Abstract
Background
Chorella is the representative taxon of Chlorellales in Trebouxiophyceae, and its chloroplast (cp) genomic information has been thought to depend only on studies concerning Chlorella vulgaris and GenBank information of C. variablis. Mitochondrial (mt) genomic information regarding Chlorella is currently unavailable. To elucidate the evolution of organelle genomes and genetic information of Chlorella, we have sequenced and characterized the cp and mt genomes of Arctic Chlorella sp. ArM0029B.
Results
The 119,989-bp cp genome lacking inverted repeats and 65,049-bp mt genome were sequenced. The ArM0029B cp genome contains 114 conserved genes, including 32 tRNA genes, 3 rRNA genes, and 79 genes encoding proteins. Chlorella cp genomes are highly rearranged except for a Chlorella-specific six-gene cluster, and the ArM0029B plastid resembles that of Chlorella variabilis except for a 15-kb gene cluster inversion. In the mt genome, 62 conserved genes, including 27 tRNA genes, 3 rRNA genes, and 32 genes encoding proteins were determined. The mt genome of ArM0029B is similar to that of the non-photosynthetic species Prototheca and Heicosporidium. The ArM0029B mt genome contains a group I intron, with an ORF containing two LAGLIDADG motifs, in cox1. The intronic ORF is shared by C. vulgaris and Prototheca. The phylogeny of the plastid genome reveals that ArM0029B showed a close relationship of Chlorella to Parachlorella and Oocystis within Chlorellales. The distribution of the cox1 intron at 721 support membership in the order Chlorellales. Mitochondrial phylogenomic analyses, however, indicated that ArM0029B shows a greater affinity to MX-AZ01 and Coccomyxa than to the Helicosporidium-Prototheca clade, although the detailed phylogenetic relationships among the three taxa remain to be resolved.
Conclusions
The plastid genome of ArM0029B is similar to that of C. variabilis. The mt sequence of ArM0029B is the first genome to be reported for Chlorella. Chloroplast genome phylogeny supports monophyly of the seven investigated members of Chlorellales. The presence of the cox1 intron at 721 in all four investigated Chlorellales taxa indicates that the cox1 intron had been introduced in early Chorellales as a cis-splice form and that the cis-splicing intron was inherited to recent Chlorellales and was recently trans-spliced in Helicosporidium.
Background
Chloroplasts and mitochondria, organelles of higher plants and algae, play important roles in energy production, photosynthesis, and metabolite production required for maintaining life. Although numerous biological functions of both organelles rely considerably on proteins imported from nuclear encoded genes, understanding the organelle genome will provide a major impact in the fields of evolution, biology, and biotechnology.
Currently, many genome projects are in progress for green microalgae. To date, more than 20 organelle genomes have been completely sequenced in green microalgae [1]. Generally, chloroplasts and mitochondria in green algae have multiple copies of a single type of circular genome. In green algae, various plastid genome sizes have been reported: 37.7 kb in the non-photosynthetic alga Helicosporidium sp. and 203.8 kb in Chlamydomonas reinhardtii[2,3]. Plastid genomes in higher plants and green algae encode 88–138 genes [4,5]. Typical plastid genomes contain a large inverted repeat (IR) region with genes for rRNA, several tRNAs, and proteins. However, plastid genomes lacking an IR region also have been reported in some species [6,7]. The size of the mitochondrial (mt) genome varies among species: 6 kb in Plasmodium to 3,000 kb in the cucumber family [8,9]. The number of mt genes also varies: 5 genes in Plasmodium and about 100 genes in Jakobid flagellates [10].
Chlorella species, one of the best-known unicellular green algae, was studied in early research on photosynthesis [11] and is now used as a model and source for biotechnology and commercial applications such as use as a food additive, feed, and bioenergy source. The Chlorella genus belongs to Trebouxiophyceae, one of the Chlorophyte groups [12]. Trebouxiophyceae, found mostly in soil and freshwater, is a large algal group including Chlorella, Oocystis, Parachlorella, Coccomyxa, and Helicosporidium. The availability of organellar genomic information in Trebouxiophyceae, however, is very limited. Plastid genomes of seven species (Chlorella vulgaris C-27, Chlorella variabilis NC64A, Coccomyxa sp. C-169., Trebouxiophyceae sp. MX-AZ01, Helicosporidium sp., Oocystis solitaria, and Parachlorella kessleri) in Trebouxiophyceae have been sequenced, and they display a wide range of genome sizes, gene content, and intron content [13,14]. An IR region is missing in the plastid genome of Chlorella vulgaris C-27 [15] and Chorella variabilis NC64A (Accession no. NC_015359) but is detected in most of the Trebouxiophyceae (Coccomyxa sp., Parachlorella kessleri, and Oocystis solitaria) group. To date, the complete mt genome sequences have been reported in four trebouxiophycean algae, and they show a limited range of genome sizes, gene repertoires, and intron content. Two of them are non-photosynthetic relatives of Chlorella—Prototheca wickerhamii[16] and Helicosporidium sp. [17]. Two others are Coccomyxa sp. C-169 of Coccomyxaceae [18] and the unclassified Trebouxiophycean alga Trebouxiophyceae sp. MX-AZ01 [14]. However, the mt genome of Chlorella species remains unknown.
In the present study, we report the chloroplast (cp) and mt sequences of Chlorella sp. ArM0029B, which was isolated from drift ice in the Arctic region and has features of high lipid accumulation and fast growth at various temperatures [19]. The plastid genome of ArM0029B is similar to that of C. variabilis NC64A except for large inversions and fewer introns. The mt sequence of ArM0029B here is the first genome to be reported for Chlorella. We compared the Chlorella sp. ArM0029B organelle genome within Trebouxiophyceae and discussed cp phylogeny and cox1 intron evolution. The unique features of both organelle genomes in Chlorella sp. ArM0029B presented here will provide an important insight into the evolution of organelle genomes within microalgal species and genetic information for biotechnology.
Results and discussion
Genomic organization and features of Arctic Chlorella sp. ArM0029B
The cp and mt genome sequences of ArM0029B were assembled as circular molecules of 119,989 bp and 65,049 bp, respectively (Figure 1). However, linear plastomes, concatenated pieces representing multiple plastomes (sometimes circular), and even branched forms were reported in many species [20,21]. The polymerase chain reaction (PCR) approach we used would not rule out linear, concatenated or branched structures of an organelle genome. Therefore, we cannot exclude other complex conformations of the organelle genome in ArM0029B. The cp genome of ArM0029B contains 114 genes excluding the non-conserved open reading frames (ORFs) encoding over 50 amino acids (Tables 1 and 2). BLASTP search against the NCBI NR database revealed that all of the 79 protein-coding genes were conserved (E value < 1E-6), while only five of them were conserved hypothetical proteins. We identified 71 additional ORFs using the Glimmer (see Additional file 1: Table S1), but they were not incorporated into the final gene set because only two of them showed homology to bacterial hypothtical proteins. ArM0029B does not carry large IRs in the plastid genome as well as C. variabilis NC64A, C. vulgaris C-27, Coccomyxa sp. C-169, and Trebouxiophyceae sp. MX-AZ01, indicating that all genes are present as a single copy. The general features and gene lists were compared (Tables 1 and 2). The overall GC content of the genome of Chlorella sp. ArM0029B is low (33.92%) similar to that of C. variabilis NC64A (33.93%) and C. vulgaris C-27 (31.6%) but in contrast to that of Coccomyxa sp. C-169 (50.71%) and Trebouxiophyceae sp. MX-AZ01 (56.25%). The length of all 114 conserved genes in the plastid genome of ArM0029B is 64,626 bp, and the genes account for a coding density of 53.8% of the total cp genome sequence. The latter value is the highest coding density among all reported Chlorella spp. to date. These results indicate that the cp genome of ArM0029B is more compact than those of the above comparable species. The mt genome of ArM0029B contains a total of 62 genes excluding the non-conserved ORFs among Trebouxioaceae (Table 1). Most of the ORFs encoding over 50 amino acids are not conserved based on NCBI BlAST search. The general features and gene list of the genome of ArM0029B were compared with four Trebouxiophyceae spp., including Prototheca wickerhamii, Helicosporidium sp., Coccomyxa sp. C-169, and Trebouxiophyceae sp. MX-AZ01 (Tables 1, 2, and 3). The gene number of the mt genome of ArM0029B is highest (62 genes) among the mt genomes of all sequenced species of Trebouxiophyceae. The overall GC content of the genome is low (28.5%) similar to that of Prototheca wickerhamii (25.8%) and Helicosporidium sp. (25.6%) but in contrast to that of two species with a high GC content, Coccomyxa sp. C-169 (53.8%) and Trebouxiophyceae sp. MX-AZ01 (53.4%). All 62 conserved genes on the mt DNA of ArM0029B cover 32,655 bp in length and account for a coding density of 50.2% of the total mt genome sequence, representing an intermediate range compared with all sequenced species of Trebouxiophyceae.
