Summary
Leukemia stem cells (LSCs) represent a biologically distinct subpopulation of myeloid leukemias, with reduced cell cycle activity and increased resistance to therapeutic challenge. To better characterize key properties of LSCs, we employed a strategy based on identification of genes synergistically dysregulated by cooperating oncogenes. We hypothesized that such genes, termed “cooperation response genes” (CRGs), would represent regulators of LSC growth and survival. Using both a primary mouse model and human leukemia specimens, we show that CRGs are comprised of genes previously undescribed in leukemia pathogenesis in which multiple pathways modulate the biology of LSCs. In addition, our findings demonstrate that the CRG expression profile can be used as a drug discovery tool for identification of compounds that selectively target the LSC population. We conclude that CRG-based analyses provide a powerful means to characterize the basic biology of LSCs as well as to identify improved methods for therapeutic targeting.
Introduction
Acute myeloid leukemia (AML) arises in hematopoietic tissues and manifests in the form of aberrant production and dissemination of immature white blood cells (Lowenberg et al., 1999). Although many therapeutic strategies have been tested, AML is generally very difficult to treat and the majority of patients ultimately succumb to disease (Burnett et al., 2011; Estey and Dohner, 2006; Lowenberg et al., 1999). While the reasons behind the failure of conventional therapy are multifaceted, one aspect of the problem appears to derive from the cellular and molecular heterogeneity that is intrinsic to leukemic disease. Indeed, primary leukemia specimens are clearly complex populations, which in at least some cases bear remnants of the developmental structure found in normal hematopoietic tissues (Bonnet and Dick, 1997; Hope et al., 2004). Hence, leukemic stem cells (LSCs) are thought to reside at the apex of a hierachical development process in which the majority of tumor cells arise from a biologically distinct LSC population. This view of leukemia pathology suggests that targeting of the LSC compartment will be critical in order to achieve lasting elimination of the disease. Notably, empirical evidence clearly suggests that LSCs are relatively quiescent and resistant to conventional chemotherapy (Costello et al., 2000; Guzman et al., 2001), thereby supporting the concept that LSCs are biologically distinct from bulk tumor cells and indicating that eradication of the malignant stem cell compartment requires the development of new therapeutic strategies.
Due to the clinical importance of targeting LSCs, investigation of the mechanisms that control their growth and survival has been an active area of research in recent years. Using mouse models of AML, several reports have described specific genes and pathways that are selectively utilized by LSCs. For example, in leukemias initiated by MLL translocations, dysregulation of β-catenin (Wang et al., 2010), GSK3-β, and Meis1 (Wong et al., 2007) have been shown to be important for survival of LSCs. Similarly, Musashi-2 was recently shown to play a key role in the pathogenesis of LSCs in BCR-ABL mediated blast crisis (Ito et al., 2010), and PML-RAR-alpha (Nasr et al., 2008) was demonstrated to have a central function in a model of acute promyelocytic leukemia. Despite these findings, the overall gamut of pathways most central to the biology of LSCs, particularly as they relate to therapeutic targeting, remains largely unknown, not least because drivers of malignant cell transformation downstream of oncogenic mutations have been very difficult to identify.
A general feature of cancers, including leukemia, is the requirement of multiple cooperating oncogenic mutations for malignant cell transformation, as for example the manifestation of acute malignancy as observed in blast crisis CML. Moreover, recent evidence from a variety of cell types shows that the process of onogene cooperation requires synergistic regulation of downstream targets at multiple levels including gene regulation (Lloyd et al., 1997; Sewing et al., 1997; Xia and Land, 2007). Notably, such targets, i.e. their corresponding genes, have been demonstrated to be essential for the cancer phenotype with high frequency and have been termed “cooperation response genes” or CRGs (McMurray et al., 2008). The approach of exploiting the consequences of the genetic interactions between oncogenes to identify CRGs thus appeared as an attractive rationale for discovery of critical pathways regulating LSC biology. In fact, McMurray et al. demonstrated in a colon cancer cell model, using activated Ras and mutant p53, that CRGs regulate a wide array of cellular processes, and included many genes not previously associated with malignancy. Thus, CRG analysis provides a powerful and relatively unbiased means by which to identify genes that contribute to oncogenic transformation.
In the present studies, we employed the CRG strategy in the context of a mouse model of blast crisis CML, a very aggressive form of acute leukemia that is usually refractory to conventional drug therapy. The system employs co-expression of the BCR-ABL and NUP98-HOXA9 translocation products, which yields a cooperative oncogenic transformation event and a well-characterized model of human blast crisis CML (Dash et al., 2002; Neering et al., 2007). Our data show that CRGs are readily evident in this system and that many of these genes modulate the in vivo growth of primitive leukemia cells. In addition, we show that the expression signature of leukemia CRGs can be utilized as a drug discovery tool to identify pharmacological agents with selective anti-leukemia activity. Finally, we extend our findings to patient-derived specimens and demonstrate a similar role for CRGs in the biology of primary human leukemic cell types. Taken together, these findings indicate that analysis of oncogene cooperation provides important insights into the biology and potential treatment of leukemic disease.
Results
Identification of cooperation response genes in primitive leukemia cells
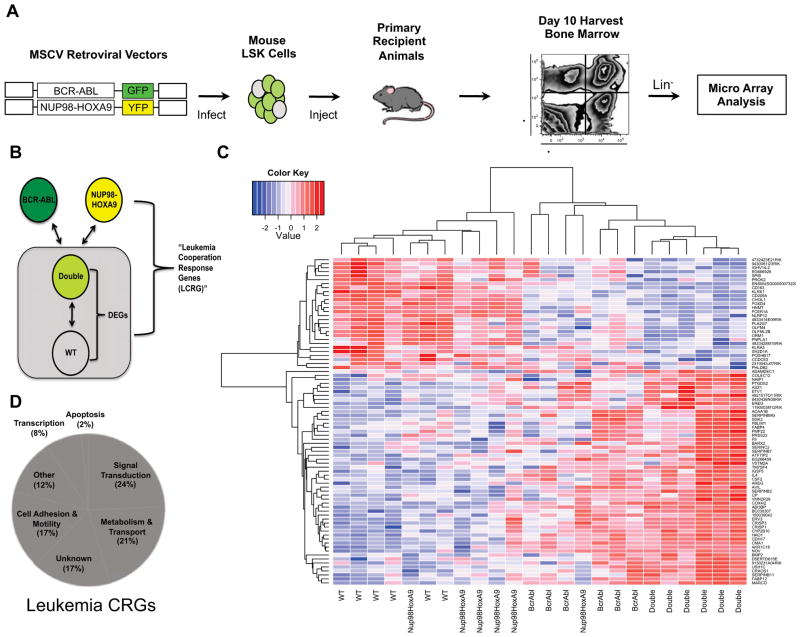
To identify leukemia cooperation response genes (CRGs), we employed a mouse model of blast crisis CML that has been shown to mimic human disease (Dash et al., 2002), and in which we have previously identified and characterized a discrete leukemia stem cell (LSC) population (Neering et al., 2007). To establish the model, hematopoietic stem cell (HSC) enriched bone marrow cells were isolated from C57Bl/6-CD45.1 congenic mice and infected with two independent retroviral vectors encoding the human translocation products BCR-ABL and NUP98-HOXA9 (Figure 1A). Each retroviral vector also expressed a distinct fluorescent protein reporter (GFP or YFP) to delineate cells transduced with one or both vectors (Neering et al., 2007). Upon transplantation into C57Bl/6-CD45.2 recipient animals, transduced cells rapidly engraft and proliferate with overt disease evident in 10–12 days. The use of two independent vectors for the initial infections results in cell populations expressing one or both vectors. These populations can be easily distinguished and isolated by flow cytometric sorting (Figure 1A). Cells representing each of the four possible genotypes (no oncogene, BCR-ABL alone, NUP98-HOXA9 alone, or both oncogenes) were sorted using a standard lineage marker panel (lin) to isolate the immature lin− compartment, as well as the CD45.1 congenic marker to ensure a pure population of donor cells. The generation and isolation of each population was repeated in six independent experiments. To identify synergistically regulated genes, genome-wide transcriptional profiling was performed on sorted cells using the Affymetrix mouse 430 2.0 microarray platform. Genes were first filtered based on differential expression between fully transformed (i.e. BCR-ABL + NUP98-HOXA9) and normal cells using the Benjamini-Hochberg correction procedure with a p-value cutoff of 0.01. This analysis yielded approximately 3500 differentially expressed genes (DEGs) representing the overall transcriptional dysregulation in primitive leukemia compared to normal cells. Next we applied a synergy filter to identify genes cooperatively regulated by BCR-ABL and NUP98-HOXA9. This was accomplished by comparing the level of gene expression in the primitive leukemia cells (BCR-ABL + NUP98-HOXA9) to that of primitive hematopoietic cells expressing each translocation alone (Figure 1B). Synergy was defined as a greater than additive fold change in gene regulation upon expression of both oncogenes in the same cell (McMurray et al., 2008). By applying the synergy criteria to the 3500 DEGs, a cooperativity response gene-set was identified in primitive leukemia. After removal of Riken cDNA and ESTs, the gene-set was comprised of 72 genes (Table S1), termed the leukemia cooperation response genes or CRGs (Figure 1C). Moreover the expression of each gene in the CRG signature was uniquely modulated in fully transformed cells (ie. BCR-ABL + NUP98-HOXA9) compared to cells bearing the single translocations (Figure 1C). To corroborate the data obtained from microarray analyses, the results were validated by quantitative real-time PCR (qRT-PCR), using freshly generated primitive (lin−) leukemic and normal bone marrow cells. Total RNA was analyzed by qRT-PCR using Taqman Low Density Arrays (TLDA), custom designed to detect expression of CRGs. A total of four independent biological replicates were evaluated. Relative gene expression was determined in primitive leukemia by comparison to paired normal controls. The analysis revealed that over 95% of CRGs identified by microarray showed a similar differential expression when assayed by TLDA (Figure S1).
