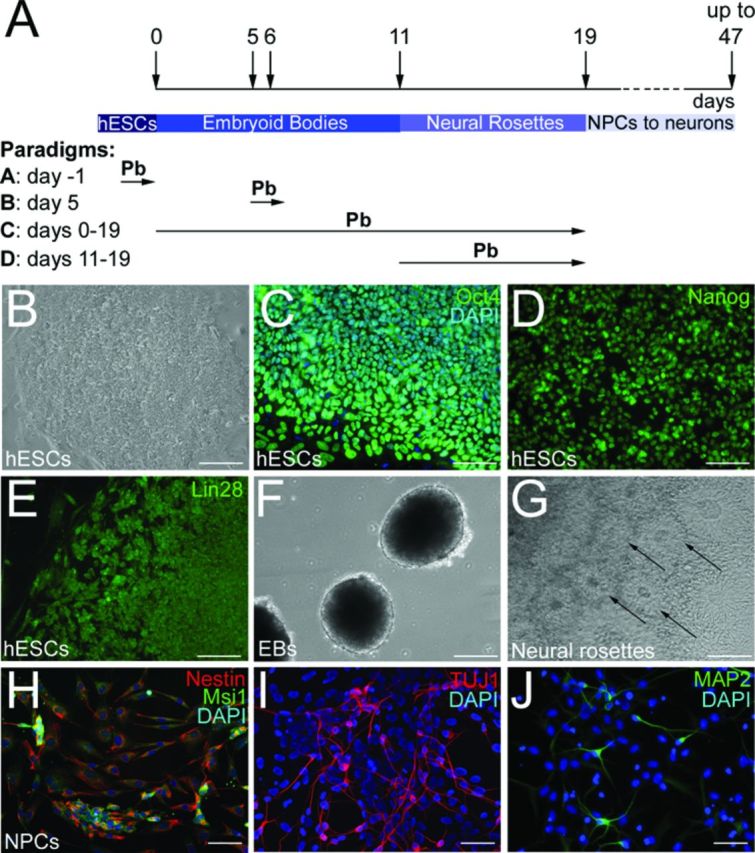
Neuronal differentiation of hESCs. (A) Schematic representation of the differentiation protocol and Pb exposure paradigms. (B) WA09 hESC cell colony on mouse embryonic fibroblast (MEF) feeder layer. hESCs are immunoreactive for the pluripotency markers Oct4 (C), Nanog (D), and Lin28 (E). (F) hESC-derived embryoid bodies (EBs) at culture day 6. (G) Formation of neural rosettes (arrows) 6 days following plating of EBs on fibronectin-coated plates. (H) hESC-derived NPCs display immunofluorescence labeling for the NPC markers Nestin (red) and Musashi1 (Msi1) (green). (I) At day 47 of the differentiation protocol, hESC-derived neurons express the neuronal-specific marker βIII-tubulin (TUJ1 antibody). (J) hESC-derived neurons also express the mature neuronal marker MAP2 at 31 days of the differentiation protocol. Nuclei are stained with DAPI. Bars: 100 μm (B)–(G); 50 μm (H)–(J).
Neuronal Differentiation of hESC-Derived NPCs
For neuronal differentiation, NPCs were passaged at a 5.105 cells/cm2 density in polyornithine/laminin-coated 12-well dishes and cultured in N2 medium supplemented with 10 ng/ml brain-derived neurotrophic factor, 10 ng/ml glial cell-derived neurotrophic factor, 10 ng/ml insulin-like growth factor (all factors from Peprotech), 200 ng/ml ascorbic acid (Sigma-Aldrich), and 100 ng/ml cAMP (Sigma-Aldrich), according to a previously published protocol (Hu and Zhang, 2010).
Pb Concentrations
Pb acetate was obtained from Sigma-Aldrich. Stock solutions (100-fold concentrated) of Pb acetate (Pb(C2H3O2)2) were prepared in sterile distilled water and dose regimens were prepared by further diluting the stock in culture medium. Control (distilled water vehicle, 0μM) and five physiologically relevant concentrations of Pb acetate were tested: 0.4μM (8 μg/dL), 0.8μM (16 μg/dL), 1.2μM (24 μg/dL), 1.5μM (32 μg/dL), and 1.9μM (40 μg/dL). Cultures were monitored daily using a Nikon Eclipse TE2000-S microscope (Nikon Instruments Inc., Melville, NY).
Viability Assay
Viability of hESC colonies and EBs was evaluated for the acute Pb exposure paradigms A and B (Fig. 1A) by Trypan blue staining for loss of membrane integrity and the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay for mitochondrial enzymatic activity.
Trypan blue staining
At the end of the 24-h Pb exposure, control and Pb-treated hESCs were harvested, stained with 0.4% Trypan blue (Life Technologies Corporation), and counted with a hemacytometer. The number of viable cells was determined by subtracting the number of dark-blue stained cells (nonviable) from the total number of cells and expressed as a percentage relative to the total number of cells. Three independent experiments were analyzed.
MTT assay
A 5 mg/ml stock solution of 3-(4,5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich) was freshly prepared in phosphate-buffered saline (PBS). Human embryonic stem cells (hESCs) were cultured in phenol red-free culture medium, and at the end of the 24-h exposure, MTT was added at a final concentration of 0.5 mg/ml to each well. Cells were then incubated in the dark at 37°C for 3 h, at which time the formazan crystals resulting from the reduction of MTT by mitochondrial enzymes were solubilized for 15 min at room temperature in dimethyl sulfoxide. Absorbance was measured at 590 nm using a spectrophotometer. Cell survival was expressed as a percentage of absorbance relative to that of control samples. Three independent experiments were analyzed.
In Vitro Differentiation Potential Studies
For evaluating the effects of Pb on the differentiation potentials of hESCs, we used a procedure involving the spontaneous differentiation of EBs (Ross et al., 2010). Briefly, hESC colonies were mechanically dissociated and maintained as EBs in FGF-2-free hESC medium in the presence of vehicle or 0.4–1.9μM Pb for 14 days, at which time quantitative reverse transcription PCR (qRT-PCR) analyses of the well-established lineage marker genes—AFP (endodermal), NCAM (ectodermal), and BRACHYURY (mesodermal)—were performed. All experiments were done in three biological replicates.
Effect of Pb on the Neural Differentiation of hESCs
The dose-response effects of Pb acetate on the generation of hESC-derived NPCs were determined in four different experimental paradigms.
Paradigm A
hESCs were acutely exposed to the different concentrations of Pb or vehicle, 24 h prior to the initiation of differentiation (day -1; Fig. 1A). The following day, control, and exposed hESC colonies were mechanically dissociated into 25–30 pieces of homogeneous size that were transferred into bacterial plates and underwent the neural differentiation protocol until day 19 (Fig. 1A).
Paradigm B
hESC colonies were mechanically dissociated into 150–200 pieces of homogeneous size that were randomly distributed into six bacterial plates. The 24-h acute exposure started at day 5 of the differentiation process, a day after initiation of neural induction with retinoic acid (day 5; Fig. 1A). As in paradigm A, cells underwent the neural differentiation protocol until day 19.
Paradigm C
hESCs were chronically exposed to Pb during the whole neural differentiation procedure up to day 19 (days 0–19; Fig. 1A).
Paradigm D
hESCs were maintained in the different Pb concentrations from days 11–19 corresponding to the phase of neural rosette formation (days 11–19; Fig. 1A).
In all paradigms A–D, the culture medium was refreshed every 2 days, and qRT-PCR analyses of the neural marker genes NESTIN, MUSASHI1 (MSI1), PAX6, and SOX2 were performed at day 19 of the differentiation process.
qRT-PCR Analysis
Total RNA was extracted from control and Pb-exposed hESC-derived cells (day 19) or EBs (day 14) using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The resulting RNA was quantified by optical density and stored at −70°C. RNA was then reversed transcribed using Superscript II Reverse Transcriptase and random primers (Life Technologies Corporation) for 50 min at 40°C followed by 15 min at 75°C. Quantitative PCR was performed using an Applied Biosystems, Inc., (ABI) PRISM 7000 Sequence Detection System and its software (Applied Biosystems) with 10 ng cDNA, 400nM of each primers and Synergy Brands (SYBR) Green PCR Master Mix (Life Technologies Corporation). Primers used for analyzing the expression of lineage specification and NPC marker genes are listed in Table 1. Data were normalized against the expression of the internal control genes Glyceraldehyde-3-phosphate dehydrogenase (GAPDH ) or Ribosomalprotein large subunit 27A (PANL27 ) (Table 1). Data analysis was performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001) and standardized by log transformation, mean centering, and autoscaling as previously described (Willems et al., 2008). Experiments were performed in at least three biological replicates.
TABLE 1. List of Primers for qRT-PCR.
| Genes | Forward | Reverse |
|---|---|---|
| AFP (Ross et al., 2010) | 5′-GAGGGAGCGGCTGACATTATT-3′ | 5′-TGGCCAACACCAGGGTTTA-3′ |
| BRACHYURY | 5′-CAGAACGAGGAGATCACAGC-3′ | 5′-GCTGGCTGTCTCCGGGTTCC-3′ |
| GAPDH | 5′-CAACGGATTTGGTCGTATTGGGC-3′ | 5′-CGTTCTCAGCCTTGACGGTG-3′ |
| MUSASHI1 | 5′-TCCTCCCCGAGCTTACAGCC-3′ | 5′-GAGCCTGTCCCTCGAACCAC-3′ |
| NCAM | 5′-CCTGATCAAGCAGGATGAC-3′ | 5′-CCAGGGACTTCAGCATGACG-3′ |
| NESTIN | 5′-CAGAGGTTGGAGGGCCGCTG-3′ | 5′-TTGAGGTGCGCCAGCTGCTG-3′ |
| PANL27 (Alicea et al., 2013) | 5′-CCATCCAGACTGAGGAAGACCCGGAAAC-3′ | 5′-GGGCAGAAGCTCTGGTTCCTC-3′ |
| PAX6 | 5′-CCCCAGCCAGAGCCAGCATG-3′ | 5′-CAGTGGCCGCCCGTTGACAA-3′ |
| SOX2 | 5′-CAAAAGGGGGAAAGTAGTTTGCTGC-3′ | 5′-CTGCCGCCGCCGATGATTG-3′ |
Antibodies and Immunofluorescence Procedures
The following antibodies were used for the characterization of hESCs and their neural/neuronal derivatives: (1) pluripotency markers: rabbit anti-Nanog, rabbit anti-Lin28, and goat anti-Oct4 (all antibodies at 1:500 and from Santa Cruz Biotechnology, Santa Cruz, CA); (2) NPC/neuronal markers: mouse anti-Nestin (1:500; EMD Millipore Corporation, Billerica, MA), rabbit anti-Msi1 (1:500; EMD Millipore Corporation), mouse anti-neuron-specific class III tubulin, βIII-tubulin (TUJ1) (1:4000; Santa Cruz Biotechnology), and mouse anti-microtubule-associated protein 2 (MAP2) (1:500; EMD Millipore Corporation). Cell cultures were processed for immunofluorescence as previously described (Senut et al., 2000). Briefly, after fixation for 10 min in 4% paraformaldehyde, cells were rinsed twice in PBS, blocked in 3% donkey serum (Sigma-Aldrich) in 0.3% Triton-X-100 (NDST3%), and incubated overnight at 4°C with primary antibodies in NDST1%. Following rinses in NDST1%, cells were incubated with secondary anti-mouse, anti-rabbit, or anti-goat antibodies conjugated to Alexa 488 (1:1000; Life Technologies Corporation), or Fast Red (1:500; Jackson ImmunoResearch Laboratories Inc., West Grove, PA) for 2 h, stained with 4’-6-diaminido-2-phenylindole (DAPI) or Hoescht dye (Sigma-Aldrich) for nuclear staining, and extensively rinsed in PBS. Specificity of the immunostaining was assessed by replacing the primary antibodies with normal serum and by omitting one immunoreagent of the immunostaining protocol. Immunostained cell culture plates were subsequently stored in the dark at 4°C until analysis.
