Abstract
Arterial spin labeling (ASL) is a non-invasive technique that can quantitatively measure cerebral blood flow (CBF). While traditionally ASL employs 2D EPI or spiral acquisition trajectories, single-shot 3D GRASE is gaining popularity in ASL due to inherent SNR advantage and spatial coverage. However, a major limitation of 3D GRASE is through-plane blurring caused by T2 decay. A novel technique combining 3D GRASE and a PROPELLER trajectory (3DGP) is presented to minimize through-plane blurring without sacrificing perfusion sensitivity or increasing total scan time. Full brain perfusion images were acquired at a 3×3×5mm3 nominal voxel size with Q2TIPS-FAIR as the ASL preparation sequence. Data from 5 healthy subjects was acquired on a GE 1.5T scanner in less than 4 minutes per subject. While showing good agreement in CBF quantification with 3D GRASE, 3DGP demonstrated reduced through-plane blurring, improved anatomical details, high repeatability and robustness against motion, making it suitable for routine clinical use.
Keywords: Arterial Spin Labeling, PROPELLER, 3D GRASE
INTRODUCTION
Arterial spin labeling (ASL) is an MRI technique that uses water molecules in the blood as an endogenous tracer to measure cerebral blood flow (CBF). It is highly effective in detecting brain lesions, cerebrovascular diseases, and brain tumors (1). Compared to traditional contrast-bolus based methods, ASL is completely non-invasive and repeatable, and offers direct measurement of quantitative perfusion values. The noninvasive nature of ASL is especially important for patients with conditions such as renal failure, or in pediatric patients where the use of external tracers may be restricted (2).
ASL has an inherently low perfusion signal-to-noise ratio (SNR) and signal averaging is necessary to obtain perfusion weighted images with sufficient SNR. As fast imaging techniques, 2D Echo Planar Imaging (EPI) and spiral imaging are commonly used for ASL acquisition to increase slice coverage. However, multi-slice 2D acquisitions result in multiple inflow times that may underestimate perfusion and cause quantification errors in certain regions. Also, both EPI and spiral imaging are highly sensitive to magnetic field inhomogeneity and susceptibility, leading to signal loss and geometric distortions. To minimize those artifacts, ASL images acquired with EPI and spiral are typically acquired with low spatial resolution. Nevertheless, higher spatial resolutions are often desirable to reduce partial volume effects and improve localization of abnormalities.
Alternatively, 3D acquisition techniques have been developed for ASL imaging. Techniques such as 3D EPI (3,4) and 3D GRASE (Gradient Echo And Spin Echo) (5) have been shown to have higher perfusion sensitivity and better spatial coverage of the brain than 2D EPI. As a gradient echo method, 3D EPI generally has better slice coverage than 3D GRASE. However, 3D GRASE has lower sensitivity to field inhomogeneity and magnetic susceptibility as a spin echo method. The robustness of 3D GRASE to magnetic field inhomogeneity is especially important for reducing image artifacts with non- Cartesian trajectories, such as PROPELLER (6). These qualities make 3D GRASE an attractive choice as an ASL imaging acquisition sequence.
One drawback has limited the widespread use of 3D GRASE. As a single-shot 3D technique, raw image data is collected after a single excitation with a long acquisition window, which results in severe through-plane blurring due to T2 decay. One apparent solution to reduce the blurring is to shorten the acquisition window by shortening the echo train length (ETL).
Two common methods for shortening the ETL are parallel imaging and multi-shot acquisitions. Parallel imaging can reduce the ETL but at the expense of SNR loss. Although the loss of SNR is partially compensated by the shorter echo time (TE) achievable with parallel imaging, at higher acceleration rates (> 2×), the SNR loss is significant (7). Multi-shot methods are another approach for reducing the ETL by splitting the full data acquisition among multiple excitations. The disadvantage of multi-shot method is the increase in scan time and susceptibility to image artifacts induced by motion. With a standard Cartesian trajectory that is commonly used in multi-shot methods, it is difficult to compensate for motion that occurs between shots.
One multi-shot technique that can reduce ETL and correct for motion error is PROPELLER (Periodically Rotated Overlapping ParallEL Lines with Enhanced Reconstruction) (6,8). Unlike the Cartesian trajectory, data acquisition in PROPELLER consists of a series of rectangular trajectories (known as blades) rotated about the center of k-space. Due to this unique rotational trajectory, one advantage of PROPELLER is the ability to perform self-referenced motion correction (6). Comparable motion correction is difficult in multi-shot methods with Cartesian trajectory without acquiring additional navigator echoes which further reduces acquisition efficiency. For long axis PROPELLER where the blade width (set by the ETL) is small compared to the blade length (the number of frequency encodes), the resolution of the final reconstructed image is determined solely by the blade length and the field of view (FOV). Hence, the original spatial resolution is fully preserved regardless of the width of the individual blade. A disadvantage of PROPELLER is an increase in scan time to fully sample k-space as a multi-shot technique. However, this disadvantage is inconsequential for ASL imaging where signal averaging is necessary to achieve adequate perfusion SNR.
