Abstract
The two North Atlantic eel species, the European eel (Anguilla anguilla) and the American eel (Anguilla rostrata), spawn in partial sympatry in the Sargasso Sea, providing ample opportunity to interbreed. In this study, we used a RAD (Restriction site Associated DNA) sequencing approach to identify species-specific diagnostic single-nucleotide polymorphisms (SNPs) and design a low-density array that combined with screening of a diagnostic mitochondrial DNA marker. Eels from Iceland (N=159) and from the neighboring Faroe Islands (N=29) were genotyped, along with 94 larvae (49 European and 45 American eel) collected in the Sargasso Sea. Our SNP survey showed that the majority of Icelandic eels are pure European eels but there is also an important contribution of individuals of admixed ancestry (10.7%). Although most of the hybrids were identified as F1 hybrids from European eel female × American eel male crosses, backcrosses were also detected, including a first-generation backcross (F1 hybrid × pure European eel) and three individuals identified as second-generation backcrosses originating from American eel × F1 hybrid backcrosses interbreeding with pure European eels. In comparison, no hybrids were observed in the Faroe Islands, the closest bodies of land to Iceland. It is possible that hybrids show an intermediate migratory behaviour between the two parental species that ultimately brings hybrid larvae to the shores of Iceland, situated roughly halfway between the Sargasso Sea and Europe. Only two hybrids were observed among Sargasso Sea larvae, both backcrosses, but no F1 hybrids, that points to temporal variation in the occurrence of hybridization.
Keywords: admixture analysis, anguilla, hybridization, RAD sequencing, SNPs
Introduction
The study of hybridization between individuals from genetically distinct populations or species is of considerable interest to understand the dynamics of speciation and adaptive divergence (Mallet, 1995; Grant and Grant, 2002; Rieseberg and Willis, 2007; Hewitt, 2011; Stolting et al., 2013). In particular, hybrid zones are widely recognized as ‘windows' or ‘natural laboratories' for evolutionary studies concerning models of speciation, selective forces involved in speciation, gene flow between species and the maintenance of species boundaries (Hewitt, 1988, 2011; Harrison, 1990; Mallet, 1995; Petit and Excoffier, 2009). A key question in many instances of hybridization concerns whether hybridization is restricted to F1 hybrids and first-generation backcrosses after which genetic incompatibilities and/or natural selection eliminate hybrid offspring, or if hybridization proceeds to the extent that introgression occurs (Allendorf et al., 2001). In the latter case, incomplete reproductive isolation may have important bearings on the evolutionary trajectories of species by decreasing divergence between species but also by allowing favourable new mutations and allelic combinations to transgress species boundaries. This holds particularly true for pelagic marine environments where few obvious physical barriers are present and speciation-with-gene-flow processes may be common (Feder et al., 2012).
A peculiar pattern of hybridization is found in the two North Atlantic eel species, the European (Anguilla anguilla) and the American eel (A. rostrata). Both species show wide distribution ranges, with the European eel being found in the eastern Atlantic from Morocco to Iceland including the Mediterranean Sea and the American eel being found in the western Atlantic from the Caribbean to Greenland. North Atlantic eels are facultatively catadromous, spawning in partial sympatry in frontal zones of the southern Sargasso Sea. After spawning, larvae (leptocephali) are transported by surface currents of the Gulf Stream to the shores of Europe/North Africa and North America, respectively. When larvae reach the continental shelf, they undergo metamorphosis into glass eels that complete the migration into fresh, brackish and coastal waters as yellow eels. After a highly variable feeding period of 5–30 years, yellow eels metamorphose into silver eels that migrate from their highly dispersed foraging areas to the common spawning ground in the Sargasso Sea, where they reproduce only once and die (van den Thillart et al., 2009). Molecular studies in both species have demonstrated that they are panmictic (Als et al., 2011; Côté et al., 2013). Remarkably, although mitochondrial DNA lineages of the two species are reciprocally monophyletic (Avise et al., 1986), low differentation is found at microsatellite loci, with FST values of 0.055 and 0.018 reported in previous studies (Mank and Avise, 2003; Wirth and Bernatchez, 2003).
It is well established that the spawning grounds of the two species overlap in the Sargasso Sea and there is also overlap in spawning time, with the American eel spawning in February–April and the European eel spawning in March–June (McCleave et al., 1987). European and American eels are known to hybridize but hybrids are observed almost exclusively in Iceland. The first evidence of hybrids in Iceland came from the study of Avise et al. (1990) based on allozymes, mitochondrial DNA markers and vertebral counts that demonstrated that Iceland populations include hybrids in low frequencies of 2–4%. The lack of discriminatory power of the markers used did not allow determining the status of hybrids and quantify the rate of introgression. This was also a problem in a later study using microsatellite loci, ascribed primarily to homoplasy (Mank and Avise, 2003). A more extensive study on the status of eels in Iceland was conducted using amplified fragment length polymorphism analysis of eels sampled at multiple Icelandic locations (Albert et al., 2006). The study revealed an overall hybrid proportion of 15.5% in Iceland, ranging from 6.7 to 100% across locations, and the occurrence of hybrids beyond the first generation. However, the markers used were not able to classify later-generation (post-F1) hybrids. Hence, the nature of introgression in Icelandic eels remains largely unresolved. Outside Iceland, only few hybrids have been identified. Albert et al. (2006) observed no hybrids out of 186 European eels and 3 later-generation hybrids out of 193 American eels. Als et al. (2011) identified a single hybrid among Sargasso Sea eel larvae and three putative hybrids among European glass eels, including one individual from Iceland. However, given the limited resolution of available markers, it is uncertain whether hybridization has been underestimated.
Here, we use a RAD (Restriction site Associated DNA) sequencing approach (Baird et al., 2008; Hohenlohe et al., 2010) to identify single-nucleotide polymorphisms (SNPs) diagnostic for European and American eels. We use a subset of these SNPs to analyze patterns, directionality and evolution of hybridization, whereas a detailed analysis of genomic divergence between the species will be presented in a forthcoming paper. Specifically, we analyze eels from Iceland, the North Atlantic Faroe Islands and the spawning region in the Sargasso Sea to address the following questions: (1) Has the level of hybridization in previous studies been underestimated? (2) Which hybrid classes are present; does hybridization lead to introgression or is it restricted to F1 hybrids and perhaps first-generation backcrosses? (3) Is the high occurrence of hybrids restricted to Iceland or can hybrids also be found in the neighbouring Faroe Islands that are the closest bodies of land to Iceland? In addition to clarifying important aspects of the biology of eels, the results will add to our understanding of speciation and hybridization processes in pelagic marine environments.
Materials and methods
Sampling
A total of 50 pure North Atlantic eels, 25 European and 25 American eels, were used for RAD sequencing and SNP discovery. All individuals were later confirmed as pure species using the developed SNP markers. European eels were collected using fyke nets at one location in the western Mediterranean, Valencia (Spain), and two locations in the eastern Atlantic, Bordeaux (France) and Burrishoole (Ireland). American eels were collected along the western Atlantic coast at Riviere Blanche (Quebec), Mira River (Nova Scotia) and St Johns River (Florida). Genomic DNA was purified using standard phenol/chloroform extraction.