Figure 1. Determining the leukemia CRG profile.
(A) Retroviruses encoding BCR-ABL and NUP98-HOXA9 were used to infect primitive hematopoietic bone marrow cells (lineage−, Sca-1+, c-kit+, termed “LSK”). Infected LSK cells were transplanted into sub-lethally irradiated mice and primitive bone marrow cells from day 10 leukemic animals were purified based on lack of expression of a standard panel of lineage markers (Ter119, B220, Gr1, and CD3e) and expression of GFP and YFP, which indicates expression of BCR-ABL or NUP98-HOXA9 respectively. Four separate bone marrow populations were purified: GFP+/YFP− (BCR-ABL alone), GFP−/YFP+ (NUP98-HOXA9 alone), GFP+/YFP+ (BCR-ABL + NUP98-HOXA9), and GFP−/YFP− (normal). RNA was isolated from each cell population and genome-wide microarray analysis performed using the Affymetrix mouse 430 2.0 array platform. (B) Gene expression analysis scheme. CRGs were identified by first determining the differentially expressed genes (DEGs) in primitive acute leukemia samples (Double = GFP+/YFP+) relative to normal cells lacking BCR-ABL and NUP98-HOXA9 expression (WT). Next, expression of DEGs was compared to cells expressing BCR-ABL or NUP98-HOXA9 alone. Leukemia CRGs (CRGs) were defined as DEGs showing differential gene expression in comparison to both BCR-ABL and NUP98-HOXA expressing cells (see methods for details). (C) Hierarchical clustering analysis of the genes representing the CRG signature in normal (WT), BCR-ABL, NUP98-HOXA9, and BCR-ABL + NUP98-HOXA9 (Double) cells from all 6 replicates. (D) Analysis of gene classes represented by CRGs. See also Figure S1 and Table S1.
Given that most of the genes identified were not previously known to be associated with blast crisis leukemia, we performed a pathway analysis and gene ontology annotation of the 72 CRGs to characterize the various cellular processes implicated. Ingenuity Pathway Analysis (IPA) revealed that the CRGs were present in a diverse set of biological functions such as signal transduction, metabolism, and cell adhesion (Figure 1D).
The CRG expression pattern is conserved amongst varying leukemic cell types
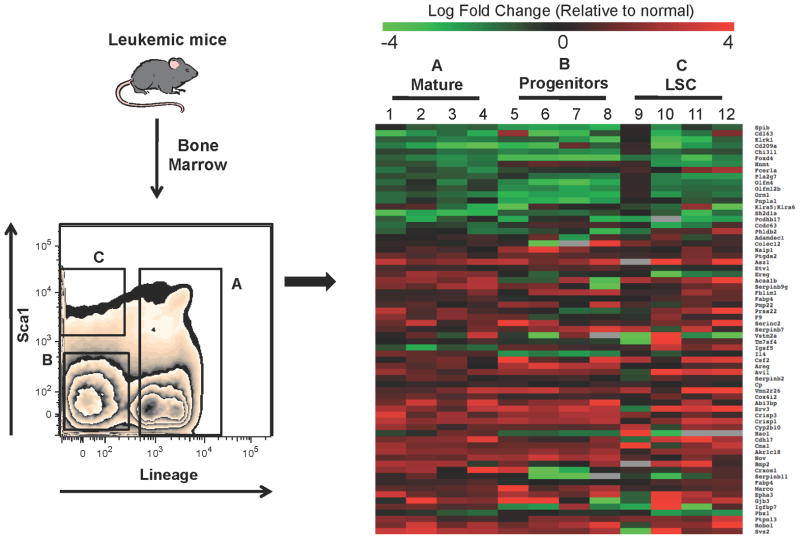
A significant challenge in understanding the biology of leukemia cells is the well-known heterogeneity found in primary malignant populations (Hope et al., 2004). Indeed, multiple discrete subpopulations are commonly evident, including the LSC compartment. While elucidating factors that control the most primitive cells is a high priority, understanding the biology of all leukemic cells is also important, since fundamental properties such as disease dissemination, growth control, drug resistance, etc. are likely to be shared amongst many cell types and are highly relevant to disease pathology. Thus, we sought to determine whether or not the CRG expression profile derived from lin− leukemia cells was conserved in more primitive as well as more mature cell types. As shown in Figure 2, we defined three distinct subpopulations based on the expression of lineage markers (lin) as well as the stem cell marker Sca-1. We have previously shown that Lin−/Sca+ leukemia cells are highly enriched for LSC content, while lin+ leukemia cells are completely devoid of LSCs (Neering et al., 2007). Thus, these two populations were isolated by flow cytometric sorting, along with the lin−/Sca1− compartment, which is enriched for leukemic progenitor cells. Total RNA was isolated from each sorted specimen, and subsequently interrogated using custom designed TLDA cards to evaluate the expression of the CRGs. A total of four independent biological replicates using paired leukemic and normal cell populations were analyzed. Intriguingly, there was a strong conservation in relative expression in the majority of leukemic populations tested (Figure 2). Over 85% of the CRG signature was maintained across all three cell populations (mature, progenitor, and LSC-enriched). The conservation of the signature across varying leukemic cell types suggests that despite readily evident cellular heterogeneity, many of the core properties that arise from oncogene cooperativity events may be conserved broadly in leukemic populations. This observation, along with the differential expression of CRGs in leukemia cells (vs. normal), may have important ramifications for the development of improved therapies.
Figure 2. Expression of leukemia CRGs is conserved in mature and primitive populations.
Leukemic animals were generated as described for Figure 1. Bone marrow was harvested from 4 independent groups of mice (n = 5 per group) and purified based on expression of BCR-ABL and NUP98-HOXA9 using GFP and YFP markers, respectively. Normal and leukemic cells were further purified based on expression of lineage markers and the stem cell antigen Sca-1. As shown in the left panel, three distinct populations were isolated: lineage positive cells (population A), lineage negative progenitors (population B), and LSC-enriched, Lin−/Sca+ (population C). RNA was isolated from each purified population and analyzed using a custom Taqman Low Density Array (TLDA) designed to interrogate the CRGs. The right panel shows a heatmap of relative expression of the CRGs in mature, progenitor, and LSC-enriched leukemia cells compared to their respective normal counterparts (Green = down-regulated, Red = up-regulated).
