Abstract
Glucosidase II (GluII) is a glycan-trimming enzyme active on nascent glycoproteins in the endoplasmic reticulum (ER). It trims the middle and innermost glucose residues (Glc2 and Glc1) from N-linked glycans. The monoglucosylated glycan produced by the first GluII trimming reaction is recognized by calnexin/calreticulin and serves as the signal for entry into this folding pathway. GluII is a heterodimer of α and β subunits corresponding to yeast Gls2p and Gtb1p, respectively. While Gls2p contains the glucosyl hydrolase active site, the Gtb1p subunit has previously been shown to be essential for the Glc1 trimming event. Here we demonstrate that Gtb1p also determines the rate of Glc2 trimming. In order to further dissect these activities we mutagenized a number of conserved residues across the protein. Our data demonstrate that both the MRH and G2B domains of Gtb1p contribute to the Glc2 trimming event but that the MRH domain is essential for Glc1 trimming.
Keywords: calnexin cycle, endoplasmic reticulum, ER quality control, glucosidase II, glycan processing
Introduction
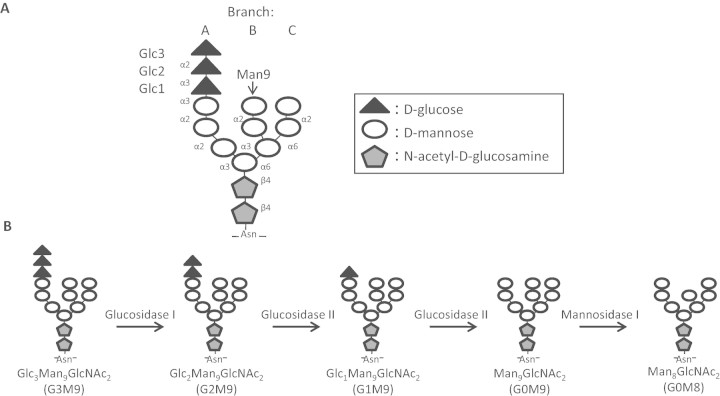
N-Linked glycosylation occurs as nascent polypeptide chains are translocated across the endoplasmic reticulum (ER) membrane. Here, a specific glycan structure (Glc3Man9GlcNAc2) (Figure 1A) is covalently attached to specific asparagines in the Asn-X-Ser/Thr sequon by the enzyme oligosaccharyltransferase (OST). This Glc3Man9GlcNAc2glycan is the target for a number of glycan-trimming enzymes whose actions play important roles in the folding and quality control of nascent glycoproteins. The outermost glucose, Glc3, is trimmed by Glucosidase I, thereby abolishing binding to the glycan by OST. Glucosidase II (GluII) is responsible for trimming both of the remaining two glucose residues, Glc2 and Glc1 (Figure 1B). These GluII-mediated reactions play a pivotal role in the calnexin/calreticulin cycle, a quality control mechanism for the correct folding of nascent glycoproteins (reviewed by Hebert and Molinari (2007)). The trimming of Glc2 creates a monoglucosylated glycan (Glc1Man9GlcNAc2; G1M9, Figure 1B) which is the substrate for the ER-resident lectin chaperones, calnexin (Cnx), and calreticulin (Crt), while the trimming of the innermost glucose residue, Glc1, means the nascent glycoprotein is no longer recognized by Cnx or Crt. If the glycoprotein is still misfolded after its release from Cnx/Crt, it can be re-glucosylated by the “folding sensor” UDP-glucose:glycoprotein glucosyltransferase (GT) (Sousa et al. 1992), recreating the monoglucosylated Glc1Man9GlcNAc2 form and the substrate for Cnx/Crt. Persistently misfolded glycoproteins are removed from the cycle and subjected to ER-associated degradation (ERAD). This process is begun with the trimming of Man9 by Mannosidase I leaving a Man8GlcNAc2glycan that is no longer recognized by GT (Molinari et al. 2003). Further mannose trimming by the newly described activity of Htm1p can subsequently generate a Man7GlcNAc2 glycan under certain conditions (Clerc et al. 2009).
Fig. 1.
N-Glycans are sequentially trimmed by a trio of glycan-trimming enzymes. (A) The enzyme OST adds this 14-sugar, tri-antennary glycan to nascent glycoproteins as they are translocated across the ER membrane. This 14-sugar glycan consists of three glucoses (triangles), nine mannoses (circles), and two N-acetylglucosamines (pentagons). The types of linkages between sugar residues are shown in abbreviated form (α2 = α1, 2, etc.). (B) The 14-sugar glycan (G3M9) is sequentially trimmed by the glucose-trimming enzymes Glucosidases I and II and by the mannose trimming Mannosidase I. It is the G1 glycan form that can be bound by the lectin chaperones calnexin (Cnx) and calreticulin (Crt). The M9 glycan form may be re-glucosylated to the G1M9 form by the “folding sensor” enzyme UDP-glucose:glycoprotein glucosyltransferase (GT) if the nascent glycoprotein is misfolded upon release from Cnx/Crt binding.