Figure 1.
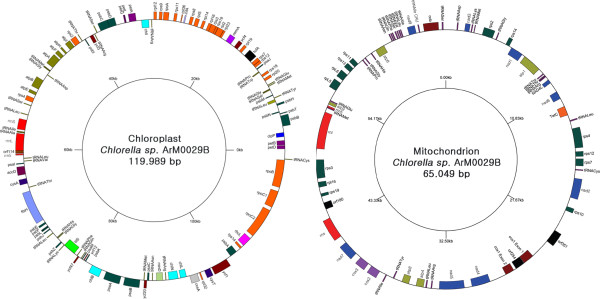
Plastid and mt genomic maps of Chlorella sp . ArM0029B.
Table 1.
General features of plastid and mt genomes in trebouxiophycean algal species
| Chloroplast | Chlorella sp. ArM0029B | C. variabilis NC64 | C. vulgaris | Coccomyxa | Trebouxiophyceae sp. |
|---|---|---|---|---|---|
| Length |
119989 |
124579 |
150613 |
175731 |
149707 |
| AT contents (%) |
66.08 |
66.07 |
68.44 |
49.29 |
43.75 |
| # genes |
114 |
114 |
115* |
115 |
115 |
| # conserved CDS |
79 |
79 |
79 |
79 |
79 |
| # tRNA |
32 |
32 |
33 |
33 |
33 |
| # rRNA |
3 |
3 |
3 |
3 |
3 |
| # introns |
1 |
3 |
3 |
1 |
5 |
| Coding density |
53.8 |
50.5 |
38.7 |
39.9/40.8 |
47.7/48.5 |
| CDS + strand |
57 |
57 |
26 |
44 |
72 |
| rRNA + srand |
3 |
3 |
3 |
3 |
3 |
| tRNA + strand |
14 |
18 |
17 |
17 |
15 |
| CDS - strand |
22 |
22 |
51 |
35 |
8 |
| rRNA - srand |
0 |
0 |
0 |
0 |
0 |
| tRNA - strand |
18 |
14 |
16 |
15 |
17 |
| Percent of +/− |
64.9 |
68.4 |
40 |
55.6 |
77.39 |
|
Mitochondria |
Chlorella
sp. ArM0029B |
Prototheca
|
Helicosporidium
sp. |
Coccomyxa
|
Trebouxiophyceae
sp. |
| Length |
65049 |
55328 |
49343 |
65497 |
74423 |
| AT contents (%) |
71.5 |
74.2 |
74.4 |
46.8 |
46.6 |
| # genes |
62 |
61 |
60 |
59 |
56 |
| # conserved CDS |
32 |
30 |
32 |
30 |
30 |
| # tRNA |
27 |
26 |
25 |
26 |
23 |
| # rRNA |
3 |
3 |
3 |
3 |
3 |
| # introns |
I (1) |
I (5) |
I (4) |
I (1) II (4) |
I (7) II (4) |
| Coding density |
50.2 |
59.8 |
64.99 |
48.38 |
42.64 |
| CDS on + strand |
18 |
13 |
9 |
29 |
29 |
| CDS on - strand |
14 |
18 |
23 |
1 |
1 |
| tRNA/rRNA on + srand |
17 |
12 |
10 |
28 |
26 |
| tRNA/rRNA on - srand |
13 |
17 |
18 |
1 |
0 |
| Percent of +/− | 56.5 | 59.3 | 68.3 | 96.7 | 98.2 |
*The asterisk indicates information without unnamed hypothetical ORFs in the cp sequence of Chlorella vulgaris C-27.
Table 2.
Gene list of the plastid genome in trebouxiophycean algal species
| |
ArM0029B |
NC64 |
C. vulgaris |
Coccomyxa |
Trebouxiophyceae sp. |
|---|---|---|---|---|---|
| MX-AZ01_NC_018569 | |||||
| accD |
o |
o |
o |
o |
o |
| atpA |
o |
o |
o |
o |
o |
| atpB |
o |
o |
o |
o |
o |
| atpE |
o |
o |
o |
o |
o |
| atpF |
o |
o |
o |
o |
o |
| atpH |
o |
o |
o |
o |
o |
| atpI |
o |
o |
o |
o |
o |
| ccsA |
o |
o |
o |
o |
o |
| cemA |
o |
o |
o |
o |
o |
| chlB |
o |
o |
o |
o |
o |
| chlI |
o |
o |
o |
o |
o |
| chlL |
o |
o |
o* |
o |
o |
| chlN |
o |
o |
o |
o |
o |
| clpP |
o |
o |
o |
o |
o |
| cysA |
o |
o |
o |
o |
o |
| cysT |
o |
o |
o |
o |
o |
| ftsH |
o |
o |
o |
o |
o* |
| infA |
o |
o |
o |
o |
o |
| minD |
o |
o |
o |
o |
o |
| petA |
o |
o |
o |
o |
o |
| petB |
o |
o |
o |
o |
o |
| petD |
o |
o |
o |
o |
o |
| petG |
o |
o |
o |
o |
o |
| petL |
o |
o |
o |
o |
o |
| psaA |
o |
o |
o |
o |
o |
| psaB |
o |
o |
o |
o |
o |
| psaC |
o |
o |
o |
o |
o |
| psaI |
o |
o |
o |
o |
o |
| psaJ |
o |
o |
o |
o |
o |
| psaM |
o |
o |
o |
o |
o |
| psbA |
o |
o* |
o |
o |
o* |
| psbB |
o |
o |
o |
o* |
o |
| psbC |
o |
o* |
o |
o |
o |
| psbD |
o |
o |
o |
o |
o |
| psbE |
o |
o |
o |
o |
o |
| psbF |
o |
o |
o |
o |
o |
| psbH |
o |
o |
o |
o |
o |
| psbI |
o |
o |
o |
o |
o |
| psbJ |
o |
o |
o |
o |
o |
| psbK |
o |
o |
o |
o |
o |
| psbL |
o |
o |
o |
o |
o |
| psbM |
o |
o |
o |
o |
o |
| psbN |
o |
o |
o |
o |
o |
| psbT |
o |
o |
o |
o |
o |
| psbZ |
o |
o |
o |
o |
o |
| rbcL |
o |
o |
o |
o |
o |
| rpl12 |
o |
o |
o |
o |
o |
| rpl14 |
o |
o |
o |
o |
o |
| rpl16 |
o |
o |
o |
o |
o |
| rpl19 |
o |
o |
o |
o |
o |
| rpl2 |
o |
o |
o |
o |
o |
| rpl20 |
o |
o |
o |
o |
o |
| rpl23 |
o |
o |
o |
o |
o |
| rpl32 |
o |
o |
o |
o |
o |
| rpl36 |
o |
o |
o |
o |
o |
| rpl5 |
o |
o |
o |
o |
o |
| rpoA |
o |
o |
o |
o |
o |
| rpoB |
o |
o |
o |
o |
o |
| rpoC1 |
o |
o |
o |
o |
o |
| rpoC2 |
o |
o |
o |
o |
o |
| rps11 |
o |
o |
o |
o |
o |
| rps12 |
o |
o |
o |
o |
o |
| rps14 |
o |
o |
o |
o |
o |
| rps18 |
o |
o |
o |
o |
o |
| rps19 |
o |
o |
o |
o |
o |
| rps2 |
o |
o |
o |
o |
o |
| rps3 |
o |
o |
o |
o |
o |
| rps4 |
o |
o |
o |
o |
o |
| rps7 |
o |
o |
o |
o |
o |
| rps8 |
o |
o |
o |
o |
o |
| rps9 |
o |
o |
o |
o |
o |
| tilS |
o |
o |
o |
o+ |
o+ |
| tufA |
o |
o |
o |
o |
o |
| ycf1 |
o |
o |
o |
o |
o |
| ycf12 |
o |
o |
o |
o |
o |
| ycf20 |
o |
o |
o |
o |
o |
| ycf3 |
o |
o |
o |
o |
o |
| ycf4 |
o |
o |
o |
o |
o |
| ycf47 |
o |
o |
o |
o |
o |
| minE |
- |
- |
o |
- |
- |
| trnA (UGC) |
o |
o |
o |
o |
o |
| TrnC (GCA) |
o |
o |
o |
o |
o |
| trnD (GUC) |
o |
o |
o |
o |
o |
| trnE (UUC) |
o |
o |
o |
o |
o |
| trnF (GAA) |
o |
o |
o |
o |
o |
| trnG (UCC) |
o |
o |
o |
o |
o |
| trnG (GCC) |
oo |
oo |
oo |
o |
o |
| trnH (GUG) |
o |
o |
o |
o |
o |
| trnI (GAU) |
o |
o |
o |
o |
o |
| trnI (CAU) |
o |
o |
o |
o |
o |
| trnK (UUU) |
o |
o |
o |
o |
o |
| trnL (CAA) |
o |
o |
o |
o |
o |
| trnL (GAG) |
o |
o |
o |
o |
o |
| trnL (UAA) |
o* |
o* |
o* |
o |
o |
| trnL (UAG) |
o |
o |
o |
o |
o |
| trnM (CAU) |
oo |
oo |
oo |
oo |
oo |
| trnN (GUU) |
o |
o |
o |
o |
o |
| trnP (UGG) |
o |
o |
o |
o |
o |
| trnQ (UUG) |
o |
o |
o |
o |
o |
| trnR (ACG) |
o |
o |
o |
o |
o |
| trnR (UCU) |
o |
o |
o |
o |
o |
| trnR (CCG) |
o |
o |
o |
o |
o |
| trnR (CCU) |
- |
- |
- |
o |
o |
| trnS (GCU) |
o |
o |
o |
o |
o |
| trnS (GGA) |
o |
o |
o |
o |
o |
| trnS (UGA) |
o |
o |
o |
o |
o |
| trnT (UGU) |
o |
o |
o |
o |
o |
| trnT (GGU) |
o |
o |
o |
o |
o |
| trnV (UAC) |
o |
o |
oo |
o |
o |
| trnW (CCA) |
o |
o |
o |
o |
o |
| trnY (GUA) |
o |
o |
o |
o |
o |
| rrnL |
o |
o |
o* |
o |
o*** |
| rrnS |
o |
o |
o |
o |
o |