Identification of CRGs that modulate leukemia growth in vivo
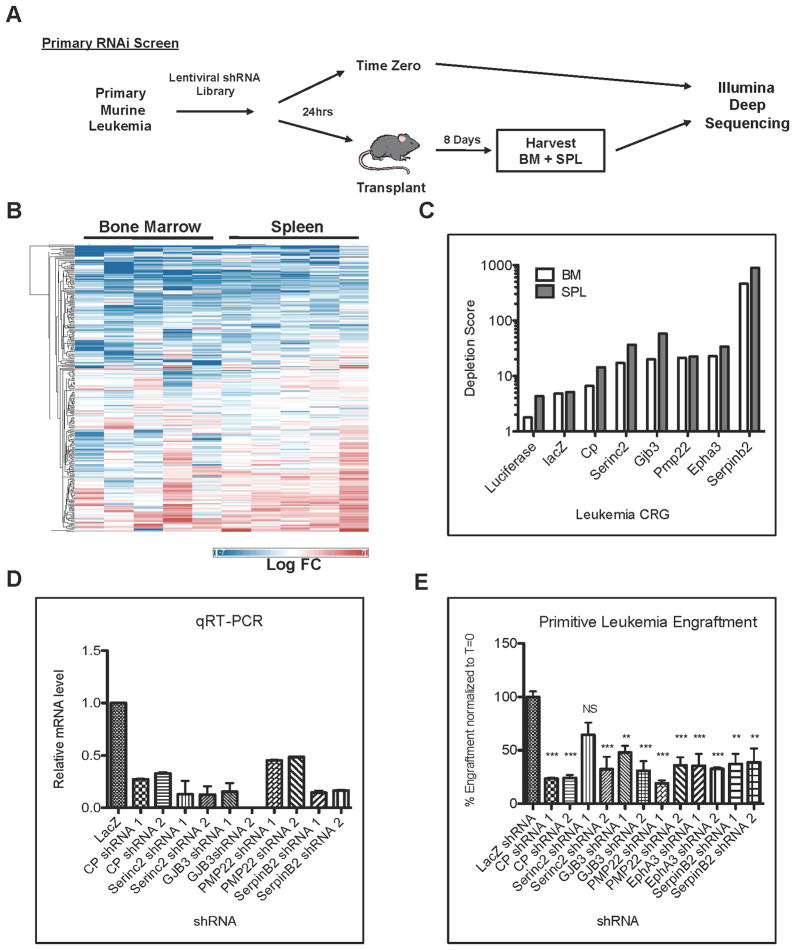
To test the hypothesis that genes comprising the CRG signature may have a functional role in the growth of leukemic cells, we designed an RNAi based approach as a means to investigate all up-regulated genes in the signature. Of the 72 genes in the CRG set, 50 were shown to be aberrantly up-regulated in leukemic cells and therefore represent potential targets for RNAi mediated knockdown (Figure S1). Primitive leukemia cells were purified from leukemic animals and transduced with a custom designed lentiviral RNAi library in which 5–10 individual shRNAs targeted each up-regulated CRG. Twenty-four hours later, infected leukemia cells were separated into two fractions. The first fraction was transplanted into recipient animals to determine the outcome of individual shRNA perturbation on leukemia growth in vivo. Genomic DNA was immediately isolated from the remaining cells to determine the time zero shRNA repertoire (Figure 3A). After eight days, bone marrow and spleen cells were harvested from recipient mice and genomic DNA was isolated. To determine the relative abundance of each shRNA before and after in vivo expansion, the DNA from all specimens was subjected to PCR-based amplification with primers flanking the shRNA sequence followed by XhoI restriction enzyme digestion to remove the hairpin structure and subsequently analyzed by Illumina deep sequencing. The entire experiment was repeated five times to ensure the validity of each gene under investigation. The fold change for each shRNA in the screen was determined relative to time zero as well as control shRNAs (luciferase, LacZ, and RFP) for both bone marrow and spleen samples (Figure 3B). Depletion was scored as a 5-fold or greater decrease in shRNA abundance relative to time zero. To minimize any background from potential off-target effects of the RNAi, only genes with 2 or more independent shRNAs achieving a 5-fold depletion were considered to be true “drop-outs” from the screen and thereby important for leukemia growth in vivo. In applying these criteria, we observed that 35 of the 50 targeted CRGs (i.e. 70%) were depleted from both the bone marrow and spleen samples (Figure S2). These findings indicate that a majority of CRGs represent genes of functional relevance to in vivo growth of primitive leukemia cells.
Figure 3. Identification of CRGs that regulate in vivo growth of leukemia cells.
(A) Primary leukemia cells were purified from leukemic mice based on lack of lineage marker expression (Ter119, B220, Gr1, and CD3e). Primitive leukemia cells were then infected with a custom designed lentiviral RNAi library targeting all up-regulated CRGs. Twenty-four hours later, infected cells were harvested and divided into two fractions. The first sample (time zero) was immediately processed to isolate genomic DNA and the second sample was transplanted into recipient animals. Five replicates were performed for each RNAi pool. Eight days post-transplant bone marrow and spleen cells were harvested from recipient mice and genomic DNA was isolated for Illumina Deep Sequencing. (B) Heatmap showing the fold change across all replicates for each shRNA in the screen relative to time zero for bone marrow and spleen samples (Blue = decreased, Red = increased). (C) Level of depletion determined for Luciferase, LacZ, CP, Serinc2, GJB3, PMP22, EphA3, and SerpinB2 shRNA from the RNAi screen shown in panel B. Next, primary leukemia progenitors were purified and infected with control or CRG shRNA lentiviruses and transplanted into recipient animals. Each CRG was targeted by 2 independent shRNA encoding lentiviruses (1, 2). (D) Gene mRNA knockdown was confirmed by qRT-PCR. (E) Ten days post-transplant leukemic bone marrow was analyzed for expression of crimson marked shRNA expressing cells by flow cytometry and engraftment was compared to time zero (T=0). Percent engraftment of LacZ control or Serinc2, CP, PMP22, EphA3, GJB3 shRNA expressing cells relative to time zero. (NS= not significant, *** p<0.001). See also Figure S2 and S3.
To corroborate the results from the RNAi screen, six individual CRGs were further characterized. The genes chosen - CP, Serinc2, GJB3, PMP22, EphA3 and SerpinB2 represent varying degrees of depletion observed from the drop out screen with CP representing the least depleted in comparison to control shRNAs and SerpinB2 representing the most depleted (Figure 3C). For these experiments, primitive leukemia cells were isolated by flow cytometry from primary animals and infected with individual shRNA lentiviruses specific for each gene or a LacZ shRNA control. The lentiviral vector used for these studies was designed to also encode the crimson fluorescent protein (CrFP), which permits identification and quantification of leukemic cells bearing the shRNA constructs (for example, see Figure S2). Twenty-four hours later, each sample was evaluated by flow cytometry to determine the percentage of CrFP+ cells (time zero), and then transplanted into recipient animals. Bone marrow was harvested from recipient animals ten days after transplant and the percentage of CrFP+ cells present in the leukemic graft was again determined by flow cytometry. As shown in Figure 3D, each of the six CRGs tested was targeted by two independent shRNA lentiviruses and knockdown of these genes led to reduced leukemia engraftment (Figure 3E). Thus, each of the genes evaluated contributes to some aspect of the in vivo behavior of primitive leukemia cells, e.g. engraftment, growth, survival, etc. To further investigate these genes, we performed parallel RNAi-mediated knock-down studies of all 6 CRGs in normal hematopoietic stem cells, and characterized the in vivo consequences using long-term bone marrow reconstitution assays. The results show that in most cases, knockdown of the CRGs had less effect in long-term engraftment of normal hematopoietic cells than their leukemia counterparts (Figure S3 and S4G).