Quantification of Nestin- and TUJ1-positive Cells
The percentage of Nestin- and TUJ1-positive cells derived from control and Pb-exposed hESCs was quantified at day 19 or day 31 of the neuronal differentiation protocol, respectively. For each cell culture well, five randomly chosen fields (X200) were captured with a CoolSNAP-Pro monochrome digital camera adapted onto a Nikon TE 2000-U inverted stage fluorescence microscope (Nikon Instruments Inc.). Exposure parameters were primarily adjusted and kept constant throughout the image collecting process. Quantifications were performed blindly to experimental conditions. For each image, the number of Nestin- or TUJ1-positive cells, as well as Hoescht/DAPI-stained nuclei, was counted. The total number of immunostained cells was then expressed as a percentage of the total number of cells, as evaluated by DAPI or Hoescht staining.
Morphological Analysis of TUJ1-Positive Cells
The morphological complexity of TUJ1-positive cells derived from control and Pb-exposed hESCs was determined using Sholl analysis (Gensel et al., 2010; Sholl, 1953). As for cell counts, all morphological analyses were conducted blindly to the experimental conditions. In each well, neurons were randomly selected and only analyzed if their neurites could be clearly identified and distinguished from adjacent cells. An average of 40–45 neurons, pooled from three independent experiments, was analyzed per each experimental condition. A template of 12.5-μm-spaced concentric circles was superimposed onto the image of each selected neuron and centered on its soma. For each neuron, we recorded: (1) the total number of times neurites intersected any given circle (sum of crossings); (2) the distance from the soma of the maximal circle crossed by a neurite (maximal distance); and (3) the total number of branchings.
Statistical Analysis
The degree of statistical significance between experimental groups was determined by one-way analysis of variance (ANOVA) followed by a post hoc Holm-Sidak test when the F value was significant, or Kruskal-Wallis one way ANOVA on ranks followed by Dunn's test using the SigmaPlot 12 software (SYSTAT Software Inc., San Jose, CA). Statistical significance was defined at p ≤ 0.05. Data from qRT-PCR experiments are expressed as means with 95% confidence intervals. All other data are expressed as the mean ± SEM.
DNA Methylation Studies and Statistical Analysis
Undifferentiated (n = 2; p28 and p30) (paradigm A in Fig. 1A) and differentiating (n = 2; p33 and p34) (paradigm B in Fig. 1A) hESCs were exposed to Pb for 24 h. In addition, hESCs (n = 3; p26, p27, and p27) were chronically exposed to Pb throughout the neural differentiation process (NPC0–19; paradigm C in Fig. 1A). At the end of Pb exposure, DNA was isolated from control, Pb-exposed hESCs, and hESC-derived NPCs with Qiagen DNA kit. The Quantifiler Human DNA Quantification Kit (Applied Biosystems) was used to determine the amount of amplifiable DNA. For genome-wide methylation analysis, we used the Infinium Human Methylation 450K BeadChip system (Illumina Inc., San Diego, CA) that allows us to quantitatively measure methylation at over 450,000 CpG dinucleotides in over 20,000 human genes, with ∼25 probes per gene. The assay covers 99% of National Center for Biotechnology Information Reference Sequence genes and 96% of CpG islands, which are regions of the genome that are enriched in CpG dinucleotides and often occur in the promoter regions of genes. Several studies have indicated that the Infinium 1 probes have a more stable signal and extended dynamic range than the Infinium 2 probes, which differ in that Infinium 1 probes have separate beads for methylated and unmethylated CpG sites whereas Infinium 2 probes consist of single beads that can detect both types of DNA (Dedeurwaerder et al., 2011; Pidsley et al., 2013; Sandoval et al., 2011; Touleimat and Tost, 2012; Wang et al., 2012). Therefore, we applied the Peak-based approach (Dedeurwaerder et al., 2011), which corrects for the Infinium 1/Infinium 2 shift by rescaling the Infinium 2 data on the basis of the Infinium 1 data density distribution. The raw data were retrieved from Genome Studio methylation module version 1.8 in the form of a complete sample methylation profile with background correction. Probe bias correction for beta values was carried out using built-in-peak-based correction and quantile normalization function in the Illumina Methylation Analyzer (Wang et al., 2012). For undifferentiated hESCs, differentiating hESCs, and NPC0–19, the calculated Pearson's correlation coefficient demonstrated high correlation of beta values between the different passages used (Supplementary table 1), justifying their use as biological replicates.
We analyzed the normalized data using the recently published Adjacent site (A)-clustering algorithm (Sofer et al., 2013). A-clustering is based on the assumption that CpG sites or probes located close to each other on the same chromosome are more likely to undergo concerted changes in methylation in response to exposure. Therefore this method groups the CpG sites into clusters based on the distance measured between methylation sites, where distance (site i, site j) = 1-correlation (site i, site j), independent of any exposure data. First, CpG sites are clustered based on the distance measure and base pair distance from each other (d-base pair merge or dbp merge). Therefore, noncorrelated CpG sites will be included in a cluster, if they are wedged between well-correlated CpG sites. Then, the distance between the CpG sites in adjacent clusters must be lesser than a predefined distance measure threshold. This will merge correlated CpG sites in adjacent clusters (≤ 1000 bps from each other on the same chromosome) to create larger clusters. To determine methylated and nonmethylated clusters, we used the optimal parameters previously described in Sofer et al. (2013), such as Spearman correlation, average clustering, distance cutoff ( = 0.25 or 25%), maximum distance for merging adjacent cluster (1000 bps), minimum cluster size of 2, paired with 999 bps dbp-merge initiation. Then, we tested the changes in methylation across the range of Pb concentrations using generalized estimating equation model (GEE; Hanley et al., 2003; Sofer et al., 2013) that calculates the average beta values for the clusters based on the weighted average of correlated observations. Firstly, because weighted average takes into consideration the weight, standard deviation, and correlation of beta values of CpG sites belonging to a cluster, it efficiently controls for outliers. Secondly, the GEE model assumes that because of the correlation structure of the data the exposure effect on all sites in a cluster is equal. The p-value estimates for the differentially methylated clusters (DMCs) from the GEE model were corrected for multiple testing using false-discovery rate (FDR). The sites were further subdivided by exposure effect size ≥0.05 or ≤−0.05 and FDR-corrected p value ≤0.05 to generate the list of significant clusters. We defined three categories of DMCs: (1) small DMCs with two CpG sites/clusters; (2) DMCs with 3–10 CpG sites/clusters; and (3) large clusters with ≥ 10 CpG sites. We annotated the significant clusters using the annotation information provided by Illumina and isolated the unique hypermethylated (+ exposure effect size) and hypomethylated (− exposure effect size) clusters mapping to genes. We performed over representation analysis (ORA) using hypergeometric testing implemented as a part of GOstat to test for association of genes with significant biological processes (Beissbarth and Speed, 2004). We isolated the significant pathway using an FDR-corrected p-value cutoff for ORA of 0.01. All analysis was done on an R platform (R > 3.0).
RESULTS
Generation and Characterization of hESC-Derived NPCs and Neurons
The procedure used to generate NPCs and neurons from hESCs is outlined in Figure 1(A). Colonies of undifferentiated hESCs (Fig. 1B) exhibited robust immunofluorescent staining for the pluripotency markers Oct4 (Fig. 1C), Nanog (Fig. 1D), and Lin28 (Fig. 1E). Two to three days following mechanical dissociation, hESCs formed spherical aggregates or EBs that continued to grow in the presence of RA and FGF-2 (Fig. 1F). Within 4–6 days of plating on fibronectin-coated plates, cells grew out of the attached EBs and organized into neural tube-like rosettes, indicating neural commitment (Fig. 1G). By culturing cells dissociated from these neural rosettes, we obtained NPCs, as evidenced by the robust staining for the NPC markers, Nestin (98.3±0.6%; Fig. 1H) and Msi1 (98.7±0.6%; Fig. 1H) observed at day 19 of the differentiation protocol. To verify that they could differentiate into neurons, hESC-derived NPCs were subsequently passaged and plated on polyornithine/laminin-coated plates in FGF-2 free medium supplemented with a cocktail of neuronal-inducing trophic factors. After 10–12 days in these culture conditions (days 29–31 of differentiation), 8.2±2% of the cells exhibited immunostaining for the early neuronal marker βIII-tubulin (TUJ1). The number of TUJ1-positive cells increased to 49.1±9.34% by day 40 of the differentiation procedure (Fig. 1I), at which time cells immunoreactive for the more mature neuronal marker MAP2 could be observed (Fig. 1J). Glial cells were rarely present at this time point.
Pb Exposure Does Not Significantly Affect the Lineage Differentiation Capabilities of hESCs
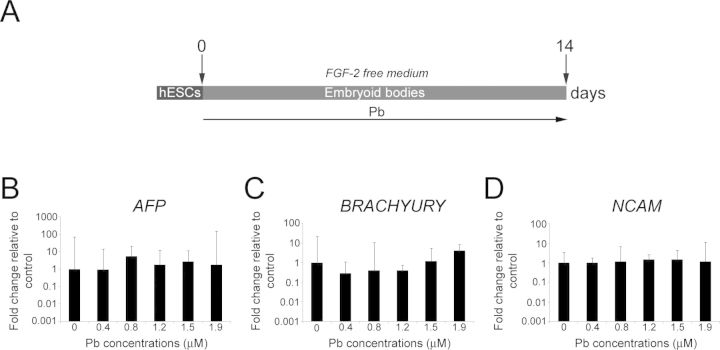
A hallmark of hESCs is their capability to differentiate into all three embryonic germ layers: mesodermal, ectodermal, and endodermal (Thomson et al., 1998). Because we plan to examine the effects of Pb exposure on the neural/neuronal differentiation of NPCs, we first verified that Pb exposure would not prevent the differentiation of hESCs, or direct them to preferentially differentiate toward a lineage other than ectodermal. To this end, we spontaneously differentiated EBs in FGF-2-free hESC medium for 14 days (Ross et al., 2010; Fig. 2A), at which time we performed qRT-PCR analyses of endodermal (AFP), mesodermal (BRACHYURY), and ectodermal (NCAM) marker genes. Exposure to 0.4–1.9μM Pb throughout the differentiation protocol neither modified the capability of hESCs to generate EBs nor altered their potentials to express markers of all three germ layers (Figs. 2B–D). Furthermore, no statistically significant differences in the expression levels of AFP (p = 0.519), BRACHYURY (p = 0.07), and NCAM (p = 0.918) were observed between control and Pb-exposed EBs, suggesting that, at the concentrations tested, Pb exposure does not markedly alter the in vitro pluripotency capabilities of hESCs (Figs. 2B–D).