Glucosidase II is a heterodimer of tightly associated GluIIα and GluIIβ subunits (Gls2p and Gtb1p, respectively, in S. cerevisiae) (Trombetta et al. 1996). The glucosyl hydrolase activity has been shown to reside in GluIIα, and this subunit alone is sufficient to hydrolyze glucose from p-nitrophenyl-α-d-glucopyranoside (p-NP-GP) in vitro (Trombetta et al. 2001). Studies in yeast have shown that Gtb1p is essential for the Glc1 trimming event that drives nascent glycoproteins out of the calnexin cycle (Wilkinson et al. 2006). Indeed, it has been reported that the GluII heterodimer competes with calreticulin for the G1M9 glycan (Totani et al. 2006) and several groups have suggested that the Mannose-6-phosphate Receptor Homology (MRH) domain within Gtb1p/GluIIβ contributes this lectin activity (Deprez et al. 2005; Totani et al. 2006; Wilkinson et al. 2006). MRH domains (Munro 2001) have been identified in a number of proteins including Yos9p and Erlectin (XTP3-B) and mutations in conserved residues in Yos9p have been shown to disrupt lectin activity (Bhamidipati et al. 2005; Szathmary et al. 2005; Quan et al. 2008). In addition to the C-terminal MRH domain, Gtb1p/GluIIβ also contains a highly conserved amino-terminal domain, hereafter described as the “G2B” domain (Glucosidase II Beta), which has been implicated in the interaction with the Gls2p/GluIIα subunit (Arendt and Ostergaard 2000; Pelletier et al. 2000).
Here, we confirm the requirement for Gtb1p in Glc1 trimming but also report a newly identified kinetic role for Gtb1p in the earlier Glc2 trimming event. Furthermore, we have used a site-directed mutagenesis approach to examine the roles of a number of conserved domains within Gtb1p in the Glc2/Glc1 trimming events. Functional analysis of these mutants in vivo indicates that both the G2B and MRH domains contribute to the Glc2 trimming event, but that the MRH domain is also essential for Glc1 trimming.
Results
Δgtb1 cells are defective in Glc2 trimming
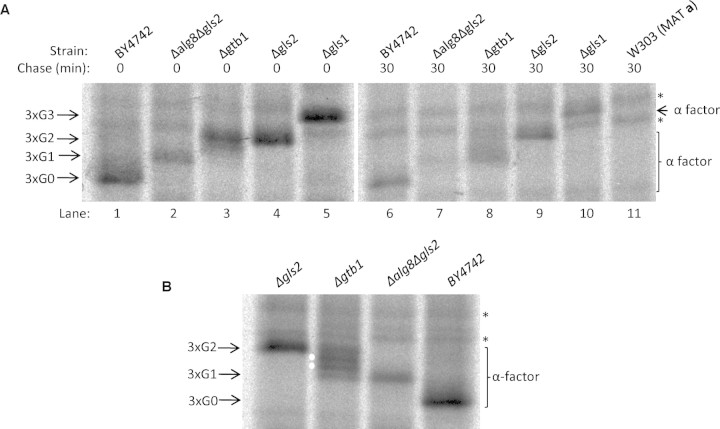
It has previously been shown that Δgtb1 mutant cells are completely deficient in Glc1 trimming of N-linked glycans on a range of glycoproteins (Wilkinson et al. 2006). We began by examining the processing of pro-α-factor, a 175 residue polypeptide which receives three N-linked glycans upon import into the ER. In order to establish a set of markers of known glycan composition, we used a series of previously characterized mutants whose defects in glycan processing are well understood. Mutant cells lacking either Glucosidase I (Δgls1), Glucosidase IIα (Δgls2), or the Δalg8/Δgls2 double mutant accumulate glycoproteins with G3, G2, and G1 glycans, respectively (Romero et al. 1997; Jakob et al. 1998a, 1998b; Simons et al. 1998). Pro-α-factor processing was analyzed in these various strains by pulse labeling followed by immunoprecipitation. The fastest migrating form of triply glycosylated pro-α-factor was observed in wildtype cells in which completion of glucose trimming yields a glycoprotein with three G0 glycans (3xG0; Figure 2A, lane 1). The slowest migrating form was then the expected 3xG3 form observed in Δgls1 (Figure 2A lane 5), while the Δgls2 and Δalg8/Δgls2 mutants produced the intermediate 3xG2 and 3xG1 forms, respectively (Figure 2A, lanes 4 and 2). The relative migration of these various forms remained similar even after a 30 min chase period (albeit with some evidence of protein turnover during this time) indicating that the various stages of glucose trimming were normally completed by our very earliest time point (Figure 2A; compare lanes 1, 2, 4, and 5 with lanes 6, 7, 9, and 10). This contrasted strikingly with the results for the Δgtb1 mutant where we observed a fuzzy band after 0 min chase which largely co-migrated with the 3xG2 form (Figure 2A; compare lanes 3 and 4) but which chased to a form more similar to 3xG1 after 30 min (Figure 2A; compare lanes 2, 3, 7, and 8). These findings suggested a slow conversion of the 3xG2 form into the 3xG1 form in Δgtb1 cells and so in order to try to capture this more clearly, we next examined processing at an intermediate time point (15 min). Under these conditions, we observed four distinct bands in Δgtb1 mutant cells (Figure 2B). The upper of these bands co-migrated precisely with the 3xG2 control (Δgls2) while the lower co-migrated with 3xG1 (Δalg8/Δgls2; Figure 2B). The intermediate bands might represent glucose trimming intermediates but might also be due to some effect on Man9 trimming. In yeast, Man9 trimming is dependent upon Mannosidase I (Mns1p) and Δmns1 cells accumulate N-linked glycans that are exclusively in the Man9 form (Camirand et al. 1991). When we analyze alpha-factor trimming in Δgtb1/Δmns1 double mutant cells, we still observe four bands (see supplementary Figure 1). Since these must all correspond to Man9 glycans, the only remaining possibility is that these four bands are glucose trimming intermediates. These findings demonstrate that while cells lacking Gtb1p can process 3xG2 glycans to the 3xG1 form, they do so only with reduced efficiency compared to wildtype cells. Slow trimming of 3xG2 to 3xG1 would be expected to yield two trimming intermediates (the larger of these having only one glycan trimmed to the G1 form and the smaller having two G1 glycans) which would be entirely consistent with the two novel forms observed in the 15 min chase (indicated by white circles in Figure 2B). A more detailed pulse-chase analysis of Δgtb1 cells (shown in Figure 4A) demonstrated that complete Glc2 trimming of pro-α-factor occurred over a period of approximately 45 min but that no further processing to the G0 form was observed. We, therefore, conclude that Gtb1p is required for efficient Glc2 trimming in yeast but that it is essential for Glc1 trimming.