SerpinB2 is an important regulator of LSC development and function
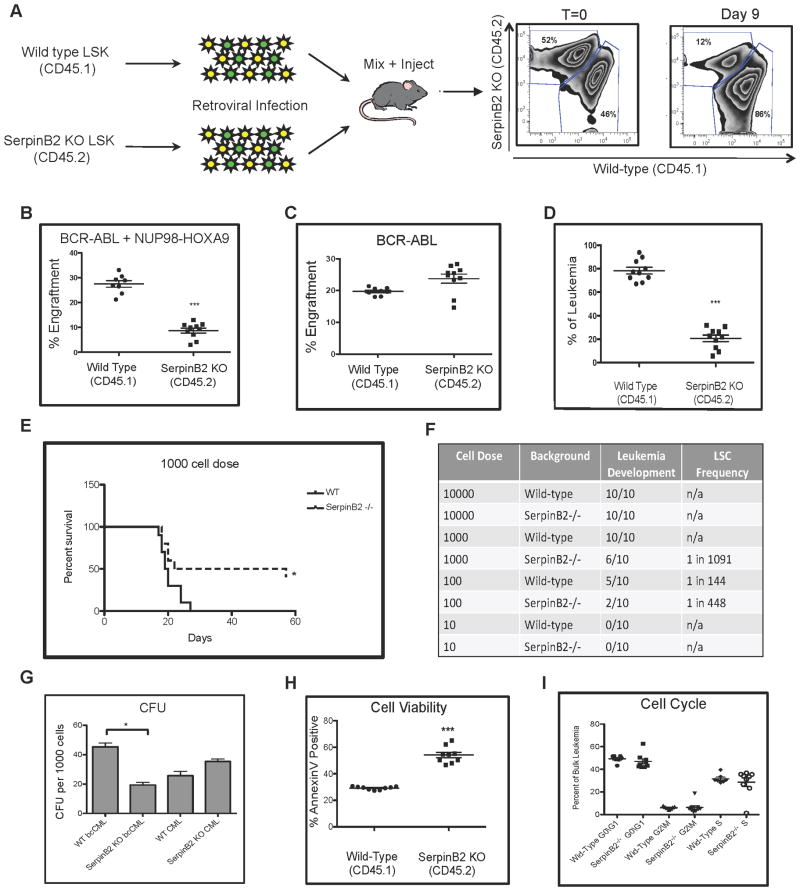
Given that SerpinB2 shRNAs induced the greatest degree of depletion in the RNAi screen, we chose to more thoroughly investigate the role of this gene in leukemia. As SerpinB2 deficient animals are available on a C57Bl/6 genetic background, we obtained SerpinB2−/− hematopoietic cells for experimental analysis. HSC-enriched cells from either SerpinB2−/− CD45.2 or wild type CD45.1 mice were infected with BCR-ABL and NUP98-HOXA9 as outlined previously (Figure 4A). Equal numbers of BCR-ABL/NUP98-HOXA9 transduced wild type or SerpinB2 KO LSK cells were transplanted into recipient animals and engraftment of wild type vs. SerpinB2 KO leukemia was determined at 9 days post transplant (Figure 4A). The analysis revealed a significant reduction of leukemia cells in bone marrow of mice transplanted with SerpinB2 KO leukemia cells compared to wild type (Figure 4B). Interestingly, engraftment of LSK cells expressing BCR-ABL alone (a model of chronic myeloid leukemia) was largely unaffected by loss of SerpinB2, with similar levels of engraftment observed between wild type and SerpinB2 KO leukemia cells (Figure 4C). This observation supports the role of SerpinB2 specifically to the cooperative gene interaction that leads to the blast crisis form of the disease. To rule out possible artifacts due to differential gene transfer efficiency, a competitive transplantation analysis of SerpinB2 KO vs. wild type leukemia cells was performed, where leukemic mice were first established with each genotype individually. Upon development of disease, primary leukemic bone marrow from each background was harvested, mixed together at equal ratios, and transplanted into secondary CD45.2 recipient mice. Nine days post transplant leukemic bone marrow was harvested from recipient animals and analyzed by flow cytometry using anti-CD45.1 and anti-CD45.2 antibodies. The relative engraftment of wild type versus SerpinB2−/− leukemia was compared to the input cell ratio (T=0). The data demonstrate that leukemia growth is significantly reduced is the absence of SerpinB2 expression (Figure 4D). We next performed limiting dilution transplantation experiments to determine the LSC frequency in the absence of SerpinB2. Those studies revealed that the LSC frequency in SerpinB2 knockout leukemia was reduced when compared to that of wild type leukemia (Figures 4E–F). Since loss of SerpinB2 may also generally impair the function of normal hematopoietic cells, we also performed control experiments in which naïve cells (i.e. non-leukemic) from wild type vs. SerpinB2−/− donors were evaluated by competitive repopulation. As shown in Figure S4, no significant changes in hematopoietic lineages, engraftment, or LSK cell frequency were evident for normal SerpinB2−/− cells, thereby further supporting a leukemia specific function for this gene. As complete loss of SerpinB2 may represent too severe of a perturbation to evaluate its role in leukemia, the phenotype was validated using two independent shRNAs to suppress but not completely eliminate SerpinB2 expression (Figure S4). The results were virtually identical to the data obtained using SerpinB2−/− cells. Taken together, the findings above demonstrate that the role of SerpinB2 is specific to leukemic populations, and that the gene plays a previously unrecognized role in growth of LSCs in vivo.
Figure 4. SerpinB2 modulates LSC activity in vivo.
(A) Primary blast crisis leukemia was generated using HSC-enriched bone marrow cells from either wild type or SerpinB2 knockout mice by co-expression of BCR-ABL and NUP98-HOXA9 as previously described. (B–C) Bone marrow from primary leukemic animals was analyzed for engraftment of cells expressing both BCR-ABL and NUP98-HOXA9, or BCR-ABL alone, using flow cytometry (*** p<0.001). (D) Primary leukemia bone marrow from wild type and SerpinB2 KO backgrounds were harvested, mixed at equal ratios, and transplanted into secondary recipient animals. Nine days post-transplant leukemic bone marrow was analyzed for engraftment of donor leukemia cells using flow cytometry and compared to time zero (T=0) (Top) (*** p<0.001). (E) Kaplan-Meier survival analysis of recipient mice transplanted with 1000 total leukemia cells from either wild type or SerpinB2 knockout primary leukemia (* p<0.01). (F) Limiting dilution results for LSC frequency between wild type and SerpinB2 knockout leukemias determined by L-CALC (Stem Cell Technologies), n=10. (G) Colony-forming ability of primary wild type or SeprinB2 KO bcCML (GFP+/YFP+) and CML(GFP+) cells (* p<0.01). (H) Percentage of apoptotic wild type or SeprinB2 KO leukemia cells analyzed by Annexin V staining (*** p<0.001). (I) Cell cycle analysis performed on primary wild type or SerpinB2 KO leukemia, n=10. See also Figure S4.
To further explore the biology underlying SerpinB2-related phenotype, we performed a series of additional experiments. First, to examine progenitor activity, the in vitro colony-forming ability of leukemia cells with or without SerpinB2 expression was analyzed. Primary wild type or SerpinB2 null leukemia cells were purified by flow cytometric cell sorting and seeded in methylcellulose cultures at varying cell doses. After seven days of culture, colonies comprised of >50 cells were scored as colony-forming units (CFUs). As shown in Figure 4G, the colony-forming potential of SerpinB2−/− leukemia cells was significantly reduced compared to wild type controls. Next, as previous reports have suggested an anti-apoptotic role of SerpinB2 expression (Antalis et al., 1998; Dickinson et al., 1995; Park et al., 2005; Tonnetti et al., 2008), studies were conducted to investigate whether SerpinB2 confers a survival advantage to leukemia cells. As shown in Figure 4H, loss of SerpinB2 expression leads to an increase in Annexin V positive cells compared to wild type controls. Finally, cell cycle analysis showed no significant difference between wild type and SerpinB2 KO leukemia cells (Figure 4I), indicating that the increased frequency of apoptotic cells was not related to overt changes in proliferative capacity. Taken together, these data suggest that SerpinB2 mediates an interaction with the in vivo microenvironment that promotes survival of leukemic stem and progenitor cells.
