Abstract
The combination of major resistance genes with quantitative resistance factors is hypothesized as a promising breeding strategy to preserve the durability of resistant cultivar, as recently observed in different pathosystems. Using the pepper (Capsicum annuum)/Potato virus Y (PVY, genus Potyvirus) pathosystem, we aimed at identifying plant genetic factors directly affecting the frequency of virus adaptation to the major resistance gene pvr23 and at comparing them with genetic factors affecting quantitative resistance. The resistance breakdown frequency was a highly heritable trait (h2=0.87). Four loci including additive quantitative trait loci (QTLs) and epistatic interactions explained together 70% of the variance of pvr23 breakdown frequency. Three of the four QTLs controlling pvr23 breakdown frequency were also involved in quantitative resistance, strongly suggesting that QTLs controlling quantitative resistance have a pleiotropic effect on the durability of the major resistance gene. With the first mapping of QTLs directly affecting resistance durability, this study provides a rationale for sustainable resistance breeding. Surprisingly, a genetic trade-off was observed between the durability of PVY resistance controlled by pvr23 and the spectrum of the resistance against different potyviruses. This trade-off seemed to have been resolved by the combination of minor-effect durability QTLs under long-term farmer selection.
Introduction
The use of resistant cultivars provides efficient, economical and environmentally friendly crop protection against plant pathogens, and many cultivars carrying major resistance genes successfully protected cultivated crops against their main pathogens. When available in genetic resources, major resistance genes are very attractive for breeding because of their simple inheritance that makes their introgression through backcrosses rapid and cheap to high-yielding but susceptible cultivars and because they are expected to confer a nearly complete resistance against the targeted pathogen. These resistances involve a gene-for-gene interaction and are usually race-specific. The widespread deployment of such resistance genes in many elite cultivars imposes a strong selection pressure on the pathogen population leading to the appearance and/or the increase in frequency of pathogen variants overcoming the resistance (or resistance-breaking (RB) variants). This adaptation of the target pathogen makes the resistance ineffective, losing its economical interest. In order to preserve the durability of cultivar resistance, management strategies were developed to handle the adaptation of the pathogen populations and limit the appearance of RB variants. When several major resistance genes are available to control a given pathogen species, pyramiding of the distinct genes into a same cultivar, alternation of resistance genes across cultivation cycles or association of cultivars with distinct resistance genes during the same cultivation cycle were proposed (Wolfe, 1985; McDonald and Linde, 2002; Mundt et al., 2002; Pink, 2002).
More recently, another strategy was suggested from experimental data showing that the durability of a major resistance gene was dependent on the plant genetic background in which it was introgressed. In three distinct pathosystems (plant–virus/fungus/nematode) the emergence and/or frequency of the pathogen variants breaking down the major resistance gene remained low or did not occur in the cultivars that combined the major resistance gene in a quantitatively resistant genetic background but increased when the resistance gene was introgressed into a plant genotype with a susceptible genetic background (Palloix et al., 2009; Brun et al., 2010; Fournet et al., 2013). These results raise the hypothesis that the selective value of allelic variation at a resistance locus depends on the genetic background. It addresses the question of how genetic background influences the effect of mutations and evolution (Chandler et al., 2013). Considering plant breeding practices it suggests that combining qualitative and quantitative resistance increases the durability of plant protection. This also raises a paradox, as the increase in resistance efficiency is expected to increase the selection pressure on the pathogen population and thus the risk to select adapted pathogen genotypes (Poland et al., 2009). However, none of the pathosystem studies cited above formally demonstrated that the increase in durability resulted from the action of quantitative resistance factors, as the durability of the major resistance was compared between pairs of genotypes with fully different genetic backgrounds. Consequently, the decrease in resistance breakdown frequency might be due to any other genes from those genetic backgrounds. Further on, Quenouille et al. (2013) showed that the breakdown frequency of the major resistance allele pvr23 controlling Potato virus Y (PVY) was correlated to the quantitative resistance level in a set of pepper genotypes carrying pvr23 and differing in their genetic background. This suggested that the breakdown frequency of the major allele and the quantitative resistance shared a common genetic basis. When very few major resistance genes are available, this would open the possibility to preserve their durability. Moreover, it would provide tools for breeders to construct resistant cultivars with enhanced durability.
In our study, we (i) examined whether resistance breakdown is a heritable trait, (ii) described the genetic architecture underlying the genetic background that affects the breakdown frequency of a major resistance gene and (iii) discussed how breeders can exploit these results to preserve the durability of major resistance genes. Using the pepper (Capsicum annuum)/PVY system, our strategy consisted of (i) detecting quantitative trait loci (QTLs) affecting the breakdown frequency of the pvr23 major resistance allele, as breakdown frequency is a key determinant of resistance durability and (ii) comparative mapping between these QTLs and QTLs affecting the quantitative resistance to PVY.
Materials and methods
Mapping population
A segregating doubled-haploid (DH) population comprising 350 lines was obtained from the F1 hybrid between two C. annuum inbred lines: ‘Perennial' carrying the PVY major resistance allele pvr23 in a partially resistant genetic background is an inbred line issued from an Indian landrace and ‘Yolo Wonder' carrying the PVY susceptibility allele pvr2+. All DH lines were genotyped with a tetra-primer ARMS–PCR (amplification refractory mutation system–polymerase chain reaction) marker that targets single nucleotide polymorphism (SNP) signatures differentiating pvr2+ from pvr23 (Rubio et al., 2008). For phenotyping, we used the 153 DH lines carrying the pvr23 allele but segregating for the genetic background.
Virus material
For evaluation of RB frequencies, we used a PVY chimera between PVY clones SON41p and LYE84.2 (Montarry et al., 2011). This PVY chimera, named ‘CI chimera', differs from SON41p only by the CI cistron, which was replaced by that of LYE84.2. The rest of the genome and more particularly the region directly involved in pvr23 RB (the VPg cistron) are strictly identical between the two PVY clones. This ‘CI chimera' was chosen because of its higher ability to break the pvr23 resistance down compared with the parental clones (Montarry et al., 2011). However, the breakdown capacity of the ‘CI chimera' was shown to be highly correlated with that of SON41p when inoculated to a large range of pepper genotypes (Quenouille et al., 2013). The ‘CI chimera' and SON41p were not infectious per se in plants carrying pvr23—that is, in these plants, the ‘CI chimera' showed very weak accumulation detected through reverse transcriptase–polymerase chain reaction but not enzyme-linked immunosorbent assay (ELISA) and suggesting that the RB mutation occurred in the resistant plant (Montarry et al., 2011), whereas mutants of the ‘CI chimera' possessing single non-synonymous substitutions in the VPg cistron could be detected at high concentration in systemically infected leaves. In addition, the CI chimera showed the same distributions of RB mutations when inoculated on plants carrying pvr23 when compared with SON41p. The level of quantitative resistance controlled by the plant genetic background was evaluated using a single-nucleotide mutant of the ‘CI chimera' carrying an amino-acid substitution at position 119 (aspartic acid to asparagine) of the VPg (named ‘CI chimera VPg-N'), which conferred the breakdown capacity towards the pvr23 resistance gene and was shown to be the most frequent in isolates from field as well as from experimental evolution experiments (Ayme et al., 2006; Palloix et al., 2009). This mutant enables the pathogen to overcome the resistance conferred by pvr23 and to reveal the quantitative resistance due to the genetic background.