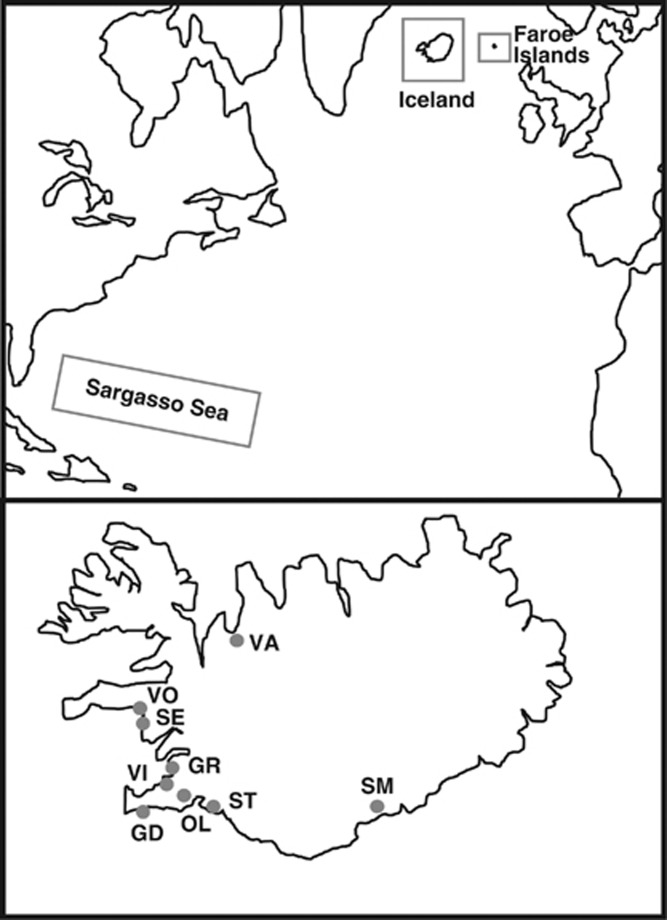
A total of 282 eels were included in the subsequent study of hybridization. Icelandic samples (N=159) were collected at 9 sites between 2000 and 2003, covering most of the known geographical distribution range of the species in Iceland, from Vatndalsa in the northern coast to Steinsmyrarfljot in the southern coast (Figure 1). The eels were not the same individuals analyzed by Albert et al. (2006) and Gagnaire et al. (2009), but they came from the same samples. Sampling locations, life stages and number of individuals sampled are detailed in Table 1. Glass eels were caught by electrofishing, whereas yellow eels were caught with traps. Samples from the Faroe Islands (N=29) were collected at two locations (Grothusvatn and Kaldbaksbotnur) in 2011 using fyke nets. The study was supplemented with the analysis of European and American eel larvae collected in the Sargasso Sea during March–April 2007 as part of the Danish Galathea 3 expedition (for details see Als et al., 2011). Samples of the two species were collected at 37 stations covering 600 km of the Subtropical Convergence frontal zone in the Sargasso Sea (64–70 °W and 24–30 °N) using a ring net of diameter 3.5 m equipped with a 25 m long net of 560 μm mesh. A subset of 94 larvae (49 European and 45 American eel) were included in the present study (Table 2). DNA from ethanol-preserved larvae was extracted using the EZNA kit (Omega Biotek, Norcross, GA, USA). Individual larvae were initially identified as European or American eel based on the analysis of the mitochondrial cytochrome b gene (Trautner, 2006).
Figure 1.
Sampling locations of European eels, including detailed locations in Iceland (labelled as in Table 1).
Table 1. Details of genetic samples for North Atlantic eels including sampling locations, code, geographic location, life stage, year of sampling and number of individuals.
| Location | Code | Location | Life stage | Year | N |
|---|---|---|---|---|---|
| Iceland | |||||
| Vatnsdalsá | VA | 65.49, −20.34 | Y | 2000 | 10 |
| Vogslækur | VO | 64.69, −22.33 | G | 2001 | 27 |
| Seljar | SE | 64.56, −22.31 | G | 2001 | 10 |
| Grafarvogur | GR | 64.15, −21.81 | Y | 2003 | 10 |
| Vifilsstadvatn | VI | 64.07, −21.87 | Y | 2002 | 10 |
| G | 2001 | 33 | |||
| Grindavik | GD | 63.83, −22.42 | Y | 2003 | 10 |
| Olfus | OL | 63.93, −21.62 | G | 2000 | 16 |
| Stokkseyri | ST | 63.81, −21.04 | G | 2001 | 10 |
| Steinsmyrarfljot | SM | 63.61, −17.92 | Y | 2000 | 23 |
| Faroe Islands | |||||
| Grothusvatn | GV | 61.84, −6.83 | Y | 2011 | 15 |
| Kaldbaksbotnur | KA | 62.06, −6.91 | Y | 2011 | 14 |
Abbreviations: G, glass eels; Y, yellow eels.
Table 2. Details of larvae sampling in the Sargasso Sea including transect, station, geographic coordinates and species sampled.
| Transect | Station | Lon. W | Lat. N | AA | AR |
|---|---|---|---|---|---|
| 1 | 6 | 63.6 | 24.3 | 0 | 1 |
| 1 | 7 | 64.0 | 25.16 | 4 | 1 |
| 1 | 8 | 64.01 | 26.01 | 4 | 1 |
| 1 | 9 | 64.01 | 26.31 | 4 | 2 |
| 1 | 10 | 63.58 | 27.01 | 1 | 0 |
| 1 | 11 | 63.6 | 27.2 | 4 | 1 |
| 1 | 12 | 64.0 | 27.42 | 1 | 1 |
| 1 | 13 | 63.59 | 28.01 | 2 | 0 |
| 2 | 16 | 67.01 | 28.29 | 2 | 0 |
| 2 | 17 | 67.0 | 27.44 | 3 | 1 |
| 2 | 19 | 66.6 | 26.3 | 0 | 1 |
| 2 | 20 | 67.0 | 25.5 | 0 | 2 |
| 2 | 21 | 66.6 | 25.37 | 2 | 2 |
| 2 | 22 | 67.0 | 25.19 | 1 | 3 |
| 2 | 23 | 67.0 | 24.6 | 3 | 4 |
| 2 | 24 | 67.05 | 24.31 | 1 | 0 |
| 3 | 25 | 69.59 | 24.6 | 0 | 3 |
| 3 | 26 | 69.58 | 25.29 | 1 | 2 |
| 3 | 27 | 69.58 | 25.27 | 7 | 2 |
| 3 | 28 | 69.59 | 26.34 | 6 | 8 |
| 3 | 29 | 69.6 | 26.6 | 1 | 1 |
| 3 | 30 | 70.05 | 27.32 | 1 | 4 |
| 3 | 31 | 70.02 | 27.59 | 0 | 4 |
| 3 | 32 | 70.02 | 26.6 | 0 | 1 |
| 3 | 33 | 70.01 | 30.0 | 1 | 0 |
Abbreviations: AA, Anguilla anguilla; AR, A. rostrata; Lat., latitude; Lon., longitude.
RAD tag sequencing, RAD data analysis and SNP identification
Genomic DNA from each individual was digested with restriction enzyme EcoRI. All 50 individuals (25 European eels and 25 American eels) were RAD-sequenced (10 individuals per lane) on an Illumina Genome Analyzer II by Beijing Genomics Institute (BGI, Hong Kong) using paired-end reads (for details see Pujolar et al., 2013).
Sequence reads from the Illumina (San Diego, CA, USA) runs were sorted according to barcode tag. Sequences were quality-filtered using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit) and reads with ambiguous barcodes/poor quality were removed from the analysis. A minimum Phred score of 10 (equivalent to 90% probability of being correct) per nucleotide position was chosen, meaning that reads were dropped if a -nucleotide position had a score of <10. This is the Phred score generally used in SNP discovery studies (Van Bers et al., 2010; Ellison et al., 2011; Scaglione et al., 2012; Wagner et al., 2012). Final read length was trimmed to 80 nucleotides in order to reduce sequencing errors present at the tail of the sequences. For subsequent analyses, only the first (left) paired-read was used. The DNA fragments created by RAD tag library preparation have a restriction site at one end and are randomly sheared at the other end that results in each instance of a restriction site sequence being sampled many times by the first reads and the genomic DNA sequence in the nearby region being randomly sampled at a lower coverage by the second paired-end reads (Etter et al., 2011), which are therefore less suitable for calling SNPs.
Sequence reads were aligned to the European eel genome draft (www.eelgenome.com) (Henkel et al., 2012) using the ungapped aligner Bowtie version 0.12.8 (Langmead et al., 2009). A maximum of two mismatches between the individual reads and the genome were allowed and alignments were suppressed for a particular read when more than one reportable alignment existed, thereby decreasing the risk of paralogous sequences in the data.