| rrn5 | o | o | o | o | o |
*gene containing one intron; **gene containing two introns; ***gene containing three introns; oo, 2 copies; +, fragmented gene.
Table 3.
Distribution of the mt protein-coding gene and trn gene among trebouxiophycean algae and chlorophycean algae
| |
Trebouxiophyceae |
Chlorophyceae |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ArM0029B | Prototheca | Helicosporidium | Coccomyxa | MX-AZ01 | Scenedesmus | Dunaliella | Gonium | Chlamydomonas | |
| atp1 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| atp4 |
o |
- |
o |
o |
o |
- |
- |
- |
- |
| atp6 |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| atp8 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| atp9 |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| cob |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| cox1 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| cox2 |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| cox3 |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| nad1 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| nad2 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| nad3 |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| nad4 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| nad4L |
o |
o |
o |
o |
o |
o |
- |
- |
- |
| nad5 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| nad6 |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| nad7 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| nad9 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rpl5 |
o |
- |
o |
o |
o |
- |
- |
- |
- |
| rpl6 |
o |
o |
o |
- |
- |
- |
- |
- |
- |
| rpl16 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps2 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps3 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps4 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps7 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps10 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps11 |
o |
o |
o |
- |
- |
- |
- |
- |
- |
| rps12 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps13 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps14 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| rps19 |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| tatC |
o |
o |
o |
o |
o |
- |
- |
- |
- |
| size |
65,049 |
55,328 |
49,343 |
65,497 |
74,423 |
42,781 |
28,331 |
15,993 |
15,758 |
|
tRNAs |
Trebouxiophyceae |
Chlorophyceae |
|||||||
| |
ArM0029B |
Prototheca
|
Helicosporidium
|
Coccomyxa
|
MX-AZ01
|
Scenedesmus
|
Dunaliella
|
Gonium
|
Chlamydomonas
|
| trnA (UGC) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnC (GCA) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnD (GUC) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnE (UUC) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnF (GAA) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnG (GCC) |
o |
o |
_ |
o |
o |
_ |
_ |
_ |
_ |
| trnG (UCC) |
o |
o |
o |
o |
_ |
o |
_ |
_ |
_ |
| trnH (GUG) |
o |
o |
o |
o* |
o* |
o |
_ |
_ |
_ |
| trnI (CAU) |
o |
o |
o |
o |
o |
_ |
_ |
_ |
_ |
| trnI (GAU) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnI (UAU) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnK (UUU) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnL (AAG) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnL (CAA) |
o |
_ |
_ |
o |
o |
_ |
_ |
_ |
_ |
| trnL (CAG) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnL (UAA) |
o |
o |
o |
o |
o |
_ |
_ |
_ |
_ |
| trnL (UAG) |
o |
o |
o |
o |
o |
_ |
_ |
_ |
_ |
| trnM (CAU) |
oo |
oo |
oo |
oo |
o |
oo |
o |
oo |
o |
| trnN (GUU) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnP (UGG) |
o |
o |
o |
o |
_ |
o |
_ |
_ |
_ |
| trnQ (UUG) |
o |
o |
o |
o |
o |
o |
o |
o |
o |
| trnR (ACG) |
o |
o |
o |
o |
_ |
o |
_ |
_ |
_ |
| trnR (CCU) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnR (UCU) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnS (GCU) |
o |
o |
o |
o* |
o* |
o |
_ |
_ |
_ |
| trnS (GGA) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnS (UGA) |
o |
o |
o |
o* |
o* |
_ |
_ |
_ |
_ |
| trnT (UGU) |
o |
o |
o |
_ |
_ |
_ |
_ |
_ |
_ |
| trnV (UAC) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| trnW (CCA) |
o |
o |
o |
o* |
o* |
o |
o |
o |
o |
| trnW (CUA) |
_ |
_ |
_ |
_ |
_ |
o |
_ |
_ |
_ |
| trnY (GUA) |
o |
o |
o |
o |
o |
o |
_ |
_ |
_ |
| Total number | 27 | 26 | 25 | 26 | 23 | 27 | 3 | 4 | 3 |
*gene containing intron; oo, 2 copies.
In the cp genome, with 74 (64.9%) conserved genes occupying one strand and 40 genes occupying the other strand, the gene distribution over the two DNA strands of ArM0029B cp genome is biased (Figure 1, Table 1). The gene contents in one strand were detected to be 68.4%, 40.5%, and 55.6% in the cp genome of NC64A, C. vulgaris C-27 and Coccomyxa C-169, respectively. These results indicate that gene distribution between the two strands of the cp genome is biased to some degree but relatively even in contrast to one of the mt genomes. In the mt genome of ArM0029B, 35 conserved genes occupy one strand, and 27 genes occupy the other strand, indicating that the genes are evenly (56.5:43.5) distributed in both strands of the ArM0029 mt genome (Table 1). The other two species, Prototheca wickerhamii and Heicosporidium sp., showed more biased occupation of the genes in one strand (59.3% and 68.3%, respectively) than Chlorella sp. ArM0029B. Furthermore, Coccomyxa and MX-AZ01 displayed a drastic biased distribution of the mt genes in one strand (96.7% and 98.2%, respectively).