Pharmacological antagonism of CRG expression results in selective inhibition of primitive leukemia cells
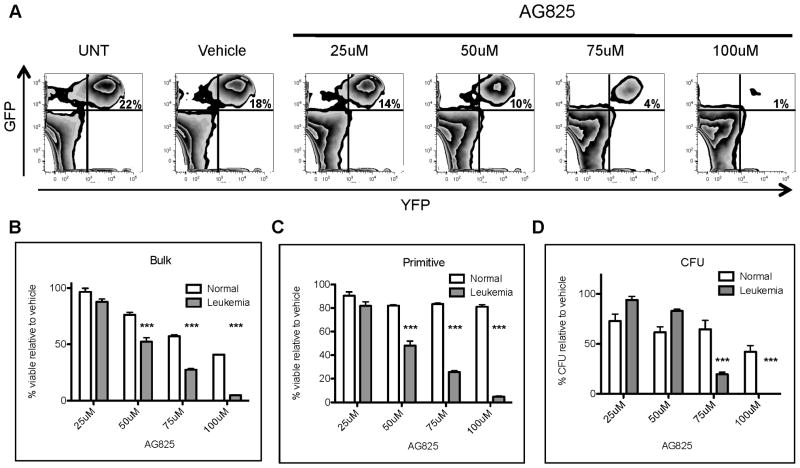
Given that specific targeting of individual CRGs resulted in inhibition of leukemic cell growth, we reasoned that simultaneous suppression of multiple CRGs could be an effective means of approaching anti-leukemia therapy. To test this concept using pharmacological agents we utilized the Broad Institutes’ Connectivity Map (CMAP) database (Lamb, 2007; Lamb et al., 2006) to identify compounds that reverse the aberrant expression of CRGs observed in leukemia cells. The CMAP database is comprised of a collection of gene expression profiles derived from small molecule compounds. The database can be queried with a gene expression signature of interest to identify those compounds that induce gene expression changes most similar and/or dissimilar to the query profile. Since we sought to reverse the gene expression pattern of genes driving pathogenesis (i.e. CRGs), we selected agents that induced expression changes most opposite to CRGs (termed “negative connectivity”). The results of this analysis are shown in Table 1, where the most negatively connected compound (defined by a maximum score of −1.0) is Tyrophostin AG825, a known ErbB2 inhibitor (Osherov et al., 1993). Based on the potential ability of this agent to suppress expression of CRGs, we hypothesized that AG825 would be selectively toxic to primary mouse leukemia cells. To test this hypothesis, primary normal and leukemia mouse bone marrow cells were treated in vitro with varying concentrations of AG825. Twenty-four hours later cell viability of total as well as more primitive cells (lin−, Sca1+) was measured using flow cytometric (Annexin V) analyses. In addition, progenitor function was assessed by methylcellulose colony forming assays (CFU). The results show that leukemia cells were eradicated by AG825 in a dose dependent fashion (Figure 5A). In contrast, comparative analyses of leukemia vs. normal cells at the bulk, primitive, and progenitor cell level demonstrate that AG825 is consistently more toxic to leukemic cell types (Figure 5B–D). Notably, normal primitive cells were only modestly affected by AG825, even at 100uM, a concentration that was highly cytotoxic to leukemia cells (Figure 5C). Further, leukemic progenitor function was completely ablated at the highest concentrations of AG825 while normal progenitors were only modestly affected (Figure 5D).
Table 1. Connectivity Map analysis of leukemia CRGs.
The 72 gene LCRG signature was used to query the Broad Institute’s Connectivity Map (CMAP). The top 49 negatively correlated compounds are shown.
| Compound | Cell Type Assayed | Array | Connectivity Score (CS) |
|---|---|---|---|
| tyrphostin AG-825 | MCF7 | u133AofAAv2 | −1 |
| 4-hydroxy-2-nonenal | RKO | hgu133plus2.mas5 | −0.944695822 |
| rosiglitazone | HL60 | hgu133a | −0.849921146 |
| yohimbine | MCF7 | u133AofAAv2 | −0.833242887 |
| deferoxamine | PC3 | hgu133a | −0.810877648 |
| staurosporine | MCF7 | hgu133a | −0.784517747 |
| LY-294002 | MCF7 | u133AofAAv2 | −0.779534308 |
| thioridazine | MCF7 | hgu133a | −0.739939595 |
| exisulind | MCF7 | hgu133a | −0.736585969 |
| 4-hydroxy-2-nonenal | RKO | hgu133plus2.mas5 | −0.723096655 |
| cobalt chloride | MCF7 | hgu133a | −0.713366654 |
| NU-1025 | MCF7 | hgu133a | −0.698630633 |
| minocycline | MCF7 | u133AofAAv2 | −0.687270265 |
| LY-294002 | MCF7 | u133AofAAv2 | −0.670194206 |
| clozapine | MCF7 | hgu133a | −0.663687044 |
| 5224221 | MCF7 | u133AofAAv2 | −0.656678467 |
| trifluoperazine | MCF7 | hgu133a | −0.643054546 |
| carbamazepine | MCF7 | u133AofAAv2 | −0.632176605 |
| 5248896 | MCF7 | u133AofAAv2 | −0.631981557 |
| Ara-C | primaryAML1 | hgu133plus2 | −0.621453076 |
| TTNPB | MCF7 | hgu133a | −0.605754727 |
| 5252917 | MCF7 | u133AofAAv2 | −0.600199581 |
| fulvestrant | MCF7 | u133AofAAv2 | −0.596498871 |
| 5182598 | MCF7 | u133AofAAv2 | −0.569870646 |
| estradiol | MCF7 | u133AofAAv2 | −0.563035483 |
| doxycycline | MCF7 | u133AofAAv2 | −0.535758294 |
| trichostatin A | MCF7 | u133AofAAv2 | −0.531625057 |
| phenanthridinone | MCF7 | u133AofAAv2 | −0.525721398 |
| W-13 | MCF7 | hgu133a | −0.524775982 |
| ikarugamycin | MCF7 | u133AofAAv2 | −0.521146751 |
| raloxifene | MCF7 | hgu133a | −0.517786179 |
| tretinoin | HL60 | hgu133a | −0.516975031 |
| prazosin | MCF7 | u133AofAAv2 | −0.506939292 |
| phenyl biguanide | MCF7 | hgu133a | −0.49527295 |
| valproic acid | MCF7 | u133AofAAv2 | −0.493871119 |
| chlorpromazine | MCF7 | hgu133a | −0.487556832 |
| colchicine | MCF7 | hgu133a | −0.447879508 |
| monensin | MCF7 | u133AofAAv2 | −0.436030095 |
| genistein | PC3 | hgu133a | −0.430373293 |
| TTNPB | PC3 | hgu133a | −0.430033398 |
| tioguanine | MCF7 | hgu133a | −0.421921152 |
| trichostatin A | MCF7 | u133AofAAv2 | −0.418453342 |
| staurosporine | MCF7 | hgu133a | −0.409294748 |
| LY-294002 | SKMEL5 | hgu133a | −0.40882847 |
| phenformin | MCF7 | hgu133a | −0.396945788 |
| haloperidol | MCF7 | u133AofAAv2 | −0.372750449 |
| 17-allylamino-geldanamycin | SKMEL5 | hgu133a | −0.366981005 |
| valproic acid | MCF7 | u133AofAAv2 | −0.360150576 |
| fulvestrant | MCF7 | hgu133a | −0.315383178 |
Figure 5. Tyrophostin AG825 is selectively cytotoxic to primitive murine leukemia cells.
(A) Leukemic bone marrow cells were treated overnight with increasing concentrations of Tyrophostin AG825 (AG825). The following day, cells were harvested and analyzed by flow cytometry to determine relative toxicity to leukemic (GFP+/YFP+) vs. normal cells (GFP−/YFP−). (B–C) Normal or leukemia cells treated with AG825 overnight were harvested and labeled with annexin V and DAPI to analyze cell death using flow cytometry. Viability of bulk and primitive cells (Lin−) exposed to AG825 are shown. (D) Normal or leukemic cells were treated with AG825 overnight followed by methylcellulose culture to measure colony formation ability, CFU (*** p< 0.001). See also Figure S5 and S6.
In addition to AG825, we evaluated the activity of 4-hydroxynonenol (4HNE), a naturally-occurring lipid peroxidation product which was also predicted by CMAP analysis to be a potent modulator of the leukemic CRG signature (Table 1). Notably, in previous independent studies of human leukemia specimens we had identified this compound to possess anti-LSC specific properties (Hassane et al., 2008). As observed with AG825, both primary bulk and primitive leukemia cells were highly sensitive to 4HNE at twenty-four hours (Figure S5). In contrast, normal bulk and primitive cells were largely unaffected by the drug with toxicity only observed at the highest concentrations. Similar results were observed with Staurosporine, another agent predicted from the CMAP analysis, on murine hematopoietic cells (Figure S5). Moreover, both AG825 and 4HNE were able to effectively reverse the CRG expression signature in murine bcCML cells (Figure S6). Taken together, these data indicate that the CRG signature can be used as a tool to predict pharmacological agents with selective toxicity to leukemia cells.
