Summary
Non-alcoholic fatty liver disease (NAFLD) affects up to 30% of the adult population in Western societies, yet the underlying molecular pathways remain poorly understood. Here, we identify the dimeric Activator Protein 1 as a regulator of NAFLD. The Fos-related antigen 1 (Fra-1) and 2 (Fra-2) prevent dietary NAFLD by inhibiting pro-steatotic PPARγ signaling. Moreover, established NAFLD and the associated liver damage can be efficiently reversed by hepatocyte-specific Fra-1 expression. In contrast, c-Fos promotes PPARγ expression, while c-Jun exerts opposing, dimer-dependent functions. Interestingly, JunD was found to be essential for PPARγ signaling and NAFLD development. This unique antagonistic regulation of PPARγ by distinct AP-1 dimers occurs at the transcriptional level and establishes AP-1 as a link between obesity, hepatic lipid metabolism and NAFLD.
Keywords: NAFLD, Steatosis, Activator Protein 1, PPARγ, Lipids, Transcription
Introduction
Given their high energy-to-weight ratio compared to carbohydrates and proteins, lipids are the most efficient energy substrate in mammals. The adipose tissue is the major lipid storage organ and it is essential for controlling metabolic homeostasis(Sethi and Vidal-Puig, 2007). In the healthy state, tissues such as muscle and liver store only minor quantities of lipids(Lara-Castro and Garvey, 2008). However, metabolic stress, as occurring in obese or alcohol-abusing patients, can cause massive ectopic lipid deposition, leading to a disease state, termed “steatosis” or “fatty liver disease”. Depending on the etiology, this disease can be further subgrouped into alcoholic or non-alcoholic fatty liver disease (AFLD and NAFLD respectively). NAFLD is the most common liver disorder in industrialized countries and it frequently leads to severe liver inflammation and damage, a disease state termed “non-alcoholic steatohepatitis”(NASH)(Browning and Horton, 2004). Moreover, NAFLD contributes to hepatic insulin resistance in diabetes(Farese et al., 2012) and is a risk factor for liver dysfunction and cancer development(Smedile and Bugianesi, 2005). Understanding the cellular and molecular mechanisms leading to NAFLD, as well as the identification of novel targets for NAFLD therapy has therefore become a priority(Cohen et al., 2011; Lazo and Clark, 2008).
The Activator Protein-1 (AP-1) (Fos/Jun)protein complex is a dimeric leucine zipper(bZIP) transcription factor. Three different Jun proteins (c-Jun, JunB and JunD) and four different Fos proteins (c-Fos, FosB, Fra-1 and Fra-2) formAP-1 dimer. Jun proteins can either form homodimers, such as c-Jun/c-Jun or c-Jun/JunB, or heterodimers, such as c-Jun/c-Fos. In contrast, Fos proteins exclusively form heterodimers(Halazonetis et al., 1988). Jun and Fos proteins also form heterodimers with other bZIP transcription factors, such as specific MAF and ATF family members(Eferl and Wagner, 2003). Thus, a vast combinatorial variety of AP-1 dimers with likely different molecular and biological functions exists(Hess et al., 2004; Verde et al., 2007; Wagner et al., 2010). Studies using genetically modified mice have unravelled essential roles of AP-1-forming proteins in development, inflammation and cancer(Eferl and Wagner, 2003). Moreover, AP-1 modulates the response to acute cellular insults, such as oxidative stress and DNA damage (Shaulian and Karin, 2002). Cellular stress typically activates AP-1 by augmenting transcription, protein stability and transactivation potential of Jun and Fos family members (Wagner and Nebreda, 2009). In the liver, the genetic inactivation of single Jun or Fos genes in hepatocytes does not compromise organ homeostasis (Bakiri and Wagner, 2013; Eferl and Wagner, 2003). However, AP-1 is critical for the liver’s response to acute stress. For example, c-Jun protects hepatocytes from injury (Fuest et al., 2012; Hasselblatt et al., 2007) and is essential for liver regeneration (Behrens et al., 2002) and carcinogenesis (Eferl et al., 2003; Machida et al., 2010; Min et al., 2012). More recently, we have documented that Fra-1, but not Fra-2, protects hepatocytes from acetaminophen overdose, a paradigm for xenobiotic-mediated acute liver failure (Hasenfuss et al., 2013). In contrast, little is known about the role of AP-1 in chronic stress conditions and the potential contribution of AP-1 to the development of hepatic metabolic disease. Here we combined system genetics with gain- and loss-of-function mouse models to study the function of AP-1 in hepatic lipid metabolism and NAFLD development. We show that, depending on dimer composition, AP-1 either represses or activates the transcription of the pro-steatotic nuclear receptor Peroxisome Proliferator-Activated Receptor γ (PPARγ), which promotes hepatic lipid uptake and lipid droplet formation. Some AP-1 proteins, such as Fra-1 and Fra-2, inhibit the PPARγ pathway and reduce hepatic lipid content. In contrast, other AP-1 proteins, such as c-Fos and JunD induce hepatic PPARγ signaling and lipid accumulation. We also show that AP-1 regulates the PPARγ pathway through direct regulation of the Pparg2 promoter. Using a mouse model for inducible hepatocyte-restricted Fra-1 expression, we demonstrate that the Fra-1-induced suppression of the PPARγ pathway can revert established NAFLD. For the first time, liver-specific single chain Jun~Fos forced dimer mice were employed, in which dimerization of a Fos protein is restricted to a single Jun partner (Bakiri et al., 2002). The analyses of these mouse models provide in vivo evidence that distinct AP-1 dimers regulate the PPARγ pathway in an antagonistic fashion. Finally, we show that JunD is essential for efficient PPARγ signalling and NAFLD formation. Overall, this study identifies AP-1 as a link between dietary obesity, hepatic lipid metabolism and NAFLD.
Results
Fra-1/AP-1 regulates hepatic lipid metabolism and NAFLD
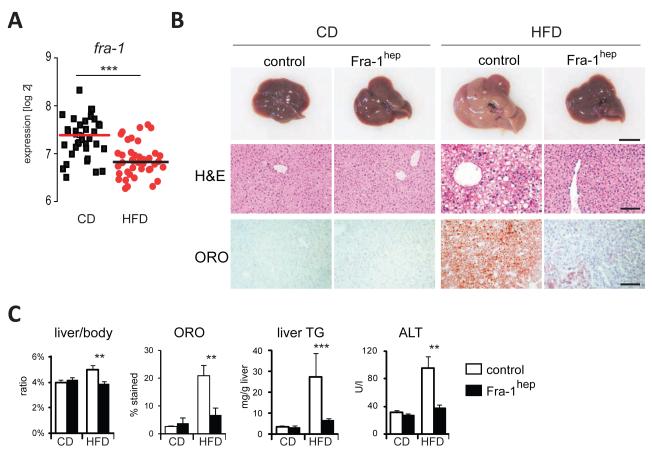
To identify a possible function of AP-1 in metabolism, we analyzed 42 genetically diverse mouse strains from the BXD mouse genetic reference population (GRP)(Peirce et al., 2004).10 animals for each strain were split evenly into two cohorts fedchow diet (CD) or high-fat diet (HFD) for five months. Hepatic AP-1 mRNA expression was then analyzed using genome-wide expression profiles from the BXD strains. Fra-1 mRNA levels were found to be significantly reduced in the HFD-fed cohort, while the expression ofc-fos, fosB, fra-2, c-jun, junb and jund were not affected by the diet(Figure 1A and FigureS1A). To explore whether Fra-1 could causally contribute to HFD-associated metabolic changes in the liver, we analyzed hepatic metabolism in Fra-1hep mice, a previously established model of Doxycycline (Dox)-controllable hepatocyte-restrictedFra-1-overexpression, which does not display any obvious phenotype under basal conditions (Hasenfuss et al., 2013)(for details on mouse strains see Table S1). After HFD feeding, the livers appeared less pale on the macroscopic level and weighed significantly lessin Fra-1hep mice compared to HFD-fed littermate controls (Figure 1B, C). Liver histology indicated a reduction in lipid droplets in mutant mice (Figure 1B), which was confirmed by the quantitation of Oil-RedO (ORO)-positive lipid droplets and liver triglyceride (TG) content analysis (Figure 1C).