Gene content and rearrangement of the cp genome
The plastid genome of ArM0029B contains 79 genes encoding proteins, 32 tRNA genes, and 3 rRNA genes similar to that of C. variabilis NC64A (Table 2). The ArM0029B plastid gene repertoire differs from that of C. variabilis NC64A except for the absence of pseudogenes similar to chlL and an intronic endonuclease in the psbC gene, and from C. vulgaris C-27 by the absence of tRNA-Val (UAC) and the minE homolog. AM0029B has a small cp genome among species although it has a similar number of genes to C. variabilis NC64A, C. vulgaris C-27, Coccomyxa sp. C-169, and Trebouxiophyceae sp. MX-AZ01 (Tables 1 and 2). The compactness of the cp genome of ArM0029B is due to a short intergenic sequence and fewer introns. The conserved gene order and rearrangement of cp genomes among ArM0029B, C. variabilis, and C. vulgaris were compared in Figure 2. The gene order in the plastid genome of Chlorella sp. ArM0029B is very similar to that of C. variabilis NC64A. Rearrangement of genomes between ArM0029B and C. variabilis was found in two regions; trnV and the 15-kb gene cluster, including “trnI-ycf20-psaC-trnN-minD-trnR1-chlN-chlL-ccsA-rpl32-cysT-ycf1-psbA”, are present in inverse orientation between Chlorella sp. ArM0029B and C. variabilis NC64A (Figure 2 and see Additional file 2: Figure S1). Marked rearrangement of gene clusters was detected between ArM0029B and C. vulgaris. Many gene clusters conserved in green algae [22] are also conserved in Chlorella sp. ArM0029B (Figure 2). Interestingly, the gene order of “trnC-rpoB-rpoC1-rpoC2-rbcL-rps14” is well conserved between ArM0029B and two Chlorella spp., C. vulgaris and C. variabilis (see Additional file 3: Figure S2) but not in related species Coccomyxa sp. C-169 and Trebouxiophyceae sp. MX-AZ01, suggesting that the gene order is well conserved and may be specific to Chlorella species. The order of psbD and psbC genes are conserved and closely linked in all sequenced Trebouxiophyceae. Interestingly, the 5′ coding region of the psbC gene seemed to be overlapped with the 3′ coding region of psbD on the same strand in ArM0029B. This phenomenon of two genes overlapping occurs frequently in the genomes of viruses, prokaryotes, mitochondria, and eukaryotes, including humans [23-25]. The overlap of psbD and psbC seemed to exist in all of the Trebouxiophyceae sequenced except for Helicosporidium sp., which lacks psbD and psbC in the plastid genome. The psbC gene in Coccomyxa sp. C-169 and Trebouxiophyceae sp. MX-AZ01 was annotated with Gly as a starting amino acid, resulting in separation of psbC from psbD. Possible Gly start codons of psbC are also found a few bases after psbD in all sequenced Trebouxiophyceae. However, the ATG or GTG start codon is also found in the 3′ coding region of psbD in those species. In other class of viridiplantae, Oltmannsiellopsis viridis and Pseudendoclonium akinetum of Ulvophyceae, Nephroselmis olivacea in Prasinophytes, and Mesostigma viride in Charophyceae also share the same feature of overlapping of psbD-psbC or a GTG start codon of psbC without overlap with psbD. However, in the case of psbC separated clearly from psbD such as in C. reinhardtii and Senedesmus, the N-terminal amino acid sequence of psbC is Met-Glu-Thr-Leu-Phe-Asn-Gly-Thr(Ser). The italic amino acids are well conserved and are encoded in all overlapped sequences of the above species, indicating that all linked genes of psbD and psbC may be overlapped in the same manner.
Figure 2.
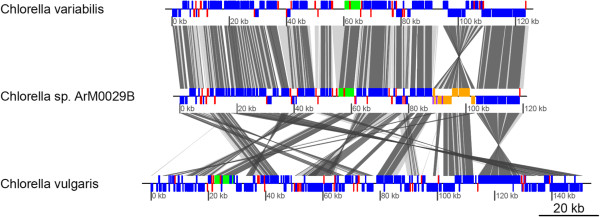
Comparison of the cp genomes of C. variabilis, Chlorellasp. ArM0029B, and C. vulgaris. Thick bars represent protein-coding genes (blue), rRNA genes (green), and tRNA genes (red). Genes in the inverted region were highlighted in orange (protein-coding genes) and purple (tRNA genes) boxes in ArM0029B. LCBs and conserved genes are connected by light and dark gray rhomboids, respectively.
Gene content of the mt genome
The mt genome of ArM0029B contains 32 mt protein coding genes, 27 tRNA genes, and 3 rRNA genes (Tables 1 and 3). The 32 protein-coding genes include 4 atp genes, 3 cox genes, 9 nad genes, 13 ribosomal protein genes, and cob and tatC genes. Helicosporidium, another trebouxiophycean alga, also has the same content of protein-coding genes. Three other trebouxiophycean algae, Prototheca, Coccomyxa, and MX-AZ01, have only 30 among 32 protein-coding genes of ArM0029B and Helicosporidium (Table 3). Two ribosomal protein genes, rpl6 and rps11, are absent in Coccomyxa and MX-AZ01. Prototheca lacks two genes, atp4 and rpl5. It is assumed that the genes in the three taxa were recently lost in the lineage of Trebouxiophyceae, possibly nuclear transferred. Compared with trebouxiophycean algae, the chlorophycean algae, including Scenedesmus, Dunaliella, Gonium, and Chlamydomonas do not have any ribosomal protein gene and tatC, which are found in Trebouxiophyceae (Table 3). Scenedesmus has the largest content of protein-coding genes among the chlorophycean algae with 13 protein-coding genes, all shared by trebouxiophycean algae. The 13 protein-coding genes include 2 atp genes, 3 cox genes, 7 nad genes, and a cob gene. Among the 13 genes, 6 are absent in the other three chlorophycean algae, Dunaliella, Gonium, and Chlamydomonas. The three chlorophycean algae contain the other seven protein-coding genes, including cob, cox1, nad1, nad2, nad4, nad5, and nad6. The ArM0029B mt genome contains 27 tRNA genes, the largest in number among trebouxiophycean algae (Table 3). The tRNA gene content of other trebouxiophycean algae ranged from 23 to 26. Both Coccomyxa and MX-AZ01 share introns in four tRNA genes. In Chlorophyceae, although Scenedesmus has 27 tRNA genes, Dunaliella, Gonium, and Chlamydomonas have only three types of tRNA genes, trnM, trnQ, and trnW. Additional file 4: Figure S3 shows the characterization of the mt tRNA genes of Chlorella sp. ArM0029B. Amongst trebouxiophycean algae, Chlorella sp. ArM0029B has the largest number of tRNA genes, and the secondary structure of tRNA genes in trebouxiophycean algal mitochondria is unknown. Thus far, the Chlorella sp. ArM0029B is known to contain the largest gene content of the mt genomes of Trebouxiophyceae.
Analysis of the conserved gene cluster in the mt genome is difficult because of limited information regarding mt genomes in trebouxiophycean algae. The mt genome of Chlorella species was not reported except for ArM0029B of this study. It has been reported that the overall gene order in mt genomes is conserved between the non-photosynthetic group “Prototheca wickerhamii and Helicosporidium sp” and the high-GC content group “Coccomyxa sp. C-169 and Trebouxiophyceae sp. MX-AZ01”, respectively. [14,17]. The overall gene order on the mt genome of Chlorella sp. ArM0029B is not conserved with any member of the non-photosynthetic group or high-GC group. Nevertheless, the gene order for “trnS-trnV-trnL” and “trnY-atp8-atp4” is well conserved in all five species of Trebouxiophyceae.
