Abstract
Background
Enzymes of the cellulose synthase (CesA) family and CesA-like (Csl) families are responsible for the synthesis of celluloses and hemicelluloses, and thus are of great interest to bioenergy research. We studied the occurrences and phylogenies of CesA/Csl families in diverse plants and algae by comprehensive data mining of 82 genomes and transcriptomes.
Results
We found that 1) charophytic green algae (CGA) have orthologous genes in CesA, CslC and CslD families; 2) liverwort genes are found in the CesA, CslA, CslC and CslD families; 3) The fern Pteridium aquilinum not only has orthologs in these conserved families but also in the CslB, CslH and CslE families; 4) basal angiosperms, e.g. Aristolochia fimbriata, have orthologs in these families too; 5) gymnosperms have genes forming clusters ancestral to CslB/H and to CslE/J/G respectively; 6) CslG is found in switchgrass and basal angiosperms; 7) CslJ is widely present in dicots and monocots; 8) CesA subfamilies have already diversified in ferns.
Conclusions
We speculate that: (i) ferns and horsetails might both have CslH enzymes, responsible for the synthesis of mixed-linkage glucans and (ii) CslD and similar genes might be responsible for the synthesis of mannans in CGA. Our findings led to a more detailed model of cell wall evolution and suggested that gene loss played an important role in the evolution of Csl families. We also demonstrated the usefulness of transcriptome data in the study of plant cell wall evolution and diversity.
Keywords: Cell wall, CesA, CslH, CslD, Transcriptome, Ferns, Liverworts, CGA, Gymnosperms
Background
Celluloses and hemicelluloses are the most abundant biopolymers in nature. In plants, they are the principal components of cell walls and the most promising renewable resources for producing biofuels [1,2]. The biosynthesis of celluloses and hemicelluloses is therefore one of the major research foci in plant biology. The past two decades have seen much progress in deciphering the molecular mechanisms of plant cell wall polysaccharide synthesis and regulation [3-8]. The identification of the cellulose synthase (CesA) gene family [9,10] and the CesA-like (Csl) gene families (collectively known as the CesA superfamily) [11] is one of the greatest achievements.
Early phylogenetic studies of CesA homologs in model plant organisms [11,12] established that there are eight Csl families: CslA, CslB, CslC, CslD, CslE, CslF, CslG and CslH, all belonging to the glycosyltransferase family 2 (GT2). Recent research in other flowering plants has added one more family (CslJ) [13]. It was proposed the Csl families might be involved in the synthesis of the backbones of hemicelluloses [11,14]. This “CSL hypothesis” has been strengthened by the functional characterization of CslA (mannan synthases) [15,16], CslC (xyloglucan synthases) [17], CslF (mixed-linkage glucan synthases) [18], and CslH (mixed-linkage glucan synthases) [19] genes. Although the functions of the other Csl families remain unknown, they are potentially involved in the synthesis of other cell wall polysaccharides or the same set of polysaccharides, e.g. through working together with other Csl or CesA genes [3].
The evolution of Csl families is also of interest to plant biologists studying the compositional diversity of cell walls [20-24]. Among the nine Csl families, CslA and CslC are distantly related to the other families; CslF and CslH are thought to be unique to monocots; CslB and CslG are confined to eudicots [25,26], and the rest of the families are found in both dicots and monocots [27]. Genomes of the lower land plants bryophyte moss Physcomitrella patens and lycophyte spike moss (Selaginella moellendorffii) only have representatives of the CesA, CslA, CslC and CslD families [27,28]. Six completed chlorophyte green algal genomes each have a single-copy CslA/C-like gene (herein named CslK), which represents the ancient CslA/C ortholog before a duplication happened in early land plants [27]. An evolutionary model was also proposed to explain the divergence order of Csl families, which has proved useful for our understanding of the cell wall diversity and evolution [22,24].
Over 40 plant genomes have been sequenced so far, including the first gymnosperm genome Picea abies (Norway spruce) [29]. However there is a lack of completed genomes for some key clades in the plant species tree: ferns, hornworts, liverworts, Streptophyta green algae (also known as advanced charophycean green algae, CGA), etc. Fortunately, the accumulation of transcriptome data in the GenBank database and the advent of the next generation sequencing have made a large amount of raw sequence data available for most of these key plants. For example, nine CGAs have significant amounts of transcriptome data recently available [30-32]. These data include ESTs (expressed sequence tags) sequenced by traditional Sanger technology, RNA-Seq data by the next generation 454 technology, as well as the pre-assembled UniGenes (mRNA contigs) in the Transcriptome Shotgun Assembly (TSA) sequence database; all of these data are available at the NCBI (National Center for Biotechnology Information) website.
Therefore, our goal in this study was to mine the transcriptomes and unfinished genomes of key plant species for Csl homologous genes in order to gain a better understanding of the evolution of the CesA/Csl superfamily. Specifically, we aimed to answer the question: when did each of the Csl families first appear in plants according to available sequence data? Answering this question will greatly improve our model of the evolution of Csl gene families and benefit the study of plant cell wall evolution and diversity.