The reference-aligned data were then used to assemble the RAD sequences into loci and identify alleles using the ref_map.pl pipeline in Stacks version 0.9995 (Catchen et al., 2011). First, exactly matching sequences are aligned together into stacks that are in turn merged to form putative loci. At each locus, nucleotide positions are examined and SNPs are called using a maximum likelihood framework. Second, a catalog is created of all possible loci and alleles. Third, each individual is matched against the catalog. A minimum stack depth of 10 reads was used, which is the number of exactly matching reads that must be found to create a stack in an individual. Finally, the program Populations in Stacks was used to process all the SNP data across individuals. Loci were processed when present in both species in at least 66.67% of the individuals.
Genome-wide measures of genetic diversity, including observed (Ho) and expected (He) heterozygosities, were calculated at each nucleotide site for all individuals. Genetic differentiation between European and American eel samples measured by FST was estimated for all individual SNPs.
SNP low-density array design and scoring
Species-diagnostic SNPs were chosen to design a 96-SNP genotyping array on the basis of the following criteria: (1) a minimum FST value of 0.95, (2) target SNP is present along with no more than five additional singleton SNPs per locus and (3) the presence of at least 100 bp flanking sequence when extending the RAD loci using the European eel genome to allow for optimal primer design. SNPs meeting those criteria were labelled as diagnostic for the purpose of the study, although not 100% fixed. All selected SNPs were distributed across different scaffolds and contigs in the eel genome in order to guarantee a good representation of the genome.
SNPs were genotyped on 96.96 Dynamic Arrays (Fluidigm Corporation, San Francisco, CA, USA) using the Fluidigm EP1 instrumentation and according to the manufacturer's protocols. The Fluidigm system uses nano-fluidic circuitry to simultaneously genotype up to 96 samples with 96 loci (see Seeb et al., 2009 for a description of the Fluidigm system methodology). Genotypes were called and the data compiled using the Fluidigm SNP Genotyping Analysis software. Each assay was assessed for plot quality and expected clustering patterns.
Hybrid identification
Hybrids were identified using STRUCTURE (Pritchard et al., 2000) and NEWHYBRIDS (Anderson and Thompson, 2002). We used STRUCTURE to estimate individual admixture proportions and their probability intervals. For this purpose we assumed an admixture model, uncorrelated allele frequencies and we did not use population priors. Given that two panmictic species were analyzed, we assumed K=2 and conducted 10 runs to check the consistency of results. A burn-in of 100 000 steps followed by 1000 000 additional Markov Chain Monte Carlo iterations were performed.
Rather than assigning individuals to a single hybrid category, NEWHYBRIDS uses a Bayesian framework to compute the posterior probability of one individual belonging to each parental and distinct hybrid class (F1, F2 and backcrosses between F1 × European eel and F1 × American eel). The genotype frequency class file was expanded to include third-generation hybrid classes originating from crosses between backcrosses and pure European eels, pure American eels and F1 hybrids. The software was run for 100 000 iterations in the burn-in, followed by 100 000 Markov Chain Monte Carlo iterations in each analysis.
Before the analysis, the power of the selected SNP markers to discriminate hybrid status was assessed using HYBRIDLAB (Nielsen et al., 2006), a program for generating simulated hybrids from population samples. Based on the observed allelic frequencies at the selected SNPs in the European and American eel baseline populations, the program generated 10 000 random genotypes of each of the following 12 categories: (1) pure European eel, (2) pure American eel, (3) F1 hybrid, (4) F2 hybrid, (5) first-generation backcross European eel × F1 hybrid, (6) first-generation backcross American eel × F1 hybrid, (7) second-generation backcross between -generation backcross (European eel × F1 hybrid) and European eel, (8) second-generation backcross between first-generation backcross (European eel × F1 hybrid) and American eel, (9) second-generation backcross between first-generation backcross (European eel × F1 hybrid) and F1 hybrid, (10) second-generation backcross between first-generation backcross (American eel × F1 hybrid) and European eel, (11) second-generation backcross between first-generation backcross (American eel × F1 hybrid) and American eel and (12) second-generation backcross between first-generation backcross (American eel × F1 hybrid) and F1 hybrid. All simulated individuals were then blindly reassigned to their most probable category using both STRUCTURE and NEWHYBRIDS.
Results
RAD sequencing
Sequencing of the RAD libraries generated an average of 8.8 million reads per individual of 90 bp, before any quality filtering, ranging from 4.4 to 13.4 million reads. After quality filtering, on average 7.3 million (82.7%) sequences per individual were retained. Retained sequences presented a mean quality score of 38.5, a median of 39.2 and a GC content of 40.6%, with no apparent differences observed between species (Table 3).
Table 3. Statistics describing the distribution of different properties of RAD sequences after each step of filtering (FASTX-Toolkit), alignment to the eel draft genome (Bowtie) and assemblage into loci (Ref_map.pl).
| FASTX | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Species |
Raw reads |
Filtered reads |
% Eliminated |
Mean Q |
Q1 |
Med |
Q3 |
%A |
%C |
%G |
%T |
| All | 8 810 898 | 7 281 642 | 17.32 | 38.45 | 37.80 | 39.24 | 40 | 29.8 | 20.5 | 20.1 | 29.6 |
| AA | 8 769 831 | 7 007 835 | 19.92 | 38.36 | 37.72 | 39.10 | 40 | 29.7 | 20.5 | 20.1 | 29.6 |
| AR | 8 851 964 | 7 555 450 | 14.72 | 38.53 | 37.88 | 39.37 | 40 | 29.8 | 20.5 | 20.1 | 29.6 |
|
Bowtie |
|||||||
|---|---|---|---|---|---|---|---|
| Species | Reads | Aligned | % Aligned | Nonaligned | % Nonaligned | Discarded | % Discarded |
| All | 7 281 642 | 4 707 002 | 64.25 | 2 504 043 | 34.15 | 117 695 | 1.60 |
| AA | 7 007 835 | 4 768 089 | 67.16 | 2 218 176 | 31.22 | 115 152 | 1.63 |
| AR | 7 555 450 | 4 645 915 | 61.33 | 2 789 910 | 37.09 | 120 238 | 1.58 |
|
Ref_map |
|||||||
|---|---|---|---|---|---|---|---|
| Species | Reads | Stacks | Loci | Loci used | % Loci used | Loci discarded | % Loci discarded |
| All | 4 707 002 | 663 555 | 322 314 | 214 178 | 66.45 | 108 136 | 33.55 |
| AA | 4 768 089 | 745 193 | 333 871 | 224 896 | 67.36 | 109 309 | 32.74 |
| AR | 4 645 915 | 581 918 | 310 756 | 203 638 | 65.53 | 107 118 | 34.47 |
Abbreviations: AA, Anguilla anguilla; AR, A. rostrata; RAD, Restriction site Associated DNA.
Retained sequences were aligned to the European eel draft genome. As expected, a higher number of European eel sequences aligned (67.16%) in comparison with American eel sequences (61.33%). The number of sequences discarded because of alternative alignments was similar for both species (1.6% Table 3).
Aligned sequences were assembled into an average of 663 555 stacks per individual and subsequently into a set of 322 314 loci. Using a minimum coverage of 10 reads, an average of 214 178 (66.45%) loci were retained and 108,136 (33.55%) loci were discarded per individual because of insufficient depth of coverage. Similar numbers of loci were retained in both species (Table 3).
A catalog of 502 526 loci was constructed using all individuals. A total of 95 884 loci (19%) shared between the two species in at least 66.67% of individuals per species were retained for SNP discovery. Using the program Populations in Stacks, a total of 767 015 SNPs were identified. When calculating FST values between the two species for each SNP detected, 3348 SNPs showed an FST>0.95.
SNP genotyping
Supplementary Table 1 shows details of 86 diagnostic SNPs for North Atlantic eels and gives the corresponding allele frequencies for European and American eel, together with the position in the European eel draft genome. We excluded 10 markers (out of the original 96; 10.4%) because of primer mismatch or incorrect/multiple product amplification. A total of 75 out of 86 SNPs (87.2%) had an FST value of 1.0 between European and American eel. The remaining 9 SNPs (12.8%) had an FST of 0.95–0.96.