FIG. 2.
Pb exposure does not affect the potentials of hESCs to passively differentiate into cells of the three germ layers. (A) Schematic representation of the differentiation protocol and Pb exposure paradigm. (B) Quantitative RT-PCR expression analysis for endodermal (AFP), (C) mesodermal (BRACHYURY), and (D) ectodermal (NCAM) lineage markers in Pb-exposed hESCs after 2 weeks of differentiation in FGF-2-free medium. Embryoid bodies generated from control and Pb-exposed hESCs express markers of different germ layers. Histogram values represent means and error bars represent 95% interval confidences (p > 0.05; n = 3, one-way ANOVA).
FIG. 3.
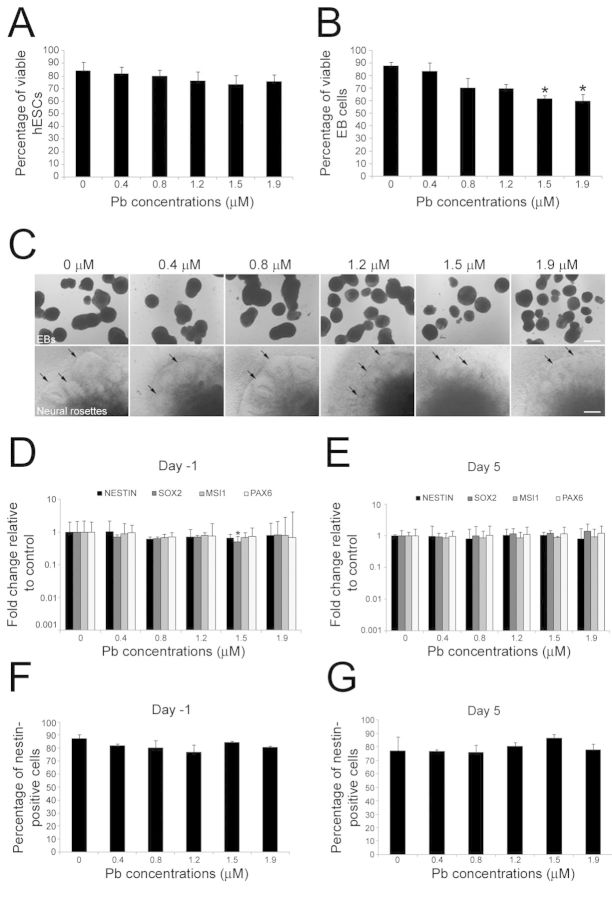
Effects of acute exposure to Pb on the neural differentiation of hESCs. (A) and (B): Analysis of cell viability by Trypan blue staining. Undifferentiated (A) and differentiating(B) hESCs were exposed to 0–1.9μM Pb for 24 h (paradigms A and B in Fig. 1A). The percentage of viable cells was calculated as follows: (total number of cells-number of nonviable dark-blue stained cells)/(total number of cells) x 100. Histogram values are means ± SEM. *Statistically significant difference (p < 0.05; n = 3, one-way ANOVA) between Pb-exposed and control hESCs. (C) Formation of embryoid bodies (EBs) and neural rosettes (arrows) from hESCs is not prevented by acute Pb exposure. (D) and (E): Quantitative RT-PCR expression analysis for the NPC marker genes NESTIN, SOX2, MSI1, and PAX6 in hESC-derived cells at day 19 of the differentiation protocol, following 24-h Pb exposure at day -1 (D) or day 5 (E). Histogram values represent means and error bars represent 95% interval confidences. *Statistically significant difference (p < 0.05; n = 3, one-way ANOVA) between Pb-exposed and control hESCs. The percentage of Nestin-positive cells generated from hESCs exposed to Pb either at day -1 (F) or day 5 (G) of the differentiation protocol was not significantly different from control percentages. Histogram values represent means ± SEM (n = 3; one-way ANOVA). Bar: 150 μm.
Acute Exposure of hESCs to Pb Does Not Prevent the Generation of NPCs
Although Pb exposure has been shown to have substantial effects on the survival and the differentiation of NPCs in animal models (see references in the Introduction section), we do not know whether it influences the directed differentiation of hESCs into the neural lineage. To determine the early effects of Pb on neural differentiation, we first examined the neural differentiation of hESCs acutely exposed to Pb (24 h) at either day -1 or day 5 of the differentiation protocol (paradigms A and B in Fig. 1A). Analysis of cytotoxicity, as assessed by Trypan blue staining at the end of the 24-h treatment, showed that there was no significant change in the viability of hESCs when they were exposed to either vehicle or Pb (0.4–1.9μM) on day -1 (p = 0.661) (Fig. 3A). In contrast, when Pb exposure was performed at day 5 of the differentiation protocol (paradigm B in Fig. 1A), a statistically significant 30.7% and 31.8% decrease in the number of viable hESCs was observed with Pb concentrations of, respectively, 1.5μM (p = 0.041) and 1.9μM (p = 0.028) compared with controls (Fig. 3B). MTT analysis of mitochondrial activity did not show changes in cell viability between control and Pb-exposed for paradigm A (p = 0.479; Supplementary fig. 1A) and paradigm B (p = 0.626; Supplementary fig. 1B). In addition, no differences in cell viability between water vehicle and a control group consisting in a molar concentration of sodium acetate solution equivalent to the 1.9 μg/dL Pb acetate were observed in the Trypan blue (p = 0.668) (not shown) or MTT assays (p > 0.05; Supplementary fig. 1). As shown in Figure 3(C), formation of EBs or neural rosettes from viable hESCs was not prevented by acute Pb exposure.
Analyses of neural marker RNAs at day 19 indicated that the expression levels for NESTIN, MSI1, SOX2, and PAX6 were similar between control hESCs and hESCs that had been acutely exposed to Pb at day -1 (paradigm A in Fig. 1A) although SOX2 expression was reduced twofold at 1.5μM of Pb (p = 0.048; Fig. 3D). No differences in the expression levels of the neural markers analyzed were observed at day 19 (Fig. 3E), when hESCs had been exposed to Pb on day 5 (Paradigm B in Fig. 1A). Quantification of Nestin-immunoreactive cells in cell cultures derived from control and Pb-treated hESCs further confirmed the presence of NPCs in our cultures, but did not reveal any differences in the percentage of positive cells between Pb and control experimental conditions (Figs. 3F and G). Taken together, these data suggest that, at the concentrations tested, acute exposure of hESCs to Pb a day prior initiation of neural differentiation or a day after initiation of neural induction does not significantly hinder their neural differentiation.
Chronic Exposure of hESCs to Pb Decreases the Expression Levels of Neural Markers in Their Derived NPCs
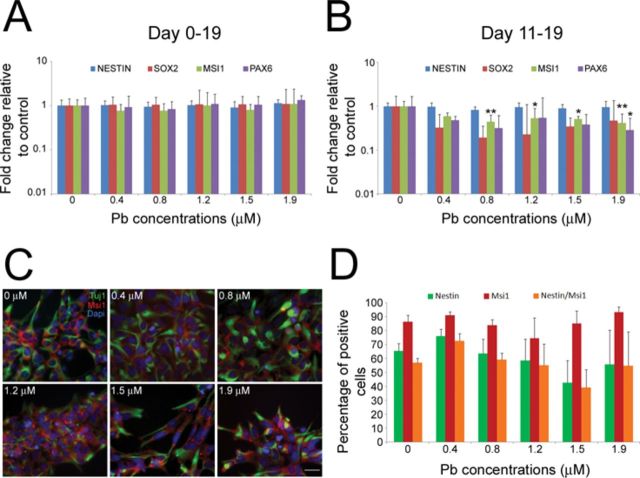
We next sought to analyze the effects that chronic exposure to Pb has on the neural differentiation of hESCs. For this purpose, 0.4–1.9μM Pb was added to the culture medium of hESCs throughout the entire differentiation protocol (day 0 to day 19; paradigm C in Fig. 1A). As shown in Figure 4(A), there were no significant differences in the expression levels of NESTIN (p = 0.144), SOX2 (p = 0.996), MSI1 (p = 0.154), and PAX6 (p = 0.964) between control and Pb-exposed hESCs. In addition, no significant changes in the number of NPCs, as assessed by Nestin immunoreactivity, were observed (data not shown). Because the organization of EBs into neural rosettes, a stage that parallels neuroectodermal development in humans (Krencik and Zhang, 2006), is a crucial step for neural commitment, we also investigated the neural differentiation capabilities of hESCs exposed to Pb during the stage of neural rosette formation (day 11 to day 19; paradigm D in Fig. 1A). Surprisingly, qRT-PCR analysis of the cells derived from control and Pb-treated hESCs revealed a statistically significant decrease in the expression levels of MSI1 and PAX6 (Fig. 4B). Specifically, a 56% (p = 0.004), 47% (p = 0.043), 49% (p = 0.024), and 50% (p = 0.002) decrease in MSI1 expression was observed following exposure to, respectively, 0.8, 1.2, 1.5, and 1.9μM Pb (Fig. 4B). Furthermore, a 72% significant decrease (p = 0.026) in PAX6 expression levels was also observed following exposure to 1.9μM Pb (Fig. 4B). Pearson correlation analysis showed a trend toward significant negative correlation between Pb concentrations and expression of MSI1 (R = −0.0753, p = 0.08) and PAX6 R = −0.778; p = 0.07). Statistically significant decreases in MSI1 and PAX6 expression levels were also observed between Pb-exposed hESCs and hESCs treated with a control sodium acetate solution (Supplementary fig. 2). To determine if changes in the number of generated NPCs could have contributed to the decrease in neural marker expression, we analyzed and compared the percentage of Nestin- and Msi1-positive cells in the cell cultures (Fig. 4C). Of the cells derived from control hESCs, about 65.6±5.2% were Nestin-positive, 86.5±4.5% were Msi1-positive, and 56.9±3.2% were stained for both Nestin and Msi1 (Fig. 4D). No statistically significant differences in the percentage of Nestin- (p = 0.712), Msi1- (p = 0.615), and double- (p = 0.666) positive cells were observed between experimental groups (Fig. 4D). Taken together, these data suggest that differentiating hESCs are more sensitive to the effects of Pb when exposed at a time of neural commitment.
FIG. 4.
Chronic exposure to Pb alters the expression levels of the neural markers MSI1 and PAX6 in hESC-derived NPCs. Quantitative RT-PCR expression analysis for the NPC marker genes NESTIN, SOX2, MSI1, and PAX6, following chronic Pb exposure at days 0–19 (A) or days 11–19 (B) of the differentiation procedure (paradigms C and D in Fig. 1A). Histogram values represent means and error bars represent 95% interval confidences. Statistically significant differences (*p < 0.05; **p < 0.01; n = 5, one-way ANOVA) between Pb-exposed hESCs and controls. (C) Immunofluorescence staining for the NPC marker Nestin (green) and Msi1 (red) in NPCs derived from hESCs exposed to 0–1.9μM Pb between day 11 and day 19 of the differentiation protocol (paradigm D in Fig. 1A). (D) The percentage of Nestin-, Msi1-, and double-stained cells obtained from hESCs differentiated in 0.4–1.9μM Pb between day 11 and day 19 of the differentiation protocol did not differ from control percentages. Histogram values represent means ± SEM (n = 3, one-way ANOVA). Bar: 20 μm.