We hypothesize that it is more advantageous to sample k-space in a non-Cartesian, multi-shot fashion with PROPELLER than simply signal averaging identical acquisitions repeatedly. In this paper, we incorporate the 3D GRASE readout sequence with PROPELLER (3DGP) to improve the spatial resolution of ASL imaging while minimizing T2 blurring and off-resonance effects. We compare the performances of 3D GRASE and 3DGP for measuring gray matter perfusion. The repeatability, CBF quantification and the perfusion SNR of 3DGP are examined in detail.
METHODS
Pulsed ASL (PASL) Implementation
ASL labeling was achieved using Q2TIPS-FAIR(9,10), a pulsed ASL implementation. Saturation bands were applied along both sides of the imaging slab to minimize aliasing artifacts in the slice encoding direction, as well as to suppress the intravascular signals carried by both arterial and venous inflows. An inversion time of 1500ms was chosen to allow blood to fully exchange with the tissue. Additional background suppression pulses were applied at 539 ms and 1345ms to null the stationary tissue signal to further improve perfusion SNR (11).
PROPELLER Acquisition
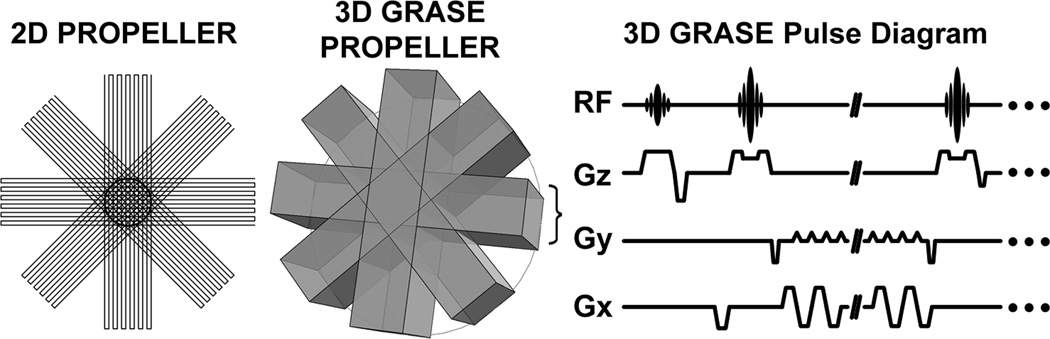
For 3DGP, a number of rectangular volumes (known as bricks (12)) are acquired at different rotation angles relative to the central kz-axis (Figure 1a). In our study, each brick was sampled with a single-shot 3D GRASE readout module (Figure 1b). 16 bricks evenly distributed from 0° to 360° were acquired per PROPELLER image by an incremental angle of 22.5°. The number of bricks acquired for each PROPELLER image satisfied both the Nyquist sampling criterion according to (6) and ensured adequate perfusion SNR. A total of four PROPELLER images were reconstructed: a control image and a label image, each acquired with a positive and a negative frequency encoding gradient. The toggling of the frequency encoding gradient was necessary to remove Nyquist ghosts using the self-referenced correction method GESTE (13). The total scan time was Tscan time = TR × NBlade × NASL × NGESTE where TR was the repetition time (3500ms), NBlade = 16 was the number of bricks per image, NASL = 2 was the number of perfusion states, and NGESTE = 2 was the number of encodes required for GESTE. The total scan time was 3 minutes 44 seconds. The acquisition pattern for PROPELLER bricks is in the following order: brick(control, GESTE+) at 0°, brick(label, GESTE+) at 0°, brick(control, GESTE−) at 0°, brick(label, GESTE−) at 0°. The order of acquisition then repeats for another 15 PROPELLER bricks evenly spaced from 22.5° to 360°.
Figure 1.
(a) The schematic illustration of k-space trajectory for 2D PROPELLER (left) and 3D GRASE PROPELLER (right). Rather than a series of rotating “blades” in the 2D PROPELLER trajectory, 3D GRASE PROPELLER consists of a series of rotating “bricks” about the central z-axis. Note the k-space is under-sampled for illustration purpose. Only 4 of 16 blades are shown. (b) 3D GRASE pulse diagram for acquiring each brick.