Results
Csl genes in fully sequenced genomes: new findings
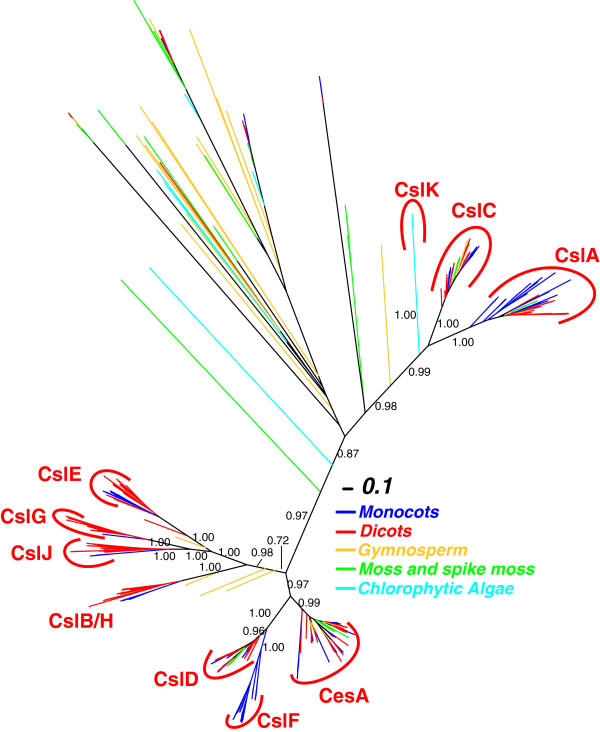
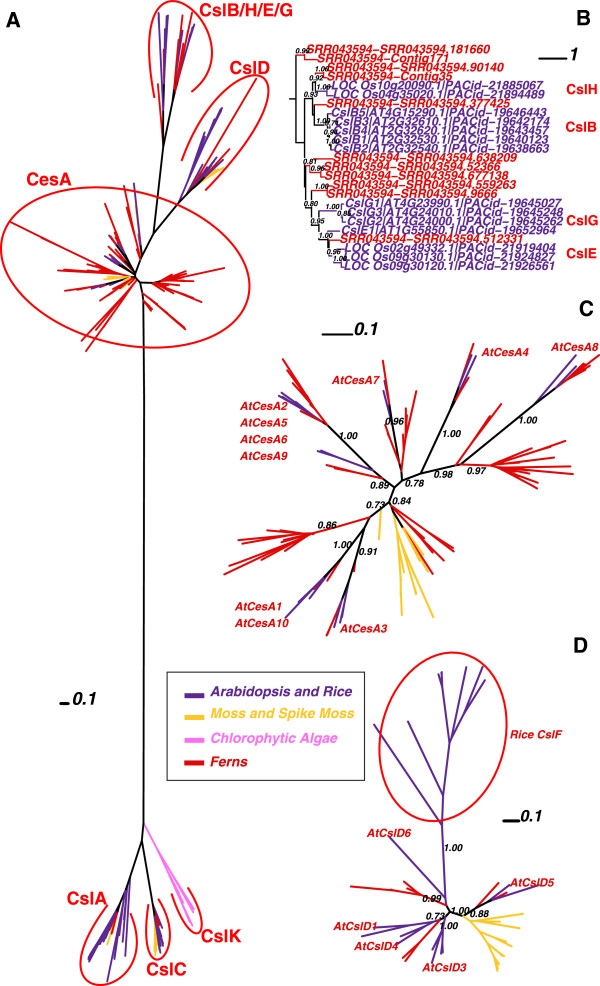
To retrieve Csl homologs, we scanned predicted protein sequences from the fully sequenced genomes of 32 land plants (23 dicots, six monocots, one gymnosperm, one moss and one spike moss), 10 CGA and two other algae (one Glaucophyta and one Rhodophyta), using two Pfam models (Cellulose_synt and Glycos_transf_2) as queries (see Additional file 1 for the list of species). The two Pfam models were used in our previous paper [27] and were able to retrieve all of the 39 documented Arabidopsis Csl genes. Figure 1 presents an unrooted phylogeny (protein IDs are provided in Additional file 2 and Additional file 3).
Figure 1.
Phylogeny of 893 GT2 proteins from 17 land plants and two green algae. The full-length protein sequences were used to build the phylogeny. The FastTree bootstrap values (1.00 = 100%) larger than 0.70 are shown beside selected nodes forming the major Csl clusters. Csl clusters are labeled according to the presence of known Csl proteins in each cluster.
Compared with our previous work [27], one of the new findings is that CslG appears to have two member genes (Pavirv00027268m and Pavirv00027269m) from P. virgatum (see also Additional file 4), ~49% identical to AtCslG2 (AT4G24000.1). The grouping of these two switchgrass genes in CslG family is strongly supported (bootstrap value = 100%), suggesting that CslG can no longer be considered a dicot-specific family.
By including the newly sequenced P. abies genome in Figure 1 (orange color), we showed that gymnosperm proteins are found in CesA, CslA, CslC, CslD families. P. abies also has proteins clustered in the large CslB/H/E/J/G clade, but these proteins’ phylogenetic groupings are not well resolved. In later sections of this paper, more gymnosperm species with transcriptomes are included to better resolve the phylogenetic clustering of the gymnosperm Csl homologs.
The other finding concerns CslJ, which is close to CslG but is very well self-clustered (bootstrap value = 100%). CslJ was thought to be unique to cereals [25,26], but here it is shown to be widely present in four (sorghum, maize, foxtail millet and switchgrass) out of six fully sequenced monocot genomes and 16 out of 23 sequenced dicot genomes (Additional file 4).
The phylogeny of CslB and CslH shown in Figure 1 (also Additional file 4) suggests that these two families are so tightly clustered that they are hardly distinguishable. Therefore it might be more appropriate to consider them as a single family.
Figure 1 also includes other GT2 proteins. Between CslA/C/K and CesA/CslD/F/B/H/E/G/J, there are some loosely clustered groups that have very long branches. The long branches suggest that proteins in these clusters are quite different from each other; clusters with long branches are usually not very stable, a sign of small sample size or rapid sequence divergence.
To explore these clusters, we built a new phylogeny with proteins using fewer flowering plants and more algae (Additional file 5). Also included in the phylogeny are published CesA/Csl protein sequences from several non-plant species including brown algae, Oomycetes, fungi and bacteria [33].
In the new phylogeny (Additional file 5) most algal homologs form a large cluster (denoted as C) including two Arabidopsis (AT2G39630.1: dolichyl phosphase β-glucosyltransferase and AT1G20575.1: dolichol phosphate mannose synthase 1), two rice, and quite a few moss and spike moss GT2 proteins. A visual examination of the multiple sequence alignments in cluster C and those of the Csl families suggests that most of these non-Csl proteins do not have the characteristic ‘D,D,D,QXXRW’ motif typically found in Csl/CesA proteins. There are also many algal homologs clustered with the non-plant CesA/Csl proteins (aqua color), suggesting that they are likely to be CesA/Csl genes of distinct origin compared to the canonical land plant CesA/Csl genes. Further study including more non-plant Csl homologs will be needed in order to gain a better understanding of their origin and evolution.
Mining for Csl homologs in short read transcriptomes/genomes
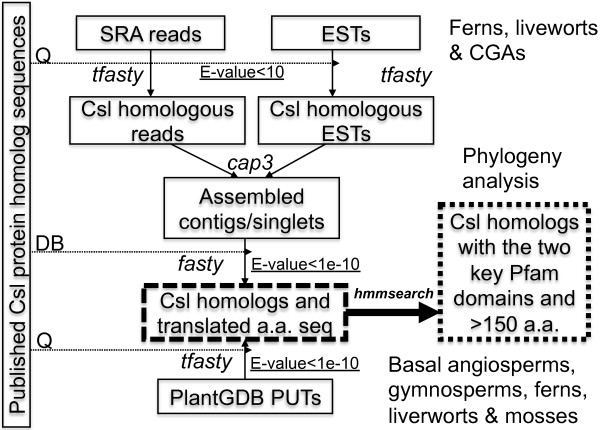
We developed a bioinformatics pipeline (Figure 2) that combined homology search and short read assembly to identify Csl homologs in: (i) transcriptome reads of nine CGAs and two ferns, (ii) genomic DNA reads of one liverwort, and (iii) pre-assembled uni-transcripts from GenBank ESTs, which consisted of PlantGDB-assembled unique transcripts (PUTs) from 26 plants, including six basal angiosperms, 16 gymnosperms (11 conifers, two cycads, one ginkgo, two gnetophyte), two ferns, one moss and one liverwort. Table 1 provides information about data for (i) and (ii) and Table 2 lists data for (iii).
Figure 2.