Expression and function of CRGs is conserved in human leukemia
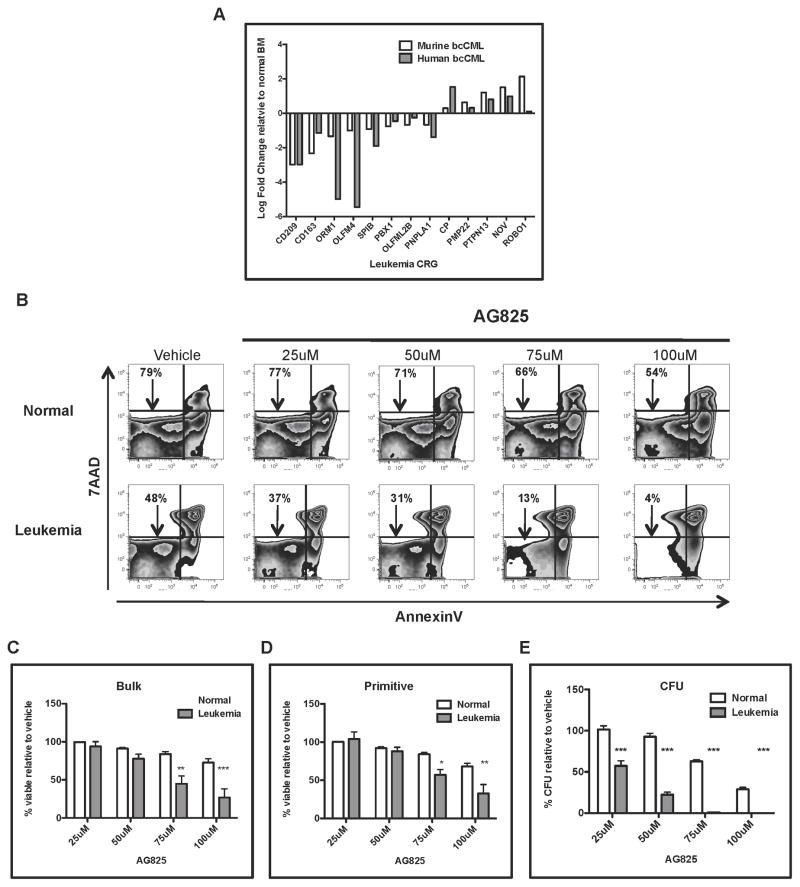
Finally, to investigate the relevance of the CRGs in human leukemia, we analyzed their expression in a panel of eight independent BCR-ABL+ blast crisis leukemia patient specimens and eight normal bone marrow control specimens. In human bone marrow, stem and progenitor cells are found within the CD34+ population, making this subpopulation roughly equivalent to murine Sca+/Lin− cells. For relative expression analyses, CD34+ cells were isolated from each leukemic or normal bone specimen using flow cytometric cell sorting. Total RNA was isolated from each specimen and analyzed by qRT-PCR using a custom TLDA containing all known human orthologs of the mouse CRGs. Of the 72 CRGs identified in mouse tissue, only 57 could be interrogated on the TLDA due to lack of a clear human counterpart. Of the 57 genes analyzed, 33 were evaluable based on successful amplification. Of these genes, analysis revealed 13 whose relative expression in comparing normal vs. leukemic cells was similar in both murine and human specimens (Figure 6A). Analysis using the Ingenuity Pathway Analysis (IPA) bioinformatics resource identified, cellular development, growth, and proliferation pathways among the known cellular functions (Table S2). The conserved 13 gene signature was then used to re-query the CMAP database for potential inhibitory compounds. As observed for murine cells, Tyrophostin AG825 and 4-HNE were identified as negatively connected compounds. To test the biological activity of AG825, leukemic patient samples (blast crisis CML) vs. normal bone marrow specimens were treated for twenty-four hours with varying concentrations followed by analysis of cell viability and colony formation ability. As observed with AG825 treatment of murine leukemia, human leukemia samples appeared to be more sensitive to the compound than normal controls (Figure 6B). Further, striking differential sensitivity to AG825 was observed for progenitor cells (denoted CFU, Figure 6E), and was also evident for bulk and phenotypically primitive populations (Figures 6C–D). Biological testing of 4HNE also demonstrated strong leukemia-specific cytotoxicity, particularly for colony-forming ability (Figure S7). Taken together, these data suggest the importance of the CRGs for growth and survival of human leukemia cells.
Figure 6. Expression and function of leukemia CRGs is conserved in human blast crisis leukemia specimens.
(A) Total RNA was isolated from CD34-enriched human or primitive murine leukemia cells and analyzed by qRT-PCR using custom Taqman Low Density Arrays (TLDA) designed to interrogate the CRGs. The relative expression in leukemia compared to normal counterparts was determined (n=8 human, n= 6 mouse). The 13 genes conserved between the two species and their relative expression is shown. (B) Normal or leukemic human specimens were treated overnight with increasing concentrations of Tyrophostin AG825 (AG825). Twenty-four hours later the cells were harvested and labeled with annexin V and DAPI for analysis of cell death using flow cytometry. (C–D) Viability of bulk and primitive (lin−) human cells treated with AG825 for twenty-four hours. (E) Normal or leukemic human cells were treated with AG825 overnight followed by methylcellulose culture to measure colony formation ability, CFU (* p<0.01), ** p< 0.01, *** p< 0.001). See also Figure S7 and Tables S2.
Discussion
Elucidating the genetic events that mediate leukemia transformation is an important step towards identifying the overall mechanisms regulating leukemia growth and survival. In the present study, we used a murine model of acute leukemia to identify genes cooperatively regulated by BCR-ABL and NUP98-HOXA9 that control in vivo growth of primitive leukemia. We demonstrated that knockdown of expression in a majority of up-regulated CRGs reduced leukemia growth upon transplantation into recipient mice. Further, we showed that the CRG expression signature was conserved in subpopulations enriched for a broad spectrum of cell types, ranging from relatively mature (lin+) to primitive (lin−/sca+). In addition, using in silico strategies, we employed the CRG profile to identify pharmacological compounds that were selectively cytotoxic to both murine and human leukemia cells. Taken together, these data identify a novel set of genes that regulate the in vivo growth and survival of primitive leukemia cells, thereby providing a foundation for future research toward elucidating central properties of LSCs.
By integrating analysis of oncogene cooperation with stem cell biology, we have begun to elucidate gene regulatory architecture underlying LSC behavior, as well as that of diverse tumor cell populations in general. The basis for the genetic studies reported here is the CRG concept that was first demonstrated in a model in which mutant Ras and p53 cooperatively mediate oncogenic transformation of colonic crypt cells (McMurray et al., 2008). Notably, these studies essentially defined “oncogene cooperativity” in molecular terms, demonstrating that the complex process of malignant cell transformation largely depends on the expression of downstream genes responding synergistically to regulation by multiple oncogenic mutations. In the present study, we demonstrate the relevance of the previously described molecular architecture of oncogene cooperation in leukemia, where CRGs were readily identified. This observation is a conceptual departure from the interpretation that leukemia arises via a “two-hit” model in which one oncogene provides a mitogenic signal (e.g. BCR/ABL) and a second oncogene serves to increase self-renewal (e.g. Nup98/HoxA9), thus assuming independence or additive behavior of the oncogene functions involved. In contrast, our data support the view that leukemogenesis is driven by highly cooperative processes yielding acute malignancy. We thus conclude that oncogene cooperativity, as defined by synergistic modulation of downstream targets, is indeed an inherent feature of leukemia biology.
The use of a primary leukemia model to study the role of CRGs has provided several novel observations. First, we were able to evaluate the expression and activity of CRGs in multiple stages of leukemia development. As reported previously, the BCR/ABL + NUP98/HOXA9 model shows considerable cellular heterogeneity, with a large proportion of cells displaying at least partial differentiation (as indicated by expression of various markers associated with mature lineages), but with several other distinct subpopulations more closely associated with stem or progenitor populations (as indicated by expression of markers such as c-kit, sca-1, CD150, Flt3, etc) (Neering et al., 2007). Notably, the expression of CRGs was well conserved across a broad spectrum of stages ranging from mature to primitive. Thus, we propose that by designing therapeutic strategies based on pathways identified by CRG analysis, it should be possible to create regimens that are potentially effective against both bulk tumor as well as more primitive stem and progenitor populations. From a conceptual standpoint, simultaneous and comprehensive targeting of heterogeneous cell populations (including the LSC compartment) represents an attractive and novel approach to improving leukemia therapy. A second notable aspect of the present CRG studies is the use of a targeted shRNA library for in vivo identification of genes that are directly relevant to leukemia pathogenesis. Using a genetic drop out screen strategy, in a single experiment we were able to simultaneously evaluate the role of aberrantly up-regulated CRGs. These studies suggest that CRGs are relevant to in vivo growth of leukemic cells at high frequency, an observation further confirmed by detailed analyses of six individual genes.