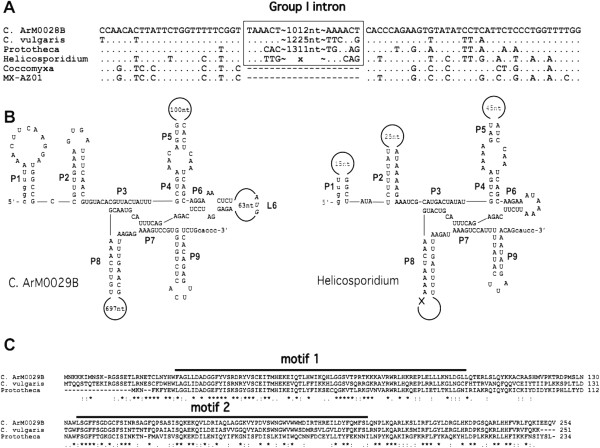
Introns in the organellar genomes of ArM0029B
Two group I introns are found in the organellar genomes of Chlorella sp. ArM0029B. One resides in trnL (UAA) of the cp genome, and the other is located in cox1 of the mt genome. The trnL (UAA) group I intron of cyanobacterial origin is an ancient self-splicing group I intron in the plastid genome that is rarely lost in some taxa [26,27]. The ArM0029B mt genome has the intron between bases 720 and 721 of cox1. Among trebouxiophycean algae, the intron with the same insertion site is also found in C. vulgaris, Prototheca, and Helicosporidium but not in Coccomyxa and MX-AZ01 (Figure 3A). The Chlorella sp. ArM0029B intron is a cis-splicing intron and has an ORF starting at Loop 6 (L6) and ending at P8-P7 (Figure 3B). The ORF has two LAGLIDADG endonuclease motifs. The endonuclease-like ORF of the group I intron is known to have two LAGLIDADG motifs [28]. The ORF with two LAGLIDADG motifs is also found in the same intron of C. vulgaris and Prototheca (Figure 3C). Unlike Chlorella and Prototheca, Helicosporidium has a trans-splicing intron without an ORF. As shown in Figure 3B, the dis-connection of the intron in Helicosporidium occurs at loop 8, which contains the ORF, assuming that the trans-splicing intron of Helicosporidium might be derived from cis-splicing by genomic rearrangement, followed by loss of the ORF. Compared with the ArM0029B intron, other related species in Trebouxiophyceae contain 3–11 introns in two to six genes of their mt genomes (Table 1), indicating that the mt genome of ArM0029B has the smallest number of introns among those reported in Trebouxiophyceae. An intron or introns split the cox1 gene into two exons in ArM0029B, four exons in Prototheca wickerhamii, three exons in Helicosporidium sp., and two exons with different cognate sites in Trebouxiophyceae sp. MX-AZ01, and no intron was found in Coccomyxa sp. C-169. The intron distribution on the plastid genome in ArM0029B is different from that of other trebouxiophycean algae: three introns in trnL3, psbA, and psbC of C. variabilis NC64A, three introns in trnL3, rrnL, and chlL of C. vulgaris, one intron in psbB of Coccomyxa sp. C-169, and five introns in ftsH, psbA, and rrnL of Trebouxiophyceae sp. MX-AZ01 (Tables 1 and 2). Fewer introns, the lack of pseudogenes, and shorter intergenic regions contributed to the more compact plastid genome of ArM0029B than that of C. variabilis NC64A and C. vulgaris C-27.
Figure 3.
The mtcox1 intron of C. ArM0028B and its distribution among trebouxiophycean algae. A. The exon and intron border sequences. B. Secondary structures of the ArM0028B cis-splicing intron and Helicosporidium trans-splicing intron. C. Comparison of the intronic ORF with two LAGLIDADG motifs.
Phylogenetic affinity of ArM0029B to other trebouxiophycean algae
Phylogenetic relationships among seven trebouxiophycean algal plastids were investigated using the aligned 10,938-base DNA sequence of six large photosystem genes—psaA, psaB, psbA, psbB, psbC, and psbD—and rbcL, which are widely used for phylogenetic studies [29-31]. Phylogenetic analysis outgrouped by four chlorophycean algae produced a single plastid maximum parsimonious (MP) tree (Figure 4A), and NJ and ML analyses showed a single tree with the similar topology (see Additional file 5: Figure S4). Four chlorophycean algae and six trebouxiophycean algae are separated into two sister clades with 100% bootstrap/Jackknife supports. Trebouxiophycean algae are separated into two clades: the Coccomyxa-[MX-AZ01] clade with 100% bootstrap/Jackknife supports and the Chlorella-Parachlorella-Oocystis clade with 84–85% bootstrap/Jackknife supports. Within a Chlorella-Parachlorella-Oocystis clade, ArM0029B-Chlorella 2 spp. formed a clade with 100% bootstrap/Jackknife support, but Parachlorella and Oocystis were clustered without bootstrap/Jackknife supports. The distance matrix of the aligned 10,938-base DNA sequence among seven trebouxiophycean algae is shown in Additional file 6: Table S2. The distance ranged from 9.193% to 10.299% between Chlorella species and ranged from 14.071% to 26.705% among Trebouxiophycean genera. The distance between ArM0029B and C. variabilis (9.193%) is smaller than the distance between ArM0029B and C. vulgaris C-27 (10.299%) or the distance between C vulgaris C-27 and C. variabilis (9.403%), indicating the close relationships of ArM0029B to Chlorella variabilis. The closer genus to Chlorella spp. was Parachlorella with 14.071 ~ 14.585% distance and Oocystis with 15.911 ~ 16.435% distance. Among other genera, Parachlorella and Oocystis had a 14.959% distance, and Coccomixa and MX-AZ01 had a 16.901% distance. Except for the genera discussed above, over 20% distance was detected among Trebouxiophycean genera. The results indicate that ArM0029B belongs to the genus Chlorella along with C. vulgaris and C. variabilis and that C. variabilis is the closer taxon to ArM0029B.The mt genome-based phylogenetic relationships among five trebouxiophycean algae were also analyzed using the translated amino acids sequences of seven genes, cob, cox1, nad1, nad2, nad4, nad5, and nad6, which are shared by trebouxiophycean and chlorophycean algal mitochondria. Phylogenetic analysis outgrouped by four chlorophycean algae produced a single MP tree (Figure 4B) and a single NJ tree with the same topology (see Additional file 5: Figure S4C). Four chlorophycean algae and five trebouxiophycean algae are separated into two sister clades with 100% bootstrap/Jackknife supports. The five-trebouxiophycean algae MP tree contained one clade, a Helicosporidium-Prototheca clade with 100% bootstrap/Jackknife support and three isolated taxa—ArM0029B, MX-AZ01, and Coccomyxa. Although the three taxa were clustered, the cluster was weakly bootstrap/Jackknife (65%/67%) supported. Although ArM0029B formed a clade with MX-AZ01 and Coccomyxa in MP and NJ trees, the mt phylogenomic affinity of ArM0029B to other trebouxiophycean algae remains to be investigated because of limited information of available mt genomes in trebouxiophycean algae. Scenedesmus, which contains the largest number of genes among Chlorophyceae, has ancient mt characteristics among green algae and is explained as a basal group in phylogenetic analysis. Green algae have lost many mt genes via gene transfer to the nucleus. ArM0029B contains more mt genes than other sequenced trebouxiophycean algae to date, suggesting that it may show ancient characteristics of its mt genome among trebouxiophycean algae. However, we cannot exclude the possibility of new integration of genes into the ancient-type trebouxiophycean alga with fewer genes in the mt genome.
Figure 4.