Measure of resistance breakdown frequency
The breakdown frequency of pvr23 was tested after inoculation of the PVY ‘CI chimera' to the 153 DH lines carrying the pvr23 allele. Throughout the test, plants were grown in a climate-controlled room at 20–22 °C, 12-h light/day. Inoculum was obtained after virus propagation in Nicotiana tabacum cv. Xanthi plants. Fifty grams of Xanthi leaves showing PVY symptoms were crushed in 200 ml of 0.03 M phosphate buffer (pH 7.0) supplemented with 2% (w:v) diethyldithiocarbamate, 4.5 g of active charcoal and 4.5 g of Carborundum. Thirty pepper seedlings with two expanded cotyledons (2–3 weeks after sowing) per genotype were inoculated mechanically on their cotyledons. Thirty-eight days post inoculation (dpi), plants were submitted to virus detection by DAS-ELISA as described by Ayme et al. (2006). Under these conditions, in each plant infected systemically, the virus population was shown to be composed of one or several VPg mutants carrying a pvr23 RB mutation (Montarry et al., 2011). For each pepper line, the RB frequency of pvr23 was assessed by the ratio of the number of systemically infected plants over the total number of inoculated plants. Two independent tests of 30 plants per DH line were performed.
Measure of quantitative resistance
To evaluate the level of the quantitative resistance due to the genetic background, we used the ‘CI chimera VPg-N' PVY mutant that overcomes the resistance conferred by pvr23 (infects all the genotypes carrying pvr23) and reveals the quantitative resistance due to the genetic background. Two different traits were assessed: the area under the disease progress curve (AUDPC) and the virus accumulation. The PVY ‘CI chimera VPg-N' was mechanically inoculated on 20 plants of each DH lines carrying pvr23. The plants were grown in a climate-controlled room as previously described. The AUDPC assessment combined both intensity of symptoms and latency period: symptom intensity was evaluated, on the 20 plants, in a 3-class scale (1=no symptom, 2=weak mosaic and 3=severe mosaic/necrosis) at 14, 21, 28 and 35 dpi. AUDPC was calculated using the formula:
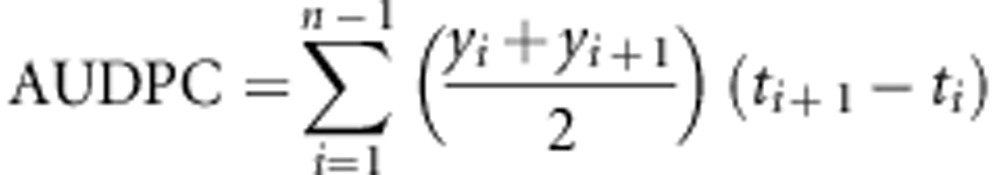 |
where ‘n' is the number of symptom assessments, ‘y' the symptom intensity and ‘t' the time in dpi.
At 36 dpi, the virus accumulation was independently evaluated on 10 individual plants per DH line by semiquantitative DAS-ELISA, using a dilution range of extracts of PVY-infected plants, and expressed relatively to a common reference sample incorporated in each ELISA plate, as in Ayme et al. (2006).
Genotyping of molecular markers and linkage map
The 350 DH progeny was an extension of the former ‘PY' mapping population (Lefebvre et al., 2002). It was genotyped with 236 molecular markers: 108 SNPs (Nicolaï et al., 2012), 29 SNPs (Jung et al., 2010), 47 simple sequence repeats (Lee et al., 2004; Minamiyama et al., 2006; Nagy et al., 2007), 33 amplified fragment-length polymorphisms (Lefebvre et al., 2002), one single-strand conformation polymorphism (SSCP-PM5, Fazari et al., 2012), two known genes and 16 new markers (Supplementary Table I) corresponding to previously mapped restriction fragment length polymorphisms and/or conserved ortholog set (COS II) sequences and designed using the Sol Genomics network (www.solgenomics.net). The two known genes were pvr2 (the major resistance locus to PVY) genotyped using the tetra-ARMS marker as in Rubio et al. (2008), and pvr6 coding for the eIF(iso)4E (eukaryotic initiation factor iso4E) protein genotyped with a new marker targeting a 82-nucleotide deletion differentiating alleles pvr6+ and pvr6 (Ruffel et al., 2006; Supplementary data I). The DH progeny was also genotyped for two phenotypic markers, the presence/absence of pungency in the fruits (locus C) and the erected/pendant fruit position (locus up).
The genetic linkage map was constructed using the Mapmaker software version 3.0b (Lander et al., 1987) with a LOD score threshold of 3.0, a maximum recombination fraction of 0.3 and distances between markers were calculated using the Kosambi mapping function. For each linkage group, marker order was verified with the ‘ripple' command and markers were conserved only if the LOD value was greater than 2.0. Three out of the 236 markers remained unlinked and 33 were deleted because they had no unique placement at LOD 2.0. The resulting map contained 200 markers distributed in 13 linkage groups (LG), which were assigned to the 12 pepper chromosomes through alignment of markers with previous pepper maps (Lefebvre et al., 2002; Wu et al., 2009; Supplementary Data II). The total length of the map was 2457.7 cm with an average length interval between markers of 12.3±12.4 cm. The increased length of this map compared with former pepper maps was mainly attributed to the larger size of the progeny (350 individuals) that allowed to declare significant a recombination rate up to 0.45 cm (slightly more than 50 Kosambi cM) and also to the assignment of the two linkage groups to the single P7 chromosome, resulting in longer chromosomes (Supplementary Data II).
Statistical analyses
Descriptive statistics was performed with the R software (http://www.r-project.org/). For each phenotypic trait, narrow-sense heritability (h2) was estimated using the formula h2=σ2G/(σ2G+σ2E/n), where σ2G corresponds to the genotypic variance, σ2E to the environment (error) variance and n to the number of replicates.
For AUDPC, the mean distributions were close to a normal distribution and we analyzed non-transformed data for QTL detection. The means of virus accumulation (VA) were non-normally distributed. Thus, for QTL detection, these data were log-transformed [ln (VA)+1] to approximate a normal distribution. Resistance breakdown frequency was a non-normally distributed trait with an excess of low values, and no transformation was efficient to improve the distribution.