Computational pipeline for data mining of short read transcriptome/genome data. Details about the mined plants and algae are provided in Table 1 and 2. SRA: sequence read archive of the NCBI; ESTs: expressed sequence tags; fasty and tfasty are two homology search commands of the FASTA package [52] (see Methods); hmmsearch is a command of the HMMER3 package [47]. The two Pfam domains include Pfam models Cellulose_synt and Glycos_transf_2. PUTs are PlantGDB-assembled unique transcripts; Q means to use as the query set in the homology search; DB means to use as the database; published Csl protein homologs are from [27].
Table 1.
Short read sequence data sets of ferns, liverwort and CGAs
| Plant clades | Species | NCBI accessions | # of reads | References |
|---|---|---|---|---|
| Fern |
Pteridium aquilinum |
SRX020701 |
730,579 |
[34] |
| Fern |
Ceratopteris richardii |
SRX154690 |
1,083,570 |
- |
| Liverwort |
Marchantia polymorpha |
SRX114614- SRX114615 |
300,372,599a |
- |
| SRX030759- SRX030787 |
22,854,396 |
- |
||
|
CGA: Charophyceae |
Chara vulgaris |
SRX041525 |
740,355 |
[32] |
|
Nitella hyalina |
SRX025843 |
949,065 |
[30,31] |
|
|
CGA: Coleochaetophyceae |
Coleochaete orbicularis |
SRX017046 |
354,659 |
|
|
Coleochaete_sp. CFD |
TSA contigs |
18,386 |
||
|
CGA: Zygnemophyceae |
Penium margaritaceum |
SRX025845 |
1,077,311 |
|
|
Spirogyra pratensis |
SRX017045 |
614,139 |
||
|
CGA: Klebsormidiophyceae |
Klebsormidium flaccidum |
SRX025847 |
994,649 |
|
|
CGA: Chlorokybophyceae |
Chlorokybus atmophyticus |
SRX025846 |
444,743 |
|
| Chaetosphaeridium globosum | SRX025844 | 884,238 |
aIllumina reads, not used in this study.
Table 2.
PlantGDB-assembled unique transcripts (PUTs)
| Plant clades | Species | # of GenBank ESTs | # of PUTs |
|---|---|---|---|
| Gymnosperm |
Cryptomeria japonica |
57,720 |
24,299 |
| Gymnosperm |
Picea abies |
14,619 |
8,715 |
| Gymnosperm |
Picea engelmannii x Picea glauca |
28,190 |
13,880 |
| Gymnosperm |
Picea glauca |
321,713 |
48,619 |
| Gymnosperm |
Picea sitchensis |
206,402 |
31,215 |
| Gymnosperm |
Pinus banksiana |
36,387 |
13,040 |
| Gymnosperm |
Pinus contorta |
40,489 |
13,570 |
| Gymnosperm |
Pinus pinaster |
35,139 |
15,648 |
| Gymnosperm |
Pinus sylvestris |
76,256 |
73,609 |
| Gymnosperm |
Pinus taeda |
329,066 |
72,829 |
| Gymnosperm |
Pseudotsuga menziesii var. menziesii |
14,354 |
9,857 |
| Gymnosperm |
Cycas rumphii |
22,000 |
10,901 |
| Gymnosperm |
Zamia vazquezii |
11,495 |
7,657 |
| Gymnosperm |
Ginkgo biloba |
21,709 |
10,210 |
| Gymnosperm |
Gnetum gnemon |
10,756 |
6,193 |
| Gymnosperm |
Welwitschia mirabilis |
10,137 |
6,606 |
| Basal angiosperm |
Amborella trichopoda |
26,403 |
15,772 |
| Basal angiosperm |
Aristolochia fimbriata |
16,454 |
7,967 |
| Basal angiosperm |
Liriodendron tulipifera |
24,146 |
14,232 |
| Basal angiosperm |
Nuphar advena |
20,601 |
13,789 |
| Basal angiosperm |
Persea americana |
16,620 |
10,928 |
| Basal angiosperm |
Saruma henryi |
10,281 |
6,754 |
| Liverwort |
Marchantia polymorpha |
33,764 |
10,959 |
| Moss |
Syntrichia ruralis |
10,010 |
7,087 |
| Fern |
Adiantum capillus-veneris |
30,561 |
16,944 |
| Fern | Ceratopteris richardii | 5,186 | 4,234 |
In Figure 2, the dashed rectangle contains all the Csl-homologous peptides in the surveyed transcriptome/genome. The peptide sequences were translated from assembled nucleotide contigs and singletons according to the fasty alignment with their best Csl hits, which are published Csl proteins previously classified into the 10 existing Csl families [27]. Note that for PlantGDB’s PUTs, the assembly step was not needed and tfasty was used to derive the translated peptide sequences.
Given that transcriptome sequencing and subsequent assembly are unlikely to recover the full-length transcripts, it was not surprising that many Csl homologs in the dashed rectangle of Figure 2 were short fragments. To clean the data, we applied the following filters to keep significant and long Csl homologs (Figure 2, dotted rectangle): (i) they had to be highly similar to known Csl proteins (E-value < 1e-10); (ii) they had to match the two characteristic Pfam domains (Cellulose_synt and Glycos_transf_2, E-value < 1e-2); and (iii) they had to be longer than 150 amino acids. These filters tend to be very stringent, so that a Csl homolog that passed all of the three filters would very likely be a true Csl gene. The filters were also helpful in reducing the impact of contamination or low quality reads on our downstream phylogenetic analyses. In particular, we found that the second filter was very critical for removing false positives. However, we were very flexible about the length filter because we did not want to miss real orthologs. When necessary, we manually inspected peptides shorter than 150 a.a. to select and include appropriate ones in the phylogenetic analyses.
For phylogenetic analysis, we combined Csl-homologous peptides of a specific plant clade (e.g. CGAs) with the known Csl proteins and then generated new phylogenies. The new phylogenies were then examined to determine if the new homologs clustered with the existing Csl families or if they formed distinct new clusters.
CGAs have representative genes from CesA, CslC and CslD families
Figure 3 presents a phylogeny with CGA homologs longer than 200 a.a. and Additional file 6 provides the sequences. It is clear that CGA homologs are found in the CslC, CslD and CesA clusters. It is surprising however that none are found in the CslA cluster, as CslA enzymes are responsible for the synthesis of mannans, which have been found in the cell walls of CGAs [35]. We investigated whether any true CslAs were removed in the stringent filtering steps by manually inspecting the fasty search results, and found no false negatives.
Figure 3.
Phylogeny with CGA homologs. Sequences of this phylogeny include 52 CGA peptides longer than 200 a.a. Other proteins are 131 pre-classified Csl proteins selected from Additional file 5.
Penium_margaritaceum-Contig85 is the only CGA peptide that has a known CslA protein (Os02g09930.1) as the best hit (identity = 24%). However, it is not clustered within the CslA clade but with another CGA peptide (Spirogyra_pratensis-Contig255) with identity = 63%. This Spirogyra_pratensis peptide has Os03g56060.1 of CslC as the best hit (identity = 27%). These two CGA peptides are further placed basal to CslA, CslC and CslK clusters (Figure 3).