Image Post-Processing
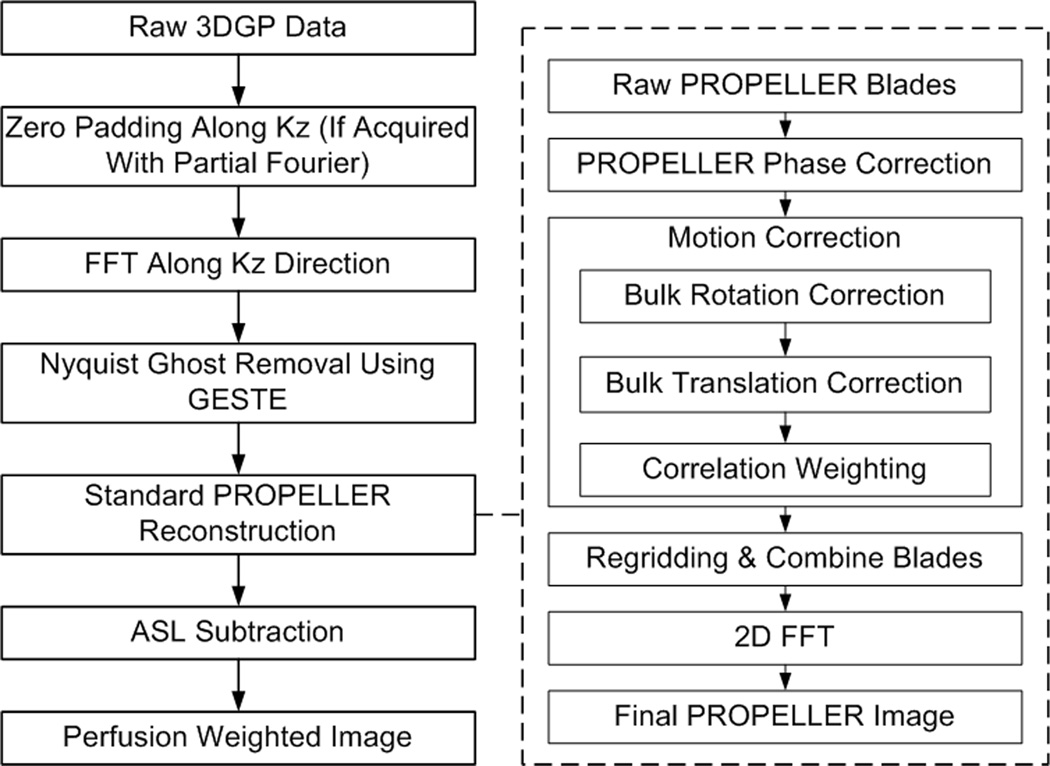
A 1D Fourier transform was first applied along kz for each raw brick, yielding the equivalent of 2D PROPELLER blades from multiple slices. GESTE (13) was applied to remove Nyquist ghosts in each blade, followed by the standard PROPELLER reconstruction (6). In the current work, motion correction (rotational and translational) was performed only along the in-plane direction. Through-plane motion was dealt with indirectly with correlation weighting (6). Note that the same motion correction reference image was used for both control and label blades during the in-plane motion correction. This prevented misalignments between the final control and label images to avoid potential subtraction errors. The corrected blades were gridded onto a Cartesian coordinate space with uniform density compensation (14). The final image for each slice was obtained by performing a 2D Fourier transform on the gridded and combined k-space data.
Each of the four PROPELLER images was reconstructed separately. The magnitude images corresponding to each GESTE encode were signal averaged to generate a single pair of control and label images. Perfusion weighted images (PWI) were then calculated by subtracting the label image from the control image. CBF quantification was done using the General Kinetic Model (10,15). The entire reconstruction process is illustrated in Figure 2.
Figure 2.
Diagram of 3DGP reconstruction process.
Experiments
Perfusion images were acquired using both 3DGP and 3D GRASE in the axial plane. In both techniques, the prescribed imaging slab was 90 mm thick consisting of 18 slices with 5 millimeter thick slices. Excitation was achieved with a 90 degree 7 lobe RF spatial spectral pulse. To further reduce blurring, partial Fourier encoding with 72% coverage was used to reduce the number of slice encodings to 13. GESTE encoding was used in both techniques for Nyquist ghost removal. The TE was minimized to optimize perfusion sensitivity by employing a center out trajectory along the slice encoding direction. The total imaging time for 3DGP and 3D GRASE was identical (3 minutes 44 seconds).
For 3DGP, 20 phase encodes and 96 frequency encodes were prescribed per brick. The in-plane FOV was 288 × 288 mm, thus achieving a voxel size of 3 × 3 × 5 mm3. A smaller FOV may be prescribed to achieve the same in-plane resolution; however, this has no effect on TE and echo spacing because a larger gradient amplitude with longer ramp times is required for the smaller FOV. A total of 16 bricks were acquired for each control/label image. Other imaging parameters included TE / TR = 17.5 / 3500 ms, total acquisition window duration = 131ms. In a separate scan, a 3DGP M0 weighted image was acquired without the ASL preparation for CBF quantification with 3 bricks and TR = 10s.