QTL analysis by regression interval mapping
For a non-normal distribution, QTL detection can be performed with a standard regression-based interval mapping approach, which is robust against deviations from normality when the threshold value is calculated by permutations (Rebaï, 1997). QTL analyses were performed using iterative composite interval mapping (iQTLm) methods implemented in the MCQTL v.5.2.4 (Jourjon et al., 2005) software. MCQTL performed QTL mapping using a linear regression model and the iQTLm scan method allows a multiple QTL model to be determined automatically (Charcosset et al., 2001). The QTL significance was determined using the MCQTL test, which is equal to –log10 of the Fisher test P-value as described in the MCQTL v.5 reference manual (http://carlit.toulouse.inra.fr/MCQTL/). The significance threshold of the MCQTL test was estimated after performing 1000 permutation tests for each trait to reach a genome-wide type-I error rate of 0.05 and 0.1 for QTLs and cofactors, respectively. This resulted in a threshold of 3.4 for [log (VA) +1] and of 3.5 for RB and AUDPC. The cofactor threshold was fixed at 3.2 for the three traits. Cofactor selection was performed by forward selection of markers. The QTL confidence intervals (CI) were defined as a two unit drop-off of the MCQTL test around the maximum MCQTL score to obtain approximately a 95% CI: CI=[max (MCQTL test)−2; max (MCQTL test)+2]. Epistasis between QTLs and the genetic background was tested using the iterative method implemented in MCQTL (threshold of 3.5). The part of phenotypic variation explained by additive and/or epistasic effects of each QTL (R2 coefficients) and the global R2 were estimated using the full QTL model in MCQTL.
QTL analysis by maximum likelihood interval mapping
For AUDPC and virus accumulation (log (VA)+1), QTL analyses were also performed using multiple QTL mapping (MQM) methods implemented in the R/qtl package of the R (Broman et al., 2003) software. We did not use this method for the RB trait because data were non-normally distributed. MQM is a maximum likelihood-based interval mapping method composed of three steps. First, missing data are ‘augmented'—that is, all genotype possibilities are modeled and their probabilities calculated from neighboring marker information. In the second step, significant markers are detected with the ‘mqmscan' function without cofactor, which corresponds to a simple interval mapping with a backward elimination. In the last step, QTLs are detected using markers detected in step two as cofactors, which corresponds to a composite interval mapping, and a backward elimination is applied to remove the nonsignificant cofactors and to establish the final QTL model. Significance LOD thresholds were estimated for each trait, with a genome-wide type-I error rate of 0.05, after performing 1000 permutation tests. The LOD threshold was estimated at 2.6 for the two traits. For each QTL, the CI was defined as a 2-LOD drop-off around the maximum LOD score to obtain approximately a 95% CI. R2 coefficients were calculated with the ‘fitqtl' function of R/qtl.
Results
Characterization of the phenotypic traits
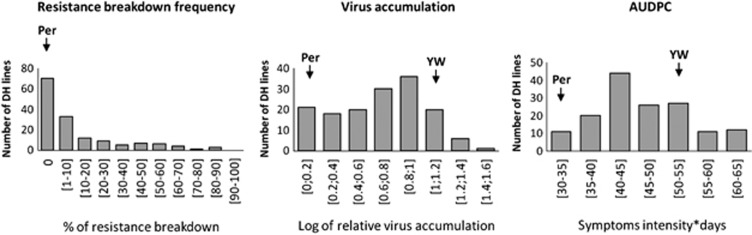
To evaluate the frequency of pvr23 resistance breakdown, the ‘CI Chimera', which is not infectious per se toward pvr23-resistant plants, was inoculated on the set of 153 DH carrying pvr23. In these plants, only RB mutants of the ‘CI Chimera' possessing single non-synonymous substitutions in the factor (VPg) conferring virulence towards the pvr23 allele were detected in previous studies (Ayme et al., 2006; Palloix et al., 2009; Montarry et al., 2011). In our tests, mosaic symptoms appeared in 100% of the susceptible (pvr2+) control Yolo Wonder 2 weeks after inoculation; however, mosaic or necrotic symptoms appeared later (4–5 weeks after inoculation) in some of the plants of some DH lines. Thirty-eight dpi, the ELISA revealed an important phenotypic variability between the 153 DH lines, with frequencies of systemic infection, corresponding to RB frequencies, varying from 0 to 93.2% with a mean (±s.d.) of 14.7% (±23.8). The correlation between the two independent tests was high (ρSpearman=0.75; P<10−7, Supplementary Data 3), and a heritability of 0.87 was estimated. The distribution of the trait was strongly skewed towards low values, with 70 DH lines displaying no resistance breakdown and 83 HD lines displaying an RB frequency between 1.7 and 93.2% (Figure 1).
Figure 1.
Histograms represent distributions of DH lines into resistance breakdown frequency, virus accumulation (ratio concentration of the sample/concentration of the control) and AUDPC classes. Arrows indicate the position of each parent in the phenotypic distribution. Per: Perennial, YW: Yolo Wonder.
The quantitative resistance was assessed using a pvr23-breaking mutant of the ‘CI Chimera', the ‘CI Chimera VPg-N'. With this virus, the pvr23-resistant lines displayed symptoms as soon as 14 dpi and 100% of the plants were infected at 35 dpi as attested by DAS-ELISA. Highly significant variations for VA and AUDPC were observed between DH lines with a heritability of 0.64 for Ln(VA+1) and 0.98 for AUDPC. For statistical and QTL analyses, VA values were log-transformed [ln(VA+1)] to approximate a normal distribution. The ln(VA+1) varied from 0 to 1.43 with a mean equal to 0.67±0.35 and AUDPC varied from 29.75 to 63 with a mean equal to 46.6±7.9 (Figure 1).
Significant correlations (P<0.05) were observed between the three traits with Spearman's coefficients equal to 0.40, 0.33 and 0.32 for RB/VA, RB/AUDPC and VA/AUDPC, respectively.
Mapping QTLs for the quantitative resistance traits
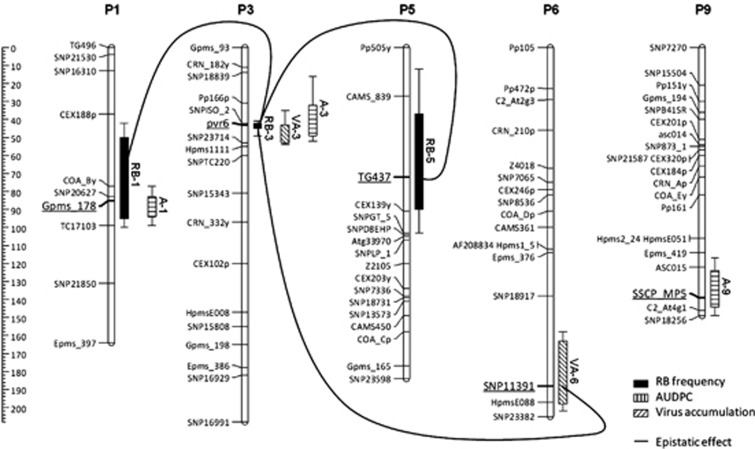
Distributions of the quantitative resistance traits AUDPC and log(VA+1) were close to normality. For these two traits, QTL detection was performed using iQTLm and MQM methods. Two significant QTLs for VA and three for AUDPC were detected by both iQTLm and MQM methods. These QTLs were named VA-3, VA-6, A-1, A-3 and A-9, according to the trait considered and the chromosome location. Hereafter, we will detail only the results from iQTLm because results from both methods are similar. The position, significance and effect of each QTL are detailed in Table 1 and Figure 2.