In terms of specific genes, these studies identified several new regulators of leukemia pathogenesis. Notably, genes in the leukemia CRG profile were generated from freshly isolated primary cells, thereby capturing expression data indicative of native in vivo signals. Subsequent analysis using the lentiviral RNAi library demonstrated almost no functional role for most of the CRGs for cells cultured in vitro over the span of eight days, which was in striking contrast to the in vivo studies (Data not shown). This observation suggests that the CRGs represent genes important for the development and survival of leukemia cells in their in vivo microenvironment. Interestingly, the genes chosen for independent validation; SerpinB2, GJB3, PMP22, Serinc2, CP, and EphA3, are all implicated in extracellular or cellular communication activity. For example, GJB3 is a gap junction protein whose cellular role is thought to be involved in cell-to-cell communication. Studies suggest that GJB3 binds to another gap junction protein, GJA1, to form a heterodimeric junction complex in ES cells (Worsdorfer et al., 2008). Moreover, GJA1 (Connexin 43) has been reported to be important for support of hematopoietic stem and progenitor cell development and survival (Cancelas et al., 2000). Our data demonstrates that knockdown of GJB3 in primitive leukemia reduces growth in vivo, suggesting that GJB3 is an important regulatory signal for pathogenesis. Similarly, both CP and SerpinB2 have extracellular functions. CP acts to maintain proper cellular iron homeostasis by influencing iron transport. Previous studies have shown that in certain cancer types iron import is elevated (Brookes et al., 2006), possibly in response to a greater need for metabolic processes. Extracellular SerpinB2 is thought to inhibit the urokinase plasminogen activator (UPA); and thereby prevent plasminogen activation, however, the actual role SerpinB2 in the plasminogen response is not well understood. Studies in endometrial cancer have reported that SerpinB2 expression is correlated with increased invasiveness and decreased long-term patient survival (Gleeson et al., 1992; Nordengren et al., 2002). Our results demonstrating that SerpinB2 expression is important for leukemia growth in vivo suggest that its activity could protect leukemia cells from apoptotic cells death in vivo. Our data further suggests that SerpinB2 is an important regulator of LSC development, in that loss of expression reduces the LSC frequency in the developing leukemia. EphA3 is a receptor tyrosine kinase that activates a signal transduction cascade when secreted ephrin molecules bind its extracellular domain. Consequently, EphA3 may represent a sensor of secreted molecules promoting leukemia growth in the marrow microenvironment. Collectively, identification of these genes as regulators of leukemia growth in vivo provides further insight on the critical role of leukemia cell extrinsic signals and niche interactions.
In principle, the genetic data derived using the CRG methodology should ultimately guide the development of pharmacological agents that selectively target leukemia cells. However, discerning how to link genomic and chemical interactions can be challenging and in many cases there are no well-defined strategies to achieve this goal. Hence, in the present studies we turned to the CMAP resource as a means to perform in silico chemo-genomic analyses of gene expression profiles derived from cooperation response studies. Our previous studies have demonstrated that both positive and negative connectivity with the CMAP can be used towards identification of agents that target leukemia stem cells (Hassane et al., 2008; Hassane et al., 2010). We adapted this approach to the expression profiles derived from CRG studies and successfully identified two agents, Tyrophostin AG825 and 4-hydroxynonenal (4HNE), that demonstrated clear activity towards primitive leukemia cells, with substantially less toxicity to normal cell types. Although AG825 was developed as an ErbB-2 inhibitor, we speculate that modulation of this pathway is not a central to leukemia-specific cell death, since subsequent experiments with the clinical ErbB-2 inhibitor Lapatinib demonstrated no difference in activity between normal and leukemia cells (data not shown). Thus, it will be of interest to further characterize the mechanism by which AG825 targets leukemia cells. Importantly, these data highlight the utility of using the CRG approach to identify compounds that would most likely never have been selected based solely on their documented mechanism of action.
While biological analyses of disease processes in mouse models can be very powerful, it is important to validate such approaches in human tissue as well. Hence, we evaluated patient-derived leukemia specimens and successfully identified human orthologs of CRGs in primary samples derived from CML blast crisis patients. Unlike the murine studies, each human specimen analyzed (8 total) was derived from an independent patient with a unique genetic background. Furthermore, while the specimens all possess the canonical BCR-ABL mutation, additional mutations resulting in blast crisis certainly differ in each patient. Thus, it is very unlikely that the murine CRG profile would be completely conserved in each human specimen. Rather, as shown, a subset of CRGs demonstrated similar changes in both murine and human leukemia cells. Importantly, the core subset of CRGs was still able to identify AG825 and 4HNE via CMAP analysis, an observation that strongly supports the functional relevance of genes conserved between murine and human tissues.
In conclusion, the findings presented in this report provide the first genetic analysis of cooperatively regulated genes in primitive leukemia cells. We report a subset of genes that influence multiple aspects of leukemia biology and the interaction of malignant cells with the surrounding microenvironment. The relevance of the CRG signature is supported by both genetic analysis of specific genes, as well as the fact that compounds with predicted inhibitory activity towards the CRG signature led to leukemia specific cytotoxicity in both mouse and human cells. These data provide intriguing insights into the synergistic nature of the LSC transformation process, and thus may yield novel strategies for effective rational drug design for the treatment of leukemia.
Experimental Methods
Generation of bcCML Mouse Model
The blast crisis CML mouse model was created as described previously (Neering et al., 2007). Briefly, six- to eight-week old naïve B6.SJL-Ptprca Pepcb/BoyJ mice were purchased from Jackson Laboratories (Bar Harbor, ME). Donor mice were sacrificed and bone marrow was harvested from the tibias and femurs. Mature lineage positive cells were depleted from the bone marrow cell suspension using the BD IMAG immunoaffinity kit (San Jose, CA) per manufacturers instructions. Purified populations of LSK cells were isolated using fluorescently conjugated antibodies specific for Sca1 and c-Kit and the BD FACS ARIA II cell sorter (San Jose, CA). Donor LSK cells were plated in one well of a 24-well cell culture plate coated with retronectin (Takara, Japan) at 1 x106 cells per ml and cultured overnight in IMDM (Invitrogen) media supplemented with 10% FBS (Invitrogen), rmIL3 (10ng/ml), rmIL6 (10ng/ml), rmFlt3 (25ng/ml), and rmSCF (50ng/ml) (Peprotech, Rocky Hill, NJ). The following day, LSK cells were infected with viral supernatant (resuspended in 50% viral supernatant and 50% 2x cytokine supplemented media) twice, once in the morning and again in the evening. The infections were repeated on day 3. On day 4, the cells were harvested and resuspended in cold PBS + 2% FBS and subsequently injected through the retro-orbital sinus into recipient sub-lethally irradiated (600 rads) six- to eight-week old C57Bl6J mice (Jackson Laboratories, Bar Harbor, ME). The LSK dose was 20,000 – 40,000 cells per mouse for all experiments and the transduction efficiencies were typically 10–15% for BCR-ABL + NUP98-HOXA9 infected cells and between 20 – 30% for BCR-ABL and NUP98-HOXA9 infected cells.
Microarray Gene Expression Analysis
Total RNA for sorted wild type cells or cells harboring BCR-ABL, NUP98-HOXA9, or BCR-ABL + NUP98-HOXA9 was harvested on the basis on fluorescent protein expression using BD FACS ARIA II flow cytometry sorter (San Jose, CA). RNA was labeled and amplified according to the NuGen procedure and hybridized to Affymetrix Mouse 430 2.0 gene expression microarrays (Affymetrix, Santa Clara, CA). Data analysis was performed using R and BioConductor (Gentleman et al., 2004). Microarray data was normalized using the dChip method of Li and Wong (Li and Wong, 2001) with mismatch probe correction. The data were log2-transformed and differentially expressed genes were identified using the limma (Smyth, 2004) with p-value correction performed according to the Benjamini-Hochberg procedure (Benjamini Y, 1995). Genes responding synergistically to the presence of both BCR-ABL and NUP98-HOXA9 only were selected according to the published cooperation response gene (CRG) identification procedure (McMurray et al., 2008). Briefly, if a corresponds to the mean expression for gene g in BCR-ABL+ cells, b corresponds to the mean expression value of g in NUP98-HOXA9+ cells, and d corresponds to the mean expression value of g in BCR-ABL+/NUP98-HOXA9+ cells, then a CRG is defined by the criterion:
The significance of the CRG scores was determined using a jackknife sub-sampling procedure to generate estimated CRG score p-values for each gene. Significant genes were selected for follow up analysis. The expression of CRGs in different cell subsets was visualized using average linkage hierarchical clustering using Pearson correlation as a distance measure. Data are deposited in Gene Expression Omnibus (GEO), accession number GSE37907.