Figure 1. Fra-1 is regulated by HFD and inhibits NAFLD and PPARγ expression.
(A) Hepatic fra-1 expression in CD and HFD (for 5 months; 60% kCal/fat) in 42 BXD inbred strains. Each data point represents the mean expression of 5 mice. (B,C), Fra-1hep mice and control littermates were on CD or HFD (for 5-9 months; 45% kCal/fat); n≥5/cohort. (B) Representative liver pictures and histology in Fra-1hep and control mice; ORO=Oil-RedO; bars=1cm and 100μm. (C) Quantitation of ORO-positive areas, liver/body ratio, liver TG content and serum ALT levels. Bar graphs are presented as mean±SEM. See also Figure S1 and Table S2 A,B.
We next addressed the effect of Fra-1 expression on NAFLD-associated liver damage and inflammation. Augmented serum levels of the liver damage marker alanine aminotransferase (ALT) and increased hepatic inflammation marker expression was observed in controls after HFD-feeding, but not in Fra-1hep mice(Figure 1C and Figure S1B). Moreover, immunohistochemistry (IHC) for the pan-lymphocyte marker CD45 and the macrophage markerF4/80 revealed a significant reduction in immune cell infiltrates in HFD-fed mutants compared to diet-matched controls (Figure S1C). In the HFD-fed state, serum IL-6 levels were also reduced in HFD-fed Fra-1hep mice compared to controls (Table S2A). We next explored the effects of hepatic Fra-1 expression on circulating metabolite and hormone levels. Serum TG and cholesterol were mildly elevated in HFD-fed Fra-1hepcompared to control mice in the fasted and/or fed states, while other serum parameters were not affected (Table S2A,B). Despite decreased NAFLD and liver damage, glucose tolerance and insulin tolerance tests (GTT and ITT) revealed that glucose metabolism was not improved, but rather slightly worsened in HFD-fed mutants as compared to controls (Figure S1D). Similar effects of Fra-1 on NAFLD development were also observed in Fra-1hep mice on a C57BL/6J background or using 60% kCal/fat HFD (Table S2B and data not shown). These data collectively suggest that hepatocyte-specific Fra-1 expression protects from dietary-induced NAFLD and secondary liver damage and inflammation, but has little impact on systemic obesity and glucose metabolism.
Fra-1 represses the PPARγ pathway
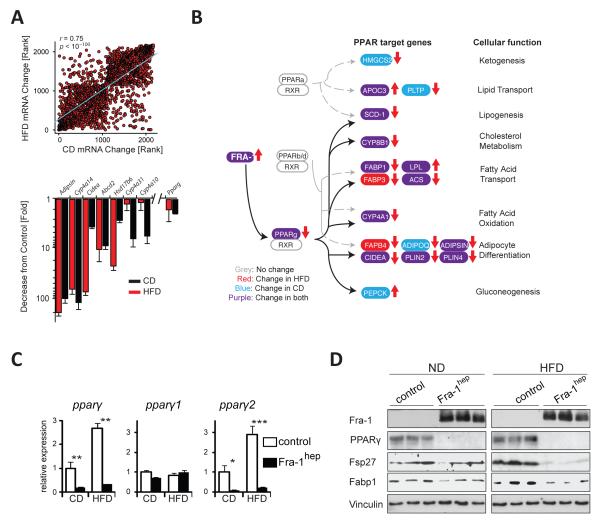
We next analyzed the molecular mechanisms underlying reduced NAFLD in Fra-1hep mice. Genome-wide hepatic gene expression analyses in CD- and HFD-fed Fra-1hep mice demonstrated that the expression of ~3000 genes was changed by at least 1.5 fold in Fra-1hep livers. The vast majority of these genes were regulated in a similar fashion in both dietary conditions and many PPARγ targets, e.g. adipsin and cidea were among the top-downregulated genes (Figure 2A). KEGG pathway analysis (Kanehisa et al., 2012) of the top 2000 most changed genes established PPARγ signaling among the most significantly affected pathways in both diet conditions (Figure 2B). A highly significant fraction of mRNAs, which were reduced in Fra-1hep mice, were encoded by genes with promoters containing putative AP-1 sites (p=9.0E-13) and PPARγ response elements (PPREs) (p=3.3E-11), as revealed by UCSC TFBS conserved tracks analyses (http://david.abcc.ncifcrf.gov/)(Huang da et al., 2009) (Table S3). qRT-PCR, immunoblot and IHC analyses confirmed decreased hepatic pparγ mRNA/PPARγ protein expression in CD- and HFD-fed Fra-1hep mice (Figure 2C,D and Figure S2A).
Figure 2. Fra-1 regulates the PPARγ pathway.
(A-D)Fra-1hep mice and control littermates on CD or HFD (for 5-9 months; 45 % kCal/fat); n≥5/condition. (A) Top: Spearman correlation of ≥1.5 fold-changed hepatic transcripts in Fra-1hep and control littermates (C57BL/6J) in CD (x-axis) and HFD (y-axis). Bottom: Top common downregulated genes in Fra-1hep livers. (B) KEGG pathway analyses for the top 2000 most changed transcripts: PPAR target genes and their cellular functions are indicated. Transcripts changed in CD, HFD or both are highlighted in blue, red and purple respectively. Arrows indicate up- or down-regulation. (C) qRT-PCR analyses of pparγ and its isoforms. (D)Immunoblot analyses in Fra-1hep and control mice. Vinculin served as loading control. Bar graphs are presented as mean±SEM. See also Figure S2 and Table S3.
Decreased pparγ mRNA levels were due to reduced expression of pparγ2mRNA, the main pparγ isoform in the liver (Figure 2C and Figure S2D)(Lee et al., 2012). Among other metabolic regulators Nr0b2, a potential PPARγ target(Kim et al., 2007)and regulator(Kim et al., 2013), which encodes the orphan nuclear receptor SHP, was also found reduced in HFD-fed Fra-1hep mice (Figure S2C). Moreover, we confirmed reduced mRNA expression for several PPARγ target genes, such as fabp1 and lpl, involved in hepatic lipid uptake, and plin2, cidea, fitm1, fitm2, g0s2, involved in lipid droplet formation (Figure S2E). Notably, the Fra-1-induced reduction of ppary2 expression was reversible, as pparγ2 levels reverted to baseline levels upon switching off the transgene (Figure S2F). Kinetic analyses of another inducible Fra-1 mouse model (Fra-1tetON mice)(Hasenfuss et al., 2013) revealed that hepatic pparγ2 mRNA decreased as early as 4 days after Fra-1 induction (Figure S2G).