Table 1. QTL detected with the iterative QTL mapping (iQTLm) method.
| Virus | Trait | QTL | Location a | Closest marker | CI b | Direction c | MCQTL test d |
Variation explained
|
Heritability | |
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Trait | |||||||||
| QTLs with additive effect: | ||||||||||
| Wild-type PVY clone (CI Chimera) | Resistance breakdown | Rb-1 | 1@87 | Gpms_178 | 42–100 | Per | 5.2 | 12.8 | 68.9 | 0.87 |
| Rb-3 | 3@43.1 | Pvr6 | 41–49 | YW | 17.4 | 39.8 | ||||
| Rb-5 | 5@57.1 | TG437 | 12–103 | Per | 3.6 | 8.6 | ||||
| QTLs with epistatic effect: | ||||||||||
| eRb-1-3 | 1@87–3@43.1 | Gpms_178/Pvr6 | 7.4 | 18.9 | ||||||
| eRb-3-5 | 3@43.1–5@57.1 | Pvr6/TG437 | 6.6 | 16.9 | ||||||
| eRb-3-6 | 3@43.1–6@188.3 | Pvr6/SNP11391 | 4.2 | 10.6 | ||||||
| Mutant PVY clone (CI Chimera VPg-N) | Virus accumulation | QTLs with additive effect: | 0.64 | |||||||
| Va-3 | 3@49.1 | SNP23714 | 35–54 | YW | 14.6 | 34.5 | 43.9 | |||
| Va-6 | 6@190.3 | SNP11391 | 158–202 | Per | 6.3 | 15.7 | ||||
| QTLs with additive effect: | ||||||||||
| Mutant PVY clone (CI Chimera VPg-N) | AUDPC | A-1 | 1@87 | Gpms_178 | 77–99 | Per | 6.3 | 16.1 | 34.1 | 0.98 |
| A-3 | 3@40.8 | SNPISO_2 | 16–52 | YW | 6.2 | 15.5 | ||||
| A-9 | 9@132 | SSCP_MP5 | 117–149 | YW | 5.9 | 14.9 | ||||
Abbreviations: AUDPC, area under the disease progress curve; PVY, Potato virus Y; QTL, quantitative trait loci; RB, resistance breakdown ; SNP, single nucleotide length polymorphism; SSCP, single-strand conformation polymorphism.
QTL locations are coded in the following way: chromosome @ position on the chromosome in cM. For example, the QTL RB-1 is localized on the chromosome 1 at 87 cM (1@87).
The confidence interval was defined as a 2-MCQTL test drop-off around the maximum MCQTL test score.
Indicates the parent who decreases the value of the trait.
The MCQTL test significance threshold was set at P=0.05 and is egal to 3.4 for Virus accumulation and 3.5 for resistance breakdown and AUDPC.
Figure 2.
Map positions on Pepper genetic map of the significant QTLs for pvr23 RB frequency, the AUDPC and the VA. On the left, the scale indicates map distances calculated by the Kosambi mapping function. For each QTL, the length of the block represents the CI of the QTL using a MCQTL test unit fall of 1 and the length of the vertical bar represents the CI of the QTL using a MCQTL test unit fall of 2. For each QTL, the closest marker is underlined. Lines indicate epistatic effects between loci.
VA-3 and VA-6 explained 34.5% and 15.7% of the VA variation, respectively. Epistasis tests did not reveal any interaction between QTLs and the genetic background. The final model combining the effects of the two significant QTLs explained 43.9% of VA phenotypic variation corresponding to 69% of the trait heritability (h2=0.64).
A-1, A-3 and A-9 explained from 14.69, 15.5 and 16.1% of the AUDPC variation. Epistasis tests did not reveal any interaction between these QTLs and the genetic background. The part of the AUDPC phenotypic variation explained by A-1, A-3 and A-9 was equal to 34.1% corresponding to 35% of the trait heritability and a large part of the variability remained unexplained (h2=0.98).
Mapping QTLs for the frequency of pvr2 3 -resistance breakdown
Owing to the skewed distribution of the RB frequency, QTL detection was performed with the standard regression-based interval mapping approach (Haley and Knott, 1992; Rebaï, 1997) using the iQTLm (Charcosset et al., 2001) method implemented in the MCQTL software (Jourjon et al., 2005). Three significant QTLs, explaining 12.8%, 39.8% and 8.6% of the pvr23 RB frequency variation, were detected on chromosomes 1, 3 and 5, respectively. QTLs were named RB-1, RB-3 and RB-5 according to the trait and the chromosome location. Table 1 and Figure 2 detail position, significance and effect of each QTL. Epistasis tests revealed that RB-3 interacts with three distinct positions in the pepper genome, on chromosomes 1, 5 and 6. The positions on chromosomes 1 and 5 interacting with RB-3 are included in the CIs of RB-1 and RB-5, respectively, suggesting interaction effects between RB-1 and RB-3 and between RB-3 and RB-5 (Figure 2). The position on chromosome 6 that interacts with RB-3 did not show any additive effect and will be further named RB-6. The final model combining additive and epistatic effects of the significant QTLs explained 68.9% of the variation of pvr23 RB frequency, which corresponds to a great part (79%) of the trait heritability (h2=0.87). Owing to the skewed segregation of the RB trait, the QTL effects (as well as heritability) may have been overestimated. Indeed, the residues of the final model did not fit a normal distribution (Shapiro–Wilk normality test W=0.87, P<10−7). The analysis was reproduced after removing the 70 lines with an RB value of 0. The same additive QTLs were detected and the final model including these three additive QTLs explained 48% of the phenotypic variance (instead of 52% with the whole population) with a normal distribution of residues (W=0.98, P=0.55). The interaction effects were not significant anymore. This very probably resulted from the fact that removing the 70 individuals with RB=0 also removed most of the individuals carrying the Yolo Wonder allele at RB-3, resulting in highly unbalanced allele frequencies at this locus (71/12), reduced population size (n=83) and decreased statistical power.