Neuronal Differentiation of NPCs Derived From hESCs Chronically Exposed to Pb Is Altered
After showing that chronic Pb exposure of differentiating hESCs may cause a decrease in the expression of NPC marker genes, we investigated whether the generating NPCs could exhibit changes in their efficiency to differentiate into neurons and/or generate neurons with altered morphology. Therefore, NPCs obtained from control hESCs and chronically Pb-exposed hESCs (day 0–19: NPC0–19 and day 11–19: NPC11–19; paradigms C and D in Fig. 1A) were cultured in neuronal differentiation conditions for 12 days (day 31 of the differentiation process), at which time we performed neuronal quantification and morphometric analyses. Although 12-day cultures do not yet contain a large number of neurons, the minimal overlap between neurons at this stage allows for the clear identification of neuritic processes which is needed for morphometric studies. Immunofluorescent analyses of the early neuronal marker TUJ1 demonstrated a statistically significant 2.9-fold increase (p = 0.05) in the number of generated neurons between NPC0–19 and control NPCs for the highest concentration of 1.9μM Pb (Fig. 5A). In contrast, no significant changes in the number of TUJ1-positive cells were observed between control NPCs and NPC11–19 (p = 0.944; Fig. 5B). Closer examination of TUJ1-positive neurons derived from the different experimental groups of NPCs revealed some morphological differences (Figs. 5C–E). Neurons differentiated from control NPCs were mostly multipolar, showing several neurites surrounding the soma and an axon-like longer process (Fig. 5C; Supplementary fig. 3). The morphology of neurons derived from NPC0–19 was overall similar to that observed in control neurons, with the exception that, at the highest Pb concentrations, the axon-like neurite extended over markedly longer distances in a subgroup of TUJ1-positive cells (Fig. 5D; Supplementary fig. 3). In contrast, at those same Pb concentrations, neurons resulting from NPC11–19 displayed less neuritic development, often adopting a unipolar shape (Fig. 5E; Supplementary fig. 3).
FIG. 5.
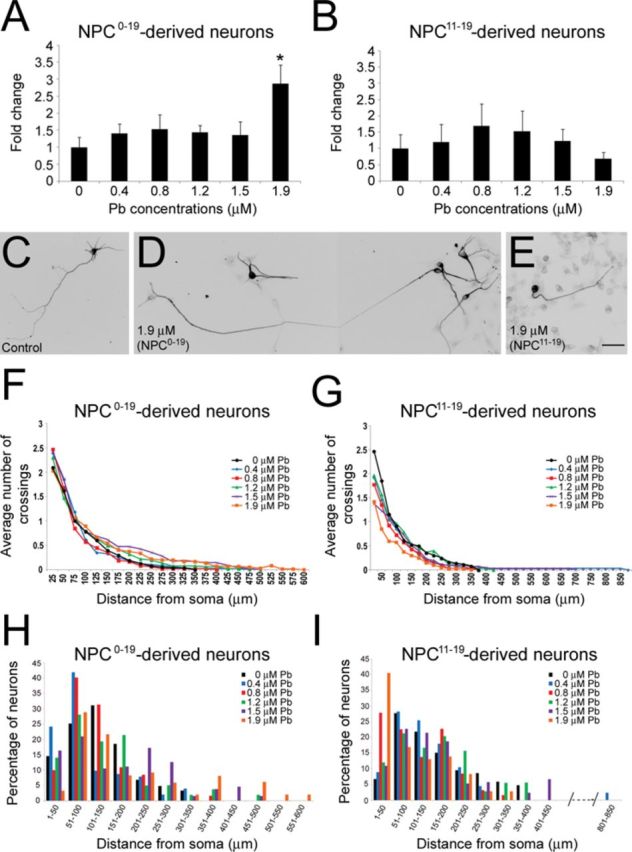
Effects of chronic exposure to Pb on the differentiation of NPCs into neurons. The number of TUJ1-positive neurons derived from control NPCs: NPC0–19 (A) and NPC11–19 (B) (paradigms C and D in Fig. 1A). Following exposure to 1.9μM Pb, the number of TUJ1-stained cells was significantly increased in neurons differentiated from NPC0–19. Histogram values represent means ± SEM. *Statistically significant difference (p = 0.05; n = 3, one-way ANOVA) between Pb-exposed and control experimental groups. Examples of TUJ1-positive neurons generated from control NPCs (C), NPC0–19 [1.9μM Pb, composite micrograph, (D)], and NPC11–19 [1.9μM Pb, (E)] at 31 days of the differentiation protocol. (F)–(I): Neurons derived from control, NPC0–19 and NPC11–19 exhibited similar overall neuritic complexity, although an increase in the percentage of neurons with longer axon-like processes was observed in the NPC0–19 group. Sholl analysis of the average number of crossings in neurons derived from control NPCs: NPC0–19 (F) and NPC11–19 (G) (n = 3). Histogram illustrating the percentage of control NPCs (H, I): NPC0–19 (H) and NPC11–19 (I) derived neurons with various lengths of axon-like processes. Bar: 30 μm.
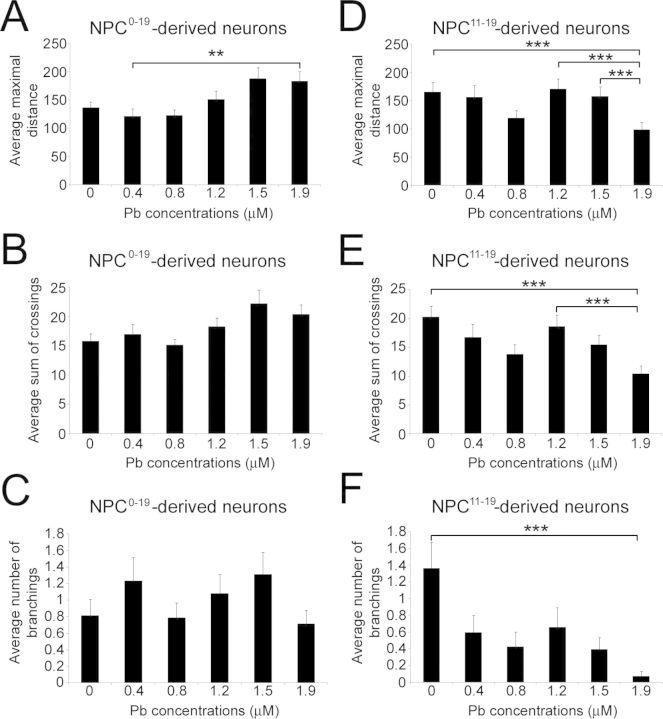
To further characterize the magnitude of the morphological changes observed between the different experimental groups, we conducted quantitative analyses of neuritic development and complexity using Sholl analysis (Figs. 5 and 6). As shown in Figures 5(F) and 5(G), neurons derived from NPC0–19 and NP11–19 exhibited similar overall neuritic complexity compared with control NPCs (0μM Pb). However, at the highest Pb concentration of 1.9μM, there was a notable increase in the percentage of NPC0–19-derived neurons with longer axon-like processes (Fig. 5H), which was not observed with NPC11–19-generated neurons (Fig. 5I). Sholl analysis of the average maximal distance reached by neurites in control and NPC0–19-derived neurons (Fig. 6A) resulted in a J-shaped dose-response curve because of a decrease in neuritic length at 0.4 and 0.8μM Pb, but an increase in neuritic length at 1.2, 1.5, and 1.9μM Pb. Statistical analysis showed a significant difference between the different median values (p = 0.007). Post hoc Dunn test identified a significant difference between the 0.4 and 1.9μM experimental groups. No significant changes in the average total number of crossings (p = 0.197; Fig. 6B) or the average number of branching (p = 0.53; Fig. 6C) were observed between NPC0–19-derived and control neurons. In contrast, Sholl analysis of neurons derived from NPC11–19 revealed a statistically significant (p ≤ 0.001) reduction in the average maximal neuritic length (Fig. 6D), average sum of crossings (Fig. 6E), and average number of branchings (Fig. 6F). Taken together, these data suggest that NPCs derived from hESCs chronically exposed to Pb display alterations in their neuronal differentiation.
FIG. 6.
Neurons generated from control NPCs, NPC0–19 and NPC11–19, exhibit different morphologies at day 31 of the differentiation protocol. Histogram representing the average maximal distance reached by the longest neurites in neurons generated from NPC0–19 (A) and NPC11–19 (B) (paradigms C and D in Fig. 1A). Histogram representing the average sum of crossings in neurons generated from NPC0–19 (C) and NPC11–1 9 (D). Histogram representing the average number of branching in neurons generated from NPC0–19 (E) and NPC11–19 (F). Histogram values represent means ± SEM. Statistically significant difference (**p < 0.01; ***p ≤ 0.001; n = 3; one-way ANOVA) between Pb exposure and control conditions.
Pb Induces Rapid Changes in the Global DNA Methylation Patterns of Undifferentiated and Differentiating hESCs
Pb-induced changes in DNA methylation may play a role in the etiology of its developmental neurotoxicity (Bihaqi and Zawia, 2012; Bihaqi et al., 2011, 2013; Dosunmu et al., 2012; Pilsner et al., 2009; Schneider et al., 2013; Wu et al., 2008a). Therefore, to determine whether and how fast Pb alters the methylation status of brain-related genes that might be relevant to the Pb-induced morphological effects we observed in neurons, we analyzed the methylation profile of undifferentiated and differentiating hESCs acutely exposed to Pb (paradigms A and B in Fig. 1A) and NPCs derived from hESCs chronically exposed to Pb throughout the neural differentiation procedure (NPC0–19; paradigm C in Fig. 1A) using the Infinium HumanMethylation450K array (GEO accession number GSE54596).