Lowering the length filter to 100 a.a. did not find any shorter peptides that clustered within the CslA clade (Additional file 7). However, three more peptides (Chara_vulgaris-Contig143, Nitella_hyalina-SRR064326.525840 and Nitella_hyalina-SRR064326.70219) clustered with the CslC clade, indicating that four out of the nine surveyed CGA species have CslC proteins.
CslD and CesA families both have CGA homologs. CslD homologs were found in Coleochaete species, while CesA homologs were found in Spirogyra pratensis and Penium margaritaceum (Figure 3). We also tried to include peptides shorter than 200 a.a. and found CesA homologs in Klebsormidium flaccidum and CslD homologs in Chaetosphaeridium globosum (Additional file 7). Deeper RNA sequencing will be needed to resolve the question of whether CslD and CesA genes are present in the other CGA species.
The presence of a major CGA-specific cluster close to the CslD clade, containing peptides from Klebsormidium flaccidum, Nitella hyaline and Chara vulgaris (Figure 3), is highly interesting. If peptides shorter than 200 a.a. were included, more CGA sequences would be clustered within this CslD-like clade (Additional file 7). It is possible, but highly speculative, that this CGA-specific clade encodes the missing CGA mannan synthases. More data are needed to determine if this clade is actually part of the CslD family or represent a new Csl family.
Liverworts have representative genes from CesA, CslA, CslC and CslD families
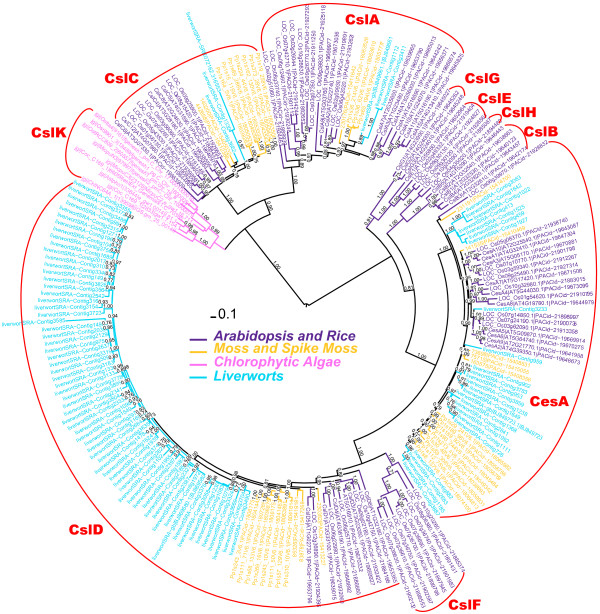
The model liverwort species Marchantia polymorpha has 33,692 ESTs and 31 genomic DNA datasets in the SRA database of NCBI. The EST data has been assembled into PUTs in the PlantGDB. Among the 31 SRA datasets, 29 are from 454 sequencing, which yields longer reads than Illumina, so we used these 29 datasets, a total of 13GB. We identified liverwort Csl homologs by combining all sequences together and using the protocol shown in Figure 2.
Figure 4 shows a phylogeny with liverwort homologs longer than 200 a.a. and a small number of selected homologs between 100 and 200 a.a. Additional file 8 provides the sequences. Similar to the fully sequenced moss and spike moss genomes, liverwort appears to have genes in CslA, CslC, CslD and CesA clusters but not in the CslB/H/E/G clusters. It is also clear that liverwort Csl homologs are often clustered with moss and spike moss sequences and ancestral to their corresponding orthologs in seed plants, suggesting that the sequence diversification of these genes happened after the split of liverworts and seed plants. Compared to other Csl families, CslD seems to have many more liverwort homologs. However, many of these homologs are very similar to each other (identity > 95% at the nucleotide level and also demonstrated by the very short branch lengths), which might be due to under-assembly. Hence the actual number of CslD homologs in liverwort is probably much lower. Since our goal is not to quantitatively but qualitatively assess the occurrence of Csl families in different plants, such under-assembly does not affect any of our conclusions. There are three expressed liverwort homologs found in the PlantGDB’s PUTs (see below), each in the CslC, CslD and CesA clusters respectively.
Figure 4.
Phylogeny with liverwort homologs. Sequences of this phylogeny include 96 liverwort peptides, most of which longer than 200 a.a. Three shorter proteins were added and clustered within the CslA cluster.
Ferns have representative genes from CesA, CslA, CslC, CslD, CslE, CslB and CslH families
Two fern species, Ceratopteris richardii and Pteridium aquilinum, have transcriptome data sequenced by 454 in the NCBI SRA database (Table 1). C. richardii and Adiantum capillus-veneris also have ESTs, which are assembled into PUTs in PlantGDB (Table 2). Following the procedure shown in Figure 2, we identified Csl homologs in the three fern species.
The phylogeny shown in Figure 5A includes fern peptides longer than 150 a.a. Additional file 9 provides the sequences. Ferns have representative genes in CslA, CslC, CslD, CesA, and even in the CslB/H/E/G clusters. Of the three fern species, C. richardii and A. capillus-veneris have homologs only in the CesA and CslD clusters while P. aquilinum has genes in all the other Csl clusters. In fact, C. richardii has many fewer Csl homologs than P. aquilinum (15 vs. 281; length > 100 a.a.), although the former has many more reads in the surveyed datasets (Table 1). This suggests that the transcriptome data of C. richardii might be very biased and does not capture the transcripts of many Csl genes, as it is unlikely that its genome does not encode CslA and CslC genes.
Figure 5.
Phylogenies with fern homologs. Sequences of these phylogenies include 182 fern peptides longer than 150 a.a. (A) The phylogeny contains all sequences; (B) The phylogeny shows only the CslB/H/E/G cluster; (C) The phylogeny shows only the CesA cluster; (D) The phylogeny shows only the CslD cluster. Sequences in (B), (C) and (D) were extracted from (A) and then re-aligned; the phylogeny was then recomputed based on the new alignments.
Most interestingly, P. aquilinum homologs are evidently found in CslE, CslB and CslH clades (Figure 5B). The fern CslB ortholog SRR043594-SRR043594.377425 has AtCslB2 (AT2G32540.1) as its best hit (sequence identity = 48% at the a.a. level) among all known Csl proteins; the CslH ortholog SRR043594-Contig35 matches OsCslH1 (Os10g20090.1) as the best hit (sequence identity = 36%); and the CslE ortholog SRR043594-SRR043594.512331 has Os09g30130.1 of the CslE family as the best hit (sequence identity = 60%). There are also fern homologs phylogenetically basal to both CslB and CslH. This suggests that the CslB/H/E clades had already diverged before ferns appeared, possibly through ancient duplications from older Csl families (i.e. CesA or CslD). Given that the completed spike moss genome does not have CslB/H/E genes, the emergence of these Csl families must have happened after spike moss split from more advanced vascular land plants but certainly before ferns.