Looking for co-localization between QTLs, the CI of A-1 proved fully included in the CI of RB-1, as was the CI of RB-3 within those of A-3 and VA-3. The major QTL affecting the RB frequency of pvr23 (RB-3) co-localizes with the major QTLs VA-3 and A-3 affecting VA and AUDPC, respectively. The QTL RB-1 also co-localizes with QTL A-1. The RB-6 locus displaying interaction effect with RB-3 is also located at the LOD peak of VA-6, indicating that RB-6 colocalizes with VA-6 (Figure 2).
Effect of parental alleles on the RB frequency
All genotypes carrying the Yolo Wonder allele at QTL RB-3 showed a very low-resistance breakdown frequency with a mean equal to 0.31%, whereas genotype carrying the Perennial allele at QTL RB-3 showed a much higher resistance breakdown frequency with a mean equal to 29.63%. The phenotypic values of the different allele combination between QTL RB-3 and epistatic QTLs are presented in Table 2. For genotypes carrying the Yolo Wonder allele at QTL RB-3, the resistance breakdown frequency was independent of the alleles at the interacting QTLs (mean of resistance breakdown frequency varies between 0.17 and 0.37%), so that these QTL effects were not significant in these genotypes. Contrarily, in genotypes carrying the Perennial allele at QTL RB-3, the RB frequency strongly depended on the alleles present at the interacting QTLs. The Yolo Wonder alleles at QTLs RB-1, RB-5 or RB-6 increased the RB frequency when combined with the Perennial allele at QTL RB-3, whereas the Perennial alleles at these QTLs strongly decreased the RB frequency (Table 2). Altogether, the combination of the Perennial alleles at the three epistatic QTLs (RB-1, RB-5 and RB-6) with the Perennial allele at RB-3 decreased the RB frequency from 69.7 to 5.3% (Table 2). The high effect of these QTLs in genotypes carrying the Perennial allele at RB3, contrarily to those carrying the Yolo Wonder allele at this QTL probably caused the significant interaction (scale effect).
Table 2. Effect of QTLs interaction on the resistance breakdown frequency of pvr2 3 .
|
Rb-1: Gpms_178 (1@85)
|
Rb-5: TG437 (5@72.3)
|
Rb-6: SNP11391 (6@188.3)
|
Combination of the 3 QTLs (Rb-1, Rb-5, Rb-6)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Yolo Wonder | Perennial | Yolo Wonder | Perennial | Yolo Wonder | Perennial | Yolo Wonder | Perennial | |
| Rb-3: (3@43.1) | ||||||||
| Yolo Wonder | 0.37% (34) | 0.30% (39) | 0.17% (39) | 0.47% (33) | 0.18% (37) | 0.36% (39) | 0.18% (9) | 0.36% (7) |
| Perennial | 42.0% (37) | 15.6% (34) | 41.8% (32) | 18.0% (31) | 36.2% (40) | 22.6% (30) | 69.7% (7) | 5.3% (6) |
This table presents the effect of the three significant epistatic interactions between loci and their combination for the resistance breakdown frequency of pvr23. The names and the positions of the markers in interaction are indicated. The numbers correspond to the average of the pvr23 resistance breakdown (RB) frequency (in %) for each allelic combination and the number between brackets corresponds to the number of lines belonging to each allelic class.
Discussion
Durable management of plant resistance to pathogen populations was mostly promoted through the spatio-temporal management of resistance genes but can also be achieved through management of resistance genes at the genotype scale: the cultivar genotype construction. In this paper we firstly succeeded to identify plant QTLs for a trait directly related to the evolution of the viral population: the frequency of emergence of viral variants adapted to the resistance. The colocation of the plant genetic factors modulating this viral evolution with the QTLs decreasing the virus accumulation further indicated that the more efficient the more durable was the resistance, which was not obvious since higher resistance levels are also expected to increase the selection pressure. Consequently, the high durability of the resistance of the Perennial pepper landrace was shown to result from a complex genetic architecture, including alleles at a major gene and at QTLs from the genetic background with complementary but also contradictory effects, which address the question of the selection processes leading to this combination in this landrace.
Heritability of the breakdown frequency of a major resistance gene
In this study, we measured the resistance breakdown (RB) frequency of a major resistance gene (pvr23) in a set of pepper DH lines carrying pvr23 but segregating for the genetic background. The observed RB frequency directly resulted from the frequency of appearance and from the accumulation dynamics of RB variants in the resistant plants, two steps of the virus evolution toward RB that are considered as major components of the resistance durability (Fabre et al., 2009). Previous studies of this pathosystem showed that the RB mutation most probably occurs in the resistant pepper host, which allowed for weak PVY multiplication and that the genetic background acts at several levels, including the mutational pathways of the virus to overcome resistance and the speed of selection of the RB variants (Montarry et al., 2011; Quenouille et al., 2013). The genetic analysis showed that the RB frequency of pvr23 was highly variable between genotypes (from 0 to 93.2%) and highly heritable (h2=0.87). Owing to the non-normal distribution of data for RB frequency, the h2 value is most probably overestimated. However, it fits the Spearman correlation value between the two repeats of the test (0.75), demonstrating the genetic control of this trait. This result is not trivial. Indeed, during the RB processes, several factors could have increased the heterogeneity of results between plants and experiments and consequently could have decreased the heritability of RB frequency. Among these factors are the stochastic nature of (i) the appearance of RB mutations in the PVY genome, (ii) the genetic drift acting on virus during systemic invasion of plants (García-Arenal et al., 2001; French and Stenger, 2003) and (iii) the fact that at least nine different VPg mutations, with contrasted effects on PVY competitiveness, can be involved in pvr23 breakdown (Ayme et al., 2006; Montarry et al., 2011). Our study demonstrated that measuring the resistance breakdown frequency with a reasonable number of plants (60 per genotype) compensates these factors of heterogeneity and allows the genetic analyses. The genetic background dependency of pvr23 durability could be attributed to four distinct genetic factors with quantitative effects: one major-QTL (RB-3) and three additional QTLs (RB-1, RB-5 and RB-6) acting additively and/or in interaction with RB-3. The discovery of such genetic factors shows that direct selection for alleles increasing the durability of a major resistance is feasible and opens new tools for durable resistance breeding. Nevertheless, the successful detection of such genetic factors affecting the RB frequency of a major resistance gene depends on the ability to generate RB strains at high frequencies through experimental evolution. For many viral diseases, the experimental conversion of an avirulent virus to an RB variant is rather easy and was commonly used to determine viral determinants of pathogenicity (Ayme et al., 2006; Margaria et al., 2007; Hajimorad et al., 2010; Lacroix et al., 2011). Moreover, the RB frequency of resistance genes is highly correlated with the durability of these genes in the field (Ayme et al., 2006; Janzac et al., 2009; Lacroix et al., 2011). In tobacco, field observations revealing that va2-RB isolates were more frequent than va0-RB isolates (Verrier and Doroszewska, 2004) were confirmed by results from experimental evolution (Lacroix et al., 2011). Moreover, the distributions of RB mutations in viral genomes are usually similar in field and laboratory observations (Moury et al., 2004; Ayme et al., 2006; Hajimorad et al., 2010). However, for some resistance genes that proved very durable in the field, it may be difficult to generate RB variants by direct inoculation of avirulent strains. In such cases, alternative protocols imposing strong inoculum pressures such as graft inoculation or inoculation through agroinfiltration can generate sufficient inoculum pressure to increase RB frequency (Bruun-Rasmussen et al., 2007; Janzac et al., 2009). These experimental procedures make it possible to measure the breakdown frequency of major resistance genes in many plant–virus pathosystems, providing powerful tools for improving the durability of virus resistance in plants. Considering other pathogens, experimental evolution may not be so relevant, due to their longer generation time, smaller populations and smaller mutation rates. However, if confirmed as a generic result, the relationship between RB QTLs and QTLs controlling the level of quantitative resistance may provide alternative selection criteria.