Fig. 2.
Δgtb1 cells exhibit delayed Glc2 trimming but are entirely deficient in Glc1 trimming. (A) Cells were pulse-labeled for 4 min with [35S] methionine/cysteine and then either snap-frozen (0 min) or chased (30 min) in the presence of NaN3 while cell extracts were prepared for immunoprecipitation studies. Glycan trimming was analyzed by 12.5% SDS–PAGE analysis of immunoprecipitated pro-α-factor. The 3xG3, 3xG2, 3xG1, and 3xG0 glycan forms are indicated. The bands indicated by asterisks are nonspecific background bands unrelated to pro-α-factor since both are evident in a MATa strain. (B) After a 4 min pulse-labeling cells were chased in the presence of NaN3 for 15 min before being snap-frozen in liquid nitrogen and processed for immunoprecipitation. Glycan trimming of pro-α-factor was analyzed by 12.5% SDS–PAGE. The 3xG2, 3xG1, and 3xG0 glycan forms are indicated. Two novel forms of pro-α-factor that migrate between 3xG2 and 3xG1 are indicated with white circles. The two nonspecific background bands are again marked by asterisks.
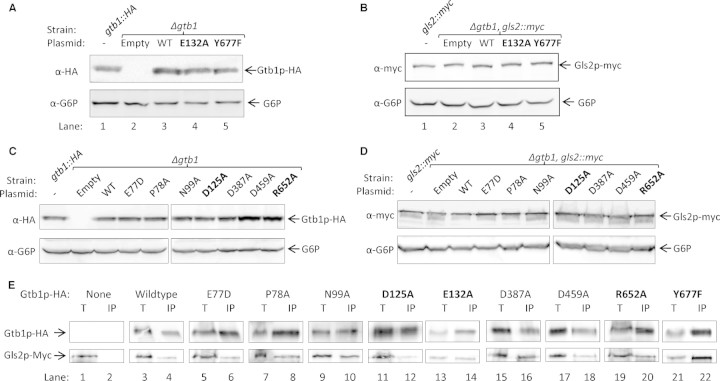
Fig. 4.
Gtb1p mutants are expressed normally and can interact with Gls2p. (A) Whole cell extracts were prepared from exponentially growing strains. After resolution by SDS–PAGE, the expression levels of Gtb1p-HA were measured by immunoblotting against the HA epitope. Glucose-6-phosphate dehydrogenase (G6P) was used as a loading control. Endogenous expression levels were measured using the gtb1::HA strain (lane 1, BWY616, (Wilkinson et al. 2006)). (B) Similarly, the expression levels of Gls2p-myc were monitored by immunoblotting against the myc epitope after resolution from whole cell extracts. Strains were made by transforming GTB1::HA plasmids into strain BWY624. Endogenous expression levels were measured using the gtb1::HA strain (lane 1, BWY618, (Wilkinson et al. 2006)). (C) The indicated mutants were processed and analyzed as in A. (D) The indicated mutants were processed and analyzed as in B. (E) The ability of the mutants to interact with Gls2p-myc was measured by immunoblotting against the HA and myc epitopes after a native immunoprecipitation of Gtb1p-HA from microsomes solubilized with 1% Triton-X100. Fifty percent of totals were loaded (T) except for mutants E132A and Y677F where 10% was loaded, IP = Immunoprecipitated. Strains used had GTB1::HA plasmids transformed into BWY624.
Mutation Analysis of Domains within Gtb1p
We next sought to determine the effects of mutations in conserved domains on the activities of Gtb1p. Point mutations were made in several conserved residues in the G2B domain, the MRH domain, and in two putative EF-hand domains (Figure 3A). The residues mutated in the G2B domain were chosen due to their high level of conservation from yeast to humans (E77D, P78A, N99A, D125A, and E132A; Figure 3B). In the MRH domain two conserved residues (R652 and Y677) (Figure 3C) were chosen that have previously been shown to be required for the function of MRH-domain-containing proteins (Bhamidipati et al. 2005; Szathmary et al. 2005; Christianson et al. 2008; Quan et al. 2008). Gtb1p also has two putative EF-hand domains at positions 376–388 and 448–460 with similarities of 82% and 88% to the EF-hand calcium binding domain signature, respectively. The EF-hand domain is involved in calcium and/or magnesium binding and conventionally consists of a 12-residue loop flanked on both sides by a 12-residue α-helical domain. EF-hand domains are usually found in tandem pairs, as they appear to be in Gtb1p, with the pairing bringing co-operativity to the Ca2+/Mg2+ binding (for review see Grabarek (2006)). In order to investigate whether either EF-hand domain plays a role in Gtb1p function, we chose to mutate residues D387 and D459, both the 12th residue of the canonical EF-hand loop. Substitution of Asp12 for alanine has previously been shown to drastically reduce the binding affinity of the EF-hand domain for its Ca2+/Mg2+ ligands (da Silva et al. 1995).