Primary RNAi Library Screen
Primary mice were generated as described previously. Spleens from 10 primary mice were harvested and pooled into a single cell suspension following alkaline RBC lysis. Leukemic spleenocytes were depleted of mature lineage positive cells using the BD IMAG immunioaffinity kit and cells expressing only BCR-ABL (GFP) and NUP98-HOXA9 (YFP) were purified using the BD FACS ARIA II cell sorter. Immature leukemic (lineage negative) cells were plated in serum-free conditions at 2×106 cells per ml and cultured overnight. The following day 3×106 cells were infected with lentiviral RNAi viral pools (Broad Institute, Boston, MA) for six hours in 800ul total volume and 4ug/ml polybrene (Millipore, Billerica, MA). The MOI for each shRNA virus was held constant at ~0.1–0.2. Each pooled infection was performed five independent times to yield 5 replicates for each pool. Post infection, 1ml of fresh media was added to the infected cells and cultured overnight. The next day (~24hours) each sample was split: one-third was resuspended in 1ml cold PBS and stored at −20C for time zero; one-third was plated in 1ug/ml of puromycin (Invitrogen) to select for transduced cells; and the remaining one-third (~1×106 cells) was injected into one recipient mouse per replicate. Eight days later, the puromycin-selected cells were harvested, resuspend in 1ml cold PBS and stored at −20C. Bone marrow and spleens were harvested from all recipient mice and processed into single cell suspensions with subsequent resuspension in 1ml cold PBS and stored at −20C. Genomic DNA was isolated from all samples using QIAamp Blood Kits (Qiagen, Valencia, CA). DNA concentration and purity (A260/A280, A260/A230) was determined using the Nanodrop 1000 (Thermo Scientific). Sequencing analysis of shRNA representation is described in detail in supplemental methods.
Drug Treatments
Human or mouse cells were cultured in serum-free media (IMDM (Gibco) +, 20% BIT (BSA, insulin, transferrin; Stem Cell Technologies, no. 09500), LDL (EMD), B-ME (Gibco, Inc), Pen/strep (Gibco, Inc), and L-glutamine (Gibco, Inc)) for twenty-four hours in presence of Tyrphostin AG825 (Sigma, St. Louis, MO) at 25, 50, 75, and 100uM or 4-hydroxy-2-nonenal (EMD, Gibbstown, NJ) at 5, 10, 20, 30uM. The following day, cells were harvested and analyzed for cell death with AnnexinV/DAPI staining using the LSRII flow cytometer (BD, San Jose, CA) or plated in methylcellulose media (Stem Cell Technologies, Vancouver, Canada) for CFU assay.
Supplementary Material
Highlights.
Downstream targets of cooperating oncogenes are identified in leukemia cells
Novel genes that regulate the in vivo behavior of leukemia stem cells are described
Genes modulating in vivo biology of leukemia stem cells are used for drug discovery
Acknowledgments
The authors gratefully acknowledge Randall Rossi, Monica Guzman, and Mark Noble for many helpful discussions and critical evaluation of the work and manuscript. We also thank Timothy Bushnell and the URMC Flow Cytometry Core for help with cell sorting experiments and Chris Proschel for supplying the SerpinB2−/− animals. The research was supported by NIHRO1-CA122206, NIHRO1-CA138249, New York State Stem Cell Foundation grant C024964, Dept of Defense grant W81XWH-07-1-0601, and University of Rochester Hematology/Oncology Training Grant (T32-HL007152).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antalis TM, La Linn M, Donnan K, Mateo L, Gardner J, Dickinson JL, Buttigieg K, Suhrbier A. The serine proteinase inhibitor (serpin) plasminogen activation inhibitor type 2 protects against viral cytopathic effects by constitutive interferon alpha/beta priming. The Journal of experimental medicine. 1998;187:1799–1811. doi: 10.1084/jem.187.11.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini YHY. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. NatMed. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55:1449–1460. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett A, Wetzler M, Lowenberg B. Therapeutic Advances in Acute Myeloid Leukemia. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- Cancelas JA, Koevoet WL, de Koning AE, Mayen AE, Rombouts EJ, Ploemacher RE. Connexin-43 gap junctions are involved in multiconnexin-expressing stromal support of hemopoietic progenitors and stem cells. Blood. 2000;96:498–505. [PubMed] [Google Scholar]
- Costello RT, Mallet F, Gaugler B, Sainty D, Arnoulet C, Gastaut JA, Olive D. Human acute myeloid leukemia CD34+/CD38− progenitor cells have decreased sensitivity to chemotherapy and Fas-induced apoptosis, reduced immunogenicity, and impaired dendritic cell transformation capacities. Cancer Res. 2000;60:4403–4411. [PubMed] [Google Scholar]
- Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J, Housman D, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JL, Bates EJ, Ferrante A, Antalis TM. Plasminogen activator inhibitor type 2 inhibits tumor necrosis factor alpha-induced apoptosis. Evidence for an alternate biological function. The Journal of biological chemistry. 1995;270:27894–27904. doi: 10.1074/jbc.270.46.27894. [DOI] [PubMed] [Google Scholar]
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson N, Gonsalves R, Bonnar J. The plasminogen activator urokinase and its inhibitor PAI-2 in endometrial cancer. Gynecol Oncol. 1992;47:58–61. doi: 10.1016/0090-8258(92)90076-u. [DOI] [PubMed] [Google Scholar]
- Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, Luger SM, Jordan CT. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- Hassane DC, Guzman ML, Corbett C, Li X, Abboud R, Young F, Liesveld JL, Carroll M, Jordan CT. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008;111:5654–5662. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassane DC, Sen S, Minhajuddin M, Rossi RM, Corbett CA, Balys M, Wei L, Crooks PA, Guzman ML, Jordan CT. Chemical genomic screening reveals synergism between parthenolide and inhibitors of the PI-3 kinase and mTOR pathways. Blood. 2010;116:5983–5990. doi: 10.1182/blood-2010-04-278044. [DOI] [PubMed] [Google Scholar]
- Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC, Obermuller F, Staddon S, Barth CF, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr R, Guillemin MC, Ferhi O, Soilihi H, Peres L, Berthier C, Rousselot P, Robledo-Sarmiento M, Lallemand-Breitenbach V, Gourmel B, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14:1333–1342. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- Neering SJ, Bushnell T, Sozer S, Ashton J, Rossi RM, Wang PY, Bell DR, Heinrich D, Bottaro A, Jordan CT. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordengren J, Fredstorp Lidebring M, Bendahl PO, Brunner N, Ferno M, Hogberg T, Stephens RW, Willen R, Casslen B. High tumor tissue concentration of plasminogen activator inhibitor 2 (PAI-2) is an independent marker for shorter progression-free survival in patients with early stage endometrial cancer. Int J Cancer. 2002;97:379–385. doi: 10.1002/ijc.1611. [DOI] [PubMed] [Google Scholar]
- Osherov N, Gazit A, Gilon C, Levitzki A. Selective inhibition of the epidermal growth factor and HER2/neu receptors by tyrphostins. J Biol Chem. 1993;268:11134–11142. [PubMed] [Google Scholar]
- Park JM, Greten FR, Wong A, Westrick RJ, Arthur JS, Otsu K, Hoffmann A, Montminy M, Karin M. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis--CREB and NF-kappaB as key regulators. Immunity. 2005;23:319–329. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tonnetti L, Netzel-Arnett S, Darnell GA, Hayes T, Buzza MS, Anglin IE, Suhrbier A, Antalis TM. SerpinB2 protection of retinoblastoma protein from calpain enhances tumor cell survival. Cancer research. 2008;68:5648–5657. doi: 10.1158/0008-5472.CAN-07-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsdorfer P, Maxeiner S, Markopoulos C, Kirfel G, Wulf V, Auth T, Urschel S, von Maltzahn J, Willecke K. Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells. 2008;26:431–439. doi: 10.1634/stemcells.2007-0482. [DOI] [PubMed] [Google Scholar]
- Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat Struct Mol Biol. 2007;14:215–223. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.