Alleles increasing quantitative resistance are favourable to the durability of major resistance genes
Using the same pepper DH progeny, we performed a QTL analysis of quantitative resistance. To get rid of the pvr23 major effect and to reveal the quantitative resistance conferred by the genetic background, we used a pvr23-RB PVY variant (the ‘CI Chimera VPg-N') that differs from the PVY variant used for the measurement of pvr23 RB frequency by a single nucleotide substitution. This mutation restores the interaction between the PVY VPg and the pepper eukaryotic translation initiation factor 4E1 (eIF4E1) encoded by pvr23, causing the breakdown of the pvr23-mediated resistance (Charron et al., 2008). Using this PVY variant, four QTLs affecting virus accumulation and/or symptom expression were detected. These four QTLs were also previously detected for the AUDPC trait with two other PVY strains, with identical allele-directed effects: resistance-favourable Perennial alleles on chromosomes P1 and P6 (LG3 and LG10 in Caranta et al., 1997) and favourable Yolo Wonder alleles on chromosomes P9 and P3 (LG9 and LG4 in Caranta et al., 1997). In this experiment, distinct natural PVY isolates with distinct pvr23 breaking down mutations in their VPg were used, indicating no differences between the QTLs detected using distinct VPg mutants. Our present results show that three of the four QTLs affecting the RB frequency of pvr23 colocate with QTLs affecting quantitative resistance (viral accumulation and/or AUDPC). These colocations may result from genetic linkage between the different traits or from a pleiotropic effect of quantitative resistance factors on the RB frequency of pvr23. Looking at the effect of parental alleles that colocate on chromosomes P1, P3 and P6, all the alleles that increased the quantitative resistance also decreased the breakdown frequency. Moreover, the QTL on chromosome P3 displayed the major effect on both virus accumulation and RB frequency. This concordance between parental allele effects for the different traits strongly suggests that colocations are due to pleiotropic effects and that alleles increasing the resistance level also increase the resistance durability. This is also consistent with the initial observations, in three pathosystems, that the durability of major genes is enhanced in partially resistant cultivars (Palloix et al., 2009; Brun et al., 2010; Fournet et al., 2013). However, it may look contradictory since higher resistance levels are expected to increase the selection pressure on the pathogen and the risk of selection of adapted pathogen populations (Poland et al., 2009). In a recent study of the different mechanisms that could account for the higher durability of polygenic resistance, Quenouille et al. (2013) showed the selection of adapted RB mutants by the polygenic resistance of Perennial was slower than by the monogenic resistance (pvr23 in a susceptible genetic background). In this same report, the additional quantitative resistance carried by the genetic background explained a large part of the variation in RB frequency. Dissecting the genetic background into QTLs indicate that breeding for quantitative resistance alleles will enhance the durability of major genes. Such a strategy is achievable for most pathosystems where breaking down variants or experimental evolution tests are not available.
Resistance durability as the result of a combination of mechanisms
At the landscape scale, it was proposed previously that diversifying the methods for pathogen control is an efficient strategy to limit adaptation of pathogen populations (Mundt et al., 2002; Pink, 2002). Diversifying resistance mechanisms at the genotype scale can also have a positive effect on durability. In accordance with Quenouille et al. (2013), our results raise the main impact of viral accumulation on the RB frequency of pvr23, since all QTLs involved in viral accumulation are also involved in the RB frequency of pvr23. Viral accumulation was measured in apical leaves and a decrease in viral accumulation can result from decreases of the efficiency of virus replication, cell-to-cell and/or systemic movement. Combining a major resistance gene with another gene involved in viral accumulation has already proved to enhance resistance durability in the PVY/tobacco system (Acosta-Leal and Xiong, 2008). In their study, Acosta-Leal and Xiong (2008) showed that the RB frequency of the va recessive gene that confers a resistance to PVY cell-to-cell movement in tobacco was reduced when va was combined with a second recessive gene (va2) conferring resistance to within-cell PVY accumulation. Two additional QTLs affected the RB frequency but had no detectable effect on viral accumulation (RB-1 and RB-5), suggesting distinct modes of action. QTL RB-1 colocates with QTL A-1 controlling symptom intensity and dynamics. In plants, a decrease in symptom intensity can result from resistance and/or tolerance mechanism. Here we consider tolerance in the sense of Schafer (1971) in which tolerant plants endure high pathogen loads without severe losses in field. Thus, QTL A-1 can be considered as a tolerance QTL since it reduces the AUDPC; however, it does not significantly affect viral accumulation. To our knowledge, mechanisms involved in plant tolerance are still unknown; however, they could also have an effect on the pvr23 RB frequency. Finally, QTL RB-5 did not colocate with any quantitative resistance QTL, suggesting that a third mechanism, independent of quantitative resistance, affects the pvr23 RB frequency. Quenouille et al. (2013) also showed that beside virus accumulation, the plant genetic background can control two additional modes of action: making the mutational pathways for viral resistance breakdown more complex and unlikely and modulating the selection pressure exerted on viral populations. QTLs RB-1 and RB-5 may be candidates for such modes of actions.