For 3D GRASE, the image matrix size was 96 × 64 with a field of view of 288 × 192mm, matching the voxel size of the 3DGP acquisition. Other imaging parameters included TE / TR = 41.2 / 3500 ms, total acquisition window duration = 379 ms. The number of control/label pairs for signal averaging was prescribed to match the 3DGP prescription exactly. The difference in the total acquisition window duration between 3D GRASE and 3DGP is solely due to the number of phase encoding lines. Reconstructed 3D GRASE images were realigned using SPM2 (Wellcome Trust Center for Neuroimage, London, UK) to remove any motion errors. A 3D GRASE M0 weighted image with TR = 10s was acquired for CBF quantification in a separate scan.
In addition to perfusion sequences, high resolution anatomical images were acquired using 3D SPGR for tissue segmentation. T1 maps were acquired using the DESPOT1 (16) sequence for CBF quantification without additional B1 correction. A total of 5 healthy volunteers of age from 24 to 30 were recruited under the supervision of the Institutional Review Board, and informed consent was obtained from each subject. All in-vivo experiments were carried out on a GE 1.5T TwinSpeed scanner (GE Healthcare, Milwaukee, WI) with an 8 channel phased array receive-only head coil (Invivo Devices, Gainesville, FL) for data collection. The pulse sequence and image reconstruction software were developed internally. Imaging reconstruction software was written in Matlab (Mathworks, Natick, MA) utilizing functions from the National Center for Image Guided Therapy (NCIGT) Fast Imaging Library (17).
Repeatability, SNR and SNR Per Pixel Calculation
To test repeatability and calculate SNR, both 3DGP and 3D GRASE acquisitions were repeated without repositioning the subject. The repeatability was tested by calculating the coefficient of repeatability, following the methods of Bland and Altman (18).