Adenoviral PPARγ delivery restores NAFLD/steatosis in Fra-1hep mice
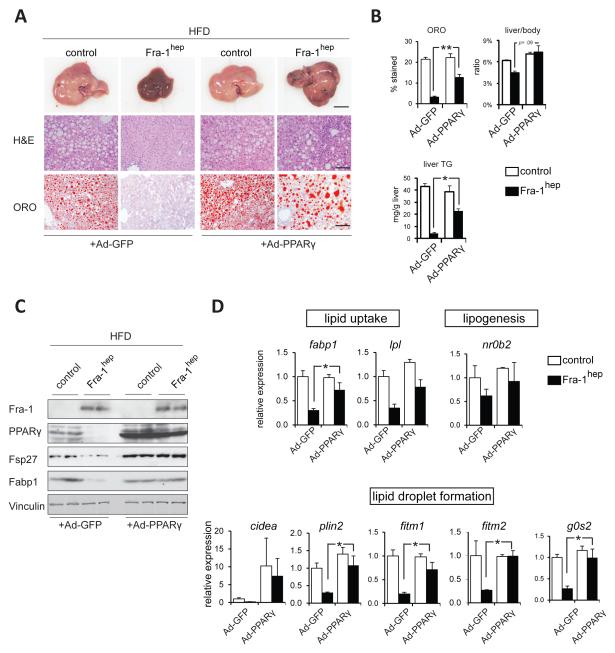
Gain- and loss-of-function studies previously established that hepatocyte PPARγ is both essential and sufficient for NAFLD formation (Gavrilova et al., 2003; Lee et al., 2012; Matsusue et al., 2003; Matsusue et al., 2008; Medina-Gomez et al., 2007; Moran-Salvador et al., 2011). To determine whether reduced NAFLD development in Fra-1hep mice is directly due to decreased PPARγ levels, HFD-fed Fra-1hep and control mice were intravenously injected with either Adeno-PPARγ or Adeno-GFP control virus 8-10 days prior to sacrifice. Adeno-PPARγ did not have any obvious effect on liver macroscopy in steatotic control mice (Figure 3A). In contrast, PPARγ expression increased ORO-positive lipid droplets, liver TG content, and liver/body weight ratio in HFD-fed Fra-1hep mutant mice (Figure3A,B). Moreover, Adeno-PPARγ increased PPARγ target gene expression in the livers of Fra-1hep mice, as compared to Adeno-GFP treated mutants (Figure 3C,D). These data demonstrate that the short-term induction of PPARγ signaling restores hepatic fat accumulation in HFD-fed Fra-1hep mice, supporting its central function in the hepatic phenotype of Fra-1hep mutant mice.
Figure 3. PPARγ delivery restores NAFLD development in Fra-1hep mice.
(A-D)Fra-1hep and control littermates on HFD (for 4-5 months, 45 % kCal/fat) were injected with Adenoviruses expressing PPARγ (Ad-PPARγ) or GFP (Ad-GFP) 8-10 days prior to sacrifice. n=3 for control genotype/cohort; n=4 for Fra-1hep mice/cohort. (A) Liver macroscopy and histology; bars=1cm and 100μm. (B)Quantitation of ORO-positive areas, liver/body ratio and liver TG. (C)Immunoblot analyses in Fra-1hep and control mice. Vinculin served as loading control.(D) qRT-PCR analyses of PPARγ target genes involved in lipid uptake and lipid droplet formation; Ad-GFP controls are set to 1. Bar graphs are presented as mean±SEM.
Reversion of NAFLD by hepatocyte-specific Fra-1 expression
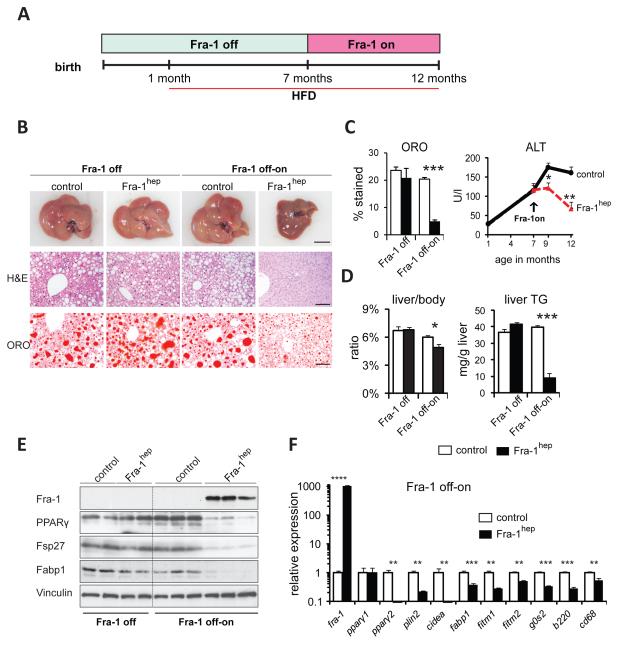
To examine whether Fra-1 induction in steatotic livers ameliorates disease symptoms, Fra-1hepand control littermates were generated in the “Fra-1 off” state and HFD-feeding was started at 1 month of age (Figure 4A). As expected, control and mutant mice were indistinguishable at 7 months of age in the absence of transgene expression(Figure 4B-D). Immunoblot analyses confirmed comparable PPARγ, Fsp27 and Fabp1 levels between control and mutant mice in the “Fra-1 off” state (Figure 4E). A cohort of Fra-1hepand control mice were kept on HFD, but Fra-1 expression was switched on in mutant mice at 7 months of age (Fra-1 off-on). After 2 months of Fra-1 induction, serum ALT was significantly lower in Fra-1hep mice than in control littermates and continued to improve 3 months later, while the mice were maintained on HFD (Figure 4C). At this point the liver was collected for macroscopy, histology, ORO-quantitation, liver/body weight ratio and liver TG content analysis, revealing an almostcomplete reversion of NAFLD in Fra-1hepmutants (Figure 4B-D). Immunoblotting and qRT-PCR analyses confirmed transgene induction, as well as the repression of PPARγ, targets of PPARγ and inflammatory markers after switching on Fra-1 expression (Figure 4E,F). These data suggest that the Fra-1-mediated repression of the PPARγ pathway efficiently reversed established NAFLD and liver damage in mice, even under continued stress of HFD feeding.
Figure 4. Fra-1 expression reverts NAFLD and liver damage.
(A)Fra-1hep and control littermates were maintained in the “Fra-1 off” state and HFD (45 % kCal/fat) was supplied from 1 month of age. Mice were analyzed at 7 months (Fra-1 off, n=2/cohort) or kept on HFD until 12 months, while transgene expression was induced (Fra-1 off-on, n=6/cohort). (B)Liver macroscopy and histology; bars=1cm and 100μm. (C) Serum ALT. (D)Quantitation of ORO-positive areas, liver/body ratio and liver TG content. (E)Immunoblot analyses in Fra-1hep and control mice. Vinculin served as loading control.(F) qRT-PCR analyses of fra-1, pparγ isoforms, PPARγ target genes and inflammation markers (Fra-1 off-on). Bar graphs are presented as mean±SEM.