Hybrid identification
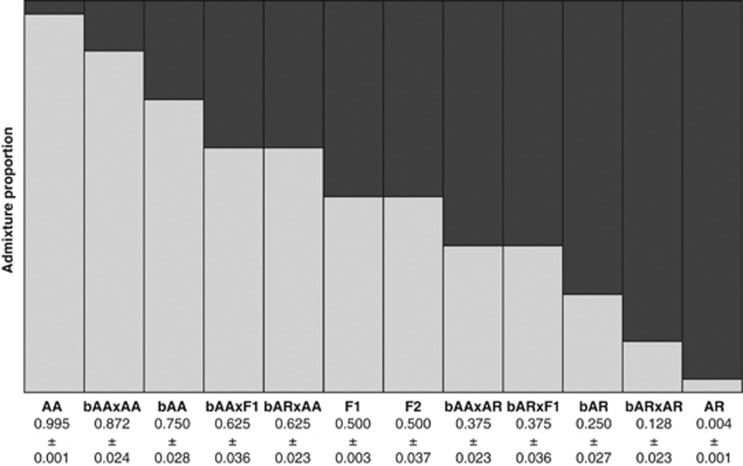
In order to test the power of the markers, a total of 120 000 individuals were simulated using 86 SNPs for 12 eel categories including first- and second-generation backcrosses and reassigned blindly. Using STRUCTURE, admixture proportion values ranged from 0.995 (pure European eel) to 0.004 (pure American eel). First-generation backcrosses showed unique admixture proportions (0.75 and 0.25). However, F1 and F2 hybrids and second-generation backcrosses could not be completely distinguished (Figure 2). Using NEWHYBRIDS, a correct assignment was made for 100% of F1 hybrids, 89% of F2 hybrids, 97% of first-generation backcrosses and 94–99% of second-generation backcrosses (Table 4). This suggests that our markers have enough discriminatory power to correctly identify up to third-generation hybrids.
Figure 2.
Admixture analysis of simulated individuals in STRUCTURE. A total of 12 categories were simulated: pure European eel (AA), pure American eel (AR), F1 hybrid, F2 hybrid, first-generation backcross European eel × F1 (bAA), first-generation backcross American eel × F1 (bAR) and second-generation backcrosses (bAA × AA, bAA × AR, bAA × F1, bAR × AA, bAR × AR and bAR × F1). Average admixture proportion value and s.d. values are presented for each simulated category.
Table 4. Assignment probability of simulated individuals in NEWHYBRIDS.
|
Assignment probability |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AR | F1 | F2 | bAA | bAR | bAA × AA | bAA × AR | bAA × F1 | bAR × AA | bAR × AR | bAR × F1 | |
| AA | 1.000 | — | — | — | — | — | — | — | — | — | — | — |
| AR | — | 1.000 | — | — | — | — | — | — | — | — | — | — |
| F1 | — | — | 1.000 | — | — | — | — | — | — | — | — | — |
| F2 | — | — | — | 0.885 | — | — | — | — | 0.064 | — | — | 0.052 |
| bAA | — | — | — | — | 0.971 | — | 0.019 | — | — | 0.010 | — | — |
| bAR | — | — | — | — | — | 0.973 | — | 0.012 | — | — | 0.014 | — |
| bAA × AA | — | — | — | — | 0.001 | — | 0.990 | — | — | — | — | — |
| bAA × AR | — | — | — | — | — | 0.012 | — | 0.988 | — | — | — | — |
| bAA × F1 | — | — | — | 0.062 | — | — | — | — | 0.937 | — | — | — |
| bAR × AA | — | — | — | — | 0.013 | — | — | — | — | 0.987 | — | — |
| bAR × AR | — | — | — | — | — | 0.009 | — | — | — | — | 0.991 | — |
| bAR × F1 | — | — | — | 0.068 | — | — | — | — | — | — | — | 0.932 |
A total of 12 categories were simulated: pure European eel (AA), pure American eel (AR), F1 hybrid, F2 hybrid, first-generation backcross European eel × F1 (bAA), first-generation backcross American eel × F1 (bAR) and second-generation backcrosses (bAA × AA, bAA × AR, bAA × F1, bAR × AA, bAR × AR and bAR × F1).
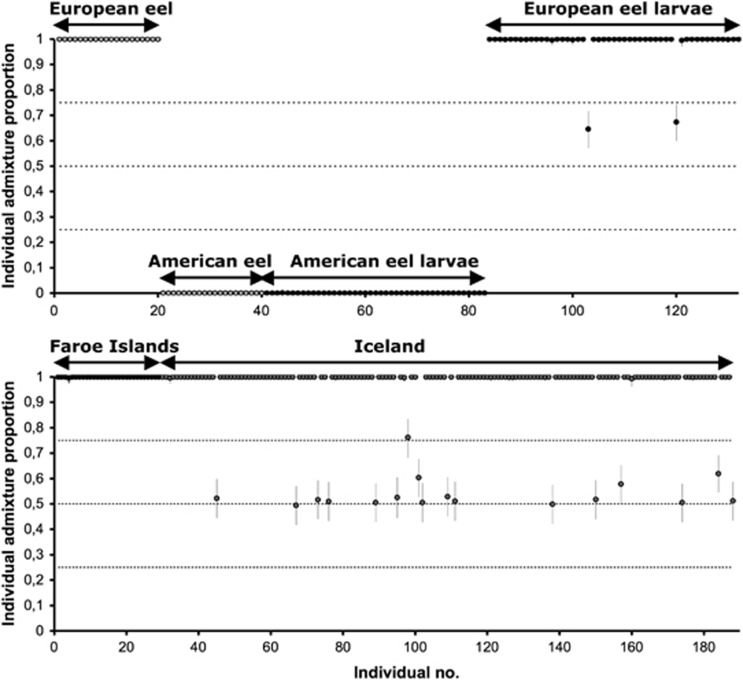
When the empirical data were examined with STRUCTURE, similar results were obtained across replicate runs, illustrating that the baseline data consisting of 25 European and American eel could be classified with high confidence (Figure 3a). All Sargasso Sea larvae identified as American eel using the cytochrome b mitochondrial marker were suggested to be nonadmixed. However, among putative European eel larvae, two individuals were clearly hybrids, with admixture proportions suggesting that they were second-generation backcrosses (Figure 3a). All 29 individuals from Faroe Islands were classified as pure European eels (Figure 3b). However, in Iceland, 142 individuals (89.3%) were pure European eels, whereas 17 individuals (10.7%) were classified as admixed with different degrees of American eel ancestry (Figure 3b). Out of the 17 admixed individuals, 13 showed a 90% probability interval overlapping 0.5, suggestive of either F1 or F2 hybrids. The remaining four individuals showed admixture proportions suggestive of backcrosses.
Figure 3.
Admixture analysis in STRUCTURE, assuming the presence of two groups (K=2): European and American eels. Individual admixture proportions along with 95% credible intervals are shown for each individual. Top panel shows baseline individuals of European eels and American eels followed by American eel larvae and European eel larvae from the Sargasso Sea. Bottom panel shows eels from the Faroe Islands and from Iceland.
A confirmation of the individual status categories obtained from STRUCTURE and a detailed classification of the hybrids was conducted in NEWHYBRIDS (Table 5). Identification of pure and hybrid individuals corresponded 100% to the results obtained with STRUCTURE. All American eel larvae were identified as pure American eels. Of the European eel larvae, 96% were pure European eels, whereas the remaining 2 individuals were identified as second-generation backcrosses between first-generation backcrosses (American eel × F1 hybrid) and pure European eels. All Faroe Islands individuals were confirmed as pure European eels. With respect to Icelandic individuals, NEWHYBRIDS confirmed the results from STRUCTURE (142 pure European eels and 17 hybrids). Out of 17 admixed individuals, 13 were F1 hybrids with a probability of P=1.00. The four later-generation hybrids identified included one first-generation backcross between pure European eel and F1 hybrid, whereas the remaining three individuals represented second-generation backcrosses between first-generation backcrosses (American eel × F1 hybrid) and pure European eels.