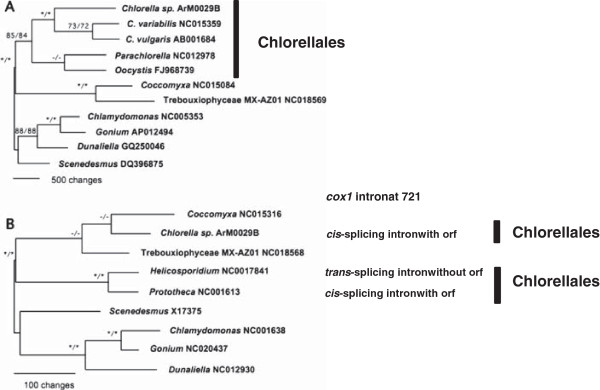
Single maximum parsimonious (MP) trees. A. The plastid MP tree from DNA sequences of rbcL and six photosystem genes. B. The mt MP tree from translated amino acid sequences of seven protein-coding genes. The distribution of cox1 intron at 721, its splicing form, and presence/absence of its orf were denoted. */*: 100% bootstrap support/100% jackknife support. −/−: bootstrap and jackknife not supported.
The Chlorellales in the phylogenies of chloroplasts and mitochondria and the meaning of cox1 intron at base position 721 of the cox1 gene
The Chlorellales is a green algal group lacking flagella whose members include Chlorella, Parachlorella, Oocystis, Prototheca, and Helicosporidium. Chlorella and Parachlorella inhabit in freshwater, marine, or land in the coccoidal form [32]. Prototheca and Helicosporidium are non-photosynthetic and parasitic coccoids. Phylogenetic relationships within the Chlorellales have been well studied, and nuclear and cp gene data have provided evidence that the Oocystaceae, including the semi-colonial Oocystis, form an early diverging clade within the Chlorellales [1]. Our cp genome phylogeny is congruent to those in previous reports and shows the formation of a strong clade containing Chlorella, Parachlorella, Oocystis, Prototheca, and Helicosporidium. The cox1 intron at base position 721 in the cox1 gene of the mt genome is found only in the members of Chlorellales in Trebouxiophyceae—i.e., Prototheca, Helicosporidium, Chlorella vulgaris and Chlorella sp. ArM0029B. The cox1 intron distribution supporting Chlorellales does not agree with mt genome phylogeny. The occurrence of the cox1 intron in free-living Arctic Chlorella sp. ArM0029B and Chlorella vulgaris, as well as in the parasitic coccoids Prototheca and Helicosporidium, indicates that the intron with the same origin had been introduced in early Chlorellales and that the trans-splicing of the intron occurred after the divergence of Prototheca and Helicosporidium in the parasitic coccoid clade. Limited taxon sampling, high variation of mt genes, and possible lateral gene transfer from other taxa might affect the topology of the phylogenetic tree in the present study. Increasing members of representative taxa in trebouxiophycean algae would help to improve the understanding of its evolution.
Conclusions
Organelle functions play an important role in maintaining an organism’s life, including energy production, photosynthesis, and metabolite biosynthesis. The chloroplast is an organelle for fatty acid/lipid biosynthesis, and the mitochondrion is an organelle for fatty acid/lipid degradation. Recently, oil-producing microalgae have been studied intensively for genetic improvement, including genomics and genetic engineering. Chlorella is an important microalgae for oil production [33]. ArM0029B is a Chlorella sp. originated from the Arctic region, which have features of fast growth at various temperatures and a high oil-accumulating trait [19].
Here, we report the 119,989-bp cp genome and 65,049-bp mt genome of Arctic Chlorella sp. ArM0029B. The plastid genome of ArM0029B lacking a large IR is close to C. variabilis NC64A: both species displayed the same content of conserved genes and almost the same gene order. However, large rearrangements are also found between ArM0029B and C. variabilis NC64A by inversion of a 15-kb gene cluster. Major structural changes were detected in introns and tRNAs in ArM0029B compared with related species of Trebouxiophyceae. The mt genome of ArM0029B contains the largest number of genes (62 genes) and smallest number of introns (one intron in cox1) among trebouxiophycean algae. Detailed information regarding the secondary structure of the tRNA genes would be obtained in a Chlorella mt genome study. Two group I introns were found in ArM0029B: a self-splicing intron in trnL (UAA) of the cp genome and another intron in cox1 of the mt genome containing an ORF encoding an endonuclease with double motifs of LAGLIDADG. Phylogenetic analysis of cp genomes suggests that three Chlorella species belong to a monophyletic group, and ArM0029B belongs to the genus Chlorella. The phylogenetic analysis of mt genomes with limited information of the available mt genome in Trebouxiophyceae could not determine the closest mt genome of ArM0029B among the four trebouxiophycean algae. The lowest number of introns in the organelle genome of ArM0029B among Chlorella spp. may be due to the limited chance of intron spreading and invasion by the isolation in the Artic environment from other related taxa. Based on the gene content, the ArM0029B organelle genomes seem to have ancient organelle characteristics with many genes and fewer introns gene in both genomes.
In the present study, cp genome phylogeny supports monophyly of the seven investigated members of Chlorellales, including three Chlorella spp., Parachlorella, Oocystis, Prototheca, and Helicosporidium. The intron distribution at base position 721 of the cox1 gene occurs in all four investigated Chlorellales taxa—Chlorella sp. ArM0029B, Chlorella vulgaris, Prototheca, and Helicosporidium—assuming that a common ancestor of the Chorellales might display the cox1 intron as a cis-splice form and that the cis-splicing intron was recently trans-spliced in Helicosporidium. When more mt genomic information is available, we will have better understanding of the mt genome phylogeny of the trebouxiophycean algae.
The unique features of Chlorella sp. ArM0029B organelle genomes presented here will provide important information to understand organellar genome evolution, including introns, gene rearrangement, and structural changes of plastids and mt genomes among species in Trebouxiophyceae and green algae.
Methods
Strain and culture conditions
Chlorella sp. ArM0029B [19] was maintained on solidified TAP medium [34] and cultured in liquid medium for analysis at 25°C with 200 rpm shaking under constant white light (40 μmol m−2 s−1).
Sequencing, assembly, and annotation of the ArM0029B organelle genomes
The ArM0029B organelle genomes were sequenced as part of the ArM0029B genome project (funded by Advanced Biomass R&D Center) using an Illumina HiSeq 2000-based whole-genome shotgun sequencing approach. The organelle sequences were obtained using the CLC Genomics Workbench version 5.5. Two large contigs (65.049 kb and 120.090 kb) with the highest average read coverages (19,505 and 7,485, respectively) were identified; the contigs displayed low GC content compared with the high GC content of the nuclear genome. Circular structures of each replicon were confirmed by polymerase chain reaction (PCR) amplification at their ends and by joining of Sanger sequence reads derived from the amplicons. The assemblies were further verified by examining paired-end distance and depth after re-mapping reads on the contig sequences. The BLAST searches of two large contigs were verified to plastid and mt genomes, respectively. For gene annotation of organelle genomes, ORFs encoding 50 amino acids or longer were identified and searched against a known protein database (NR). Genes encoding proteins homologous to known short cp peptides were manually identified. Glimmer (ver. 3.02) was used to predict additional putative protein-coding genes [35]. tRNA and rRNA genes were respectively detected using ARAGORN [36] and RNAmmer 1.2. The rrn5 in the mt genome was detected based on BLAST search and the 5S rRNA data bank [37]. The complete sequences of the ArM0029B chloroplast and mitochondrion were deposited in GenBank under the accession numbers KF554427 and KF554428, respectively.
For comparison of the mt genomes of ArM0029B, we used all four mt genomes to date reported in Trebouxiophyceae: Prototheca wickerhamii (NC_001613), Helicosporidium sp. ex Simulium jonesi (NC_017841), Coccomyxa sp. C-169 (NC_015316), and Trebouxiophyceae sp. MX-AZ01 (NC_018568). We skipped Parachlorella minor because of the very low gene content and arguable placement to be included in Trebouxiophyceae. For the plastid genomes of ArM0029B, we selected four reported species, including C. variabilis NC64A (NC_015359), C. vulgaris C-27 (NC_001865), Coccomyxa sp. C-169 (NC_015084), and Trebouxiophyceae sp. MX-AZ01 (NC_018569). We did not include non-photosynthetic Trebouxiophyceae species to compare plastid genomes because they do not contain many photosynthetic genes.