The CslD family seems to have already diversified before ferns split from seed plants, as AtCslD1/4 and AtCslD5 have clear orthologs in ferns (Figure 5D). Similarly, different CesA sub-clusters containing the Arabidopsis genes CesA3, CesA4, CesA7, CesA8, CesA1/10 and CesA2/5/6/9, respectively, all have orthologs in ferns (Figure 5C), suggesting that their divergence occurred as early as in the last common ancestor of ferns and later evolved land plants. It is most interesting to observe that, for the three major components of the cellulose synthase complex of secondary cell walls in Arabidopsis: CesA4, CesA7 and CesA8, their common ancestral genes had already diversified in ferns, in contrast to the earliest vascular plant spike moss, whose CesAs are all clustered into one monophyletic group (yellow color).
There are also additional fern-specific CesA clusters, e.g. the large red clusters in the CesA circle of Figure 5A and also the red sub-clusters beside AtCesA8 and beside AtCesA1/10 of Figure 5C. All the members of these clusters have known CesA proteins as the best hit, but future experimental studies are needed to verify whether they truly have cellulose synthase activity or not.
Gymnosperms have Csl genes basal to CslB/H and to CslE/G respectively
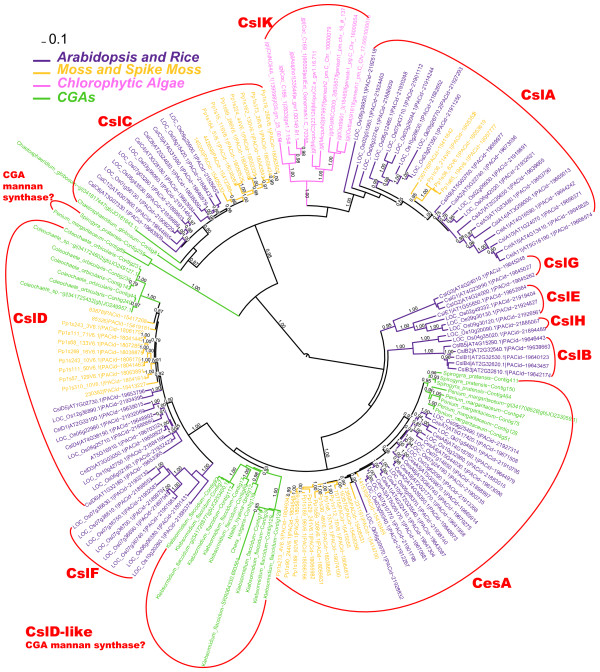
Although there is one gymnosperm genome P. abies available, Figure 1 suggests that more gymnosperm sequences are needed to resolve the uncertain clustering of P. abies homologs in CslB/H/E/J/G clusters. Therefore, we selected from PlantGDB six basal angiosperms, 16 gymnosperms, two ferns, one moss and one liverwort (Table 2). The basal angiosperms include plant species that are neither eudicotyledons nor monocotyledons, such as magnoliids, which are ancestral to both dicot and monocot plants. Following the procedure in the bottom part of Figure 2, we identified Csl homologs in these plants.
Figure 6 shows a phylogeny with all of the Csl homologs longer than 200 a.a. from the 26 surveyed plant transcriptomes and Additional file 10 provides the sequences. We mainly looked at gymnosperms and basal angiosperms. In agreement with what we found in Figure 1, no gymnosperm homolog (lighter red) is found inside the individual cluster of CslB/H/E/G. More precisely, the CslE family clustered with a large gymnosperm cluster with a bootstrap value lower than 70%. So it appears that each individual family does not have clear orthologs in gymnosperms. Instead, there are gymnosperm-specific clades basal to CslE/G families and CslB/H families, respectively. Basal angiosperm homologs however were found in all of the individual families including CslG. Interestingly, all of the six basal angiosperms have genes in the CslB/H/E/G clusters. Particularly, Aristolochia fimbriata has six genes found in all of the four Csl families.
Figure 6.
Phylogeny with PUT homologs. Sequences of this phylogeny include 167 PUT peptides longer than 200 a.a.
Discussion
To our best knowledge, this is the first comprehensive and large-scale data mining of Csl homologs in the transcriptomes of various plants and algae. Prior to our study, Sorensen et al. searched the EST data of C. nitellarum (CGA species) and found CslD orthologs [35]. Richmond built a web resource (http://cellwall.stanford.edu) in 2000 to collect Csl genes in plant genomes and ESTs [11], but that web resource is no longer available. Publications of transcriptome/genome data of diverse plants and algae (Tables 1 and 2) in the past few years have made our comprehensive search possible.
Here we categorized CesA/Csl genes residing in 44 fully sequenced plant and algal genomes (Additional file 11), as well as 38 transcriptomes of CGAs, ferns, gymnosperms, and basal gymnosperms, and raw genomic DNA reads from liverworts (Additional file 6, Additional file 8, Additional file 9, Additional file 10). We studied their distribution in 10 different Csl protein families using phylogenetic analyses, which not only offer cell wall polysaccharide and bioenergy researchers with a list of Csl genes in bioenergy-related crops, but also provide new insights into the evolution and function of the CesA/Csl families in different plants.
About mannans in CGAs
As shown in Figures 3 and 7, CslA appears to be absent in CGAs. There are many possible reasons for why these genes are missing in CGAs, but we believe the following are the most likely: (i) CGA mannan synthase genes were not captured by the transcriptome data that we mined due to low expression, or (ii) CGA mannan synthases are not encoded by the canonical CslA gene family. In other words, convergent evolution may have given gene families other than CslA the ability to synthesize mannans in CGAs, e.g. the CGA-specific clusters found in Figure 3. Notably, these clusters are close to the CslD family in the phylogeny. The literature contains discussions of the possibility of CslD proteins are glucomannan synthases [3,36,37]. It is therefore tempting to speculate that the CslD and/or CslD-like genes (Figure 3 and Additional file 7) are responsible for the synthesis of CGA mannans, given that (i) CGAs have genes only in the CesA, CslA, CslD and CslD-like clusters, (ii) the cell walls of CGAs contain both xyloglucans and mannans, (iii) the CslA family encodes xyloglucan synthases and (iv) the function of the CslD family is still unknown.
Figure 7.