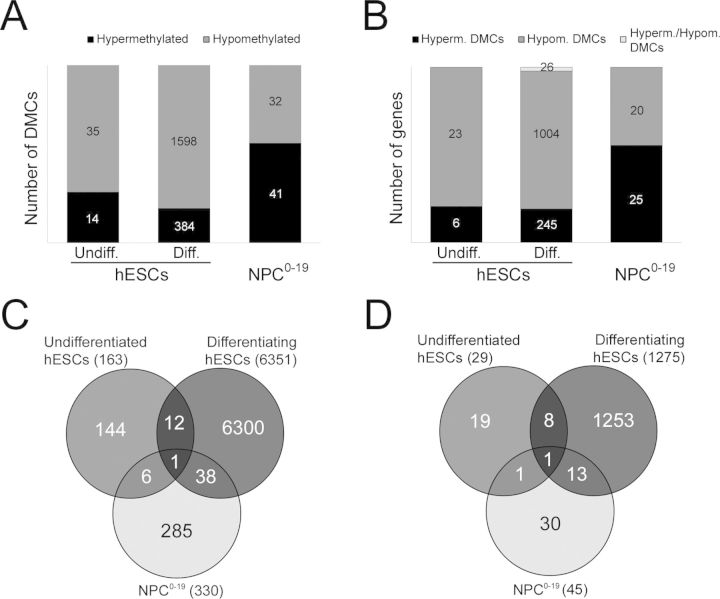
Because methylation changes of a single CpG dinucleotide in a specific gene region are unlikely to affect gene transcription, we identified all adjacent CpG sites by chromosomes and clustered them based on the correlation between their beta values and base pair distance from each other using the A-clustering approach. GEE analysis of linearly Pb-dependent methylation changes allowed us to isolate DMCs that exhibited statistically significant (p ≤ 0.05; FDR corrected) positive or negative exposure, which we subsequently subdivided into hypermethylated and hypomethylated groups. As shown in Table 2, a large number of DMCs were identified in acutely exposed undifferentiated and differentiating hESCs, and to a lesser degree in chronically exposed NPC0–19. Following FDR correction and filtering for exposure effect (≥0.05 or ≤−0.05), we identified 49 DMCs mapping to 29 genes for undifferentiated hESCs (Table 2; Figs. 7A and B). About 30% of the CpG sites within DMCs were located in promoter-associated regions and 80.4% localized to CpG resort (CpG islands and the surrounding shores and shelves). Compared with undifferentiated hESCs, differentiating hESCs exhibited a greater number of DMCs (n = 1982) that mapped to 1275 genes (Table 2; Figs. 7A and B), suggesting that differentiating hESCs are more vulnerable to methylation changes in response to acute Pb exposure. It also showed a decrease in percentage of CpG sites mapping to CpG resort (59.6%) and CpG sites mapping to promoters (11.7%) for all identified DMCs. Analysis of NPC0–19 generated from chronically exposed hESCs revealed significant methylation changes in 73 clusters mapping to 45 unique genes, with 83.9% of the CpG sites locating in CpG resorts and 31.5% of the CpG sites associating with promoters (Table 2; Figs. 7A and B). Interestingly, NPC0–19 displayed a larger percentage (13.7%) of DMCs with ≥ 10 CpG compared with undifferentiated (4.1%) and differentiating (1.9%) hESCs (Table 2). Analysis of the DMC-related CpG sites in the three experimental groups showed that a single CpG site—cg04778274—mapping to a CpG island in the LGI4 (Leucine-Rich Repeat, member 4) gene was common to all three groups and consistently showed Pb-induced hypomethylation (Fig. 7C; Group A in Supplementary table 2). In addition, we identified 39 CpG sites shared between differentiating hESCs and NPC0–19 (Fig. 7C; Group B in Supplementary table 2), 13 CpG sites common to undifferentiated and differentiating hESCs (Fig. 7C; Group C in Supplementary table 2), and 7 CpG sites common to undifferentiated hESCs and NPC0–19 (Fig. 7C; Group D in Supplementary table 2). The respective genomic regions are detailed in Supplementary table 3.
TABLE 2. A-Clustering Analysis of Pb-Induced Methylation Changes in Undifferentiated hESCs, Differentiating hESCs, and NPC0–19 (Paradigms A, B, and C in Fig. 1A). Table Gives the Summary of the Methylation Profiles for Each Experimental Group After Determining Differentially Methylated Clusters (DMCs) Using GEE (See the Materials and Methods section).
| Undifferentiated hESCs | Differentiating hESCs | NPC0–19 | |
|---|---|---|---|
| Number of clusters (before filtering) | 12785 | 11519 | 7167 |
| Significant clusters (after filtering) | 49 | 1982 | 73 |
| Total number of CpG sites mapped to DMCs | 163 | 6351 | 330 |
| Unique genes associated with DMCs | 29 | 1275 | 45 |
| DMCs with 2 CpG sites (%) | 59.2 | 53.6 | 41.1 |
| DMCs with 3 to 10 CpG sites (%) | 36.7 | 44.5 | 45.2 |
| DMCs with ≥ 10 CpG sites (%) | 4.1 | 1.9 | 13.7 |
| CpG sites associated with promoters (%) | 30.1 | 11.7 | 31.5 |
| CpG sites mapping to CpG resort (%) | 80.4 | 59.6 | 83.9 |
FIG. 7.
Differential methylation analysis following Pb exposure in undifferentiated hESCs, differentiating hESCs, and NPC0–19 (paradigms A, B, and C in Fig. 1A). CpG sites were assigned to clusters based on correlation of beta values and base pair distance from each other and the effects of exposure to Pb were tested using the generalized estimating equation (GEE) model. (A) The differentially methylated clusters (DMCs) were filtered based on estimated p value ≤ 0.05 (FDR corrected) and minimum exposure effect size ≥0.05 or 5% or ≤0.05 or −5% to obtain a list of significant clusters. The significant clusters were further classified into hypermethylated (+ exposure effect size) and hypomethylated (− exposure effect size) DMCs for undifferentiated hESCs and differentiating hESCs acutely exposed to Pb, and NPCs derived from hESCs chronically exposed to Pb. (B) The significant DMCs were mapped to genes, using the publicly available Human methylation 450K annotation information and classified into hypermethylated (+ exposure effect size) and hypomethylated (− exposure effect size) genes. Notice that some genes were mapped to both hypermethylated and hypomethylated clusters and were assigned to a separate group. (C) Venn diagram illustrating the overlap between the CpG sites/probes mapped to all significant DMCs irrespective of their exposure effect for the three experimental groups. (D) Venn diagram of the overlap between unique genes mapped to significant DMCs irrespective of their exposure effect for the three experimental groups.
A total of 29, 1275, and 45 genes eliciting dose-dependent methylation effects to Pb were identified in undifferentiated hESCs, differentiating hESCs, and NPC0–19, respectively (Table 2; Figs. 7B and D). Whereas 26 genes contained both hyper- and hypo-methylated DMCs in differentiating hESCs, no such genes could be found in undifferentiated hESCs or NPC0–19 (Fig. 7B). As shown in a Venn diagram (Fig. 7D), and in addition to the LGI4 gene common to all three experimental groups, we identified eight genes common to undifferentiated and differentiating hESCs, one gene common to undifferentiated hESCs and NPC0–19, and 13 genes common to differentiating hESCs and NPC0–19 (Supplementary table 4). Gene ontology (GO) analysis of the Pb-related differentially methylated genes in the differentiating hESCs revealed that, at the Pb concentrations tested, the top hit categories were “calcium ion import” (p = 0.0008) and “neuron projection development” (p = 0.009) for hypermethylated genes (Table 3), and “neurological system processes” (p = 0.0012), “calcium ion import” (p = 3.31E-05), and “actin cytoskeleton organization (p = 1.02E-05) for hypomethylated genes (Table 3). For example, genes key to neurogenesis such as PLXNA4 (Waimey et al., 2008) and NEUROG1 (Sun et al., 2001) (Table 4) displayed Pb-induced hypermethylation and hypomethylation, respectively (Supplementary table 4). The overrepresented GO categories are decided based on the FDR-corrected p values as well as their relevance to the study, i.e., they must be involved in either well-known signaling pathways within neuron or involved in maintaining neural differentiation or growth. Due to the small number of genes exhibiting significant Pb-induced methylation changes in undifferentiated hESCs and NPC0–19, GO analysis was not meaningful and we instead analyzed target genes individually. Interestingly, hypermethylation changes were observed in genes known to influence brain development and functions such as EFNA2 (Holmberg et al., 2005; Jiao et al., 2008), GRIK4 (Knight et al., 2012), and LXH3 (Lee and Pfaff, 2003) in NPC0–19 (Table 4). These genes also displayed Pb-induced hypomethylation in differentiating hESCs (Table 4; Supplementary table 4).
TABLE 3. GO (Gene Ontology) Analysis of Genes Exhibiting Linearly Pb-Induced Methylation Changes (Hypermethylation and Hypomethylation) in Differentiating hESCs (Paradigm B in Fig. 1A).
| Methylation status | GO category | p value |
|---|---|---|
| Hypermethylated | GO:0070509 Calcium ion import (4/32) | 0.0008 |
| Hypermethylated | GO:0090279 Regulation of calcium ion import (3/19) | 0.002 |
| Hypermethylated | GO:0048172 Regulation of short-term neuronal synaptic plasticity (2/7) | 0.0036 |
| Hypermethylated | GO:0031175 Neuron projection development (18/726) | 0.009 |
| Hypomethylated | GO:0030029 Actin filament-based process (50/488) | 8.9E-06 |
| Hypomethylated | GO:0030036 Actin cytoskeleton organization (46/437) | 1.02E-05 |
| Hypomethylated | GO:0070509 Calcium ion import (9/32) | 3.31E-05 |
| Hypomethylated | GO:0050877 Neurological system processes (93/1267) | 0.0012 |
TABLE 4. Examples of Genes Exhibiting Pb-Induced Methylation Changes (GEE Analysis, See Methods) in Differentiating (Diff.) hESCs and NPC0–19 (Paradigms B and C in Fig. 1A).
| Cells | Cluster | Pb effect size | Pb effect standard error | p value | p adjusted | CpG sites/clusters | CpG sites | Genes | Methylation type | Gene location |
|---|---|---|---|---|---|---|---|---|---|---|
| Diff. hESCs | 3227 | −0.052 | 0.017 | 0.003 | 0.0127 | 5 | cg17830308 | NEUROG1 | chr5:134870740–134872051 | TSS1500 |
| cg11863728 | ||||||||||
| cg08451582 | ||||||||||
| cg02604503 | ||||||||||
| cg13336665 | ||||||||||
| Diff. hESCs | 5099 | 0.083 | 0.020 | 3.32E-05 | 0.0003 | 2 | cg24469977 | PLXNA4 | non CpG | TSS200, 1st Exon |
| cg24091954 | ||||||||||
| Diff. hESCs | 5842 | −0.088 | 0.02 | 9.94E-06 | 8.59E-05 | 3 | cg15833797 | LHX3 | chr9:139090420–139095472 | Body |
| cg21172615 | ||||||||||
| cg08967938 | ||||||||||
| Diff. hESCs | 10227 | −0.136 | 0.028 | 1.54E-06 | 1.58E-05 | 2 | cg15490703 | EFNA2 | chr19:1295513–1295891 | Body |
| cg22676470 | ||||||||||
| Diff. hESCs | 7121 | −0.100 | 0.040 | 0.013 | 0.040498 | 5 | cg25113360 | GRIK4 | non CpG | TSS1500TSS200 |
| cg24226238 | ||||||||||
| cg27179693 | ||||||||||
| cg10961484 | ||||||||||
| cg25316569 | ||||||||||
| NPC0–19 | 6415 | 0.106 | 0.016 | 1.05E-10 | 5.8E-08 | 2 | cg15490703 | EFNA2 | chr19:1295513–1295891 | Body |
| cg22676470 | ||||||||||
| NPC0–19 | 4442 | 0.112 | 0.025 | 1.02E-05 | 0.0018 | 4 | cg18247436 | GRIK4 | chr11:120856725–120857174 | Body |
| cg15174564 | ||||||||||
| cg24255125 | ||||||||||
| cg12122877 | ||||||||||
| NPC0–19 | 3581 | 0.065 | 0.013 | 2.62E-07 | 0.00007 | 2 | cg00522018 | LHX3 | Chr9:139090420–139095472 | Body |
| cg14381623 |
DISCUSSION
Accumulating evidence that any level of Pb may have deleterious effects on the neurological outcome in exposed children (Kuehn, 2012; Bellinger, 2013) has prompted the need for a better understanding at a cellular level of the mechanisms of Pb toxicity in the developing human brain. The problem of shortage of available human cell lines and tissues has been revolutionized by the derivation of hESCs (Thomson et al., 1998). Using this well-defined cellular model, we show that the directed neural/neuronal differentiation of hESCs, although not prevented, is significantly altered by Pb exposure. We found that Pb had distinct effects depending on the differentiation stage at which hESCs were exposed. Thus, if Pb exposure occurred at the stage of neural rosette formation, it induced a decrease in the expression of the NPC marker genes MSI1 and PAX6 and led to the generation of neurons with shorter processes and reduced branching. In contrast, when Pb treatment started at the time of EB formation, it resulted in an increased number of neurons and a propensity to longer neurites at the highest concentration. Furthermore, our data show that Pb rapidly induces aberrant patterns of DNA methylation in genes key to brain development.