Comparative analysis of cp genomes
The complete cp genomes of ArM0029B, C. variabilis NC64A, and C. vulgaris C-27 were compared using the MAUVE alignment tool [38] to identify rearrangement-free LCBs (locally collinear blocks) among genomes, yielding 25 LCBs with a minimum weight of 170. The genome sequence of the C. variabilis chloroplast was artificially rearranged prior to the MAUVE alignment so that the genome-level alignments could be maximally shown. Conserved genes among the three cp genomes were identified using the BLASTN search. genoPlotR [39] was then used to visualize conserved genes in the context of genomes and LCBs.
Secondary structure analyses of intron and mt trn genes
The secondary structure of the group I intron was constructed based on the methods of Burke et al.[40] and Michel and Westhoff [41]. For the secondary structure of mt trn genes, the method of Chuang et al.[42] was consulted.
Phylogenetic analyses
The phylogenetic relationships of ArM0029B among green algae were investigated using both chloroplast and mitochondrion genomic information. As an ingroup, all reported trebouxiophycean organellar genomic information was included. Currently, information concerning six cp genomes is available in Trebouxiophyceae: Chlorella vulgaris C-27 (AB_001684.1), Chlorella variabilis NC64A (NC_015359), Parachlorella kessleri (NC_012978), Coccomyxa sp. C-169 (NC_015084), Oocystis solitaria (FJ968739), and Trebouxiophyceae sp. MX-AZ01 (NC_018569). Four reported trebouxiophycean algal mt genomes are Coccomyxa sp. C-169 (NC_015316), Helicosporidium sp. ex Simulium jonesi (NC_017841), Prototheca wickerhamii (NC_001613), and Trebouxiophyceae sp. MX-AZ01 (NC_018568). A partial clone (AB011523) of the cox1 gene for Chlorella vulgaris was also used. To avoid bias by taxon sampling, four chlorophycean algae, known both as cp and mt genomes, were used as an outgroup. These include Chlamydomonas reinardtii (NC_005353 for cp; NC_001638 for mt), Gonium pectoral (AP_012494 for cp; NC_020437 for mt), Dunaliella salina (GQ_250046 for cp; NC_012930 for mt), and Scenedesmus obliquus (DQ_396875 for cp; X17375 for mt).
DNA sequences of seven cp protein genes, including psaA, psaB, psbA, psbB, psbC, psbD, and rbcL were used for the cp phylogenetic MP, NJ and ML tree using Paup ver. 6.0. Bootstrap and jackknife analyses of MP tree were also performed with 1,000 replication. Shared gene contents among chlorophycean and trebouxiophycean algal mitochondria were very limited, and the DNA sequence variation of protein-coding genes was highly variable for successful alignment. Translated amino acid sequences of seven protein-coding genes, including cob, cox1, nad1, nad2, nad4, nad5, and nad6 were used for the mt MP tree. In the analysis, gapped sequences were not included. Bootstrap and jackknife analyses were also performed with 1,000 replication.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
W-JJ and JL designed the research and wrote the paper. HJ, JML, JP, YMS, and H-GC performed the research. All of the authors read and approved the manuscript.
Supplementary Material
Additional putative CDSs predicted by Glimmer ver. 3.02. Additional 71 ORFs (>50 aa) were identified from the cp genome of ArM0029B using the Glimmer gene prediction tool. These ORFs were not added to the finalized gene set because ab initio gene prediction that uses a short-sized genome for self-training and prediction itself could result in many false genes. ORFs overlapping tRNA or rRNA genes were excluded.
Confirmation of a 15-kb gene cluster inversion in the plastid genome of Chlorella sp. ArM0028B compared with C. variabilis NC64A. (A) Diagram of the inverted gene cluster in the plastid genomes of C. variabilis NC64A and Chlorella sp. ArM0029B. PCR primers are marked as psaBF, ycf20R, psbAF, and rps14R with arrows. (B) PCR confirmation of a 15-kb gene cluster inversion in the plastid genome of Chlorella sp. ArM0028B. Lane 1: primer set (psaBF and ycf20R); Lane 2: primer set (psbAF and rps14R); Lane 3: primer set (psaBF and psbAF); Lane 4, primer set (ycf20R and rps14R). The expected sizes of PCR products in lanes 1 and 2 are 801 bp and 855 bp, respectively. The primers sequences used for PCR are follows. 5′-TATGTTTTAACTTATGCGGCATTCTT-3′ for psaBF; 5′-AACATTGAATTGCAAAAATGTTCC-3′ for ycf20R; 5′-CAACCGATGTATAAACGGTTTTCA-3′ for psbAF; 5′-TCTTCAAGGTCTTTTACCTGGT-3′ for rps14R. Total genomic DNA purified from ARM0029B was used for PCR reactions. PCR amplifications in only lanes 1 and 2 with the expected sizes indicating that a 15-kb gene cluster in the plastid genome of ArM0029B exists in the inverse orientation compared with the plastid genome of C. variabilis NC64A.
Conserved plastid genome gene clusters among trebouxiophycean algae. The arrangement of cp genes, including trnC to rpoC2 and rps14, are compared in ArM0029B and related species. The red-boldface indicates genes with a conserved order. The direction of the box arrow denotes sense orientation of transcription of the gene.
Secondary structure of mt trn genes in Chlorella sp. ArM0029B.
Single ML (A, HYK85 + G + I model) and NJ (B) trees from the DNA sequences of seven cp genes and a NJ tree (C) from translated amino acid sequences of seven mt protein-coding genes. */*: 100% bootstrap support/100% jackknife support. −/−: bootstrap and jackknife not supported.
Distance matrix of seven cp gene sequences in Trebouxiophyceae.
Contributor Information
Haeyoung Jeong, Email: hyjeong@kribb.re.kr.
Jong-Min Lim, Email: jmlim114@kribb.re.kr.
Jihye Park, Email: park@greenplant.re.kr.
Young Mi Sim, Email: ymsim@kribb.re.kr.
Han-Gu Choi, Email: hchoi82@kopri.re.kr.
Jungho Lee, Email: jlee@greenplant.re.kr.
Won-Joong Jeong, Email: wonjoong@kribb.re.kr.
Acknowledgments
This work was supported by a Grant from the Advanced Biomass R&D Center (ABC) of the Global Frontier Project funded by the Ministry of Education, Science and Technology of Korea (ABC-2011-0031343).