Fig. 3.
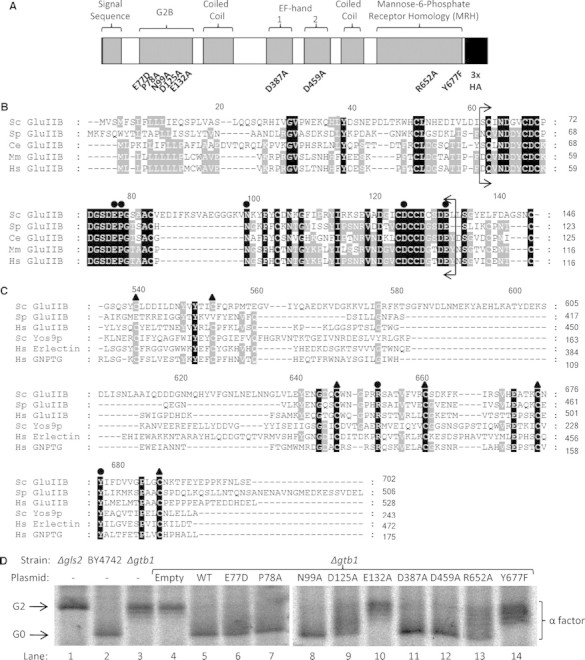
Mutation analysis of Gtb1p. (A) Domain architecture of Gtb1p with sites of mutagenesis shown. The protein has been tagged at its C-terminus with a HA epitope. (B) The protein sequences of the N-terminal region of GluIIβ orthologs from the species Saccharomyces cerevisiae (Sc), Saccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Mus musculus (Mm), and Homo sapiens (Hs) were aligned using ClustalW2 software (Larkin et al. 2007). The residues bracketed (Q63-E132 in Gtb1p) form the highly conserved G2B domain. Residues chosen for mutagenesis, E77, P78, N99, D125, and E132, are indicated by black circles. (C) The MRH domains of several GluIIβ orthologs and also those from Yos9p (Sc), Erlectin/XTP3-B (Hs, domain 2), and GlcNAc-phosphotransferase (GNPTG, Hs) were aligned. The six conserved cysteine residues of the MRH domain are indicated by black triangles and the residues mutated, R652 and Y677, are indicated by black circles. Black shading represents 100% conservation while gray shading represents 60% conservation across sequences chosen. All numbering is for Sc GluIIβ (Gtb1p). (D) Mutant plasmids were transformed into the Δgtb1 strain and a 5 min radio-labeling was carried out before immunoprecipitation of pro-α-factor. After resolution by 12.5% SDS–PAGE, radio-labeled pro-α-factor was visualized by autoradiography. Once again Δgls2 and wild-type (BY4742) strains provide references for the 3xG2 and 3xG0 forms, respectively.
Mutants E132A and Y677F are Defective in Glc1 Trimming
Various mutant alleles were transformed into a Δgtb1 strain and then tested for their ability to rescue the Δgtb1 phenotype. Glycan trimming was analyzed by SDS–PAGE analysis of radio-labeled pro-α-factor using the same short pulse labeling described earlier. As before, we used pro-α-factor from Δgls2 and BY4742 strains as markers for the 3xG2 and 3xG0 glycan forms, respectively (Figure 3D, lanes 1 and 2). As expected, the glycosylated pro-α-factor found in extracts from Δgtb1 cells migrated more slowly than that from cells carrying the functional HA-tagged copy of GTB1 (GTB1::HA, (Wilkinson et al. 2006)) (Figure 3D, compare lanes 4 and 5). Cells expressing GTB1::HA containing either the E132A or Y677F point mutations exhibited severe mutant phenotypes with no evidence of fully trimmed 3xG0 under these conditions (Figure 3D, compare lanes 2 and 4 with lanes 10 and 14). In all other mutants trimming to the 3xG0 form was apparent, but in two cases, D125A and R652A, there appeared to be significant levels of various intermediates between the 3xG2 and 3xG0 forms (Figure 3D, compare lanes 2 and 4 with lanes 9 and 13) and so these mutants were also chosen for further investigation. None of the other mutants tested, including those in the putative EF-hand domains, showed any significant defect in this assay but we do not exclude the possibility that one or more might exert some subtle effect.