To the best of our knowledge, this is the first study characterizing the morphological and epigenetic effects of Pb exposure on the neuronal differentiation of hESCs. Hence, we corroborate earlier reports (Wobus and Loser, 2011) indicating that the neuronal differentiation capabilities of hESCs make them a suitable cell-based model to gain useful insights into the effects of heavy metals on neuronal development and their underlying mechanisms. An advantage in using hESCs compared with other (e.g., animal-derived) cellular systems is that they preclude species-specific differences in development and response to environmental toxicants, making them relevant for human safety issues (Wobus and Loser, 2011). The use of ESCs for exploring the developmental neurotoxicity of Pb, and heavy metals in general, is in its early stages and the reports so far available are focused mostly on mouse (Baek et al., 2011; Sanchez-Martin et al., 2013; Visan et al., 2012; Zimmer et al., 2011) and more recently human (Zimmer et al., 2012).
We chose to test concentrations of Pb ranging from 0.4 to 1.9μM (8–40 μg/dL) because we wanted to be close to the range of Pb concentrations detected in the blood of exposed children. The current CDC threshold for initiation of treatment in young children is 5 μg/dL (about 0.25μM) and the vast majority of exposed ≤3-year-old children exhibit BLLs between 5 and 14 μg/dL (0.25–0.7μM) (2011 data; www.cdc.gov/nceh/lead/data). Furthermore, at such BLLs, exposed children manifest measurable deficits in various cognitive and intellectual skills (Canfield et al., 2003; Lanphear et al., 2005; Nelson and Espy, 2009), that are of unknown mechanisms .
Here, we report that acute exposure to physiological levels of Pb does not affect the viability of undifferentiated hESCs, which concurs with previous reports with mouse ESCs (Visan et al., 2012) or human amniotic fluid-derived fetal stem cells (Gundacker et al., 2012). The lack of effects of acute Pb exposure on the undifferentiated hESCs could be due to their relative resistance to hypoxia and oxidative stress (Bolin et al., 2006). Indeed, exposure to Pb has been associated with increased lipid peroxidation, decreased expression, or changes in the activity of antioxidant enzymes, and reduction of nitric oxidative synthase, factors that contribute to increased oxidative stress and conditions mimicking hypoxia (see Nava-Ruiz et al., 2012 for review). Decreasing oxygen or nitric oxide levels have been shown to improve the proliferation of mouse ESCs (Tejedo et al., 2010). In contrast, differentiating hESCs exhibited a dose-dependent decrease in viability in response to acute Pb exposure, reaching a significant 31–32% decrease at 1.5–1.9μM Pb. This greater vulnerability of differentiating versus undifferentiated state of hESCs to acute Pb exposure is similar to a previous report demonstrating that whereas differentiating mouse ESCs viability was reduced by 15% at doses as low as 0.1μM Pb, a similar effect was only observed with undifferentiated cells at levels of about 70 μm Pb (Visan et al., 2012). Using the MTT assay, we did not detect any remarkable changes in the mitochondrial enzymatic activity of Pb-exposed hESCs. The discrepancy between the Trypan blue and MTT data for differentiating hESCs could be due to the difficulty to control cell numbers in growing EBs and/or interferences between MTT and Pb, because Pb has been shown to influence mitochondrial activity (Sanders et al., 2009). We also found that, at the concentrations tested, Pb does not modify the in vitro pluripotency potentials of exposed hESCs, because they could differentiate into cells that expressed markers of all three embryonic lineages. However, we cannot exclude that their further differentiation into lineage-specific cells may be altered, warranting further investigation.
In all exposure paradigms tested, Pb exposure did not prevent hESCs from generating true NPCs, as assessed by expression/immunostaining of the neural markers Msi1, Nestin, Sox2, and Pax6, and their capability to differentiate into TUJ1-positive neurons. However, and in contrast to acute treatments, we found that chronic Pb exposure induced a significant reduction in the expression levels of the neural marker genes MSI1 and PAX6, while maintaining the percentage of NPCs generated stable. These effects were observed only when hESCs had been exposed to Pb at the stage of neural rosettes formation (days 11–19), suggesting an increased sensitivity of the differentiating hESCs at this particular time window. Because both MSI1 and PAX6 genes are essential for neural stem/progenitor self-renewal, proliferation, and differentiation of the NPCs (Okano et al., 2005; Osumi et al., 2008), it is likely for some of these functions to be altered in NPC11–19. We were however surprised that a similar decline in neural marker expression did not occur in cells derived from hESCs treated with Pb throughout the differentiation protocol, which includes the neural rosette formation stage. It is possible that Pb exposure prior to this window of Pb sensitivity provides some form of protection to the differentiating hESCs. Although the mechanisms of such protection are unclear, they could be partially related to the capability of Pb to interfere with calcium (Ca2+)-sensitive pathways. Indeed, Pb can substitute to Ca2+ and regulate the activity of various key cellular enzymes, including protein kinase C (Lidsky and Schneider, 2003), a protein involved in neuroprotection (Maher, 2001). Furthermore, calcium preconditioning has recently been shown to confer protection to pig retinal ganglion cells neurons against glutamate-induced excitotoxicity (Brandt et al., 2011). Interestingly, preconditioning with low doses of the heavy metal cobalt chloride was shown to improve infarct size and functional deficits induced in the neonatal rat brain following hypoxia (Jones et al., 2008).
Our data show that NPC0–19 obtained under 1.9μM Pb generated almost three times more TUJ1-positive neurons than control NPCs. Furthermore, whereas the longest neurites of control neurons did not extend beyond a distance of 300–350 μm, about 18% of NPC0–19-derived neurons reached a maximal distance ranging from 400 to 600 μm. This increase in neuronal generation observed in NPC0–19-derived neurons suggests that prior treatment with the highest doses of Pb may have promoted neuronal differentiation and/or improved survival of the resulting neurons. These data are reminiscent of previous in vitro studies reporting that 3–30μM concentrations of Pb exhibited neurotrophic effects, increasing both neuronal numbers and neuritic length in cultures of developing rat primary dorsolateral cortical cells (Davidovics and DiCicco-Bloom, 2005). Similar neurotrophic effects of Pb on the generation of MAP2-positive neurons were recently reported following continuous direct exposure of mouse ESCs to Pb(II) acetate concentrations of 30–1000 μg/dL (Baek et al., 2011). However, in our study the “trophic” effects were a delayed rather than a direct response to Pb, indicating subtle but long-lasting effects of Pb.
Although we observed no remarkable changes in their overall number compared with control conditions, neurons generated from NPC11–19 clearly demonstrated a significant decrease in neuritic average maximal length, average number of crossings and branchings. We observed a significant decrease in the average maximal length of axon-like neurites in neurons derived from NPC11–19 generated under exposure to 1.9μM Pb. This could be related in part to the decreased expression of PAX6 observed at this concentration in NPC11–19, gene that was recently shown to increase axonal growth in cultures of mouse post-mitotic neurons (Sebastian-Serrano et al., 2012). The decreased MSI1 expression may also be involved in the changes in neuronal morphology observed in NPC11–19. Msi1 is an RNA-binding protein that is enriched in the embryonic and adult central nervous system (CNS) (Kaneko et al., 2000; Sakakibara et al., 1996) and binds to multiple targets, including Numb, a regulator dendritic branching, axonal growth and synaptogenesis (Franklin et al., 1999; Imai et al., 2001; Nishimura et al., 2006; Shen et al., 2002). Therefore, because Msi1 downregulates Numb expression, it is possible that in response to the reduction of Msi expression in NPC11–19, Numb expression is upregulated, subsequently leading to shorter processes. However, the fact that we did not observe a marked increase in axonal length, which is also induced by Numb via antagonization of the Notch signaling pathway (Franklin et al., 1999; Imai et al., 2001), suggests that other developmental mechanisms are involved. Further research is needed to better understand the involvement of MSI1 and PAX6 in the effects of Pb exposure on human neuronal growth.
Alterations in epigenetic marks help define the transcriptional pathways that contribute to the differentiation of hESCs into NPCs (Xie et al., 2013). DNA methylation regulates gene expression and is most commonly associated with gene silencing (Elango and Yi, 2011). As a first step in understanding the role of epigenetic determinants in Pb-induced neurotoxicity, we first analyzed the genome-wide methylation patterns of undifferentiated and differentiating hESCs exposed to Pb for 24 h (acute exposure) and in hESC-derived NPCs (chronic exposure). The rationale behind the acute exposure paradigms is that we wanted to determine how fast Pb exposure could trigger DNA methylation changes in hESCs and that the limited number of epidemiological studies looking at the effects of Pb exposure on the human epigenome relate to long-term exposure paradigms. The latter studies demonstrated that changes in DNA methylation may play a role in the etiology of Pb neurotoxicity (Schneider et al., 2013; Senut et al., 2012). Our observation of changes in the methylation status of numerous CpG sites occuring within 24 h of exposure shows that Pb can rapidly modify the epigenetic landscape of hESCs. This is consistent with the dynamism of epigenetic determinants and the rapid turnover of DNA methylation (Yamagata et al., 2012). Compared with undifferentiated hESCs and NPC0–19, Pb exposure dramatically increased the number of hypomethylated and hypermethylated DMCs in the differentiating hESCs. This not only reflects the active regulation of gene expression taking place during cellular differentiation (Meissner et al., 2008; Fisher and Fisher, 2011), but also suggests that the epigenome of hESCs at the early stages of neuronal induction is extremely susceptible to DNA methylation changes in response to acute Pb exposure. Furthermore, once engaged in the neural differentiation pathway, a majority of DMCs showed Pb-associated hypomethylation. This “hypomethylation” effect is in accordance with previous human epidemiological studies showing an inverse relationship between bone/BLLs and methylation of LINE-1 (Pilsner et al., 2009; Wright et al., 2008) or collagen type1 alpha2 (Hanna et al., 2012). Unexpectedly, we did not observe this “hypomethylation” effect in NPCs derived from hESCs chronically exposed to Pb throughout the entire neural differentiation protocol. Indeed, the observation that chronic exposure to Pb induced methylation changes at a much more reduced scale than that observed in differentiating hESCs may reflect an adaptation to prolonged Pb exposure, substantiating our observations of a lack of changes in neural gene expression in NPC0–19. In the three experiment groups analyzed, our DNA methylation studies did not show methylation changes for MSI1 and PAX6, which suggests that Pb-induced decreased expression of these genes in NPC11–19 may be mediated by mechanisms other than DNA methylation (i.e., histone modifications) (references in Senut et al., 2012) or other than epigenetics (post-transcriptional) (Mata et al., 2005). Additional studies are necessary to further characterize the methylation profiles of these genes in NPC11–19. Interestingly, genes that showed linear changes of methylation profile with increasing Pb concentrations included genes involved in neurogenesis, such as EFNA2 (Holmberg et al., 2005; Jiao et al., 2008), GRIK4 (Knight et al., 2012), and the motor neuron specification factor LHX3 (Lee and Pfaff, 2003), which were hypomethylated in differentiating hESCs and hypermethylated and NPC0–19. In addition, the axonal guidance PLXN4 (Waimey et al., 2008) and the neuronal differentiation factor NEUROG1 (Sun et al., 2001) were found to be hypermethylated and hypomethylated, respectively, in Pb-exposed differentiating hESCs. Interestingly, an increase in NEUROG1 expression was recently reported in mouse brains exposed to Pb during gestation and in neurons derived from Pb-treated mouse ESCs (Sanchez-Martin et al., 2013). Because methylation changes close to the transcription start site may impact gene expression, hypomethylation of NEUROG1 in Pb-exposed differentiating cells could be in part involved in the morphological effects we observed in the generated neurons. As a whole, these data show that acute and chronic Pb exposure of hESCs can epigenetically alter genes key to neuronal development.