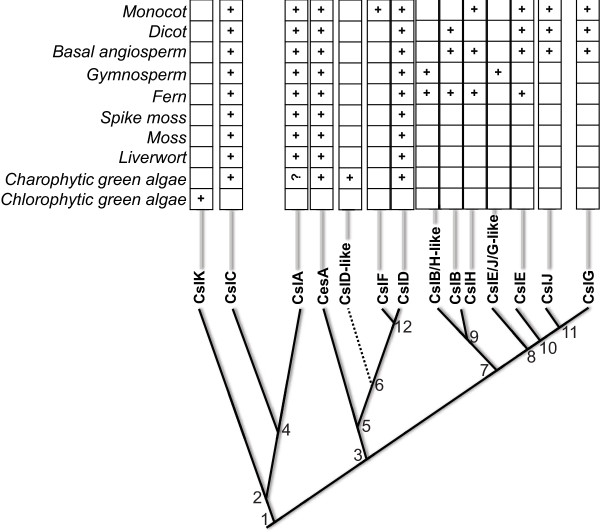
An evolutionary model of Csl gene families. The top panel shows the occurrence of Csl families. “+” means the family is found; “?” means it is uncertain. In the bottom panel, the numbers 1 to 12 are used to label each divergence node, which are detailed in the main text. The dotted line means it is uncertain if the CslD-like family truly exists.
The first possibility could be validated by deeper RNA sequencing of CGAs that have experimental evidence of mannans, e.g. Coleochaete nitellarum and Spirogyra spI[35]. However, we noticed that two closely related species, Coleochaete orbicularis and Spirogyra pratensis, were included in our data mining and only one relevant sequence (Spirogyra_pratensis-Contig255) was found in either species; this sequence is most similar to a known CslC protein. For this reason, the possibility that mannan synthesis genes are missing from the data for artifactual reasons seems unlikely.
About MLG in horsetail (Equisetum arvense)
In NCBI’s taxonomy database, the two fern species, Ceratopteris richardii and Pteridium aquilinum, both belong to Polypodiopsida under Moniliformopses (ferns). Interestingly, horsetails (Equisetopsida) that are also of Moniliformopses have been shown to have mixed-linkage glucans (MLGs) in their cell walls [38-41]. Since MLGs are only narrowly found in the plant kingdom, in Poaceaes, horsetails and some algae, it has been proposed that horsetails and algae might have independently acquired their abilities to synthesize MLGs by using enzymes of the Csl families. Here we found that CslH has orthologs in the fern species Pteridium aquilinum. It is likely that these CslH genes also encode MLGs in Pteridium aquilinum, although experimental evidence is needed to prove this. On the other hand, this suggests that horsetails probably also have CslH orthologs that are responsible for the synthesis of MLGs in their cell walls. Therefore, our finding supports the hypothesis that CslH genes were in the common ancestor of ferns and seed plants but later lost in gymnosperms. Such gene loss event might be fairly prevalent, as Ceratopteris richardii, which has more reads than Pteridium aquilinum (Table 1), appears to have no CslH genes.
Given that both CslF and CslH encode MLG synthases and CslF is strictly confined to monocots, we conclude that CslH is the more ancient MLG synthase family [41].
It was suggested that leafy liverwort Lophocolea bidentata might have MLG-like polysaccharides [42], but our search in the liverwort Marchantia polymorpha genome did not find any CslH orthologs. Spike moss and CGAs were also suggested to have MLG [35], but the MLG is unlikely to be synthesized by CslH as no CslH orthologs were found in the completed spike moss genome or the surveyed CGA transcriptomes. In this case, the convergent evolution hypothesis is still a plausible explanation for the synthesis of MLGs in these organisms that do not have CslH and CslF families.
Evolution of Csl families
Many of our previous views about the Csl families were changed in light of our new findings, which led to a revised evolutionary model with more details (Figure 7). Twelve nodes were labeled to represent the speculated evolutionary events that might have led to the divergence of CesA/Csl gene families.
About CslA/C/K
Node 1 represents the endosymbiosis event(s) that gave rise to the earliest plant cell. Two distinct ancestral genes were passed to the earliest plant cell, which shared an even earlier GT2 ancestor in ancient prokaryotes. One gene was the ancestor of extant CslA/C/K families and the other was the ancestor of the rest of the Csl families. A larger scale analysis including non-plant GT2 proteins will be useful to disentangle the different origins of the two groups of families.
After node 2, the ancestral gene became the current CslK family in chlorophytes, while in CGA, it evolved into the CslA and CslC families through duplication (node 4). This duplication event should have occurred after the split of CGAs and chlorophytes. We did not find CslA genes in CGAs, suggesting that CslA might have been lost in evolution.
About CesA/CslD/F
The other ancestral gene that the earliest plants inherited was very likely to be a CesA gene, which might be from some ancestral cyanobacteria [43,44]. Node 3 represents an early gene duplication that occurred, probably in ancient algal species, where one gene later evolved to be the latest common ancestor of CesA/CslD/F families, while the other evolved to be the latest common ancestor of CslB/H/E/J/G.
Node 5 implies that the ancestor of CesA/CslD duplicated and diverged into the CesA and CslD clades. As both families are present in CGAs, their divergence must have happened before CGAs appeared. Afterwards, the evolution of the CslD genes seem to be enigmatic in that some CGAs (Coleochaete orbicularis and Chaetosphaeridium globosum) have CslD homologs while others have CslD-like homologs (Figure 3 and Additional file 7). It is therefore uncertain if there was an additional divergence (node 6) that gave rise to the CslD-like homologs in CGAs. Completed CGA genomes will be needed to reach a conclusive answer.
It is worthy of mentioning that different CesA subfamilies, including the AtCesA4, AtCesA7 and AtCesA8 subfamilies, diversified after spike moss but before ferns appeared (Figure 5C). This suggests that the secondary cell wall cellulose synthase protein complex has been in existence since ferns.
CslF genes are only present in monocots, and they have long branches in the phylogenies (Figure 1), suggesting a rapid divergence after splitting from CslD family through duplication (node 12). Therefore, among all Csl families, CslF was the last one to arise.
About CslB/H/E/J/G
The divergence of CslB/H and CslE/J/G (node 7) should be much later than node 5, because CslB/H/E appeared since ferns. The common ancestor of CslB/H/E/J/G might be very ancient, but the diversification of this ancestral gene into each of the individual families seems to have occurred much later.
The most exciting findings of this paper are about these families. Prior to our study, it was believed that: i) CslB and CslG are dicot-specific; ii) CslH and CslJ are found only in cereals; iii) CslE is only found in angiosperms. As shown in Figure 8, these views are subject to modifications: 1) CslB, CslH and CslE have orthologs in ferns and basal angiosperms; 2) CslG is found in switchgrass and basal angiosperms (also see Figure 1 and Additional file 4); Additional file 3) CslJ is found in most dicots (Figure 1 and Additional file 4) and in basal angiosperms too.