SNR was calculated from the repeated acquisition (19). The noise for the SNR calculation was estimated from the difference image of the two repeated PWIs. The mean signal and the standard deviation of the noise were obtained from the gray matter region of the PWIs and the difference image, respectively. Both gray matter and white matter were identified from tissue segmentation maps, obtained from the high resolution anatomical images processed by SPM2.
Image reconstruction algorithms can affect the final SNR of the reconstructed image. Since 3D GRASE and 3DGP use very different image reconstruction algorithms, the average raw SNR per pixel was calculated for both methods by integrating the raw MRI signal as described by Liang and Lauterbaur (Equation 8.48 of (20)). The standard deviation of the noise for this calculation was measured from a single acquisition with all of the RF pulses turned off.
Results
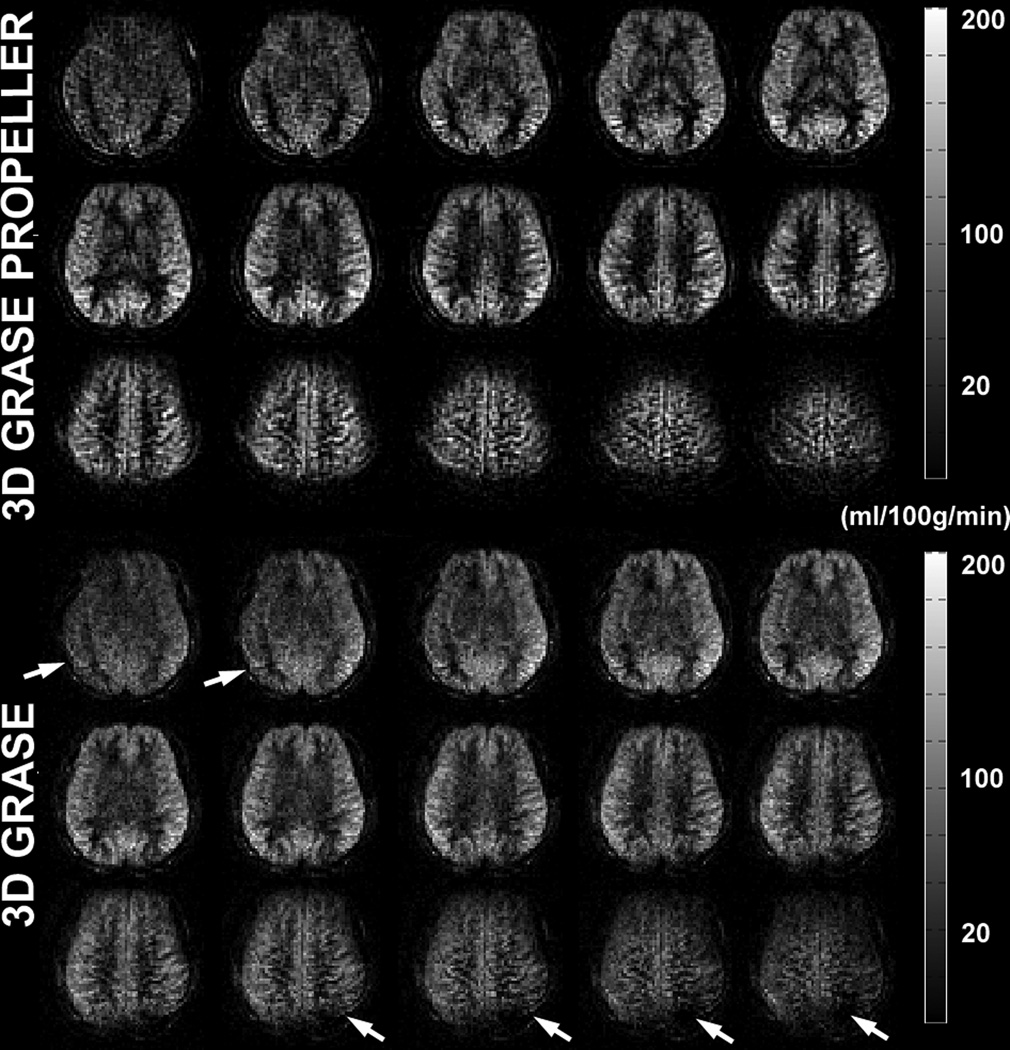
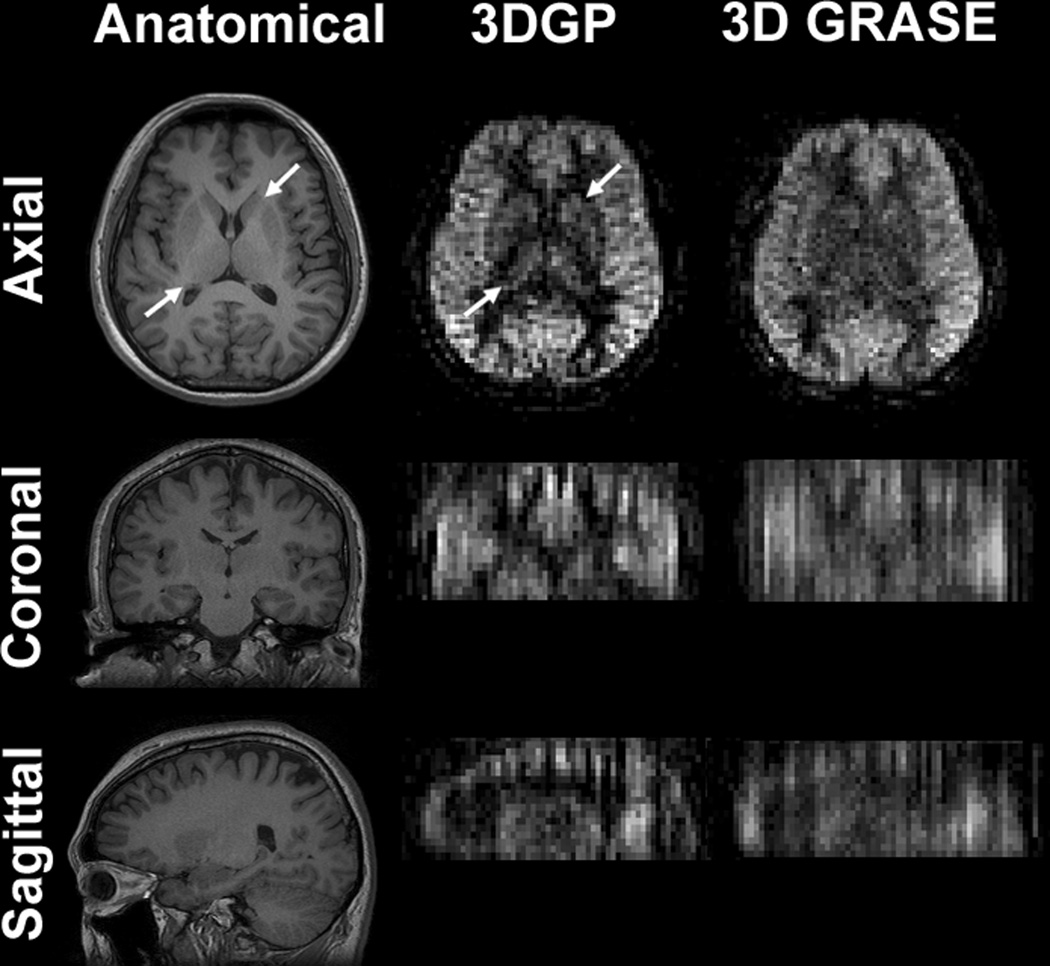
Figure 3 shows the absolute CBF maps of slices 2 through 16 obtained in one of the five subjects from 3DGP and 3D GRASE acquisitions. Both sets of images showed full brain coverage and SNR of good diagnostic quality. Through-plane blurring artifacts were reduced in 3DGP comparing to 3D GRASE, most apparent in the superior and inferior slices. The improvements on blurring can be better seen in the reformatted coronal and sagittal views, shown in Figure 4. Anatomical details such as the thalamus and the basal ganglia were better revealed in 3DGP (indicated by the arrows in row one of Figure 4) while the same structures were hard to identify in corresponding 3D GRASE slices.