PPARγ links AP-1 to lipid metabolism
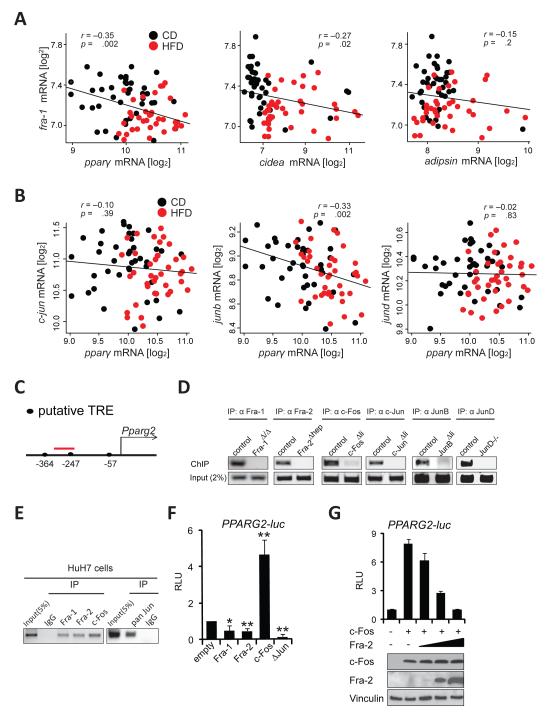
We next searched for correlations between AP-1 genes, PPARγ and PPARγ targets in gene expression arrays from the BXD family of wild-type inbred mouse strains. This analysis revealed that hepatic pparγ expression, assessed with a probe detecting both pparγ isoforms, is significantly upregulated in HFD-fed cohorts (Figure S3A). As expected, pparγ mRNA levels strongly correlated with the expression of PPARγ targets, such as cidea and adipsin (Figure S3B). Notably, and consistent with our findings in Fra-1hepmice, a significant inverse correlation was found between fra-1 and pparγ and fra-1 and cidea regardless of the diet (Figure 5A). Other PPARγ targets, such as adipsin followed a similar trend without reaching statistical significance (Figure 5A). Interestingly, junb, a potential dimerization partner for Fra-1 also negatively correlated with pparγ (Figure 5B), indicating that several AP-1 proteins consistently regulate PPARγ signaling in genetically diverse populations.
Figure 5. Several AP-1 proteins regulate the PPARγ pathway.
Correlation plots for fra-1 with pparγ,cidea and adipsin (A) and for Jun members with pparγ(B) in CD and HFD (5months; 60 % kCal/fat) in the BXD inbred family. Each data point represents the average expression from 5 pooled mice. Pearson’s r was used to analyze correlations and p-values are indicated. (C) Proximal murine Pparg2 promoter: Position of the putative AP-1 binding TPA-responsive element (TRE) is indicated relative to the transcription start. The ChIP-PCR amplicon is depicted in red. (D)ChIP assays using hepatic chromatin from AP-1-deficient mice. Endpoint PCR products representative of 3 independent experiments are shown. (E) ChIP assays in Huh7 cells; primers amplifying a region homologous to (C) were used. Data are representative of 3 independent experiments. (F,G) Human PPARG2 reporter assays in HuH7 cells. Data are mean±s.e.m of 4 independent experiments in (F). Technical replicates of one representative experiment (n=2) is shown and ectopic c-Fos and Fra-2 expression is confirmed by immunoblot in (G). RLU: relative luminescence units. ΔJun: truncated c-Jun. Control (empty vector) is set to 1. Bar graphs are presented as mean±SEM. See also Figure S3.
Antagonistic regulation of Pparg2 by AP-1
The proximal promoter of the mouse Pparg2 gene (encoding PPARγ2), in which we identified several putative AP-1 sites (Figure 5C), is conserved across species (Figure S3D). Thus, we assessed the binding of various Jun and Fos proteins to the Pparg2 promoter. Chromatin samples were prepared from liver tissue of mice deficient for individual AP-1 genes (Table S1) and their respective littermates to control for antibody specificity. Chromatin immunoprecipitation (ChIP) assays revealed that Fra-1, Fra-2, c-Fos, c-Jun, JunB and JunD all efficiently bound to the proximal Pparg2 promoter fragment in liver tissue (Figure 5D). α-Flag ChIP assays also revealed a significant enrichment for the same promoter fragment in livers from Fra-1hep and c-Foshep mice (Figure S3E), a mouse model for inducible c-Fos expression in the liver (Table S1). No enrichment was observed for an unrelated genomic fragment in the same α-Flag ChIP samples (Figure S3E). Fra-1, Fra-2, c-Fos and Jun proteins also bound to the human PPARG2 promoter, as α-Fra-1, α-Fra-2, α-c-Fos, α-pan-Jun, but not control IgG ChIP samples were enriched for the proximal PPARG2 promoter region in human HuH7 hepatoma cells (Figure5E). These data indicate that Jun and Fos proteins are functionally involved in the direct regulation of the mouse Pparg2/human PPARG2 promoter. We therefore analyzed the effects of Fos and Jun proteins on PPARG2 promoter activity. Reporter assays revealed that the activity of a human PPARG2 luciferase reporter was inhibited by transfecting Fra-1 and also by Fra-2 in HuH7 and 293T cells (Figure 5F and Figure S3F). In contrast, the PPARG2 luciferase reporter was activated by c-Fos and inhibited by a dominant-negative Δc-Jun construct, which lacks a transactivation domain (Figure 5F and Figure S3F). Finally, PPARG2 reporter activation by c-Fos was efficiently inhibited by co-transfecting increasing amounts of Fra-2 (Figure 5G). These data demonstrate that c-Fos and Fra-1/2 regulate the PPARG2 promoter in an antagonistic manner.
AP-1 dimer-specific regulation of PPARγ signaling
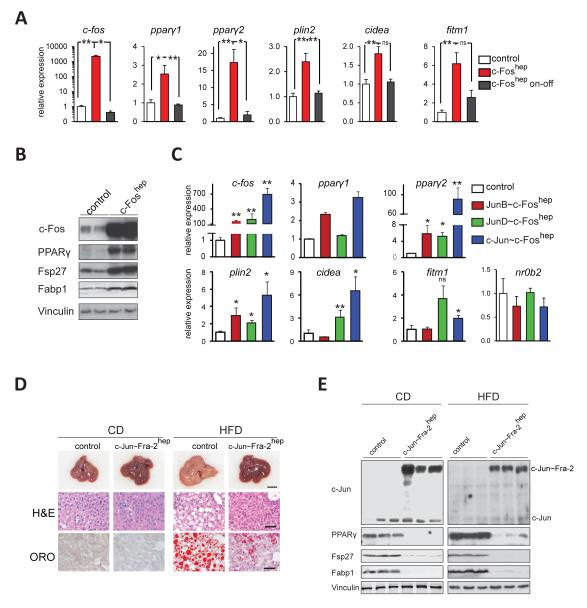
The consequences of hepatic c-Fos expression were analyzed in c-Foshep mice. As early as 1 week after c-Fos induction, a strong increase in ppary2 mRNA, PPARγ protein and PPARγ target gene expression was observed in c-Foshep mice, while ppary1 was only mildly affected and nr0b2 unchanged (Figure 6A,B and Figure S4A,B). After switching off transgene expression, c-fos, ppary2 and PPARγ target genes reverted to baseline levels (on-off, Figure 6A). The reversible induction of ppary2expression was confirmed in c-Foshep mice after 8 weeks of transgene induction (Figure S4B). Next, we studied the effect of Jun~c-Fos forced dimers(Bakiri et al., 2002) on hepatic PPARγ signaling using transgenic mice, in which dimerization of c-Fos is restricted to c-Jun, JunB or JunD (Table S1). The expression of all Jun~c-Fos dimers, such as c-Jun~c-Fos, JunB~c-Fos and JunD~c-Fos, increased ppary2 mRNA and PPARγ target gene expression in the livers of mutant mice, with c-Jun~c-Fos causing the strongest induction (Figure 6C). As the expression of c-Fos or Jun~c-Fos forced dimers rapidly caused lethal liver dysplasia (Bakiri et al., unpublished), the long-term consequences of increased PPARγ signaling on lipid metabolism and NAFLD could not be further investigated.