In conclusion, our study demonstrates that Pb exposure during the neural differentiation of hESCs alters the number and morphology of generated neurons and induces aberrant patterns of DNA methylation of genes involved in neurodevelopmental pathways. Further studies are now in progress to determine whether genes eliciting Pb-induced changes in their methylation status exhibit changes in expression that may partly explain the morphological features observed in hESC-derived neurons following Pb exposure. Cross comparison with genetic databases for neurological disorders will allow us to identify key pathways affected by Pb and to better understand the mechanisms behind the neurological deficits observed in Pb-exposed children.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences (1R21ES021983 and ES012933 to D.M.R.); NIH Cancer Center Support Grant (NIH P30 CA22453).
Supplementary Material
Acknowledgments
The authors are grateful to Dr Jose Cibelli and all the members of the Cellular Reprogramming Laboratory (CRL) at Michigan State University for the use of CRL facilities, stem cell expertise, and insightful discussions. Particularly, we thank Dr Eun Ah Chang for expert training in stem cell cultures and Dr Steven Suhr for skillful assistance with primer design, qRT-PCR techniques, and helpful comments. Methylation data were deposited under the GEO accession number GSE54596.
REFERENCES
- Alicea B., Murthy S., Keaton S. A., Cobbett P., Cibelli J. B., Suhr S. T. Defining the diversity of phenotypic respecification using multiple cell lines and reprogramming regimens. Stem Cells Dev. 2013;22:2641–2654. doi: 10.1089/scd.2013.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D. H., Park S. H., Park J. H., Choi Y., Park K. D., Kang J. W., Choi K. S., Kim H. S. Embryotoxicity of lead (II) acetate and aroclor 1254 using a new end point of the embryonic stem cell test. Int. J. Toxicol. 2011;30:498–509. doi: 10.1177/1091581811416522. [DOI] [PubMed] [Google Scholar]
- Basha M. R., Wei W., Bakheet S. A., Benitez N., Siddiqi H. K., Ge Y. W., Lahiri D. K., Zawia N. H. The fetal basis of amyloidogenesis: Exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J. Neurosci. 2005;25:823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T., Speed T. P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bellinger D. C. Prenatal exposures to environmental chemicals and children's neurodevelopment: An update. Saf. Health Work. 2013;4:1–11. doi: 10.5491/SHAW.2013.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi S. W., Bahmani A., Subaiea G. M., Zawia N. H. Infantile exposure to lead and late-age cognitive decline: Relevance to AD. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.02.012. doi: 10.1016/j.jalz.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi S. W., Huang H., Wu J., Zawia N. H. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: Implications for Alzheimer's disease. J. Alzheimers Dis. 2011;27:819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Bihaqi S. W., Zawia N. H. Alzheimer's disease biomarkers and epigenetic intermediates following exposure to Pb in vitro . Curr. Alzheimer Res. 2012;9:555–562. doi: 10.2174/156720512800617964. [DOI] [PubMed] [Google Scholar]
- Bolin C. M., Basha R., Cox D., Zawia N. H., Maloney B., Lahiri D. K., Cardozo-Pelaez F. Exposure to lead and the developmental origin of oxidative DNA damage in the aging brain. FASEB J. 2006;20:788–790. doi: 10.1096/fj.05-5091fje. [DOI] [PubMed] [Google Scholar]
- Brandt S. K., Weatherly M. E., Ware L., Linn D. M., Linn C. L. Calcium preconditioning triggers neuroprotection in retinal ganglion cells. Neuroscience. 2011;172:387–397. doi: 10.1016/j.neuroscience.2010.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker C. J., Schmithorst V. J., Haynes E. N., Dietrich K. N., Egelhoff J. C., Lindquist D. M., Lanphear B. P., Cecil K. M. Altered myelination and axonal integrity in adults with childhood lead exposure: A diffusion tensor imaging study. Neurotoxicology. 2009;30:867–875. doi: 10.1016/j.neuro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield R. L., Henderson C. R., Jr, Cory-Slechta D. A., Cox C., Jusko T. A., Lanphear B. P. Intellectual impairment in children with blood lead concentrations below 10 microg per deciliter. N. Engl. J. Med. 2003;348:1517–1526. doi: 10.1056/NEJMoa022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil K. M., Brubaker C. J., Adler C. M., Dietrich K. N., Altaye M., Egelhoff J. C., Wessel S., Elangovan I., Hornung R., Jarvis K., et al. Decreased brain volume in adults with childhood lead exposure. PLoS Med. 2008;5:e112. doi: 10.1371/journal.pmed.0050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E. A., Beyhan Z., Yoo M. S., Siripattarapravat K., Ko T., Lookingland K. J., Madhukar B. V., Cibelli J. B. Increased cellular turnover in response to fluoxetine in neuronal precursors derived from human embryonic stem cells. Int. J. Dev. Biol. 2010;54:707–715. doi: 10.1387/ijdb.092851ec. [DOI] [PubMed] [Google Scholar]
- Davidovics Z., DiCicco-Bloom E. Moderate lead exposure elicits neurotrophic effects in cerebral cortical precursor cells in culture. J. Neurosci. Res. 2005;80:817–825. doi: 10.1002/jnr.20539. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S., Defrance M., Calonne E., Denis H., Sotiriou C., Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- Dosunmu R., Alashwal H., Zawia N. H. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 2012;133:435–443. doi: 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N., Yi S. V. Functional relevance of CpG island length for regulation of gene expression. Genetics. 2011;187:1077–1083. doi: 10.1534/genetics.110.126094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C. L., Fisher A. G. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Gen. Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Franklin J. L., Berechid B. E., Cutting F. B., Presente A., Chambers C. B., Foltz D. R., Ferreira A., Nye J. S. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr. Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- Gensel J. C., Schonberg D. L., Alexander J. K., McTigue D. M., Popovich P. G. Semi-automated Sholl analysis for quantifying changes in growth and differentiation of neurons and glia. J. Neurosci. Methods. 2010;190:71–79. doi: 10.1016/j.jneumeth.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddabasappa A., Hamilton W. R., Chaney S., Xiao W., Johnson J. E., Mukherjee S., Fox D. A. Low-level gestational lead exposure increases retinal progenitor cell proliferation and rod photoreceptor and bipolar cell neurogenesis in mice. Environ. Health Perspect. 2011;119:71–77. doi: 10.1289/ehp.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. E., Kelly M. E., Samsam T. E., Goodman J. H. Chronic developmental lead exposure reduces neurogenesis in adult rat hippocampus but does not impair spatial learning. Toxicol. Sci. 2005;86:365–374. doi: 10.1093/toxsci/kfi156. [DOI] [PubMed] [Google Scholar]
- Guilarte T. R., Opler M., Pletnikov M. Is lead exposure in early life an environmental risk factor for Schizophrenia? Neurobiological connections and testable hypotheses. Neurotoxicology. 2012;33:560–574. doi: 10.1016/j.neuro.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundacker C., Scheinast M., Damjanovic L., Fuchs C., Rosner M., Hengstschlager M. Proliferation potential of human amniotic fluid stem cells differently responds to mercury and lead exposure. Amino Acids. 2012;43:937–949. doi: 10.1007/s00726-011-1154-1. [DOI] [PubMed] [Google Scholar]
- Hanley J. A., Negassa A., Edwardes M. D., Forrester J. E. Statistical analysis of correlated data using generalized estimating equations: An orientation. Am. J. Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- Hanna C. W., Bloom M. S., Robinson W. P., Kim D., Parsons P. J., vom Saal F. S., Taylor J. A., Steuerwald A. J., Fujimoto V. Y. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum. Reprod. 2012;27:1401–1410. doi: 10.1093/humrep/des038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J., Armulik A., Senti K. A., Edoff K., Spalding K., Momma S., Cassidy R., Flanagan J.G., Frisen J. Ephrin-A2 reverse signaling negatively regulates neural progenitor proliferation and neurogenesis. Genes Dev. 2005;19:462–471. doi: 10.1101/gad.326905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Schneider J. S. Effects of lead exposure on proliferation and differentiation of neural stem cells derived from different regions of embryonic rat brain. Neurotoxicology. 2004;25:1001–1012. doi: 10.1016/j.neuro.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Hu B. Y., Zhang S. C. Directed differentiation of neural-stem cells and subtype-specific neurons from hESCs. Methods Mol. Biol. 2010;636:123–137. doi: 10.1007/978-1-60761-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaako-Movits K., Zharkovsky T., Romantchik O., Jurgenson M., Merisalu E., Heidmets L. T., Zharkovsky A. Developmental lead exposure impairs contextual fear conditioning and reduces adult hippocampal neurogenesis in the rat brain. Int. J. Dev. Neurosci. 2005;23:627–635. doi: 10.1016/j.ijdevneu.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Jiao J.-W., Feldheim D. A., Chen D. F. Ephrins are negative regulators of adult neurogenesis in diverse regions of the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8778–8783. doi: 10.1073/pnas.0708861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe E. M., McQuate A. L., Zhao X. Crosstalk among epigenetic pathways regulates neurogenesis. Front. Neurosci. 2012;6:59. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. M., Kardashyan L., Callaway J. K., Lee E. M., Beart P. M. Long-term functional and protective actions of preconditioning with hypoxia, cobalt chloride, and desferrioxamine against hypoxic-ischemic injury in neonatal rats. Pediatr. Res. 2008;63:620–624. doi: 10.1203/PDR.0b013e31816d9117. [DOI] [PubMed] [Google Scholar]
- Kaneko Y., Sakakibara S., Imai T., Suzuki A., Nakamura Y., Sawamoto K., Ogawa Y., Toyama Y., Miyata T., Okano H. Musashi1: An evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- Kermani S., Karbalaie K., Madani S. H., Jahangirnejad A. A., Eslaminejad M. B., Nasr-Esfahani M. H., Baharvand H. Effect of lead on proliferation and neural differentiation of mouse bone marrow-mesenchymal stem cells. Toxicol. In Vitro. 2008;22:995–1001. doi: 10.1016/j.tiv.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Knight H. M., Walker R., James R., Porteous D. J., Muir W. J., Blackwood D. H. R., Pickard B. S. GRIK4/KA1 protein expression in human brain and correlation with bipolar disorder risk variant status. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;22:21–29. doi: 10.1002/ajmg.b.31248. [DOI] [PubMed] [Google Scholar]
- Kovatsi L., Georgiou E., Ioannou A., Haitoglou C., Tzimagiorgis G., Tsoukali H., Kouidou S. p16 promoter methylation in Pb2+ -exposed individuals. Clin. Toxicol. 2010;48:124–128. doi: 10.3109/15563650903567091. [DOI] [PubMed] [Google Scholar]
- Krencik R., Zhang S. C. Stem cell neural differentiation: A model for chemical biology. Curr. Opin. Chem. Biol. 2006;10:592–597. doi: 10.1016/j.cbpa.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kuehn B. M. Panel advises tougher limits on lead exposure. JAMA. 2012;307:445. doi: 10.1001/jama.2012.50. [DOI] [PubMed] [Google Scholar]