We next examined steady-state expression levels of the various mutant proteins in Western blots. The levels observed for both the Y677F and E132A mutant forms of Gtb1-HA were comparable to those seen for either the unmutated plasmid control or expressed from a genomic copy of GTB1::HA (Figure 4A). From these data, we can conclude that the loss of Gtb1p function is not due to any gross perturbation in protein expression levels. Similar results were also obtained for the D125A and R652A mutants (Figure 4C). Next, we tested for any indirect effect on the expression levels of the Gtb1p's binding partner Gls2p. To this end, we transformed the Gtb1p-HA constructs into a Δgtb1 strain carrying a myc-tagged genomic allele of GLS2 (BWY624, made as described for BWY625 in Wilkinson et al. (2006)). As previously reported, we found the expression level of Gls2p-myc to be unaffected in the Δgtb1 mutant (Figure 4B, compare lanes 2 and 3; (Wilkinson et al. 2006)), and our data further demonstrate that Gls2p-myc levels are similar in the E132A and Y677F mutant cells compared to wildtype indicating that neither mutation has any dominant-negative effect on Gls2p-myc stability (Figure 4B). Once again similar results were obtained for all other mutants (Figure 4D). It has previously been shown that Gtb1p interacts with Gls2p (Wilkinson et al. 2006), and so we next tested whether the mutations might affect heterodimer formation. Native immunoprecipitations of Gtb1p-HA were performed using HA-specific antibodies. Immunoprecipitates were resolved by SDS–PAGE, and then both Gtb1p-HA and Gls2p-myc were detected by immunoblotting with the relevant antibodies (Figure 4E). As expected, we found that Gls2p-myc could be co-immunoprecipitated with Gtb1p-HA from extracts prepared from wildtype control cells (Figure 4D, lane 4). In all mutants tested, we found evidence of co-immunoprecipitation of Gls2p-myc with Gtb1-HA indicating that none of the mutations abolished heterodimer formation (Figure 4E). Nonetheless, we did note in the case of D125A that the proportion of Gls2-myc which could be co-immunoprecipitation was reproducibly lower than in wild type suggesting that D125 might contribute to heterodimer stability (Figure 4E, compare lanes 3 and 4 with lanes 11 and 12).
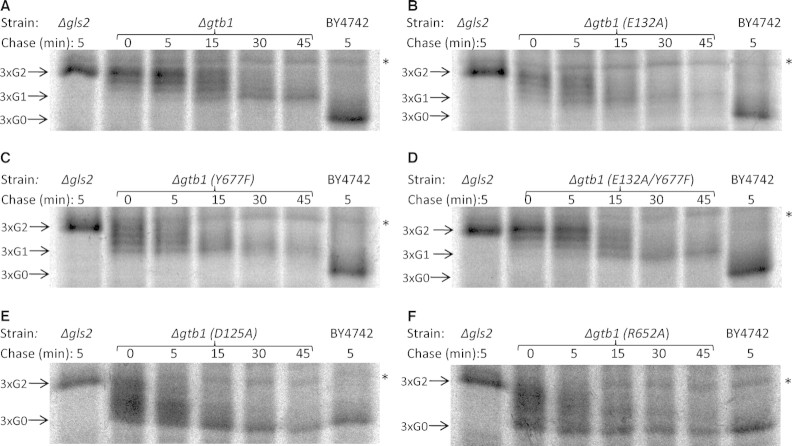
The E132A and Y677F Mutations Exert Differential Effects on Glc2 versus Glc1 Trimming
We have shown that Glc2 trimming is delayed in the absence of Gtb1p but this effect appears distinct from the role of Gtb1p in Glc1 trimming which is abolished in the null mutant (Wilkinson et al. 2006). Given these two distinct phenotypes, we next sought to determine which of these were affected in the E132A, Y677F, D125A, and R652A mutants. To this end, we next performed a more thorough pulse-chase analysis. Once again, we found the 3xG2 form present in Δgtb1 cells after an initial short pulse but that this subsequently chased into a spectrum of four distinct bands after 15 min, with the larger three then reducing in levels over time until only the lower molecular weight 3xG1 form remained after some 45 min (Figure 5A). This compares strikingly to the wildtype control (BY4742) in which pro-α-factor chases to the 3xG0 form within 5 min (Figure 5A). The same four bands are observed in both the E132A and Y677F mutants but the chase profiles are different, with the lower (3xG1) band being evident from the earliest time point indicating more rapid completion of Glc2 trimming than in the null mutant (Figure 5, compare panel A with panels B and C). Moreover, we detected a trace of the fully trimmed (3xG0) form in E132A cells after 15 min of chase (Figure 5B). The effects of E132A and Y677F on Glc2 trimming appear additive, and thus distinct, since the double mutant (E132A/Y677F) phenotype is indistinguishable from the null (Figure 5, compare panels A, B, C, and D). From these results, we conclude that the Y677F mutation in the MRH domain abolishes the Glc1 trimming activity of Gtb1p and further contributes to the Glc2 trimming activity of this protein. In contrast, the E132A mutation exerts a distinct effect on Glc2 trimming but remains competent for Glc1 trimming albeit very inefficiently. The D125A and R652A mutations have more subtle effects with both being delayed in Glc2 trimming when compared to the wildtype control, but both being able to complete trimming to the 3xG0 form within the course of the experiment (Figure 5, panels E and F).
Fig. 5.
Pulse chase analysis to monitor progression of glucose trimming. Cells from Δgtb1 (A) and each of the mutant strains, E132A (B), Y677F (C), E132A/Y677F (D), D125A (E), and R652A (F), were pulse-chased in the presence of 2mM NaN3 after radio-labeling for 5 min and were processed for immunoprecipitation as before. Glycan trimming of pro-α-factor was monitored by 12.5% SDS–PAGE and autoradiography. Δgls2 and BY4742 strains were chased for 5 min and provide reference points for the 3xG2 form and fully trimmed 3xG0 forms, respectively. A nonspecific background band that migrated slightly slower than the 3xG2 form is apparent in each strain (marked by asterisk).