Figure6. Antagonistic regulation of PPARγ signaling by AP-1.
(A) qRT-PCR analyses of c-fos (endogenous+ectopic), pparγ1/2 and PPARγ targets in c-Foshep and control mice. c-Fos expression was induced for 1 week (c-Fos on) or induced for 1 week and switched off for 1 week (c-Fos on-off). n=8 for c-Foshepmice (on), n=10 for controls (on), n=3 for c-Foshep (on-off). (B) Immunoblot analyses in c-Foshep (c-Fos on for 1 week) and control mice. Vinculin served as a loading control. Blots are representative of 3 controls and 4 mutants.(C)qRT-PCR analyses in JunB~c-Foshep(n=6), JunD~c-Foshep(n=5), and c-Jun~c-Foshep(n=4)(transgene on for 1 month) and control (n=5) mice. Liver macroscopy and histology (D) and immunoblot analyses (E) of c-Jun~Fra-2hep mice (c-Jun~Fra-2 switched on at 1 month) and control littermates on CD or HFD (for 4 months;60 % kCal/fat). Vinculin served as loading control; n≥5/condition. Bar graphs are presented as mean±SEM. See also Figure S4.
We next explored the effect of Fra-2 and Fra-2/AP-1 dimers on PPARγ signaling and hepatic lipid metabolism. In contrast to c-Fos and Jun~c-Fos dimers, hepatocyte-restricted Fra-2 monomer expression in Fra-2hep mice inhibited the PPARγ pathway and prevented NAFLD development (Figure S4C-G). c-Jun~Fra-2 forced dimers in c-Jun~Fra-2hep mice suppressed PPARγ signaling and NAFLD development to a similar extent as Fra-2 monomers(Figure 6D,E and Figure S4H-J). In contrast to most established PPARγ target genes, nr0b2levels were unaffected in Fra-2hepmice (Figure S4G), while a mild up-regulation of nr0b2 mRNA was observed in CD-fed c-Jun~Fra-2hep mice (Figure S4J). These data collectively suggest an antagonistic regulation of the PPARγ pathway and lipid metabolism by c-Fos/AP-1 and Fra-2/AP-1 dimersin vivo.
JunD is essential for NAFLD development
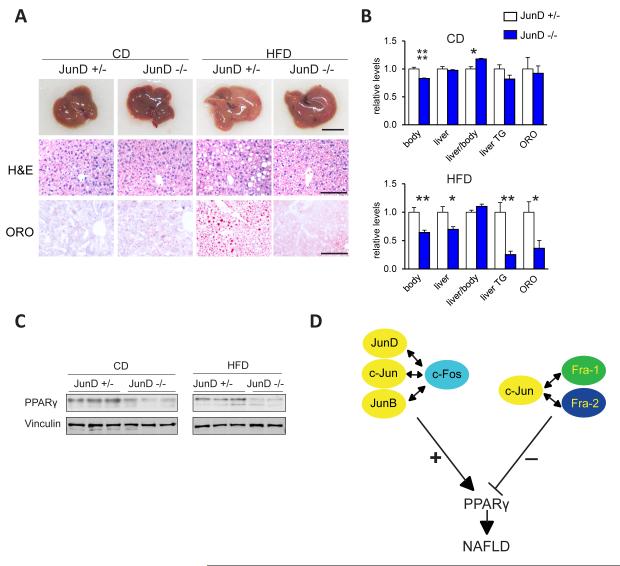
To investigate whether individual Jun or Fos members are essential for hepatic PPARγ expression and lipid metabolism, we employed loss-of-function mutant mice. Individual gene inactivation of Fra-1, Fra-2 or c-Fos had no effect on hepatic pparγ2 expression nor on HFD-induced NAFLD (Figure S5A and data not shown). Similarly, the single inactivation of c-Jun or Junb did not affect hepatic pparγ2 expression (Figure S5A), while JunD−/− mice displayed decreased pparγ2levels in the liver under basal conditions (Figure S5A). We therefore analyzed HFD-induced NAFLD development in JunD-deficient mice. Liver macroscopy, histology, liver TG content analyses and ORO-quantitation revealed decreased HFD-induced NAFLD in JunD−/− livers (Figure 7A,B). qRT-PCR, immunoblot and IHC analysis confirmed decreased pparγ2 mRNA/PPARγ protein levels in HFD fed JunD−/− mice compared to controls in both diet conditions (Figure 7C and Figure S5B). qRT-PCR analyses revealed reduced expression of the PPARγ targets cidea and fitm1in JunD−/−livers, whereas nr0b2 expression was not affected (Figure S5C).
Figure7. JunD is essential for PPARγ expression and steatosis formation.
(A-C)Analyses of JunD−/− and JunD+/− control littermates (males and females) on CD or HFD (for 9 months; 45 % kCal/fat); n≥7/condition. (A) Representative liver macroscopy and histology; bars=1cm and 100μm. (B) Macroscopic parameters, serum ALT, liver TG content and quantitation of ORO-positive areas; relative levels are plotted and sex matched controls set to 1. Bar graphs are presented as mean±SEM. (C)Immunoblot analysis for PPARγ and Vinculin.(D) Antagonistic regulation of PPARγ expression by different AP-1 dimers: c-Fos induces PPARγ as a dimer with any Jun protein, while Fra-1 and Fra-2 repress PPARγ, likely by dimerizing with c-Jun, thereby affecting hepatic lipid metabolism and NAFLD. See also Figure S5 and Table S4.
As previously reported(Thepot et al., 2000), JunD−/− mice had a reduced body weight and this effect was maintained in HFD (Figure7B). Total liver and fat pad weights were specifically decreased after HFD-feeding in JunD−/− mice compared to littermate controls (Figure 7B and Table S4). Interestingly, reduced pparγ2 mRNA was also observed in heart tissue of HFD-fed JunD−/− mice (Figure S5D), indicating that AP-1 might also regulate the PPARγ pathway in other organs.