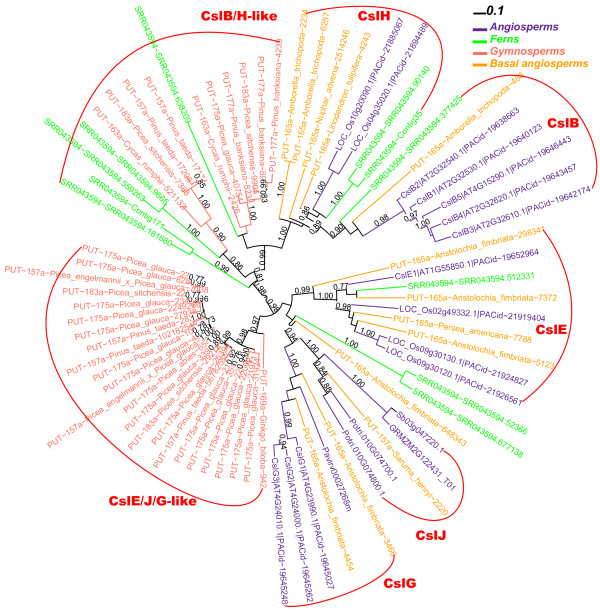
Figure 8.
Phylogeny with CslB/H/E/J/G homologs from selected organisms. Sb03g047220.1, GRMZM2G122431_T01, Potri.010G074700.1, Potri.010G074800.1 of CslJ family and Pavirv00027268m of CslG family are selected from Figure 1. The rest proteins are selected from Figures 3 to 6.
No gymnosperm genes are found in any of the individual families of CslB/H/E/J/G. However the presence of CslB/H-like cluster (Figure 8) suggests that CslB and CslH might have evolved in ferns and then lost in gymnosperms. Similarly the gymnosperm-specific CslE/J/G-like gene cluster contains expressed genes from almost all surveyed gymnosperms including the ginkgo species, suggesting that it is functionally very important and conserved (short branches in Figure 8).
It remains a mystery why and how gymnosperms lost CslB, CslH and CslE genes but retained the apparently more ancestral CslB/H-like and CslE/J/G-like families. However, all of the CslB/H/E/J/G families are no longer narrowly distributed and they appear to be much older than previously thought. It has been suggested that gymnosperms have lower substitution rate in their genomes [45], which should be considered for the future study of the evolution of CslB/H/E/J/G families.
About the divergence order of these families, node 8 and 9 must precede the occurrence of ferns. CslE then diverged from CslJ/G (node 10), probably also before ferns. CslG might have evolved in early angiosperms through gene duplication from the CslJ family (node 11).
Our study suggests that gene duplication and gene loss (e.g. loss of CslB/H/E in gymnosperms) occurred very often throughout plant genome evolution, and together they have played a significant role in shaping the expansion and diversification of the Csl families.
Conclusions
In summary, the following major contributions were made in this paper: 1) we demonstrated that the toolkits for the study of the plant cell wall evolution and diversity could be complemented by bioinformatics data mining of the transcriptomes of plant clades that do not have completed genomes; 2) we found that fern transcriptomes have expressed genes of the CslB/H/E families so these families are much older than we thought; 3) we predicted that CslH genes might also exist and encode MLG synthases in horsetails; 4) we speculated that the mannan synthases in CGAs might be encoded by Csl families other than CslA as it is missing in all surveyed CGA transcriptomes; and 5) we proposed a more complete model for the evolution of Csl families and suggested that gene loss following duplication played a significant role in the evolution of Csl gene families.
Methods
Sequence data
Previously categorized Csl protein sequences were downloaded from the supplemental data of [27].
The fully sequenced plant and algal genomes were downloaded from Phytozome and JGI [46], except for Picea abies, downloaded from http://congenie.org. The HMMER3 package [47] was used to search the two Pfam domains (Cellulose_synt and Glycos_transf_2) against the above genomes, following our previous papers [27,48,49]. The fern, liverwort and CGA short read data sets were downloaded from the NCBI SRA database. The pre-assembled PUT data sets were downloaded from PlantGDB [50].
The owners of the unpublished fern (Ceratopteris richardii) transcriptome data and the unpublished liverwort (Marchantia polymorpha) genome data agreed with the use of these data in this study and were acknowledged in the Acknowledgement. The liverwort genome sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community. All other SRA sequence data that have been published were properly cited in Table 1.
Data mining pipeline
The pipeline was depicted in Figure 2. For the assembly of 454 transcriptomes/genomes of ferns, liverwort and algae, we used cap3[51] with overlap length > 60 bp and overlap percent identity > 97% (-o 60 and -p 97). Because short read assembly is well known to be computationally intense when the data size is large, we did a pre-screening homology search prior to the assembly and only assembled the reads of a same species that are homologous to known Csl proteins.
The FASTA package [52] was used for all homology searches. Specifically, fasty and tfasty commands were used, which have the advantage that they can tolerate sequence errors and tend to yield longer alignments by including stop codons and frame shifts, as compared to the common BLAST searches.
After fasty/tfasty search, the peptide sequences were translated from assembled nucleotide contig/singleton sequences according to the alignment with their best Csl hits. Symbols of frame shifts (“/” and “\”) and stop codons (“*”) in the fasty/tfasty alignments were removed before multiple sequence alignment (MSA).
Phylogenetic analysis
MSAs were generated using MAFFT v6.935b with the L-INS-i method [53], which is among the most accurate sequence alignment algorithms. Phylogenies were made using the FastTree program version 2.1.3 [54]. FastTree implements an ultrafast and fairly accurate approximate maximum likelihood method. The accuracy of FastTree is considered to be slightly better than PhyML version 3.0, with minimum-evolution nearest neighbor interchanges moves, and is 100 to 1,000 times faster and requires much less computer memory.
FastTree analyses were conducted with default parameters; specifically, the amino acid substitution matrix was JTT, the number of rate categories of sites (CAT model) was 20, and the local support values of each node were computed by resampling the site likelihoods 1000 times and performing the Shimodaira Hasegawa test. Based on our previous work [27,48,49], FastTree performs sufficiently well for protein family evolution studies.
We also tried the much slower but more accurate PhyML program to build all of the phylogenies and the tree topology does not differ much and does not change any of our findings. The iTOL server was used to generate the phylograms [55].
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YY conceived this study, carried out all the analysis and wrote the paper. MAJ, HC, and MR helped in the data analysis and the paper writing. All authors read and approved the final manuscript.
Supplementary Material
44 fully sequenced plants and algae.
Circular view of Figure 1.
Sequences included in Figure 1.
Zoomed-in view of CslB/H/E/J/G clusters in Figure 1.
Phylogeny with plant and algal GT2 homologs forming three large clusters (the inset shows the radial view).
Csl homologs found in CGAs and sequences.
Phylogeny with CGA homologs longer than 100 a.a.
Csl homologs found in Marchantia polymorpha and sequences.
Csl homologs found in ferns and sequences.
Csl homologs found in PUTs of surveyed plants and sequences.
Csl homologs of 44 plant and algal genomes.
Contributor Information
Yanbin Yin, Email: yyin@niu.edu.
Mitrick A Johns, Email: rjohns@niu.edu.
Huansheng Cao, Email: hcao@niu.edu.
Manju Rupani, Email: z1694921@students.niu.edu.