Discussion
The glucose trimming action of Glucosidase II (GluII) along with the G1-specific lectin chaperones calnexin and/or calreticulin (Cnx/Crt) and the re-glucosylation action of UDP-glucose:glycoprotein glucosyltransferase (GT) comprise the Cnx/Crt cycle, an important folding and quality control pathway for glycoproteins (reviewed by Moremen and Molinari (2006)). It was previously reported that Gtb1p (GluIIβ) was required for Glc1 trimming (Wilkinson et al. 2006), the terminal glucose trimming step that releases nascent glycoproteins from Cnx/Crt binding. In this work, by using a rapid freezing technique that traps trimming intermediates, we have uncovered a further novel role for Gtb1p in enhancing the kinetics of Glc2 trimming. Furthermore, by mutagenizing conserved residues across the protein, we have shown that the G2B and MRH domains play important roles in facilitating Glc2 trimming and that a functional MRH domain is an absolute requirement for Glc1 trimming.
Mutation of residue Y677 in Gtb1p's MRH domain had a major effect on Glc2 trimming but completely abolished Glc1 trimming. Similarly, mutation of the other conserved residue within the MRH domain, R652A, led to a reduction in Gtb1p-dependent trimming efficiency. Interestingly, the equivalent mutation to R652A in the MRH domain of Yos9p has been shown to disrupt lectin activity (Quan et al. 2008). Initially, we considered two possible activities for this lectin domain; a role in interacting with Gls2p through binding of one of its glycans (Wilkinson et al. 2006) or a role in binding glycan substrate during glucose trimming of nascent glycoproteins. Co-immunoprecipitation experiments allowed us to rule out the possibility that a functional MRH domain was required for the interaction with Gls2p and thus left the likely explanation that this domain binds the glycan substrate during glucose trimming as has been suggested previously (Deprez et al. 2005; Totani et al. 2006; Wilkinson et al. 2006). Totani et al. (2006) report that GluII recognizes the terminal mannose on branch C of the glycan (see Figure 1A) and we speculate that it is the MRH domain of Gtb1p that binds this residue. The requirement for a functional MRH domain differs markedly between the Glc2 and Glc1 trimming events, and this reflects more general differences between the two reactions.
The substrates for Glc2 and Glc1 cleavage differ being Glcα1-3Glc and Glcα1-3Man, respectively, and the rates of the two reactions are different with Glc2 cleavage occurring more rapidly than Glc1 cleavage both in vivo (Hubbard and Robbins 1979) and in vitro (Totani et al. 2006). Although GluII has a single glucosyl hydrolase site (on GluIIα) (Pelletier et al. 2000), kinetic data suggest two distinct substrate binding affinities given the fact that it has both high and low affinity binding sites for p-NP-GP (Alonso et al. 1991) and that the low affinity site can be selectively inhibited by bromoconduritol (Alonso et al. 1993). We propose a model in which the MRH domain of GluIIβ binds the glycan substrate and by doing so increases GluIIα's effective affinity for the substrate. Given that GluIIα has a lower affinity for the G1 glycan compared to the G2 glycan, it follows that the abolition of MRH binding would have a greater effect on Glc1 trimming than on Glc2 trimming. This is indeed the case as the Y677F mutation had a major effect on Glc2 trimming but completely abolished Glc1 trimming.
The role of the G2B domain (Q63-E132) is largely unknown although it is the most highly conserved region of Gtb1p, having a 61% similarity to human GluIIβ compared to 21% similarity for the whole protein (Pelletier et al. 2000). Two mutations in this domain, D125A and E132A, produced defects in Gtb1p-dependent trimming. Interestingly, this domain has been implicated in the interaction with GluIIα (Arendt and Ostergaard 2000), and the reduced interaction observed for I125A in co-immunoprecipitation studies with Gls2p appears to support this idea. However, the E132A mutant protein still interacted with Gls2p (Figure 4E) and yet has a very severe Glc1 trimming defect implying a further role for this domain. The function of the G2B domain appears distinct from that of the MRH domain as mutating both domains together has an additive effect on Glc2 trimming and given the different effect each domain has on Glc1 trimming. While no sign of Glc1 trimming was apparent in the Y677F mutant, after a 45 min chase the E132A mutant appears to remain competent for Glc1 trimming albeit less efficiently than the wildtype Gtb1p. This region of the protein bears similarity to the C-type lectin domain and although this domain is not thought to exist in lower eukaryotes (Zelensky and Gready 2005), we cannot discount a lectin activity for this region of Gtb1p. Perhaps a separate lectin activity from the G2B domain in combination with that of the MRH domain would serve to align Gtb1p precisely on the glycan, potentiating the glucose trimming reaction. The specificity of the lectin activity of Gtb1p will be revealed by analysis of binding to different high-mannose glycans and it will be interesting to discover the relative specificities and contributions of the G2B and MRH domains.
Material and methods
Yeast strains, media, and growth conditions
Yeast strains were grown at 30°C with rotation in the YP medium (2% peptone, 1% yeast extract) containing 20 mg/L adenine and 2% glucose (YPAD) or in minimal medium (0.67% yeast nitrogen base) with 2% glucose plus appropriate supplements for selective growth. All growth media were obtained from Difco Labs (MD, USA). Standard techniques of mating haploid strains, sporulation, and tetrad analysis were employed to construct double mutant strains. The genotypes of yeast strains used are detailed in Table I. Null mutant strains were obtained from the systematic knockout series in strain BY4742 (Winzeler et al. 1999).
Table I.