Discussion
Combined forward and reverse genetic approaches have a strong potential for discovering new regulators of metabolism. The initial identification of Fra-1as a potential obesity-related gene in livers from the BXD population of inbred mouse strains prompted us to further explore the role of AP-1 proteins. Subsequent mechanistic studies led to the discovery that AP-1 can function as a molecular link between obesity and liver metabolism. First, this study established AP-1 as a potent regulator of lipid metabolism and NAFLD development. Second, gene pathway analysis and BXD population genetics highlighted the AP-1 complex as a novel regulator of hepatic PPARγ signaling. Third, we demonstrate that Fra-1 repressed the PPARγ-dependent expression of genes involved in lipid uptake/lipid droplet formation, thereby efficiently improved established steatosis, liver damage and inflammation. Fourth and maybe most intriguingly, the mouse Pparg2 and the human PPARG2 promoter were found to be regulated in an antagonistic fashion by distinct AP-1 dimers (Figure 7D).
PPARγ promotes lipid uptake by increasing the expression of lipid transporters, such as fatty acid binding proteins (Fabps), and by promoting lipid storage in lipid droplets. Lipid droplet proteins (LDPs) inhibit TG lipolysis thereby preventing lipid-droplet breakdown (Fujimoto et al., 2008; Puri et al., 2008; Sun et al., 2012). Several LDPs are regulated by PPARγ at the transcriptional level(reviewed in Tontonoz and Spiegelman, 2008) and promote NAFLD in mice, including Cidea, Plin2, and Fsp27(Chang et al., 2006; Dalen et al., 2004; Matsusue et al., 2008; Sun et al., 2012; Varela et al., 2008; Zhou et al., 2012). Similarly, deletion of the fatty acid transporter Fabp1reduced the dietary induction of NAFLD (Newberry et al., 2003; Newberry et al., 2006). Previous studies suggested that hepatic PPARγ also promotes hepatic lipogenesis(Matsusue et al., 2003; Medina-Gomez et al., 2007). In Fra-1hep mice, which display a dramatic reduction in PPARγ levels, decreased expression of the Stearoyl-CoA desaturase-1 (SCD-1), a key enzyme in the generation of unsaturated fatty acids, was observed. However, no consistent changes in the expression of SREBP-1/2, the main transcriptional regulators of de novo lipogenesis, nor in the SREBP-1/2 targets FAS and ACC, were observed. Since FAS and ACC catalyze the rate-limiting steps in fatty acid synthesis, altered lipogenesis likely does not play a major role in the Fra-1-mediated repression of NAFLD. Instead, decreased hepatic lipid uptake and lipid droplet formation is most likely the primary cause for reduced steatosis formation in Fra-1hep, Fra-2hep and c-Jun~Fra-2hep mice. Previous reports have shown that PPARγ induced the expression of Nr0b2 (Kim et al., 2007). In line with this, HFD-fed Fra-1hep mice displayed reduced pparγ2 and nr0b2 levels, which appeared normalized after Adeno-PPARγ treatment. More recently, Nrob2 was shown to be required for hepatic PPARγ expression and NAFLD(Kim et al., 2013). However, we did not observe a consistent correlation between pparγ2 andnr0b2 expression across dietary conditions in our AP-1 mutant mouse models. Therefore, AP-1 likely regulates pparγ2 and NAFLD independently of Nr0b2.
Hepatocyte-specific Pparg deletion, like Fra-1 overexpression, has been shown to reduce liver TG and to increase serum TG levels in the obese ob/ob mice and the AZIP lipodystrophy model, likely due to decreased hepatic lipid uptake(Gavrilova et al., 2003; Matsusue et al., 2003). Moreover, PPARγ2-dependent hepatic steatosis has been suggested to buffer systemic TG levels (Medina-Gomez et al., 2007). Similar to mice with hepatocyte-restricted Pparg deletion, Fra-1hep mice displayed worsened glucose metabolism after HFD-feeding, despite a reduction in NAFLD. Thus, hepatic Fra-1 overexpression largely phenocopies the effects of hepatocyte-specific Ppargdeletion on lipid and glucose metabolism. As elevated serum TG levels are associated with diabetes development, increased serum TG levels likely contribute to the deterioration of glucose metabolism in HFD-fed Fra-1hep and Pparg-deficient mice.
Our data suggest a functional antagonism between activating c-Fos/AP-1 and repressing Fra/AP-1 dimers. Interestingly, c-Jun/c-Fos dimers increased, whereas c-Jun/Fra-2 dimers reduced PPARγ2 expression, suggesting a partner-dependent effect of c-Jun on Pparg2 promoter activity. Previous studies in other organs suggested overlapping functions of c-Fos and Fra-1/2(Fleischmann et al., 2000; Matsuo et al., 2000). Thus, we here identify Pparg as the first gene to be antagonistically regulated by different Fos proteins. This finding raises the intriguing question: How do structurally similar protein complexes, such as c-Fos/AP-1 and Fra/AP-1 dimers, have opposite effects on the same promoter? While further research is required to address this question, several AP-1 co-repressors, such as Sirt1 (Purushotham et al., 2009)and HDAC3 (Feng et al., 2011; Knutson et al., 2008; Sun et al., 2012) are involved in hepatic lipid metabolism. We speculate that specifically Fra/AP-1 dimers might interact with such co-repressors to inhibit Pparg2 promoter activity.
Among Jun proteins, JunD was found to be important for NAFLD development in the liver. As JunD−/− mice are leaner and display reduced adiposity, the possibility that extrahepatic functions of JunD contribute to decreased NAFLD development in JunD−/− mice cannot be excluded. However, JunD is essential for normal pparγ2 mRNA/PPARγ protein expression and bound to the Pparg2 promoter in liver tissue, suggesting JunD as a novel physiologically relevant regulator of hepatic PPARγ signaling. Given the key function of PPARγ in NAFLD development, reduced PPARγ signaling in the liver likely contributes to NAFLD resistance in JunD−/− mice.
The described data from gain-and loss-of-function mouse models, together with the correlations between AP-1 components and the PPARγ pathway in the BXD cohort, establish AP-1 as an important regulator of PPARγsignaling and NAFLD. HFD affects a plethora of cellular signaling cascades, such as the Insulin(Kim and Kahn, 1994), the JNK(Hibi et al., 1993) and the PKC(Boyle et al., 1991) pathways. As these pathways are also known regulators of AP-1 expression and activity, exploring how they affect AP-1 levels and dimerization during obesity is certainly an important challenge for future experiments. Moreover, extensive cross-talk between AP-1 and transcription factors of the NF-κB(Fujioka et al., 2004) and the nuclear receptor family(Glass and Saijo, 2010; Ricote and Glass, 2007; Wan et al., 2007) has been described. Future studies should reveal the molecular interplay of these pathways with AP-1 signaling in the context of NAFLD in both mice and human.
Experimental procedures
Animal procedures
Mice were maintained in a 12 hour light/12 hour dark cycle with food and water ad libitum. Chow (D8604, Harlan), 45% kCal/fat HFD (D12451, Research diets), 60%kCal/fat HFD (D12492, Research diets) were used as specified in the figure legends. If not indicated otherwise, male mice were used and HFD feeding was started between 4-8 weeks of age. Dox (1g/l) was supplied in sucrose-containing (100g/l) drinking water. The BXD mice were sacrificed after overnight fasting, while in other experiments, the mice were sacrificed in CO2-chambers between 2pm and 5pm in the fed state. Fasted Cholesterol and TG measurements were performed using serum from overnight fasted mice. For intra-peritoneal glucose tolerance (GTT) and insulin tolerance tests(ITT), mice were fasted for 6 hours (GTT) or 8 hours (ITT) and intra-peritoneally injected with 1 mg glucose/kg body weight (GTT) or 0,5U Insulin/kg body weight (ITT). Glucose and insulin were diluted in PBS to an injectable volume. Blood glucose was determined by tail puncture for all time points. All mouse experiments were performed in accordance with local and institutional regulations. Details on mouse strains can be found in Table S1.