Acknowledgements
This work was supported the start-up funds and the Research & Artistry Award from Northern Illinois University to Y.Y. We acknowledge all of the individual groups worldwide who have made their genomic and transcriptomic data available to the public. In particular, we acknowledge Dr. John L. Bowman for allowing us to use the Marchantia polymorpha genomic DNA data and Dr. Stanley J. Roux for the Ceratopteris richardii RNA-Seq data, which are all available at the SRA database before publication.
References
- Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ Jr, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T. The path forward for biofuels and biomaterials. Science. 2006;15(5760):484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;15(4):559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;15:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye ML, Bernard S, Kousar S, Chevalier L, Vicre-Gibouin M, Lerouxel O. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front Plant Sci. 2012;15:79. doi: 10.3389/fpls.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvskov P. In: Annual Plant Reviews: Plant Polysaccharides, Biosynthesis and Bioengineering. Ulvskov P, editor. Oxford, UK: Wiley-Blackwell; 2011. [Google Scholar]
- Carpita NC. Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr Opin Biotechnol. 2012;15(3):330–337. doi: 10.1016/j.copbio.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15(11):625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Dixon RA. On-off switches for secondary cell wall biosynthesis. Mol Plant. 2012;15(2):297–303. doi: 10.1093/mp/ssr098. [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci U S A. 1996;15(22):12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. CELLULOSE BIOSYNTHESIS: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;15:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;15(2):495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD. Cellulose synthase-like genes of rice. Plant Physiol. 2002;15(2):336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB. Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J. 2006;15(2):145–167. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides - a complex process. Curr Opin Plant Biol. 2006;15(6):621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, Anderson P. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;15(5656):363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci U S A. 2005;15(6):2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci U S A. 2007;15(20):8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;15(5769):1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A. 2009;15(14):5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA. Evolution and diversity of green plant cell walls. Curr Opin Plant Biol. 2008;15(3):286–292. doi: 10.1016/j.pbi.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Fangel JU, Ulvskov P, Knox JP, Mikkelsen MD, Harholt J, Popper ZA, Willats WG. Cell wall evolution and diversity. Front Plant Sci. 2012;15:152. doi: 10.3389/fpls.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper Z, Michel G, Herve C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB. Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol. 2011;15:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- Sorensen I, Domozych D, Willats WG. How have plant cell walls evolved? Plant Physiol. 2010;15(2):366–372. doi: 10.1104/pp.110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Tuohy MG. Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol. 2010;15(2):373–383. doi: 10.1104/pp.110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino F, Bacic A. Plant cell walls: the skeleton of the plant world. Funct Plant Biol. 2010;15(5):357–381. doi: 10.1071/FP09279. [DOI] [Google Scholar]
- Fincher GB. Revolutionary times in Our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 2009;15(1):27–37. doi: 10.1104/pp.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Huang J, Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009;15(1):99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT. The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol. 2007;15(2):207–219. doi: 10.1007/s11103-006-9083-1. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hällman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Käller M, Luthman J, Lysholm F, Niittylä T, Olson A, Rilakovic N, Ritland C, Rosselló JA, Sena J. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;15(7451):579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010;15:96. doi: 10.1186/1471-2229-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;15(1):e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodniok S, Brinkmann H, Glockner G, Heidel AJ, Philippe H, Melkonian M, Becker B. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 2011;15:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010;15(1):82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- Der JP, Barker MS, Wickett NJ, De Pamphilis CW, Wolf PG. De novo characterization of the gametophyte transcriptome in bracken fern. Pteridium aquilinum. BMC Genomics. 2011;15:99. doi: 10.1186/1471-2164-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O’Neill MA, Fei Z, Rose JK, Domozych DS, Willats WG. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011;15(2):201–211. doi: 10.1111/j.1365-313X.2011.04686.x. [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y, Yin L, Oikawa A, Scheller HV. Mannan synthase activity in the CSLD family. Plant signaling & behavior. 2011;15(10):1620–1623. doi: 10.4161/psb.6.10.17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WG, Scheller HV. The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant. 2011;15(6):1024–1037. doi: 10.1093/mp/ssr026. [DOI] [PubMed] [Google Scholar]
- Sorensen I, Pettolino FA, Wilson SM, Doblin MS, Johansen B, Bacic A, Willats WGT. Mixed-linkage (1 - > 3), (1 - > 4)-beta-D-glucan is not unique to the poales and is an abundant component of Equisetum arvense cell walls. Plant J. 2008;15(3):510–521. doi: 10.1111/j.1365-313X.2008.03453.x. [DOI] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR. Mixed-linkage (1 - > 3,1 - > 4)-beta-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 2008;15(1):104–115. doi: 10.1111/j.1469-8137.2008.02435.x. [DOI] [PubMed] [Google Scholar]
- Burton RA, Fincher GB. (1,3;1,4)-Beta-D-glucans in cell walls of the poaceae, lower plants, and fungi: a tale of Two linkages. Mol Plant. 2009;15(5):873–882. doi: 10.1093/mp/ssp063. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Exploring the evolution of (1,3;1,4)-beta-D-glucans in plant cell walls: comparative genomics can help! Curr Opin Plant Biol. 2009;15(2):140–147. doi: 10.1016/j.pbi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Primary cell wall composition of bryophytes and charophytes. Ann Bot. 2003;15(1):1–12. doi: 10.1093/aob/mcg013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles DR, Brown RM. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose. 2004;15(3–4):437–448. [Google Scholar]
- Nobles DR, Romanovicz DK, Brown RM. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;15(2):529–542. doi: 10.1104/pp.010557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo E, Ritland C, Bohlmann J, Ritland K. Slow but not low: genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol Biol. 2012;15:8. doi: 10.1186/1471-2148-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;15(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;15(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Chen H, Hahn MG, Mohnen D, Xu Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant physiol. 2010;15(4):1729–1746. doi: 10.1104/pp.110.154229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Huang J, Gu X, Bar-Peled M, Xu Y. Evolution of plant nucleotide-sugar interconversion enzymes. PLoS One. 2011;15(11):e27995. doi: 10.1371/journal.pone.0027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong QF, Lawrence CJ, Schlueter SD, Wilkerson MD, Kurtz S, Lushbough C, Brendel V. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;15(2):610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Madan A. Cap3: a DNA sequence assembly program. Genome Res. 1999;15(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;15(1):24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;15(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;15(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;15(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
44 fully sequenced plants and algae.
Circular view of Figure 1.
Sequences included in Figure 1.
Zoomed-in view of CslB/H/E/J/G clusters in Figure 1.
Phylogeny with plant and algal GT2 homologs forming three large clusters (the inset shows the radial view).
Csl homologs found in CGAs and sequences.
Phylogeny with CGA homologs longer than 100 a.a.
Csl homologs found in Marchantia polymorpha and sequences.
Csl homologs found in ferns and sequences.
Csl homologs found in PUTs of surveyed plants and sequences.
Csl homologs of 44 plant and algal genomes.