A genetic trade-off between broad-spectrum resistance and resistance durability that can be solved by breeding strategy
Among the four additive and/or epistatic QTLs affecting the RB frequency, the alleles from Perennial were shown to decrease the risk of pvr23 breakdown at three of these QTLs (RB-1, RB-5 and RB-6), whereas the Perennial allele increased strongly this risk for the major QTL RB-3. This result looks surprising since the pvr23 allele was previously shown to be highly durable in the Perennial landrace (Palloix et al., 2009). It is noteworthy that the peak of QTL RB-3 was localized at the pvr6 locus that encodes the eIF(iso)4E, an isoform of eIF4E1 encoded by pvr2. It was previously shown that the Perennial allele at pvr6 included a deletion of 82 nucleotides followed by a premature stop codon, compared with the pvr6+ allele from Yolo Wonder. The eIF(iso)4E protein encoded by the Perennial pvr6 allele was therefore non-functional (Ruffel et al., 2006) and can be considered as a deleted allele of pvr6+. When combined with different pvr2 alleles, the Perennial pvr6 allele was shown to enlarge the spectrum of resistance to additional potyviruses, including Chilli veinal mottle virus (ChiVMV) and Pepper veinal mottle virus (Caranta et al., 1997; Moury et al., 2005; Rubio et al., 2009). Hence, the Perennial allele at the pvr6/RB-3 locus contributes to a gain of resistance against Pepper veinal mottle virus and ChiVMV but also to a decrease in the durability of the pvr23 resistance to PVY together with a decrease in quantitative resistance traits VA and AUDPC. The Perennial inbred line was selected from a landrace issued from farm selection in India, where several potyviruses including ChiVMV are prevalent in pepper crops, and it is highly likely that pvr23 and pvr6 have been selected locally to enlarge the spectrum of resistance against these potyviruses. Whether pvr6 is the functional gene for the RB-3 QTL remains to be determined through functional validation. However, the tight colocation between pvr6 and RB-3 already reveals a trade-off between a large resistance spectrum against potyviruses when combined with a pvr2 resistance allele and the durability of the pvr23 PVY resistance in the Perennial genotype.
Such a trade-off looks unusual since resistance gene pyramiding and broad-spectrum resistance are often considered favourable to durability (McDonald and Linde, 2002; Kou and Wang, 2010). This statement was recently reinforced by Le Van et al. (2013) who showed that broad-spectrum resistance QTLs in apple did not exert any differential selection pressures on a mixed Venturia inaequalis inoculum, whereas narrow-spectrum QTLs did. In pepper, the combination of pvr2 resistance alleles (particularly pvr21 and pvr22 alleles) with pvr6 is already used in breeding programs to create resistant varieties against a large range of potyviruses (Rubio et al., 2009) and such cultivars may lead to a premature breakdown of pvr2-mediated resistance by PVY. More generally, translation initiation factor-mediated virus resistance is widespread (Robaglia and Caranta, 2006) and combination between mutated or knockout alleles at different genes coding for translation initiation factors is expected to provide a way to breed for broad-spectrum resistance. In tomato, the narrow-range resistance conferred by a knockout allele of eIF4E1 was enlarged when combined with a knockout of eIF4E2 (Mazier et al., 2011). Our study indicates combining mutant alleles from genes belonging to the same multigenic family should be used carefully, since the trade-off observed in pepper between large spectrum and durability of resistance may also occur in other plant species.
Our previous observations indicate introgressing resistance alleles from the plant germplasm into new elite cultivars with susceptible genetic backgrounds can endanger the long-term use of these genes as well as provide an evolutionary springboard to the pathogen for resistance breakdown (Palloix et al., 2009). Investigating the background dependence of the durability of pvr23 led to an improved understanding of how selection acted on this gene/genetic background combination (Chandler et al., 2013). In mass selection of plant populations by farmers, only the seeds from the healthiest and most productive individuals participate to the next cultivated generation. In Northwest India where Perennial originated, plants combining pvr23 and pvr6 certainly gain a selective advantage in presence of ChiVMV. However, the rapid adaptation of PVY and/or other potyviruses would have counterselected this allelic combination in a few cultivation seasons. Over a few plant generations, and in heterogeneous plant populations, this multipathogen selection pressure promoted the recombinant individuals carrying favorable alleles at additional QTLs (RB-1, RB-5 and RB-6) which, in interaction with RB-3, decreased the risk of pvr23 resistance breakdown. Such a polygenic combination of co-adapted alleles contributed to increase resistance in efficiency, spectrum and durability.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.s4n76
Acknowledgments
We thank B Mangin (INRA-Toulouse) for help in the use of the MCQTL software, G Girardot, P Mistral, G Nemouchi, K Nozeran, B Savio and V Simon for technical assistance, AM Sage-Palloix for providing the C genetic resources, J-P Bouchet for bioinformatic assistance and K Lamour (Tennessee Univ., USA) for critical reading of the manuscript. This work was financially supported by the Comité Technique Permanent de la Sélection (CTPS, French Ministry of Agriculture and Fisheries) by the Agence Nationale de la Recherche (Project VirAphid ANR-2010STRA-001-01) and by the Région Provence Alpes Côte d'Azur (PACA).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Acosta-Leal R, Xiong Z. Complementary functions of two recessive R-genes determine resistance durability of tobacco ‘Virgin A Mutant' (VAM) to Potato virus Y. Virology. 2008;379:275–283. doi: 10.1016/j.virol.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Ayme V, Souche S, Caranta C, Jacquemond M, Chadoeuf J, Palloix A, et al. Different mutations in the genome-linked protein vpg of potato virus y confer virulence on the pvr23 resistance in pepper. Mol Plant Microbe Interact. 2006;19:557–563. doi: 10.1094/MPMI-19-0557. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Brun H, Chèvre A-M, Fitt BD, Powers S, Besnard A-L, Ermel M, et al. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010;185:285–299. doi: 10.1111/j.1469-8137.2009.03049.x. [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Møller IS, Tulinius G, Hansen JKR, Lund OS, Johansen IE. The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol Plant Microbe Interact. 2007;20:1075–1082. doi: 10.1094/MPMI-20-9-1075. [DOI] [PubMed] [Google Scholar]
- Caranta C, Lefebvre V, Palloix A. Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol Plant Microbe Interact. 1997;10:872–878. [Google Scholar]
- Chandler CH, Chari S, Dworkin I. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 2013;29:358–366. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charcosset A, Mangin B, Moreau L, Combes L, Jourjon M-F, Gallais A. Heterosis in maize investigated using connected RIL populations. Quantitative genetics and breeding methods: the way ahead. Les colloques n°96. INRA Editions: Paris; 2001. [Google Scholar]