Blood analyses
Blood was collected from the submandibular vein, by tail puncture or by cardiac puncture at experimental endpoints. Unless indicated otherwise specified, parameters were analyzed in the fed state. Serum ALT, TG and cholesterol levels were determined using a Reflovet blood chemistry analyzer and glucose using an Accucheck glucose analyzer (Aviva). Serum Leptin, Resistin, Adiponectin and IL-6 were measured using Quantikine ELISA kits (R&D) and serum Insulin was determined with an ultrasensitive ELISA (Mercodia). Serum ß-HB and FFA were measured using enzymatic assays (Cayman Chemicals).
qRT-PCR/immunoblot analysis
qRT-PCR was performed using the GoTaq® qPCR Master Mix and an Eppendorf light cycler. Expression levels were calculated using the ΔCt-method. Data were normalized to a housekeeping gene (rps27 or rpl0). Primer sequences are available upon request. Immunoblot analysis was performed using standard protocols and following antibodies: ACC, PPARγ, CEBPβ, c-Jun, phospho-CREBP, total CREB (Cell Signaling), Vinculin (Sigma), PPARγ, Parp-1, CEBPα, c-Fos, Fra-1 (Santa Cruz), HNF4, FAS, Fabp1 (Abcam), and Fsp27 (Novus Biologicals), SREBP-1/2 (BD Bioscience). Nuclear extracts from liver tissue were obtained using the NE-PER Nuclear Protein Extraction Kit (Pierce).
Histology
H&E- and ORO-staining were performed using standard procedures. ORO-positive areas were quantified as previously described(Mehlem et al., 2013). IHC was performed as described (Hasenfuss et al., 2013) using following antibodies: PPARγ (Cell Signaling), CD45 (Abcam), F4/80 (AbD Serotec).
RNA microarray
RNA was isolated using the RNEasy Midi kit (Qiagen) and RNA integrity was evaluated using an Agilent 2100 Bioanalyzer. Samples of RNA integrity score above 7.8 were used for microarray analysis. 100ng of RNA was labeled with Cy3 (RNA pool from at least 5 control mice, which were either fed CD or HFD) or Cy5 (RNA samples from individual mutants, which were either fed CD or HFD) using the Low Input Quick Amp Labeling Kit Version 6.5 (Agilent). Labeled RNAs were purified using RNeasy spin columns (Qiagen) and hybridized to a mouse gene expression array G3 8×60K (Agilent microarray design ID 028005, P/N G4852A). On each array, the Cy3-labeled control pool and one Cy5 labeled mutant sample were hybridized at 65°C for 17 hours. The microarray was scanned on a 2505C DNA microarray scanner (Agilent) and images were analyzed using the Feature Extraction Software Version 10.7 (Agilent). Multiple testing correction was performed using the Benjamin-Hochberg procedure. Data are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE52275 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52275). Hepatic gene expression of the BXD strains was analyzed using Mouse Gene 1.0 ST Arrays (Affymetrix) and are accessible on http://www.genenetwork.org.Standard array analysis methods were used, e.g. RMA normalization, as described elsewhere(Irizarry et al., 2003).
Gene pathway analysis
Microarray data were analyzed separately in CD and HFD conditions by comparing control to mutant livers. All nominally significant changes with fold change ≥ 1.5 were retained. Gene sets were then winnowed using multiple testing correction (Benjamin-Hochberg) and entered independently into Web Gestalt (http://bioinfo.vanderbilt.edu/webgestalt/). Enriched pathways were generated based on KEGG gene ontology annotations. The PPARγ signaling pathway was found significantly modulated in both dietary conditions. The two independently-generated pathways were then overlaid and redrawn to generate the pathway diagram.
Cell culture and reporter assay
HuH7 and 293T cells were cultured in DMEM/10%FCS at 37°C and 5% CO2. For reporter assays, 0.8× 105 HuH7 or 293T cells were plated per well of a 24-well plate. 24 hours later, 0.01μg Renilla vector, 0.2μg PPARG2-luc vector(Saladin et al., 1999)and 0.6μg pCMV-AP-1 or pCMV-empty control vector were transfected using Lipofectamine 2000 (Invitrogen). Cells were harvested 48h after transfection and luciferase activity was analyzed using the Dual-Glo Luciferase Assay (Promega).
Liver TG content analysis
Frozen liver tissue (25-75mg) was homogenized in chloroform/methanol (8:1 v/v; 500ul per 25 mg tissue) and shaken at RT for 8-16 hours. H2SO4 was added to a final concentration of 0.28M. After centrifugation, the lower phase was collected, dried, and TG content was measured using an enzymatic assay (Caymen Chem).
Chromatin immunoprecipitation (ChIP)
ChIP was performedusing following antibodies: Flag (F3165, Sigma), Fra-1 (SC-183, Santa Cruz), Fra-2 (rat, CNIO polyclonal), c-Fos (PC-05, Calbiochem), c-Jun (BD), JunB (SC-73, Santa Cruz), JunD (CS5000, Cell Signaling). Pan Jun ChIP with HuH7 cells has been performed with a mixture of2 antibodies raised against an epitope present in all Jun proteins. For details on the ChIP protocol, see also Supplementary Experimental Procedures.
Statistical analysis
Statistical significance was calculated using Student’s two-tailed t-test if not indicated otherwise, *: p<0.05; **: p<0.01; ***: p<0.001, ****: p<0.0001.
Supplementary Material
Highlights.
- AP-1 is a regulator of hepatic lipid metabolism.
- Fra-1prevents and importantly, fully reverts NAFLD in vivo.
- c-Fos/c-Jun dimers activate, while Fra/c-Jun dimers repress the Pparg2 promoter.
-JunD deficiency impairs PPARγ signaling and inhibits NAFLD.
Acknowledgements
We thank Drs. N. Djouder, M. Perez-Moreno, R. Ricci, M. Serrano and G. Sumara for critical reading of the manuscript and valuable suggestions; the CNIO Transgenics Unit and G. Luque and G. Medrano for technical help with mouse procedures. J.A. is the Nestlé Chair in Energy Metabolism and his laboratory is supported by grants from the Ecole Polytechnique Fédérale de Lausanne, the EU Ideas program (AdG-231138), the Swiss National Science Foundation (31003A-140780) and NIH (R01AG043930). Erwin F. Wagner laboratory is supported by the Banco Bilbao Vizcaya Argentaria Foundation (F-BBVA), a grant from the Spanish Ministry of Economy (BFU2012-40230) and an ERC-Advanced grant ERC-FCK/2008/37. Martin K. Thomsen was supported by a Juan de la Cierva postdoctoral fellowship. Sebastian C. Hasenfuss received a Boehringer Ingelheim Fonds (BIF) PhD fellowship and an EMBO short term fellowship (ASTF 198–2012).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest
For procedure details, see Extended Experimental Procedures.