Table 5. Assignment probability in NEWHYBRIDS of all Icelandic and Sargasso Sea hybrids.
| Location | Individual |
Assignment probability |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AR | F1 | F2 | bAA | bAR | bAA × AA | bAA × AR | bAA × F1 | bAR × AA | bAR × AR | bAR × F1 | ||
| VA | VADA-1 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VA | VADA-2 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VO | VOGG-11 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VO | VOGG-5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| VO | VOGG-9 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SE | SSG-10 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SE | SSG-7 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VI | VFG-9 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VI | VIFIG-2 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| VI | VIFIG-5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| OL | OLFG-5 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| ST | STG-2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| ST | STG-3 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SM | STEG-1 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SM | STEG-31 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SM | STEG-38 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| SM | STEG-40 | 0.000 | 0.000 | 0.000 | 0.000 | 0.983 | 0.000 | 0.000 | 0.000 | 0.017 | 0.000 | 0.000 | 0.000 |
| Sargasso | L-125 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| Sargasso | L-211 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | 0.000 | 0.000 | 0.000 | 0.000 | 0.995 | 0.000 | 0.000 |
Categories include pure European eel (AA), pure American eel (AR), F1 hybrid, F2 hybrid, first-generation backcross European eel × F1 (bAA), first-generation backcross American eel × F1 (bAR) and second-generation backcrosses (bAA × AA, bAA × AR, bAA × F1, bAR × AA, bAR × AR and bAR × F1). Locations are labelled as in Table 1.
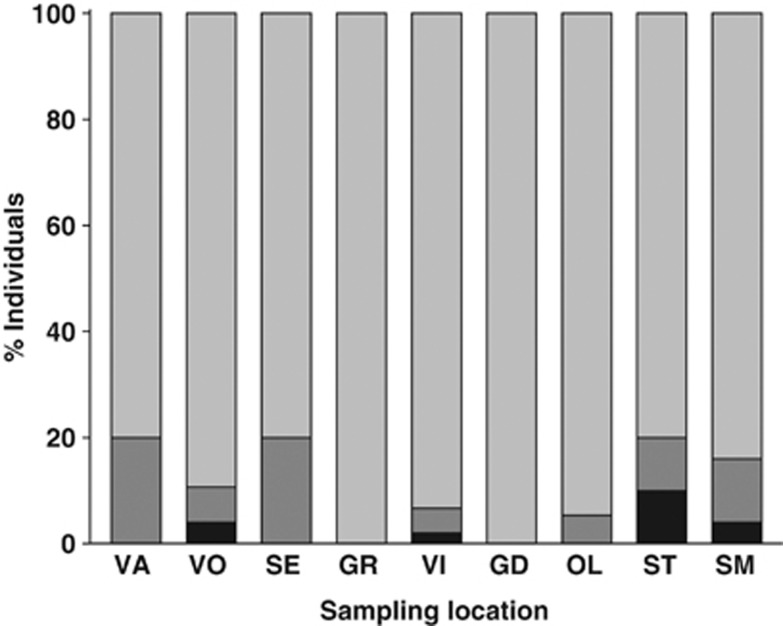
Across locations in Iceland, hybrids were detected in 7 out of 9 locations (Figure 4), including the northernmost (Vatnsdalsá) and southernmost (Steinsmyrarfljot) locations, with a proportion ranging from 5 to 20%. Only at two locations (Grafarvogur and Grindavik) were all individuals identified as pure European eels. F1 hybrids were found in all locations in which hybrids were present, whereas backcrosses were found at four locations (Vogslækur, Vifilstadvatn, Stokkseyri and Steinsmyrarfljot). Across life stages, the proportion of hybrids ranged from 9 to 20% in glass eels and from 0 to 20% in yellow eels.
Figure 4.
Proportion of pure European eels (light grey), F1 hybrids (dark grey) and later-generation hybrids (black) in all Icelandic locations, labelled as in Table 1.
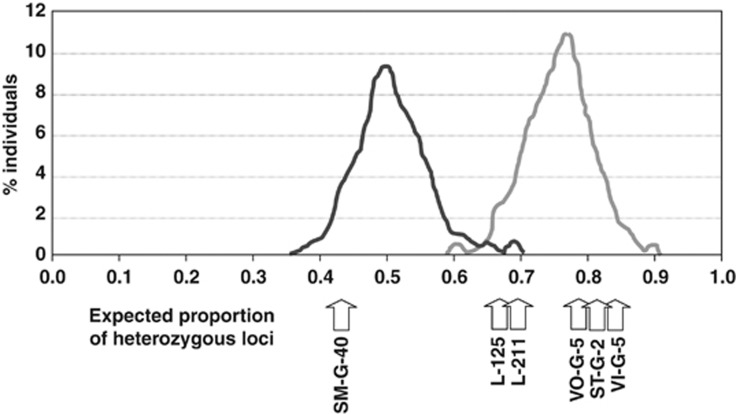
We further explored the nature of the six individuals identified as later-generation hybrids (four Icelandic individuals and two Sargasso Sea larvae) by comparing the observed proportion of heterozygote loci at each individual with the expected proportion in first-generation backcrosses (European eel × F1) and second-generation backcrosses between first-generation backcrosses (American eel × F1) and European eels (Figure 5), using the simulated data from HYBRIDLAB. In accordance with NEWHYBRIDS, only one individual from Iceland clearly presented an observed heterozygosity within the expected values for first-generation backcrosses (European Eel × F1). The remaining three individuals from Iceland presented much higher heterozygosity values than first-generation backcrosses, concordant with second-generation backcrosses between first-generation backcrosses (American eel × F1) and pure European eels. Observed heterozygosities for the two Sargasso larvae fitted the expectation for second-generation backcrosses but were also within the 5% uppermost distribution of first-generation backcrosses.
Figure 5.
Comparison of observed and expected proportion of heterozygous loci in (1) first-generation backcrosses European eel × F1 (dark grey) and (2) second-generation backcrosses between first-generation backcrosses American eel × F1 hybrid and European eels (light grey) based on 10 000 simulated individuals per category using HYBRIDLAB. Arrows indicate observed proportion of heterozygous loci.
In addition to SNP genotyping, all individuals were identified as European or American eel on the basis of the mitochondrial cytochrome b gene. All hybrid individuals (2 European eel larvae from the Sargasso Sea and 17 Icelandic individuals) showed the European eel mitochondrial haplotype. Surprisingly, 1 individual out of the 142 Icelandic individuals identified as pure European eels (on the basis of the SNP array with a 100% probability using both STRUCTURE and NEWHYBRIDS) presented the American eel mitochondrial haplotype.
Discussion
Species-diagnostic markers for detecting hybridization
The greater capacity to analyze genome-wide patterns of variation using genotyping-by-sequencing approaches has improved the means of identifying hybrids and examining the genetic and evolutionary consequences of species hybridization. Using an array of diagnostic RAD-generated SNPs with highest FST values between European and American eel, we were able to reliably identify hybrids up to the third generation. Simulations showed a high discriminatory power of the markers, as we were able to correctly identify 100% of F1 hybrids and 99% of backcrosses with pure parental species. Our results confirm that SNPs are effective markers to study hybridization and detect introgression (Hohenlohe et al., 2011, 2013; Amish et al., 2012; Nussberger et al., 2013). Because of their relatively low mutation rate and consequently low number of alleles, SNPs are more likely to be diagnostic loci than highly polymorphic markers such as microsatellites that present a higher probability of allele sharing between related species due to homoplasy (Balloux and Goudet, 2002).
Has hybridization in North Atlantic eels been underestimated?
The high discriminatory power of the set of diagnostic loci allows us to address the question of whether previous studies have underestimated the number of hybrids between the two species. This seems only to a minor extent to be the case. Als et al. (2011) identified a single hybrid among larvae collected in the Sargasso Sea, whereas the present study identified two hybrids among a subsample of the same larvae, both second-generation backcrosses. An eel larvae survey conducted in the Sargasso Sea in March–April 2007 during the Danish Galathea 3 marine expedition, from which the larvae included in our study are derived, showed both species cooccurring at most of the 37 sampling stations distributed along the longitudes 64 °W, 65 °W, 67 °W and 70 °W between 24 and 30° latitude, with American eel being predominant in the West and gradually being replaced by European eel towards the East (Als et al., 2011). Nevertheless, the observation in our study of only 2 putative hybrids out of 96 individuals (2.06%) does not support the existence of a distinct marine hybrid zone where the two North Atlantic eel species admix. Furthermore, the two larvae identified as hybrids were sampled at stations >600 km apart: one hybrid was collected at longitude 64 °W/ latitude 27.42 °N in the mid-eastern Sargasso Sea, whereas the other hybrid was collected at longitude 69.6 °W/latitude 26 °N in the western Sargasso Sea.