List of S. cerevisiae strains used
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | (Brachmann et al. 1998) |
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | (Brachmann et al. 1998) |
| W303 | MATa, ade2–1, can1–100, his3–11,15, leu2–3, 112, trp1–1, ura3–1 | (Thomas and Rothstein 1989) |
| BWY616 | As BY4742 except gtb1::HA-kanMX4 | (Wilkinson et al. 2006) |
| BWY618 | As BY4741 except gls2::myc-kanMX4 | (Wilkinson et al. 2006) |
| BWY624 | MATα, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gls2::myc-kanMX4, Δgtb1::kanMX4 | This study |
| BWY629 | MATα, his3Δ1, leu2Δ0, ura3Δ0, Δalg8, Δgls2 | (Wilkinson et al. 2006) |
| BWY635 | MATα, his3Δ1, leu2Δ0, ura3Δ0, Δgtb1, Δmns1 | (Wilkinson et al. 2006) |
| BWY636 | MATα, his3Δ1, leu2Δ0, ura3Δ0, Δgls2, Δmns1 | (Wilkinson et al. 2006) |
Molecular biology
The GTB1::HA sequence was amplified by PCR from the yeast strain BWY616 with the forward primer GGACTAGTGGGTAGGTGCACAAGCG and reverse primer ACGCGTCGACCACATCATGCCCCTGAGC which incorporated SpeI and SalI restriction enzyme sites at the respective ends of the amplified sequence. The PCR product was cloned into a TOPO-TA vector (Invitrogen, USA) according to the manufacturer's instructions. The resulting plasmid was used as a template for the generation of point mutants using a QuickChange SDM kit (Stratagene, La Jolla, CA) as per the manufacturer's instructions. DNA sequence analysis was carried out to verify that the correct mutations had been made. The wildtype GTB1::HA cassette and all point mutant versions were then subcloned into the single-copy yeast expression vector pRS316 (Sikorski and Hieter 1989) after excision from TOPO-TA using SpeI and SalI. Individual plasmids were then transformed into the required yeast strain by lithium acetate transformation.
Yeast cell extract preparation
Yeast whole cell extracts were prepared from 5.0 A600 nm units of exponentially growing cells by suspension in the SDS sample buffer containing 5% β-mercapto-ethanol (β-ME) and 0.5 mm glass beads followed by disruption at 6.5 M/s for 30 s (Hybaid Ribolyser, Fisher Scientific, USA) and incubation for 5 min at 95°C.
Antibodies and immunoblot analysis
Antibodies raised against pro-α-factor (Tyson and Stirling 2000) and ppCPY (Young et al. 2001) have been previously described. Monoclonal antibodies against the myc (9E10) and HA (12CA5) epitopes were obtained from Sigma-Aldrich (St. Louis, MO) as was the antibody against glucose-6-phosphate dehydrogenase (G6P). After migration on SDS–PAGE gels, proteins were electroblotted to PVDF membranes (Immobilon-P, Millipore, Boston, MA), and antibody incubations were performed as previously described (Wilkinson et al. 1997). Immunodetection was carried out using enhanced chemi-luminescence (Perkin-Elmer, Boston, MA) with HRP-conjugated IgGs (Sigma-Aldrich, St. Louis, MO).
Native immunoprecipitation
For native immunoprecipitation, 5.0 A280 nm units of microsomes, prepared as previously described (Wilkinson et al. 1997), were pelleted at 17,000 × g for 15 min at 4°C and re-suspended with vortexing in 500 μL of solubilization buffer (20 mM Tris-HCl pH 7.4, 5 mM Mg(OAc)2, 10 μg/mL leupeptin, 5 μg/mL chymostatin/pepstatin, 10 μg/mL aprotinin, 1 mM AEBSF, 12% glycerol) containing 1% Triton X-100 and 200 mM NaCl. The mixture was incubated on ice for 30 min prior to centrifugation at 10,000 × g for 10 min. For anti-HA precipitation, 50 μL of anti-HA affinity matrix (rat monoclonal antibody, clone 3F10, Roche Diagnostics, IN, USA), pre-washed in the solubilization buffer, was added followed by incubation at 4°C for 2 h. Antigens were dissociated from the beads by the addition of SDS sample buffer containing 5% β-ME and heating at 95°C for 5 min.
Radio-labeling and denaturing immunoprecipitation
Yeast strains were grown overnight to 0.3 A600 nm in selective YNB media without any ammonium sulfate but containing, in addition to the required supplements, 10 mM ammonium chloride. Typically, 7.5 A600 nm equivalents of cells were pelleted before re-suspension in 0.5 mL of the appropriate media and incubation at 30°C for 5 min. Cells were then radio-labeled with 10 μCi/A600 nm [35S] methionine/cysteine (Met/Cys) (Perkin-Elmer, Boston, MA) and incubated at 30°C for 4 min. Cells were pelleted upon completion of radio-labeling, the s/n was removed and the cell pellet was immediately snap-frozen in liquid N2. Cell pellets were subsequently boiled for 5 min at 95°C to destroy enzymatic activity before being re-suspended in the 0.5 mL spheroplast buffer (1.4 M sorbitol, 50 mM Tris-HCl pH7.4, 2 mM MgCl2, 10 mM NaN3) and incubated at 30°C for 30 min with 12 U/A600 nm of yeast lytic enzyme (Sigma-Aldrich, St. Louis, MO) in order to digest the cell wall and form spheroplasts. Spheroplasts formed were pelleted and re-suspended in the 200 μL lysis buffer (50 mM Tris-HCl pH7.4, 1 mM EDTA, 1% SDS). Samples were then boiled at 95°C for 5 min followed by 2 min on ice. One milliliter of immunoprecipitation (IP) buffer (187.5 mM NaCl, 62.5 mM Tris-HCl pH8.0, 6.25 mM EDTA, 1.25% Triton X-100) was added to each sample, followed by the addition of 50 μL insoluble protein A suspension (Sigma-Aldrich, St. Louis, MO). Samples were cleared by rotation at room temperature (RT) for 30 min, after which insoluble protein A was pelleted. The resulting s/n was transferred to a clean tube and 2 μL/A600 nm of antibody was added. Samples were rotated at RT for 2 h. Following this, 50 μL of Protein A-Sepharose CL4B beads (as a 20% w/v suspension in IP buffer containing 0.1% SDS) was added and rotated for a further hour at RT. Beads were then pelleted by centrifuging and washed three times with 1 mL of IP buffer. Antigens were dissociated from beads by the addition of SDS sample buffer containing 5% β-ME and heating to 95°C for 5 min. Samples were resolved by SDS–PAGE (resolving over 20 cm) before being processed for autoradiography.