References
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakiri L, Wagner EF. Mouse models for liver cancer. Molecular oncology. 2013;7:206–223. doi: 10.1016/j.molonc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Sibilia M, David JP, Mohle-Steinlein U, Tronche F, Schutz G, Wagner EF. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M, Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BH, Li L, Paul A, Taniguchi S, Nannegari V, Heird WC, Chan L. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol Cell Biol. 2006;26:1063–1076. doi: 10.1128/MCB.26.3.1063-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalen KT, Schoonjans K, Ulven SM, Weedon-Fekjaer MS, Bentzen TG, Koutnikova H, Auwerx J, Nebb HI. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes. 2004;53:1243–1252. doi: 10.2337/diabetes.53.5.1243. [DOI] [PubMed] [Google Scholar]
- Eferl R, Ricci R, Kenner L, Zenz R, David JP, Rath M, Wagner EF. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Farese RV, Jr., Zechner R, Newgard CB, Walther TC. The problem of establishing relationships between hepatic steatosis and hepatic insulin resistance. Cell Metab. 2012;15:570–573. doi: 10.1016/j.cmet.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hafezi F, Elliott C, Reme CE, Ruther U, Wagner EF. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 2000;14:2695–2700. doi: 10.1101/gad.187900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuest M, Willim K, MacNelly S, Fellner N, Resch GP, Blum HE, Hasselblatt P. The transcription factor c-Jun protects against sustained hepatic endoplasmic reticulum stress thereby promoting hepatocyte survival. Hepatology. 2012;55:408–418. doi: 10.1002/hep.24699. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, Uwagawa T, Li Z, Evans DB, Abbruzzese JL, Chiao PJ. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–7819. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman ML. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Hasenfuss SC, Bakiri L, Thomsen MK, Hamacher R, Wagner EF. The AP-1 transcription factor Fra-1 is dispensable for murine liver fibrosis, but modulates xenobiotic metabolism. Hepatology. 2013 doi: 10.1002/hep.26518. doi: 10.1002/hep.26518. [DOI] [PubMed] [Google Scholar]
- Hasselblatt P, Rath M, Komnenovic V, Zatloukal K, Wagner EF. Hepatocyte survival in acute hepatitis is due to c-Jun/AP-1-dependent expression of inducible nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:17105–17110. doi: 10.1073/pnas.0706272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HI, Koh YK, Kim TH, Kwon SK, Im SS, Choi HS, Kim KS, Ahn YH. Transcriptional activation of SHP by PPAR-gamma in liver. Biochemical and biophysical research communications. 2007;360:301–306. doi: 10.1016/j.bbrc.2007.05.171. [DOI] [PubMed] [Google Scholar]
- Kim SC, Kim C, Axe D, Cook A, Lee M, Li T, Smallwood N, Chiang JY, Hardwick JP, Moore DD, et al. All-trans-retinoic acid ameliorates hepatic steatosis in mice via a novel transcriptional cascade. Hepatology. 2013 doi: 10.1002/hep.26699. doi: 10.1002/hep.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kahn CR. Insulin stimulates phosphorylation of c-Jun, c-Fos, and Fos-related proteins in cultured adipocytes. J Biol Chem. 1994;269:11887–11892. [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Castro C, Garvey WT. Intracellular lipid accumulation in liver and muscle and the insulin resistance syndrome. Endocrinol Metab Clin North Am. 2008;37:841–856. doi: 10.1016/j.ecl.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo M, Clark JM. The epidemiology of nonalcoholic fatty liver disease: a global perspective. Semin Liver Dis. 2008;28:339–350. doi: 10.1055/s-0028-1091978. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Ko EH, Kim JE, Kim E, Lee H, Choi H, Yu JH, Kim HJ, Seong JK, Kim KS, et al. Nuclear receptor PPAR gamma-regulated monoacylglycerol O-acyltransferase 1 (MGAT1) expression is responsible for the lipid accumulation in diet-induced hepatic steatosis. Proc Natl Acad Sci U S A. 2012;109:13656–13661. doi: 10.1073/pnas.1203218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Tsukamoto H, Liu JC, Han YP, Govindarajan S, Lai MM, Akira S, Ou JH. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology. 2010;52:480–492. doi: 10.1002/hep.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24:184–187. doi: 10.1038/72855. [DOI] [PubMed] [Google Scholar]
- Matsusue K, Haluzik M, Lambert G, Yim SH, Gavrilova O, Ward JM, Brewer B, Jr., Reitman ML, Gonzalez FJ. Liver-specific disruption of PPAR gamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest. 2003;111:737–747. doi: 10.1172/JCI17223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ. Hepatic steatosis in leptin-deficient mice is promoted by the PPAR gamma target gene Fsp27. Cell Metab. 2008;7:302–311. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez G, Gray SL, Yetukuri L, Shimomura K, Virtue S, Campbell M, Curtis RK, Jimenez-Linan M, Blount M, Yeo GS, et al. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS Genet. 2007;3:e64. doi: 10.1371/journal.pgen.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nature protocols. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- Moran-Salvador E, Lopez-Parra M, Garcia-Alonso V, Titos E, Martinez-Clemente M, Gonzalez-Periz A, Lopez-Vicario C, Barak Y, Arroyo V, Claria J. Role for PPAR gamma in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 2011;25:2538–2550. doi: 10.1096/fj.10-173716. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology. 2006;44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V, Ranjit S, Konda S, Nicoloro SM, Straubhaar J, Chawla A, Chouinard M, Lin C, Burkart A, Corvera S, et al. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A. 2008;105:7833–7838. doi: 10.1073/pnas.0802063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M, Glass CK. PPARs and molecular mechanisms of transrepression. Biochim Biophys Acta. 2007;1771:926–935. doi: 10.1016/j.bbalip.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladin R, Fajas L, Dana S, Halvorsen YD, Auwerx J, Briggs M. Differential regulation of peroxisome proliferator activated receptor gamma1 (PPAR gamma1) and PPAR gamma2 messenger RNA expression in the early stages of adipogenesis. Cell Growth Differ. 1999;10:43–48. [PubMed] [Google Scholar]
- Sethi JK, Vidal-Puig AJ. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007;48:1253–1262. doi: 10.1194/jlr.R700005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Smedile A, Bugianesi E. Steatosis and hepatocellular carcinoma risk. Eur Rev Med Pharmacol Sci. 2005;9:291–293. [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepot D, Weitzman JB, Barra J, Segretain D, Stinnakre MG, Babinet C, Yaniv M. Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development. 2000;127:143–153. doi: 10.1242/dev.127.1.143. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPAR gamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Varela GM, Antwi DA, Dhir R, Yin X, Singhal NS, Graham MJ, Crooke RM, Ahima RS. Inhibition of ADRP prevents diet-induced insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2008;295:G621–628. doi: 10.1152/ajpgi.90204.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle. 2007;6:2633–2639. doi: 10.4161/cc.6.21.4850. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Schonthaler HB, Guinea-Viniegra J, Tschachler E. Psoriasis: what we have learned from mouse models. Nat Rev Rheumatol. 2010;6:704–714. doi: 10.1038/nrrheum.2010.157. [DOI] [PubMed] [Google Scholar]
- Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- Zhou L, Xu L, Ye J, Li D, Wang W, Li X, Wu L, Wang H, Guan F, Li P. Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology. 2012;56:95–107. doi: 10.1002/hep.25611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.