It is surprising that the two hybrids from the Sargasso Sea were second-generation backcrosses, whereas no F1 hybrids or first-generation backcrosses were observed. One possible explanation could be that the occurrence of hybridization differs between years, a pattern that has previously been suggested to explain temporally varying proportions of hybrids observed in Iceland (Avise et al., 1990; Albert et al., 2006). This could be because of differences in temporal overlap of spawning time between the species across years, possibly mediated by a more southern or northern displacement of thermal fronts across years. Indeed, the thermal fronts where eels are assumed to spawn show considerable differences in intensity and geographical position between years (Munk et al., 2010).
No hybrids were detected in the Faroe Islands. In the case of Icelandic eels, the set of diagnostic markers confirms that hybrids are common. The proportion of hybrid individuals found in our study (10.7%) is lower than the values reported by Albert et al. (2006) using amplified fragment length polymorphism markers (15.5%). However, the higher proportion of hybrids in the latter study might be because of the different locations sampled across studies. The two locations with the highest percentage of hybrids in the study of Albert et al. (2006) were not included in our study: the northeasternmost location, Saudarkrokur, constituted by 100% hybrids, and the northwesternmost location, Reykholar, constituted by 45% hybrids. When comparing locations across studies, hybrids were found in all locations shared, although hybrid proportions were only similar at one location, Vogslaekur (11.1% in our study and 11.9% in Albert et al., 2006). Only at one location, Stokkseyri, the proportion of hybrids was higher in our study (20%) in comparison with the Albert et al. (2006) study (8.3%). At the remaining locations, hybrid proportions were higher in Albert et al. (2006). However, the overall hybrid proportion in both studies was well higher than the values reported by the earlier study of Avise et al. (1990) that suggested that ∼2–4% of the gene pool of Icelandic eels is derived from American eel ancestry. The lower hybridization detected might be attributable to the low resolution of the markers used (allozymes and mitochondrial DNA) in comparison with amplified fragment length polymorphisms (Albert et al., 2006) or SNPs (this study).
Patterns of hybridization
The increased resolution of our array of diagnostic RAD-generated SNP markers sheds new light on the status of hybrids and the nature of introgression in Icelandic eels. According to our SNP survey, pure European eels made up the majority of Icelandic eels but there is also an important influx of individuals of admixed ancestry. In our study hybrids encompassed: F1 hybrids resulting from crosses between European eel females and American eel males, first-generation backcrosses between pure European eels and F1 hybrids (one individual) and second-generation backcrosses between first-generation backcrosses (pure American eel × F1 hybrid) and pure European eels (three individuals). None of the admixed individuals were F2 hybrids resulting from crosses between F1 hybrids. Although the hybrid classes observed were restricted to the first three generations, the finding of American eel mtDNA in an individual with a seemingly pure European nuclear genome suggests that successful hybridization over several generations is possible, although rare. In this sense, later hybrids might have been missed because of low sample sizes. It should also be noted that SNPs that are fixed between species are likely to mark chromosomal regions under strong directional selection or genomic incompatibilities (Gagnaire et al., 2012). During subsequent generations of backcrossing, selection could remove introgressed alleles at diagnostic markers. The number of individuals representing multigeneration backcrosses could therefore in principle be underestimated.
Although F1 hybrids are likely to outperform either pure parental species in surviving due to hybrid vigor (Templeton, 1986; Johnson et al., 2010), in many species F1 hybrids have been shown to be infertile, often regarded as an evolutionary dead end when hybrids are unable to produce viable progeny. This is not the case of the European eel, as our study shows the presence of first- and second-generation backcrosses in Iceland that not only indicates that F1 hybrids are viable and can survive in Iceland, but also that they can successfully migrate back to the Sargasso Sea as silver eels and reproduce as part of the spawning stock. Our study also shows that it is possible for a backcross originated from a cross between an F1 and an American eel to return to the Sargasso and interbreed with a pure European eel, with the resulting second-generation backcross eventually settling down in Iceland.
Alternatively, those individuals identified as second-generation backcrosses might be first-generation backcrosses between European eel × F1 hybrid showing higher-than-expected heterozygosities (Figure 5). One mechanism that could explain this is intrinsic outbreeding depression (Templeton, 1986; Allendorf et al., 2001) thought to be because of genetic incompatibilities between species involving deleterious epistatic interactions caused by the combination of incompatible alleles from different loci in hybrids (Dobzhansky, 1936; Muller, 1942). It is often found that F2 hybrids show reduced fitness because of disruption of sets of co-adapted genes by recombination that occurs in the second or later generations when assortment of alleles at different loci takes place (Gharrett and Smoker, 1991; Johnson et al., 2010). Eels are highly fecund, with an estimated mean fecundity of 3.6 million of eggs per female (MacNamara and McCarthy, 2012); therefore, even though the expected proportion of heterozygous loci in first-generation backcrosses is 50%, there will be variance around this number. In a large number of offspring from a single cross, there would consequently be some individuals that show high heterozygosity for diagnostic alleles at loci that presumably are under selection. These individuals might be functionally closer to F1 hybrids, with more sets of co-adapted alleles from each of the parental species relative to average backcrosses, where pronounced disruption of co-adapted gene complexes is expected. Finally, a higher-than-expected heterozygosity could also be explained by production of partly unreduced gametes in F1 hybrids given rise to triploid progeny when backcrossed to either parental species (Johnson and Wright, 1986; Castillo et al., 2007), although heterozygosity values should vary randomly and not necessarily fit the observed pattern of deviating backcrosses.
In total, we find it most likely that the deviating backcrosses are indeed second-generation backcrosses, as they were identified as such with high probability using NewHybrids and as the observed heterozygosity matched expectations for this type of cross. The finding that F1 hybrids can backcross to one species (American eel) and in the subsequent generation backcross to the other species (European eel) is remarkable. At present, the underlying mechanisms cannot be identified. However, as European and American eels have different peak spawning time (McCleave et al., 1987), this is likely to be an important prezygotic reproductive isolation mechanism. This may be broken up in hybrids leading to intermediate spawning time and a higher probability of interbreeding with either of the parental species.
Why are hybrids primarily found in Iceland?
Although genetic markers have shown the occurrence of hybrids in Iceland (Avise et al., 1990; Albert et al., 2006), the geographic range limit of hybrids and their presence in other European locations has remained uncertain. The observation in our study of 100% pure European eels in the Faroe Islands, which are the closest bodies of land to Iceland at a distance of 450 km, suggests that hybrids do not spread much towards the East and are limited to Iceland. Early indications that hybrids might occur in the Faroe Islands came from the study of Boëtius (1980) based on total number of vertebrae, previously regarded as the best distinguishing character between European and American eels. In a comprehensive survey of over 15 000 individuals comprising all of Europe and North Africa, Northern European locations showed 1–4% individuals with relatively low vertebrae counts, as is typical of American eel. Although vertebrae counts in Italy (N=1287) ranged between 111 and 119, individuals showing vertebrae counts of <110 were observed in both Iceland (108–110) and Faroe Islands (106–110) that pointed to the presence of either pure American eels or hybrids in Northern Europe and a relatively high degree of mixing between the two species. However, the observation in our study of 100% pure European eel individuals with no signatures of admixed ancestry in the Faroe Islands suggests that the shift towards American values in vertebrae counts observed by Boëtius (1980) is not because of the presence of hybrids and might be the result of a high variability in this trait.