Figure 3.
A comparison of CBF maps acquired with 3DGP and 3D GRASE from one of the five subjects. Fifteen slices of a total of 18 are shown at inflow times TI = 1500ms. Severe blurring can be observed in the most superior and inferior slices of 3D GRASE, indicated by the arrows.
Figure 4.
Reformatted views of ASL images acquired in the axial plane. Severe blurring can be seen in the 3D GRASE images along the slice encoding direction. The blurring effect was significantly reduced in the 3DGP images. In-plane image details, such as thalamus and basal ganglia indicated by the arrows, can be easily identified in the 3DGP images whereas the same region was less recognizable in the 3D GRASE images.
The mean measured CBF values for 3D GRASE were 50 ± 14 ml/100g/min for gray matter and 23 ± 7 ml/100g/min for white matter. The average calculated CBF values for 3DGP were 52 ± 16 ml/100g/min for gray matter and 19 ± 7 ml/100g/min for white matter. The difference in white matter perfusion between 3DGP and 3D GRASE was significant according to a paired t-test (p < 0.0003) while it was not for gray matter (p > 0.28). The increase in white matter perfusion in 3D GRASE might be the result of severe through-plane blurring where the perfusion values were falsely increased by the contributions from other slices. This was also reported in the mean gray matter and white matter perfusion ratio: 2.7 for 3DGP and 2 for 3D GRASE. The individual CBF values for all subjects are shown in Table 1.
Table 1.
Mean CBF measurements for all subjects.
| Subject | Type | Gray Matter CBF (ml/100g/min) |
White Matter CBF (ml/100g/min) |
Ratio |
|---|---|---|---|---|
| 1 | 3DGP | 69.56 ± 3.12 | 28.1 ± 0.94 | 2.5 |
| GRASE | 60.49 ± 1.07 | 29.22 ± 1.16 | 2.1 | |
| 2 | 3DGP | 56.01 ± 0.25 | 22.80 ± 0.63 | 2.5 |
| GRASE | 60.58 ± 2.8 | 28.27 ± 1.39 | 2.1 | |
| 3 | 3DGP | 64.91 ± 0.95 | 21.24 ± 0.44 | 3.0 |
| GRASE | 60.48 ± 1.96 | 27.2 ± 1.31 | 2.2 | |
| 4 | 3DGP | 38.81 ± 1.83 | 15.09 ± 1.72 | 2.6 |
| GRASE | 39.27 ± 0.4 | 19.55 ± 0.59 | 2.0 | |
| 5 | 3DGP | 30.34 ± 2.39 | 10.25 ± 1.47 | 3.0 |
| GRASE | 28.56 ± 1.83 | 12.99 ± 0.58 | 2.1 |
Both techniques showed good repeatability in perfusion measurements. The maximum difference in CBF between repeated scans was less than 5 ml/100g/min for whole gray matter and white matter measurements. The coefficients of repeatability of 3D GRASE and 3DGP were 5.6 and 6.2 ml/100g/min for gray matter, and 3.6 and 3.3 ml/100g/min for white matter, respectively. This means the difference between two perfusion measurements for the same subject is expected to be less than the calculated coefficients of repeatability in 95% of pairs of observations. The SNR results across all subjects were 7.2 ± 1.82 for 3D GRASE and 4.9 ± 1.37 for 3DGP. The raw SNR per pixel values for all subjects were 9.4 ± 0.63 for 3D GRASE and 28.8 ± 1.96 for 3DGP. A detailed SNR analysis is included in the discussion section.
Discussion
This paper described the use of a non-Cartesian trajectory PROPELLER to minimize through-plane blurring in a 3D GRASE readout sequence for ASL imaging. PROPELLER was chosen due to a shorter ETL and its robustness against motion. This new technique, 3DGP, was demonstrated to show improved overall image quality in terms of reduced through-plane blurring, less off-resonance artifacts and improved image resolution.
Compared to a single-shot 3D GRASE readout, each 3DGP brick is less susceptible to off-resonance effects as a result of the reduced ETL. PROPELLER has demonstrated strong insensitivity to local off-resonance related distortion (21), and the final reconstructed 3DGP image exhibited few image distortions. In comparison, off-resonance induced distortion in 3D GRASE was more apparent and remained coherent across the time series. As a result of averaging, the final perfusion image was presented with the same distortion; whereas such artifact was absent in the 3DGP images.