- Lanphear B. P., Hornung R., Khoury J., Yolton K., Baghurst P., Bellinger D. C., Canfield R. L., Dietrich K. N., Bornschein R., Greene T., et al. Low-level environmental lead exposure and children's intellectual function: An international pooled analysis. Environ. Health Perspect. 2005;113:894–849. doi: 10.1289/ehp.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Pfaff S. L. Synchronization of neurogenesis and motor neuron specification bydirect coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–645. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lidsky T. I., Schneider J. S. Lead neurotoxicity in children: Basic mechanisms and clinical correlates. Brain. 2003;126:5–19. doi: 10.1093/brain/awg014. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maher P. How protein kinase C activation protects nerve cells from oxidative stress-induced cell death. J. Neurosci. 2001;21:2929–2938. doi: 10.1523/JNEUROSCI.21-09-02929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Marguerat S., Bähler J. Post-transcriptional control of gene expression: A genome-wide perspective. Trends Biochem. Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Mazumdar M., Xia W., Hofmann O., Gregas M., Sui S. H., Hide W., Yang T., Needleman H. L., Bellinger D. C. Prenatal lead levels, plasma amyloid beta levels, and gene expression in young adulthood. Environ. Health Perspect. 2012;120:702–707. doi: 10.1289/ehp.1104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava-Ruiz C., Mendez-Armenta M., Rios C. Lead neurotoxicity: Effects on brain nitric oxide synthase. J. Mol. Histol. 2012;43:553–563. doi: 10.1007/s10735-012-9414-2. [DOI] [PubMed] [Google Scholar]
- Neal A. P., Guilarte T. R. Molecular neurobiology of lead (Pb(2+)): Effects on synaptic function. Mol. Neurobiol. 2010;42:151–160. doi: 10.1007/s12035-010-8146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. M., Espy K. A. Low-level lead exposure and contingency-based responding in preschoolers: An exploratory study. Dev. Neuropsychol. 2009;34:494–506. doi: 10.1080/87565640902964565. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yamaguchi T., Tokunaga A., Hara A., Hamaguchi T., Kato K., Iwamatsu A., Okano H., Kaibuchi K. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol. Biol. Cell. 2006;17:1273–1285. doi: 10.1091/mbc.E05-07-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H., Kawahara H., Toriya M., Nakao K., Shibata S., Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Osumi N., Shinohara H., Numayama-Tsuruta K., Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- Peng S., Hajela R. K., Atchison W. D. Characteristics of block by Pb2+ of function of human neuronal L-, N-, and R-type Ca2+ channels transiently expressed in human embryonic kidney 293 cells. Mol. Pharmacol. 2002;62:1418–1430. doi: 10.1124/mol.62.6.1418. [DOI] [PubMed] [Google Scholar]
- Pidsley R., Wong C. C. Y., Volta M., Lunnon K., Mill J., Schalkwyk L. C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner J. R., Hu H., Ettinger A., Sanchez B. N., Wright R. O., Cantonwine D., Lazarus A., Lamadrid-Figueroa H., Mercado-Garcia A., Tellez-Rojo M. M., et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ. Health Perspect. 2009;117:1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Brew B. J., Guillemin G. J. Lead dysregulates serine/threonine protein phosphatases in human neurons. Neurochem. Res. 2011;36:195–204. doi: 10.1007/s11064-010-0300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. J., Suhr S. T., Rodriguez R. M., Chang E. A., Wang K., Siripattarapravat K., Ko T., Cibelli J. B. Human-induced pluripotent stem cells produced under xeno-free conditions. Stem Cells Dev. 2010;19:1221–1229. doi: 10.1089/scd.2009.0459. [DOI] [PubMed] [Google Scholar]
- Sakakibara S., Imai T., Hamaguchi K., Okabe M., Aruga J., Nakajima K., Yasutomi D., Nagata T., Kurihara Y., Uesugi S., et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin F. J., Fan X., Lindquist D. M., Xia X., Puga A. Lead induces similar gene expression changes in brains of gestationally exposed adult mice and in neurons differentiated from mouse embryonic stem cells. PLoS ONE. 2013;8:e80558. doi: 10.1371/journal.pone.0080558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T., Liu Y., Buchner V., Tchounwou P. B. Neurotoxic effects and biomarkers of lead exposure: A review. Rev. Environ. Health. 2009;24:15–45. doi: 10.1515/reveh.2009.24.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M. A., Bibikova M., Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Schneider J. S., Kidd S. K., Anderson D. W. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol. Lett. 2013;217:75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian-Serrano A., Sandonis A., Cardozo M., Rodriguez-Tornos F. M., Bovolenta P., Nieto M. Palphax6 expression in postmitotic neurons mediates the growth of axons in response to SFRP1. PLoS ONE. 2012;7:e31590. doi: 10.1371/journal.pone.0031590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut M. C., Cingolani P., Sen A., Kruger A., Shaik A., Hirsch H., Suhr S. T., Ruden D. Epigenetics of early-life lead exposure and effects on brain development. Epigenomics. 2012;4:665–674. doi: 10.2217/epi.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut M. C., Suhr S. T., Kaspar B., Gage F. H. Intraneuronal aggregate formation and cell death after viral expression of expanded polyglutamine tracts in the adult rat brain. J. Neurosci. 2000;20:219–229. doi: 10.1523/JNEUROSCI.20-01-00219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Zhong W., Jan Y. N., Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Sholl D. A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Sofer T., Schifano E. D., Hoppin J. A., Hou L., Baccarelli A. A. A-clustering: A novel method for the detection of co-regulated methylation regions, and regions associated with exposure. Bioinformatics. 2013;29:2884–2891. doi: 10.1093/bioinformatics/btt498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes L., Letz R., Gerr F., Kolczak M., McNeill F. E., Chettle D. R., Kaye W. E. Neurotoxicity in young adults 20 years after childhood exposure to lead: The Bunker Hill experience. Occup. Environ. Med. 1998;55:507–516. doi: 10.1136/oem.55.8.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan F. A., Day J. J. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vincens M., Misono S., Lin M. Z., Zubiaga A., Hua X., Fan G., Greenberg M. E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Neuron. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tejedo J. R., Tapia-Limonchi R., Mora-Castilla S., Cahuana G. M., Hmadcha A., Martin F., Bedoya F. J., Soria B. Low concentrations of nitric oxide delay the differentiation of embryonic stem cells and promote their survival. Cell Death Dis. 2010;1:e80. doi: 10.1038/cddis.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Toscano C. D., Guilarte T. R. Lead neurotoxicity: From exposure to molecular effects. Brain Res. Brain Res. Rev. 2005;49:529–554. doi: 10.1016/j.brainresrev.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Touleimat N., Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- Verina T., Rohde C. A., Guilarte T. R. Environmental lead exposure during early life alters granule cell neurogenesis and morphology in the hippocampus of young adult rats. Neuroscience. 2007;145:1037–1047. doi: 10.1016/j.neuroscience.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visan A., Hayess K., Sittner D., Pohl E. E., Riebeling C., Slawik B., Gulich K., Oelgeschlager M., Luch A., Seiler A. E. Neural differentiation of mouse embryonic stem cells as a tool to assess developmental neurotoxicity in vitro. Neurotoxicology. 2012;33:1135–1146. doi: 10.1016/j.neuro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Waimey K. E., Huamng P.-H., Chen M., Cheng H.-J. Plexin-A3 and plexin-A4 restrict the migration of sympathetic neurons but not their neural crest precursors. Dev. Bio. 2008;315:448–458. doi: 10.1016/j.ydbio.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yan L., Hu Q., Sucheston L. E., Higgins M. J., Ambrosone C. B., Johnson C. S., Smiraglia D. J., Liu S. IMA: An R package for high-throughput analysis of Illumina's 450K Infinium methylation data. Bioinformatics. 2012;28:729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L. D., Cory-Slechta D. A., Gilbert M. E., Tiffany-Castiglioni E., Zawia N. H., Virgolini M., Rossi-George A., Lasley S. M., Qian Y. C., Basha M. R. New and evolving concepts in the neurotoxicology of lead. Toxicol. Appl. Pharmacol. 2007;225:1–27. doi: 10.1016/j.taap.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Willems E., Leyns L., Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Wobus A. M., Loser P. Present state and future perspectives of using pluripotent stem cells in toxicology research. Arch. Toxicol. 2011;85:79–117. doi: 10.1007/s00204-010-0641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. P., Dietrich K. N., Ris M. D., Hornung R. W., Wessel S. D., Lanphear B. P., Ho M., Rae M. N. Association of prenatal and childhood blood lead concentrations with criminal arrests in early adulthood. PLoS Med. 2008;5:e101. doi: 10.1371/journal.pmed.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. O., Schwartz J., Wright R. J., Bollati V., Tarantini L., Park S. K., Hu H., Sparrow D., Vokonas P., Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Basha M. R., Brock B., Cox D. P., Cardozo-Pelaez F., McPherson C. A., Harry J., Rice D. C., Maloney B., Chen D., et al. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): Evidence for a developmental origin and environmental link for AD. J. Neurosci. 2008a;28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Basha M. R., Zawia N. H. The environment, epigenetics and amyloidogenesis. J. Mol. Neurosci. 2008b;34:1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- Xie W., Schultz M. D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J. W., Tian S., Hawkins R. D., Leung D., et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y., Szabo P., Szuts D., Bacquet C., Aranyi T., Paldi A. Rapid turnover of DNA methylation in human cells. Epigenetics. 2012;7:141–145. doi: 10.4161/epi.7.2.18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. C., Wernig M., Duncan I. D., Brustle O., Thomson J. A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zimmer B., Kuegler P. B., Baudis B., Genewsky A., Tanavde V., Koh W., Tan B., Waldmann T., Kadereit S., Leist M. Coordinated waves of gene expression during neuronal differentiation of embryonic stem cells as basis for novel approaches to developmental neurotoxicity testing. Cell Death Differ. 2011;18:383–395. doi: 10.1038/cdd.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer B., Lee G., Balmer N. V., Meganathan K., Sachinidis A., Studer L., Leist M. Evaluation of developmental toxicants and signaling pathways in a functional test based on the migration of human neural crest cells. Environ. Health Perspect. 2012;120:1116–1122. doi: 10.1289/ehp.1104489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurich M. G., Eskes C., Honegger P., Berode M., Monnet-Tschudi F. Maturation-dependent neurotoxicity of lead acetate in vitro: Implication of glial reactions. J. Neurosci. Res. 2002;70:108–116. doi: 10.1002/jnr.10367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.