Why are hybrids between European and American eel found primarily in Iceland? Hybrids could exhibit a genetically defined intermediate duration of the larval phase synchronized with an intermediate development time (Avise et al., 1990). This would explain the presence of hybrids in Iceland that is somewhat situated halfway en route from the Sargasso Sea to Europe. However, the migration duration of European and American eel larvae from the Sargasso Sea to their respective continents remains unresolved and highly debated, although it is obvious that migration distance is much longer for European than for American eels. For European eels, migration time estimates range from 7 to 9 months to 2 years depending on the assumptions and methods used, whereas estimates for American eels range between 6 and 12 months (Bonhommeau et al., 2009). Different larval migration duration is also supported by a transcriptome study of larvae of both eel species collected in the Sargasso Sea (Bernatchez et al., 2011). Larvae showed very similar transcriptome profiles at different age stages, but there was a significant delay of up- and down-regulation in European as opposed to American eels, consistent with the assumption of a shorter larval stage and migration time in the latter.
It is possible to speculate that hybrids might show an intermediate migratory behaviour and larval developmental (metamorphosis) between the two North Atlantic parental species that ultimately brings hybrid larvae to the shores of Iceland. Several studies have shown an intermediate migratory behaviour in hybrids. Experimentally produced hybrids of an exclusively migratory European population and a partially migratory African population of the blackcap Sylvia atricapilla showed intermediate migratory restlessness and an intermediate percentage of birds displaying restlessness compared with the two parental stocks (Berthold and Querner, 1981). Similarly, hybrids between common redstarts Phoenicurus phoenicurus and black redstarts P. ochruros showed intermediate time course of migratory activity relative to the parental species (Berthold and Querner, 1995). In fish, hybrids between coastal cutthroat trout (Oncorhynchus clarki clarki) and steelhead trout (O. mykiss) showed both intermediate swimming ability and intermediate migratory behaviour to that of either species (Moore et al., 2010). Hence, it is plausible that hybrids between European and American eels would also show intermediate migratory behaviour, although this remains to be investigated.
The question still remains of why hybrid eels are abundant in Iceland but not in the neighbouring Faroe Islands. Here it should be noted that the North Atlantic Current (the northeastward extension of the Gulf Stream) transporting the eel larvae actually separates into two branches ∼1000 km southwest of Iceland. The one branch passes the Faroese shelf and continues into the North Sea and Norwegian Sea towards the coast of Norway. The other branch, the Irminger Current, shifts in a northwestern direction towards and along the west coasts of Iceland. Hence, in terms of migratory routes, eels from Iceland and the Faroe Islands are expected to be separated by >1000 km, even though the shortest direct waterway distance is considerably shorter.
Biological implications of patterns of hybridization
European and American eel represent extreme life histories and a peculiar hybridization scenario involving two panmictic species and yet a very localized occurrence of hybrids. Nevertheless, even very low gene flow between the species may be biologically significant and could explain the surprisingly low genetic differentiation between North Atlantic eels observed in previous studies (Mank and Avise, 2003; Wirth and Bernatchez, 2003). At equilibrium, FST should be approximately equal to 1/(1+4Nem) (Wright, 1931), where Ne denotes effective population size and m is migration rate per generation. Estimates of long-term Ne in Atlantic eels derived from microsatellite markers are on the order of 4000–10 000 (Wirth and Bernatchez, 2003; Pujolar et al., 2011), whereas a recent study employing RAD sequencing suggests that historical Ne in European eel may be as high as ca. 100 000 to 1 000 000 (Pujolar et al., 2013). If we assume that FST between the species is ca. 0.05 (roughly reflecting the estimates by Mank and Avise, 2003; Wirth and Bernatchez, 2003) then at Ne values of 4000 to 10 000 this would correspond to migration rates of 1.2 × 10−3 and 4.8 × 10−4, whereas for Ne values of 100 000 and 1 000 000 m it would be as low as 4.8 × 10−5 and 4.8 × 10−6. Hence, depending on which of the Ne estimates best reflects the true Ne, it would only take a low to very low level of gene flow to maintain overall low genetic differentiation between the species.
We predict that the occurrence of low genetic differentiation between closely related and occasionally hybridizing species may be a common occurrence among marine fishes and invertebrates exhibiting large effective population sizes, even if gene flow is quantitatively very low. In the Northern Hemisphere, plaice (Pleuronectes platesa) and flounder (Platichthys flesus) represent a good example of hybridizing species that could be used for further studying the generality of such patterns (Kijewska et al., 2009).
Data archiving
All sequencing files (.fastq) can be found on NCBI's Sequence Read Archive under project number PRJNA195555 (European eel) and PRJNA230782 (American eel).
Acknowledgments
We thank Russel Poole, Javier Lobon, Eric Feunteun and participants in the Galathea 3 Eel Project for provision of samples, Karen-Lise D Mensberg, Dorte Meldrup and Annie Brandstrup for technical assistance and three anonymous reviewer for constructive comments on the manuscript. We acknowledge funding from the Danish Council for Independent Research, Natural Sciences (Grant 09-072120).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- Albert V, Jónsson B, Bernatchez L. Natural hybrids in Atlantic eels (Anguilla anguilla, A. rostrata): evidence for successful reproduction and fluctuating abundance in space and time. Mol Ecol. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- Als TD, Hansen MM, Maes GE, Castonguay M, Riemann L, Aarestrup L, et al. All roads lead to home: panmixia of European eel in the Sargasso Sea. Mol Ecol. 2011;20:1333–1346. doi: 10.1111/j.1365-294X.2011.05011.x. [DOI] [PubMed] [Google Scholar]
- Amish SJ, Hohenlohe PA, Painter S, Leary RF, Muhfeld C, Allendorf FW, et al. RAD sequencing yields a high success rate for westslope cutthroat and rainbow trout species-diagnostic SNP assays. Mol Ecol Res. 2012;12:653–660. doi: 10.1111/j.1755-0998.2012.03157.x. [DOI] [PubMed] [Google Scholar]
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Helfman GS, Saunders NC, Hales LS. Mitochondrial DNA differentiation in North Atlantic eels: population genetic consequences of an unusual life history pattern. Proc Natl Acad Sci USA. 1986;83:4350–4354. doi: 10.1073/pnas.83.12.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Nelson WS, Arnold J, Koehn RK, Williams GC, Thorsteinsson V. The evolutionary genetic status of Icelandic eels. Evolution. 1990;44:1254–1262. doi: 10.1111/j.1558-5646.1990.tb05229.x. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux F, Goudet J. Statistical properties of population differentiation estimators under stepwise mutation in a finite island model. Mol Ecol. 2002;11:771–783. doi: 10.1046/j.1365-294x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, St-Cyr J, Normandeau E, Maes GE, Als TD, Kalujnaia S, et al. Differential timing of gene expression regulation between leptocephali of the two Anguilla species in the Sargasso Sea. Ecol Evol. 2011;1:459–467. doi: 10.1002/ece3.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P, Querner U. Genetic basis of migratory behaviour in European warblers. Science. 1981;212:77–79. doi: 10.1126/science.212.4490.77. [DOI] [PubMed] [Google Scholar]
- Berthold P, Querner U. Microevolutionary aspects of bird migration based on experimental results. Isr J Zool. 1995;41:377–385. [Google Scholar]
- Boëtius J. Atlantic Anguilla: a presentation of old and new data of total number of vertebrae with special reference to the occurrence of Anguilla rostrata in Europe. Dana. 1980;1:93–112. [Google Scholar]
- Bonhommeau S, Blanke B, Tréguier AM, Grima N, Rivot E, Vermand Y, et al. How fast can the European eel (Anguilla anguilla) larvae cross the Atlantic Ocean. Fish Oceanogr. 2009;18:371–385. [Google Scholar]