Since the in-plane resolution of 3DGP depends primarily on the blade length, the image resolution can be improved by increasing the resolution along the frequency encode direction. This imposes only a minor increase in TE if the Nyquist sampling criterion is satisfied. However, if the Nyquist sampling criterion is not satisfied, the total number of PROPELLER bricks and/or the ETL need to be increased accordingly. Increasing the number of PROPELLER bricks will increase scan time, while increasing the ETL will increase TE and geometrical distortion. Therefore, it is desirable to acquire 3DGP images with the minimum ETL possible while maximizing the number of the bricks acquired for a given scan time. Given that a short ETL is desired, a minimum ETL of 16 is needed for adequate motion correction (22). However, thicker blades ensure the full capture of the k-space center to prevent signal loss during rotation and provide better motion correction(8). In our experience, an ETL of 20 is a good compromise between signal capture and motion correction. In perfusion imaging, signal averaging is necessary to achieve perfusion images with adequate SNR. One often needs to acquire more bricks than the Nyquist sampling criterion to achieve adequate SNR, thus providing the opportunity to adjust the ETL without increasing scan time. Nonetheless, this makes 3DGP a good candidate for high resolution ASL imaging.
GESTE is chosen for Nyquist ghost correction due to its robustness against system instability and higher efficiency than reference scan based techniques (13). In the presence of background suppression where tissue signal was reduced by 90%, the magnitude image is still dominated by residual tissue signal, and the performance of GESTE is not affected. GESTE encoding is executed separately for the control and label image in the current implementation. However, it may be possible to merge GESTE and ASL encoding by coupling the readout gradient polarity change with the blood magnetization labeling state, which would eliminate the need for acquiring two GESTE images for each control/label image. This is currently under investigation.
The SNR measurements of 3D GRASE for the perfusion weighted images were consistently higher than 3DGP according to in-vivo experiments. This was unexpected for three reasons. First, 3DGP has a shorter TE than 3D GRASE, which minimizes signal loss due to T2 relaxation. Second, the time between refocusing RF pulses is shorter for 3DGP (about 1/3 of the time of 3D GRASE), which minimizes signal loss due to magnetic field inhomogeneities and reduces T2 blurring. Third, the long acquisition duration required by single-shot 3D GRASE allows more noise to be introduced into the final image than signal due to T2 decay (23). While longer acquisition duration of 3D GRASE does result in more through-plane blurring, the SNR advantage of 3D GRASE cannot be attributed to this. For a concentric-out trajectory along kz, the T2 relaxation attenuates signal acquired at higher spatial frequencies but does not affect the noise. This is different from applying an exponential filter to the k-space data during post-processing, where both high frequency data and noise in k-space are attenuated leading to an overall increase in SNR.
We suspected that the SNR loss of 3DGP compared to 3D GRASE was occurring during image reconstruction. To further investigate this hypothesis, the average image SNR per pixel was calculated directly from the raw k-space data before image reconstruction (20). This revealed that the SNR of 3DGP images was 3 times greater than 3D GRASE (average SNR per pixel for 3DGP: 28.8, 3D GRASE: 9.4) despite the fact that 3D GRASE acquires 3 times as much data. The two SNR measurements (before and after reconstruction) revealed the signal loss of 3DGP was occurring during image reconstruction. We attributed this loss of SNR to geometrical and intensity distortions from magnetic field inhomogeneities during PROPELLER acquisition. As the 3DGP brick rotates, the geometric distortion changes direction for each brick. Combining these individual bricks without correcting for the distortion would result in incoherent signal averaging leading to signal loss. Although the geometrical distortion in 3D GRASE is larger than 3DGP due to the longer ETL, it has minimal effect on SNR since the distortion pattern is consistent throughout the entire experiment and is averaged coherently. We believe the SNR of 3DGP can be recovered by correcting for those residual off-resonance errors as demonstrated in (24), which we are currently investigating.
Conclusions
In this paper, we have shown that through-plane blurring present in 3D GRASE can be significantly reduced by incorporating a PROPELLER trajectory for ASL imaging. This technique, 3D GRASE PROPELLER or 3DGP, has demonstrated robustness against motion, off-resonance effects, and improved in-plane and through-plane image resolution, while inheriting the usual merits of 3D GRASE. By incorporating additional off-resonance correction, 3DGP has a great potential for higher resolution ASL imaging at higher field strength.
Acknowledgment
The authors thank the Center of Biomedical Imaging (CBI) at Wake Forest University School of Medicine (Winston-Salem, NC) for unfettered access to the MRI scanner and support from the Functional Neuroimaging Lab at Brigham and Women's Hospital (Boston, MA). The authors also thank Dr. Brian Hargreaves for publicly posting his gridding code and Dr. Jing Yuan at the BWH/National Center for Image Guided Therapy for providing the code for the spatial spectral pulses. This work was supported in part by NIH U41 RR-019703 and 5R01AA016748-02. This work was also in part funded by the German Ministry of Research and Education (BMBF) by grant 01EV0702.