Pulse-chase
Cells were prepared for radio-labeling as described above but a 5 min labeling was carried out. A sample was taken prior to chase initiation and this constituted the time zero sample. Chases were performed by adding cold nonradioactive Met and Cys and sodium azide (NaN3) (final concentration of 2 mM each) to the sample upon completion of the labeling period and returning the sample to agitation at 30°C. NaN3 was used to block ER-to-Golgi transport so that glycan trimming could be monitored for extended periods. At various time points after this, an equal amount of media were removed, the cells were pelleted and after removal of supernatant were snap-frozen in liquid N2 and processed for denaturing immunoprecipitation as described above.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust.
Conflict of interest statement
None declared.
References
- Alonso JM, Santa-Cecilia A, Calvo P. Glucosidase II from rat liver microsomes. Kinetic model for binding and hydrolysis. Biochem J. 1991;278:721–727. doi: 10.1042/bj2780721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Santa-Cecilia A, Calvo P. Effect of bromoconduritol on glucosidase II from rat liver. A new kinetic model for the binding and hydrolysis of the substrate. Eur J Biochem. 1993;215:37–42. doi: 10.1111/j.1432-1033.1993.tb18004.x. [DOI] [PubMed] [Google Scholar]
- Arendt CW, Ostergaard HL. Two distinct domains of the beta-subunit of glucosidase II interact with the catalytic alpha-subunit. Glycobiology. 2000;10:487–492. doi: 10.1093/glycob/10.5.487. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Camirand A, Heysen A, Grondin B, Herscovics A. Glycoprotein biosynthesis in Saccharomyces cerevisiae. Isolation and characterization of the gene encoding a specific processing alpha-mannosidase. J Biol Chem. 1991;266:15120–15127. [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AC, Kendrick-Jones J, Reinach FC. Determinants of ion specificity on EF-hands sites. Conversion of the Ca2+/Mg2+ site of smooth muscle myosin regulatory light chain into a Ca(2+)-specific site. J Biol Chem. 1995;270:6773–6778. doi: 10.1074/jbc.270.12.6773. [DOI] [PubMed] [Google Scholar]
- Deprez P, Gautschi M, Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell. 2005;19:183–195. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Hubbard SC, Robbins PW. Synthesis and processing of protein-linked oligosaccharides in vivo. J Biol Chem. 1979;254:4568–4576. [PubMed] [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, te Heesen S, Aebi M, Roth J. Genetic tailoring of N-linked oligosaccharides: The role of glucose residues in glycoprotein processing of Saccharomyces cerevisiae in vivo. Glycobiology. 1998;8:155–164. doi: 10.1093/glycob/8.2.155. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Molinari M. N-Linked glycan recognition and processing: The molecular basis of endoplasmic reticulum quality control. Curr Opin Struct Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr Biol. 2001;11:R499–R501. doi: 10.1016/s0960-9822(01)00302-5. [DOI] [PubMed] [Google Scholar]
- Pelletier MF, Marcil A, Sevigny G, Jakob CA, Tessier DC, Chevet E, Menard R, Bergeron JJ, Thomas DY. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology. 2000;10:815–827. doi: 10.1093/glycob/10.8.815. [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Dijkgraaf GJ, Shahinian S, Herscovics A, Bussey H. The yeast CWH41 gene encodes glucosidase I. Glycobiology. 1997;7:997–1004. doi: 10.1093/glycob/7.7.997. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JF, Ebersold M, Helenius A. Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 1998;17:396–405. doi: 10.1093/emboj/17.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Totani K, Ihara Y, Matsuo I, Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high-mannose-type glycans. J Biol Chem. 2006;281:31502–31508. doi: 10.1074/jbc.M605457200. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Fleming KG, Helenius A. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry. 2001;40:10717–10722. doi: 10.1021/bi010629u. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Simons JF, Helenius A. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem. 1996;271:27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BM, Esnault Y, Craven RA, Skiba F, Fieschi J, Kepes F, Stirling CJ. Molecular architecture of the ER translocase probed by chemical crosslinking of Sss1p to complementary fragments of Sec61p. EMBO J. 1997;16:4549–4559. doi: 10.1093/emboj/16.15.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BM, Purswani J, Stirling CJ. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J Biol Chem. 2006;281:6325–6333. doi: 10.1074/jbc.M510455200. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Young BP, Craven RA, Reid P, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.