- Charron C, Nicolaï M, Gallois J, Robaglia C, Moury B, Palloix A, et al. Natural variation and functionnal analyes provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008;54:56–68. doi: 10.1111/j.1365-313X.2008.03407.x. [DOI] [PubMed] [Google Scholar]
- Fabre F, Bruchou C, Palloix A, Moury B. Key determinants of resistance durability to plant viruses: insights from a model linking within- and between-host dynamics. Virus Res. 2009;141:140–149. doi: 10.1016/j.virusres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Fazari A, Palloix A, Wang LH, Sage-Palloix AM, Djian-Caporalino C. The root-knot nematode resistance N-gene co-localizes in the Me-genes cluster on the pepper (Capsicum annuum L.) P9 chromosome. Plant Breeding. 2012;131:665–673. [Google Scholar]
- Fournet S, Kerlan MC, Renault L, Dantec JP, Rouaux C, Montarry J. Selection of nematodes by resistant plants has implications for local adaptation and cross-virulence. Plant Pathol. 2013;62:184–193. [Google Scholar]
- French R, Stenger DC. Evolution of wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu Rev Phytopathol. 2003;41:199–214. doi: 10.1146/annurev.phyto.41.052002.095559. [DOI] [PubMed] [Google Scholar]
- García-Arenal F, Fraile A, Malpica JM. Variability and genetic structure of plant virus populations. Annu Rev Phytopathol. 2001;39:157–186. doi: 10.1146/annurev.phyto.39.1.157. [DOI] [PubMed] [Google Scholar]
- Hajimorad MR, Wen RH, Eggenberger AL, Hill JH, Maroof MAS. Experimental adaptation of an rna virus mimics natural evolution. J Virol. 2010;85:2557–2564. doi: 10.1128/JVI.01935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Janzac B, Fabre MF, Palloix A, Moury B. Phenotype and spectrum of action of the Pvr4 resistance in pepper against potyviruses, and selection for virulent variants. Plant Pathol. 2009;58:443–449. [Google Scholar]
- Jourjon MF, Jasson S, Marcel J, Ngom B, Mangin B. MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics. 2005;21:128–130. doi: 10.1093/bioinformatics/bth481. [DOI] [PubMed] [Google Scholar]
- Jung J, Park SW, Liu WY, Kang B-C. Discovery of single nucleotide polymorphism in Capsicum and SNP markers for cultivar identification. Euphytica. 2010;175:91–107. [Google Scholar]
- Kou Y, Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Lacroix C, Glais L, Verrier J-L, Jacquot E. Effect of passage of a Potato virus Y isolate on a line of tobacco containing the recessive resistance gene va2 on the development of isolates capable of overcoming alleles 0 and 2. Eur J Plant Pathol. 2011;130:259–269. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lee JM, Nahm SH, Kim YM, Kim BD. Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet. 2004;108:619–627. doi: 10.1007/s00122-003-1467-x. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Pflieger S, Thabuis A, Caranta C, Blattes A, Chauvet JC, et al. Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome. 2002;45:839–854. doi: 10.1139/g02-053. [DOI] [PubMed] [Google Scholar]
- Le Van A, Caffier V, Lasserre-Zuber P, Chauveau A, Brunel D, Cam B, et al. Differential selection pressures exerted by host resistance quantitative trait loci on a pathogen population: a case study in an apple 9 Venturia inaequalis pathosystem. New Phytol. 2013;197:899–928. doi: 10.1111/nph.12086. [DOI] [PubMed] [Google Scholar]
- Margaria P, Ciuffo M, Pacifico D, Turina M. Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol Plant Microbe Interact. 2007;20:547–558. doi: 10.1094/MPMI-20-5-0547. [DOI] [PubMed] [Google Scholar]
- Mazier M, Flamain F, Nicolaï M, Sarnette V, Caranta C. Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS One. 2011;6:e29595. doi: 10.1371/journal.pone.0029595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- Minamiyama Y, Tsuro M, Hirai M. An SSR-based linkage map of Capsicum annuum. Mol Breeding. 2006;18:157–169. [Google Scholar]
- Montarry J, Doumayrou J, Simon V, Moury B. Genetic background matters: a plant-virus gene-for-gene interaction is strongly influenced by genetic contexts. Mol Plant Pathol. 2011;12:911–920. doi: 10.1111/j.1364-3703.2011.00724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Morel C, Johansen E, Guilbaud L, Souche S, Ayme V, et al. Mutations in Potato virus Y Genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol Plant Microbe Interact. 2004;17:322–329. doi: 10.1094/MPMI.2004.17.3.322. [DOI] [PubMed] [Google Scholar]
- Moury B, Palloix A, Caranta C, Gognalons P, Souche S, Selassie KG, et al. Serological, molecular, and pathotype diversity of Pepper veinal mottle virus and Chili veinal mottle virus. Phytopathology. 2005;95:227–232. doi: 10.1094/PHYTO-95-0227. [DOI] [PubMed] [Google Scholar]
- Mundt C, Cowger C, Garrett K. Relevance of integrated disease management to resistance durability. Euphytica. 2002;124:245–252. [Google Scholar]
- Nagy I, Stágel A, Sasvári Z, Röder M, Ganal M. Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.) Genome. 2007;50:668–688. doi: 10.1139/g07-047. [DOI] [PubMed] [Google Scholar]
- Nicolaï M, Pisani C, Bouchet JP, Vuylsteke M, Palloix A. Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum) Genet Mol Res. 2012;11:2295–2300. doi: 10.4238/2012.August.13.3. [DOI] [PubMed] [Google Scholar]
- Palloix A, Ayme V, Moury B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009;183:190–199. doi: 10.1111/j.1469-8137.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Pink D. Strategies using genes for non-durable disease resistance. Euphytica. 2002;124:227–236. [Google Scholar]
- Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Quenouille J, Montarry J, Palloix A, Moury B. Farther, slower, stronger: how the plant genetic background protects a major resistance gene from breakdown. Mol Plant Pathol. 2013;14:109–118. doi: 10.1111/j.1364-3703.2012.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebaï A. Comparison of methods for regression interval mapping in QTL analysis with non-normal traits. Genet Res. 1997;69:69–74. [Google Scholar]
- Robaglia C, Caranta C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Rubio M, Caranta C, Palloix A. Functional markers for selection of potyvirus resistance alleles at the pvr2-eIF4E locus in pepper using tetra-primer ARMS-PCR. Genome. 2008;51:767–771. doi: 10.1139/G08-056. [DOI] [PubMed] [Google Scholar]
- Rubio M, Nicolaï M, Caranta C, Palloix A. Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E and pvr6-eIF(iso)4E alleles for resistance to pepper veinal mottle virus. J Gen Virol. 2009;90:2808–2814. doi: 10.1099/vir.0.013151-0. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Gallois J-L, Moury B, Robaglia C, Palloix A, Caranta C. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J Gen Virol. 2006;87:2089–2098. doi: 10.1099/vir.0.81817-0. [DOI] [PubMed] [Google Scholar]
- Schafer JF. Tolerance to plant disease. Annu Rev Phytopathol. 1971;9:235–252. [Google Scholar]
- Verrier JL, Doroszewska T.2004. The ‘va' resistance to PVYN in Nicotiana tabacum: an assessment of the frequency of ‘va' breaking PVYN strains based on seven years of field survey on a worldwide basis. In: Proceedings of the 12th European Association for Potato Research (Virology Section) Meeting 2004, 13–19 June 2004, Rennes, France.86.
- Wolfe M. The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annu Rev Phytopathol. 1985;23:251–273. [Google Scholar]
- Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, et al. A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet. 2009;118:1279–1293. doi: 10.1007/s00122-009-0980-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.