- Castillo AGF, Beall E, Moran P, Martinez JL, Ayllon F, Garcia-Vazquez E. Introgression in the genus Salmo via allotriploids. Mol Ecol. 2007;16:1741–1746. doi: 10.1111/j.1365-294X.2007.03257.x. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe PA, Cresko WA, Postlethwait JH. Stacks: building and genotyping loci de novo from short-read sequences. G3 Genes Genom Genet. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté C, Gagnaire PA, Bourret V, Verrault G, Castonguay M, Bernatchez L. Population genetics of the American eel (Anguilla rostrata). FST=0 and North Atlantic Oscillation effects on demographic fluctuations of a panmictic species. Mol Ecol. 2013;22:1763–1776. doi: 10.1111/mec.12142. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Location of sterility factors on Drosophila pseudoobscura. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, Glass NL, et al. Population genomics and local adaptation in wild isolates of a model microbial organism. Proc Natl Acad Sci USA. 2011;107:2831–2837. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter PD, Preston JL, Bassham S, Cresko WA, Johnson EA. Local de novo assembly of RAD paired-end contigs using short sequencing reads. PLoS One. 2011;6:e18561. doi: 10.1371/journal.pone.0018561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Ecol Evol. 2012;28:342–349. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Albert V, Jónsson B, Bernatchez L. Natural selection influences AFLP intraspecific genetic variability and introgression patterns in Atlantic eels. Mol Ecol. 2009;18:1678–1691. doi: 10.1111/j.1365-294X.2009.04142.x. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Bernatchez L. Comparative genomics reveals adaptive protein evolution and a possible cytonuclear incompatibility between European and American eels. Mol Biol Evol. 2012;29:2909–2919. doi: 10.1093/molbev/mss076. [DOI] [PubMed] [Google Scholar]
- Gharrett AJ, Smoker WW. Two generations of hybrids between even– and odd-year pink salmon (Oncorhynchus gorbuscha): a test for outbreeding depression. Can J Fish Aquat Sci. 1991;48:1744–1749. [Google Scholar]
- Grant BR, Grant PR. Simulating secondary contact in allopatric speciation: an empirical test of premating isolation. Biol J Linn Soc. 2002;76:545–556. [Google Scholar]
- Harrison RG. Hybrid zones: windows on the evolutionary process. Oxford Surv Evol Biol. 1990;7:69–128. [Google Scholar]
- Henkel CV, Burgerhout E, de Wijze DL, Dirks RP, Minegishi Y, Jansen HJ, et al. Primitive duplicate Hox clusters in the European eeĺs genome. PLoS One. 2012;7:e32231. doi: 10.1371/journal.pone.0032231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. Hybrid zones: natural laboratories for evolutionary studies. Trends Ecol Evol. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Quaternary phylogeography: the roots of hybrid zones. Genetica. 2011;139:617–638. doi: 10.1007/s10709-011-9547-3. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:23. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Amish SJ, Catchen JM, Allendorf FW, Luikart G. Next-generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westlope cutthroat trout. Mol Ecol. 2011;11:117–122. doi: 10.1111/j.1755-0998.2010.02967.x. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Day MD, Amish SJ, Miller MR, Kamps-Hugues N, Boyer MC, et al. Genomics patterns of introgression in rainbow and westlope cutthroat trout illuminated by overlapping paired-end RAD sequencing. Mol Ecol. 2013;22:3002–3013. doi: 10.1111/mec.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Wright JM. Female brown trout x male Atlantic salmon hybrids produce gynogens and triploids when backcrossed to male Atlantic salmon. Aquaculture. 1986;57:345–358. [Google Scholar]
- Johnson JR, Fitzpatrick BM, Shaffer HB. Admixture dynamics of tiger salamanders: fitness of early-generation hybrids and the retention of low-fitness genotypes in contemporary populations. BMC Evol Biol. 2010;10:147. doi: 10.1186/1471-2148-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijewska A, Burzynski A, Wenne R. Molecular identification of European flounder (Platichthys flesus) and its hybrids with European plaice (Pleuronectes platessa) ICES J Mar Sci. 2009;66:902–906. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleave JD, Kleckner RC, Castonguay M. Reproductive sympatry of American and European eels and implications for migration and taxonomy. Am Fish Soc Symp. 1987;1:286–297. [Google Scholar]
- MacNamara R, McCarthy TK. Size-related variation in fecundity of European eel (Anguilla anguilla) ICES J Mar Sci. 2012;69:1333–1337. [Google Scholar]
- Mallet J. A species definition for the modern synthesis. Trends Ecol Evol. 1995;10:294–299. doi: 10.1016/0169-5347(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Microsatellite variation and differentiation in North Atlantic eels. J Hered. 2003;94:30–34. doi: 10.1093/jhered/esg062. [DOI] [PubMed] [Google Scholar]
- Moore ME, Goetz FA, Van Doornik DM, Tezak EP, Quinn TP, Reyes-Tomassini JJ, et al. Early marine migration patterns of wild coastal cutthroat trout (Oncorhynchus clarki clarki), steelhead trout (Oncorhynchus mykiss) and their hybrids. PLoS One. 2010;5:e12881. doi: 10.1371/journal.pone.0012881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Isolation mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Munk P, Hansen MM, Maes GE, Nielsen TG, Castonguay M, Riemann L, et al. Oceanic fronts in the Sargasso Sea control the early life and drift of Atlantic eels. P Roy Soc B Biol Sci. 2010;277:3593–3599. doi: 10.1098/rspb.2010.0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen EE, Bach LA, Kotlicki P. HYBRIDLAB (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Res. 2006;6:971–973. [Google Scholar]
- Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P. Development of NSP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Res. 2013;13:447–460. doi: 10.1111/1755-0998.12075. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujolar JM, Bevacqua D, Cappocioni F, Ciccotti E, De Leo GA, Zane L. No apparent genetic bottleneck in the demographically declining European eel using molecular genetics and forward-time simulations. Conserv Genet. 2011;12:813–825. [Google Scholar]
- Pujolar JM, Jacobsen MW, Frydenberg J, Als TD, Larsen PF, Maes GE, et al. A resource of genome-wide single-nucleotide polymorphisms generated by RAD tag sequencing in the critically endangered European eel. Mol Ecol Res. 2013;113:706–714. doi: 10.1111/1755-0998.12117. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione S, Acquadro A, Portis E, Tirone M, Knapp S, Lanteri S. RAD tag sequencing as a source of SNP markers in Cynara cardunculus. BMC Genomics. 2012;13:3. doi: 10.1186/1471-2164-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeb JE, Pascal CE, Ramakrishnan R, Seeb LW. SNP genotyping by the 5'-nuclease reaction: advances in high-throughput genotyping with nonmodel organisms. Method Mol Biol. 2009;578:277–292. doi: 10.1007/978-1-60327-411-1_18. [DOI] [PubMed] [Google Scholar]
- Stolting KN, Nipper R, Lindtke D, Caseys C, Waeber S, Castiglione S, et al. Genomic scan for single nucleotide polymorphisms reveals patterns of divergence and gene flow between ecologically divergent species. Mol Ecol. 2013;22:842–855. doi: 10.1111/mec.12011. [DOI] [PubMed] [Google Scholar]
- Templeton AR.1986Coadaptation and breeding depressionIn: Soulé ME (ed.) Conservation Biology: The Science of Scarcity and Diversity Sinauer: Sunderland; 105–116. [Google Scholar]
- Trautner J. Rapid identification of European (Anguilla anguilla) and North American eel (Anguilla rostrata) by polymerase chain reaction. Inf Fischereiforsch. 2006;53:49–51. [Google Scholar]
- Van Bers NEM, Van Oers K, Kerstens HHD. Genome-wide SNP detection in the great tit Parus major using high throughtput sequencing. Mol Ecol. 2010;19:89–99. doi: 10.1111/j.1365-294X.2009.04486.x. [DOI] [PubMed] [Google Scholar]
- van den Thillart G, Rankin JC, Dufour S. Spawning Migration of the European Eel: Reproduction Index, a Useful Tool for Conservation Management. Springer: Dordecht: The Netherlands; 2009. [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwalko S, Greuter L, et al. Genome-wide RAD sequence data provide unprecedented resolution in species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol. 2012;22:787–798. doi: 10.1111/mec.12023. [DOI] [PubMed] [Google Scholar]
- Wirth T, Bernatchez L. Decline of Atlantic eels: a fatal synergy. P Roy Soc B Biol Sci. 2003;270:681–688. doi: 10.1098/rspb.2002.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.