References
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and Molecular Evolution of the Green Algae. Crit Rev Plant Sci. 2012;15(1):1–46. [Google Scholar]
- de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;15:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, de Pamphilis CW, Miller W, Harris EH, Stern DB. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. The Plant cell. 2002;15(11):2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M. A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol. 2007;15:2. doi: 10.1186/1741-7007-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol. 2006;15(6):1324–1338. doi: 10.1093/molbev/msk018. [DOI] [PubMed] [Google Scholar]
- Manhart JR, Hoshaw RW, Palmer JD. Unique chloroplast genome in spirogyra maxima (chlorophyta) revealed by physical and gene mapping1. J Phycol. 1990;15(3):490–494. [Google Scholar]
- Manhart JR, Kelly K, Dudock BS, Palmer JD. Unusual characteristics of Codium fragile chloroplast DNA revealed by physical and gene mapping. MGG Mol Gen Genet. 1989;15(2–3):417–421. doi: 10.1007/BF00334385. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Gardner MJ, Williamson DH, Wilson RJ. The putative mitochondrial genome of Plasmodium falciparum. J protozool. 1991;15(3):243–245. doi: 10.1111/j.1550-7408.1991.tb04436.x. [DOI] [PubMed] [Google Scholar]
- Ward BL, Anderson RS, Bendlich AJ. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae) Cell. 1981;15(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria Of Protists. vol. 38. 2004. pp. 477–524. [DOI] [PubMed] [Google Scholar]
- Benson AA. Following the path of carbon in photosynthesis: a personal story. Photosynth Res. 2002;15(1–3):29–49. doi: 10.1023/A:1020427619771. [DOI] [PubMed] [Google Scholar]
- Friedl T. Inferring taxonomic positions and testing genus level assignments in coccoid green lichen algae: a phylogenetic analysis of 18 s ribosomal rna sequences from dictyochloropsis reticulata and from members of the genus myrmecia (chlorophyta, trebouxiophyceae cl. Nov.)1. J Phycol. 1995;15(4):632–639. [Google Scholar]
- Lang BF, Nedelcu A. In: Genomics of Chloroplasts and Mitochondria. vol. 35. Bock R, Knoop V, editor. Netherlands: Springer; 2012. Plastid Genomes of Algae; pp. 59–87. [Google Scholar]
- Servín-Garcidueñas LE, Martínez-Romero E. Complete mitochondrial and plastid genomes of the green microalga Trebouxiophyceae sp. strain MX-AZ01 isolated from a highly acidic geothermal lake. Eukaryotic Cell. 2012;15(11):1417–1418. doi: 10.1128/EC.00244-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Nagai T, Kapoor M, Sugita M, Ito M, Ito S, Tsudzuki J, Nakashima K, Tsudzuki T, Suzuki Y, Hamada A, Ohta T, Inamura A, Yoshinaga K, Sugiura M. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc Natl Acad Sci U S A. 1997;15(11):5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Plante I, Lang BF, Kuck U, Burger G. Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii. Gene content and genome organization. J Mol Biol. 1994;15(1):75–86. doi: 10.1006/jmbi.1994.1210. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Keeling PJ. The mitochondrial genome of the entomoparasitic green alga helicosporidium. PLoS One. 2010;15(1):e8954. doi: 10.1371/journal.pone.0008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Burki F, Yamada T, Grimwood J, Grigoriev IV, van Etten JL, Keeling PJ. The GC-rich mitochondrial and plastid genomes of the green alga coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS One. 2011;15(8):e23624. doi: 10.1371/journal.pone.0023624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JW, Hwangbo K, Lee SY, Choi HG, Park YI, Liu JR, Jeong WJ. A new Arctic Chlorella species for biodiesel production. Bioresour Technol. 2012;15:340–343. doi: 10.1016/j.biortech.2012.09.026. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant cell. 2004;15(7):1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol. 2004;15(4):953–970. doi: 10.1016/j.jmb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae pedinomonas minor, parachlorella kessleri, and oocystis solitaria reveal a shared ancestry between the pedinomonadales and chlorellales. Mol Biol Evol. 2009;15(10):2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Chisholm SW. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 2004;15(11):2268–2272. doi: 10.1101/gr.2433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S, Bergström S, Edlund T, Grundström T, Jaurin B, Lindberg FP, Olsson O. Overlapping genes. Annu Rev Genet. 1983;15:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni V, Makałowski W, Galdzicki M, Sood R, Makałowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;15(2):280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besendahl A, Qiu YL, Lee J, Palmer JD, Bhattacharya D. The cyanobacterial origin and vertical transmission of the plastid tRNA(Leu) group-I intron. Curr Genet. 2000;15(1):12–23. doi: 10.1007/s002940050002. [DOI] [PubMed] [Google Scholar]
- Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;15(4987):1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Lee J, Manhart JR. Three ORF-Containing Group I Introns in Chloroplast SSU of Caulerpa sertularioides (Ulvophyceae) and Their Evolutionary Implications. Algae. 2003;15:183–190. [Google Scholar]
- Daugbjerg N, Andersen RA. Phylogenetic analyses of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol Biol Evol. 1997;15(12):1242–1251. doi: 10.1093/oxfordjournals.molbev.a025733. [DOI] [PubMed] [Google Scholar]
- Freshwater DW, Fredericq S, Butler BS, Hommersand MH, Chase MW. A gene phylogeny of the red algae (Rhodophyta) based on plastid rbcL. Proc Natl Acad Sci U S A. 1994;15(15):7281–7285. doi: 10.1073/pnas.91.15.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart JR. Phylogenetic analysis of green plant rbcL sequences. Mol Phylogenet Evol. 1994;15(2):114–127. doi: 10.1006/mpev.1994.1014. [DOI] [PubMed] [Google Scholar]
- Aslam Z, Shin W, Kim MK, Im WT, Lee ST. Marinichlorella Kaistiae gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) based on polyphasic taxonomy. J Phycol. 2007;15(3):576–584. [Google Scholar]
- Larkum AWD, Ross IL, Kruse O, Hankamer B. Selection, breeding and engineering of microalgae for bioenergy and biofuel production. Trends Biotechnol. 2012;15(4):198–205. doi: 10.1016/j.tibtech.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic; 1989. p. 780. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;15(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;15(1):11–16. doi: 10.1093/nar/gkh152. ARAGORN, tRNA (and tmRNA) detection. http://130.235.46.10/ARAGORN/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Barciszewski J, Erdmann VA. 5S ribosomal RNA database Y2K. Nucleic Acids Res. 2000;15(1):166–167. doi: 10.1093/nar/28.1.166. 5S RIBOSOMAL RNA DATABASE. http://rose.man.poznan.pl/5SData/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;15(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SG. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;15(18):2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Belfort M, Cech TR, Davies RW, Schweyen RJ, Shub DA, Szostak JW, Tabak HF. Structural conventions for group I introns. Nucleic Acids Res. 1987;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;15(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Chuang LY, Lin YD, Yang CH. PtRNAss: Prediction of tRNA secondary structure from nucleotide sequences. IAENG Int J Comput Sci. 2010;15(3):204–209. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional putative CDSs predicted by Glimmer ver. 3.02. Additional 71 ORFs (>50 aa) were identified from the cp genome of ArM0029B using the Glimmer gene prediction tool. These ORFs were not added to the finalized gene set because ab initio gene prediction that uses a short-sized genome for self-training and prediction itself could result in many false genes. ORFs overlapping tRNA or rRNA genes were excluded.
Confirmation of a 15-kb gene cluster inversion in the plastid genome of Chlorella sp. ArM0028B compared with C. variabilis NC64A. (A) Diagram of the inverted gene cluster in the plastid genomes of C. variabilis NC64A and Chlorella sp. ArM0029B. PCR primers are marked as psaBF, ycf20R, psbAF, and rps14R with arrows. (B) PCR confirmation of a 15-kb gene cluster inversion in the plastid genome of Chlorella sp. ArM0028B. Lane 1: primer set (psaBF and ycf20R); Lane 2: primer set (psbAF and rps14R); Lane 3: primer set (psaBF and psbAF); Lane 4, primer set (ycf20R and rps14R). The expected sizes of PCR products in lanes 1 and 2 are 801 bp and 855 bp, respectively. The primers sequences used for PCR are follows. 5′-TATGTTTTAACTTATGCGGCATTCTT-3′ for psaBF; 5′-AACATTGAATTGCAAAAATGTTCC-3′ for ycf20R; 5′-CAACCGATGTATAAACGGTTTTCA-3′ for psbAF; 5′-TCTTCAAGGTCTTTTACCTGGT-3′ for rps14R. Total genomic DNA purified from ARM0029B was used for PCR reactions. PCR amplifications in only lanes 1 and 2 with the expected sizes indicating that a 15-kb gene cluster in the plastid genome of ArM0029B exists in the inverse orientation compared with the plastid genome of C. variabilis NC64A.
Conserved plastid genome gene clusters among trebouxiophycean algae. The arrangement of cp genes, including trnC to rpoC2 and rps14, are compared in ArM0029B and related species. The red-boldface indicates genes with a conserved order. The direction of the box arrow denotes sense orientation of transcription of the gene.
Secondary structure of mt trn genes in Chlorella sp. ArM0029B.
Single ML (A, HYK85 + G + I model) and NJ (B) trees from the DNA sequences of seven cp genes and a NJ tree (C) from translated amino acid sequences of seven mt protein-coding genes. */*: 100% bootstrap support/100% jackknife support. −/−: bootstrap and jackknife not supported.
Distance matrix of seven cp gene sequences in Trebouxiophyceae.