References
- 1.Calamante F, Thomas DL, Pell GS, Wiersma J, Turner R. Measuring cerebral blood flow using magnetic resonance imaging techniques. J Cereb Blood Flow Metab. 1999;19(7):701–735. doi: 10.1097/00004647-199907000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Pollock JM, Tan H, Kraft RA, Whitlow CT, Burdette JH, Maldjian JA. Arterial spin-labeled MR perfusion imaging: clinical applications. Magn Reson Imaging Clin N Am. 2009;17(2):315–338. doi: 10.1016/j.mric.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gai ND, Talagala SL, Golay X, Hoogenraad FG, Butman JA. Evaluation of 3D-EPI PULSAR with and without Background Suppression Inversion Recovery Pulse. Berlin, Germany: 2007. p. 3486. [Google Scholar]
- 4.Talagala SL, Ye FQ, Ledden PJ, Chesnick S. Whole-brain 3D perfusion MRI at 3.0 T using CASL with a separate labeling coil. Magn Reson Med. 2004;52(1):131–140. doi: 10.1002/mrm.20124. [DOI] [PubMed] [Google Scholar]
- 5.Gunther M, Oshio K, Feinberg DA. Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med. 2005;54(2):491–498. doi: 10.1002/mrm.20580. [DOI] [PubMed] [Google Scholar]
- 6.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med. 1999;42(5):963–969. doi: 10.1002/(sici)1522-2594(199911)42:5<963::aid-mrm17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Wang J, Connick TJ, Wetmore GS, Detre JA. Continuous ASL (CASL) perfusion MRI with an array coil and parallel imaging at 3T. Magn Reson Med. 2005;54(3):732–737. doi: 10.1002/mrm.20574. [DOI] [PubMed] [Google Scholar]
- 8.Pipe JG, Zwart N. Turboprop: improved PROPELLER imaging. Magn Reson Med. 2006;55(2):380–385. doi: 10.1002/mrm.20768. [DOI] [PubMed] [Google Scholar]
- 9.Kim SG. Quantification of relative cerebral blood flow change by flow-sensitive alternating inversion recovery (FAIR) technique: application to functional mapping. Magn Reson Med. 1995;34(3):293–301. doi: 10.1002/mrm.1910340303. [DOI] [PubMed] [Google Scholar]
- 10.Luh WM, Wong EC, Bandettini PA, Hyde JS. QUIPSS II with thin-slice TI1 periodic saturation: a method for improving accuracy of quantitative perfusion imaging using pulsed arterial spin labeling. Magn Reson Med. 1999;41(6):1246–1254. doi: 10.1002/(sici)1522-2594(199906)41:6<1246::aid-mrm22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magn Reson Med. 2000;44(1):92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Holdsworth SJ, Skare S, Nordell A, Newbould RD, Bammer R. 3D SAP-EPI for Self-Navigated T1w Spoiled Gradient Echo Imaging. Toronto, Canada: 2008. [Google Scholar]
- 13.Hoge WS, Tan H, Kraft RA. Robust Elimination of EPI Nyquist Ghosts via Spatial and Temporal Encoding (EPI-GESTE) Magn Reson Med. doi: 10.1002/mrm.22564. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson J, Meyer CH, Nishimura DG, Macovski A. Selection fo a convolution function for Fourier inversion using gridding. IEEE Transactions on Medical Imaging. 1991;10(3):473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- 15.Buxton RB, Frank LR, Wong EC, Siewert B, Warach S, Edelman RR. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magn Reson Med. 1998;40(3):383–396. doi: 10.1002/mrm.1910400308. [DOI] [PubMed] [Google Scholar]
- 16.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003;49(3):515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- 17.Hoge WS. NCIG Fast Imaging Library. Boston, MA: 2010. [Google Scholar]
- 18.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 19.Sijbers J, den Dekker AJ, Van Audekerke J, Verhoye M, Van Dyck D. Estimation of the noise in magnitude MR images. Magn Reson Imaging. 1998;16(1):87–90. doi: 10.1016/s0730-725x(97)00199-9. [DOI] [PubMed] [Google Scholar]
- 20.Liang Z-P, Lauterbur PC. Principles of magnetic resonance imaging: a signal processing perspective. New York: Wiley-IEEE Press; 2000. [Google Scholar]
- 21.Wang FN, Huang TY, Lin FH, Chuang TC, Chen NK, Chung HW, Chen CY, Kwong KK. PROPELLER EPI: an MRI technique suitable for diffusion tensor imaging at high field strength with reduced geometric distortions. Magn Reson Med. 2005;54(5):1232–1240. doi: 10.1002/mrm.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamhane AA, Arfanakis K. Motion correction in periodically-rotated overlapping parallel lines with enhanced reconstruction (PROPELLER) and turboprop MRI. Magn Reson Med. 2009;62(1):174–182. doi: 10.1002/mrm.22004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mugler J, Brookeman J. The optimum data sampling period for maximum signal-to-noise ratio in MR imaging. Rev Magn Reson Med. 1988;3(1):1–51. [Google Scholar]
- 24.Skare S, Andersson J. Calibration free distortion correction for PROPELLER EPI. Proceedings 16th ISMRM Scientific Meeting. 2008:417. [Google Scholar]